Abstract
Natural killer cell immunoglobulin-like receptor (KIR) haplotype-specific DNA fragments were sequenced to identify centromeric and telomeric allele level haplotype structures and their frequencies from 76 unrelated individuals with European ancestry. Analysis was simplified by redefining the 5’ boundary of the centromeric KIR gene cluster to include only exons 7-9 of KIR3DL3. Three consensus allele level haplotypes were identified for a centromeric gene presence/absence structure designated as Cen-A1. KIR3DL3*00201 (ex 7-9)—KIR2DL3*001—KIR2DL1*00302 was the most frequent (37.5%) centromeric structure. Single consensus haplotypes were observed for haplotype structures Cen-B1 and Cen-B2. Six Tel-A1 and two Tel-B1 consensus haplotypes were observed; the most prevalent (23.0%) was KIR2DL4*00102—KIR3DL1*002—KIR2DS4*00101—KIR3DL2*002. A small number of nucleotide substitutions (≤3) in the coding regions of the functional KIR genes created microvariants of the consensus haplotypes. Eight less common haplotype structures were also detected. Four carried hybrid genes formed during gene deletion events, two carried an insertion with a 2DL5/3DP1 fusion gene, and two included a very large insertion. These data show that the KIR gene complex is composed of a limited number of conserved allele level centromeric and telomeric haplotypes that have diversified by mutation, recombination within a locus, and unequal crossing over.
Keywords: Natural killer cell, killer immunoglobulin-like receptor, haplotype, population study
Introduction
The recognition of malignant or infected cells by natural killer (NK) cells is controlled by both inhibitory and stimulatory receptors (1;2). Of the 14 members of the killer cell immunoglobulin-like receptor (KIR) family, many (KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL5, KIR3DL1, KIR3DL2, and KIR3DL3) are inhibitory receptors that prevent NK destruction of normal cells. Some of these receptors recognize subsets of human leukocyte antigens (HLA) as their ligands (3-6). Stimulatory receptors that may initiate NK destruction of damaged cells include KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5, and KIR3DS1; some of these receptors recognize HLA ligands but the ligands of others are not yet known (7;8) (9). KIR2DL4 has the capacity to be both stimulatory and inhibitory (10); it recognizes HLA-G as its ligand (11). Together, KIR and other NK receptors, like the NKG2 and the natural cytotoxicity receptors, control NK reactivity.
The genes encoding KIR are found in two adjacent clusters within the human leukocyte receptor complex on chromosome 19 (12). Within each cluster, KIR genes, lying in a head to tail orientation, are separated by about 2 Kb. Studies of KIR gene presence or absence in various individuals have noted that not all 14 KIR genes are found in a single haplotype (13-15). KIR2DL2 and KIR2DL3 appear to be encoded as allotypes; individual copies of chromosome 19 carry one or the other but not both genes. This is also true for KIR3DL1 and KIR3DS1. Three KIR genes (KIR2DL5, KIR2DS3, KIR2DS5) appear to be duplicated and may appear in the centromeric and/or the telomeric cluster (16-18). The complex also includes two pseudogenes, KIR2DP1 and KIR3DP1. Framework genes, found in most KIR haplotypes, flank each cluster:KIR3DL3 and KIR3DP1 flank the centromeric cluster while KIR2DL4 and KIR3DL2 flank the telomeric cluster.
DNA sequencing of single copies of chromosome 19 (18;19) and family segregation (15;20) (21) have defined three common centromeric (Cen-A1, Cen-B1, Cen-B2) and two common telomeric (Tel-A1, Tel-B1) haplotype structures based on the presence and absence of specific KIR genes. As examples, Cen-A1 is comprised of KIR3DL3—KIR2DL3—KIR2DP1—KIR2DL1—KIR3DP1 in this gene order while Tel-A1 includes KIR2DL4—KIR3DL1—KIR2DS4—KIR3DL2 (Table 1). Recombination within the region separating the two KIR gene clusters generates various centromeric--telomeric combinations. Less common KIR haplotypes appear to have been generated by unequal crossing over in this multigene family. Some haplotypes carry duplicated KIR genes (22); others carry large deletions (23). Some are marked by the presence of chimeric genes generated by the splicing of two KIR genes to form, for example, an expressed form of the KIR3DP1 pseudogene (24) or a hybrid KIR3DL1/KIR3DL2 gene (25). Overall, the KIR gene complex is a model for genetic complexity.
Table 1.
Genes carried by centromeric and telomeric KIR haplotypes.
| Haplotype Structure1 | Genes Included (centromere to telomere) |
|---|---|
| Cen-A1 | KIR3DL3-KIR2DL3-KIR2DP1-KIR2DL1-KIR3DP1 |
| Cen-B1 | KIR3DL3-KIR2DS2-KIR2DL2-KIR2DL5B-KIR2DS3 (or KIR2DS5)-KIR2DP1-KIR2DL1-KIR3DP1 |
| Cen-B2 | KIR3DL3-KIR2DS2-KIR2DL2-KIR3DP1 |
| Tel-A1 | KIR2DL4-KIR3DL1-KIR2DS4-KIR3DL2 |
| Tel-B1 | KIR2DL4-KIR3DS1-KIR2DL5A-KIR2DS3 (or KIR2DS5)-KIR2DS1-KIR3DL2 |
In addition to variation in the presence or absence of genes, the KIR genes are also polymorphic (26). This structural variation can impact levels of surface expression, ligand binding and NK cell activity (27-30). Family segregation, linkage disequilibrium, and haplotype isolation have defined allelic associations for segments of some KIR haplotypes (16;18;20;21;31;32). This study extends those observations to define the frequency and allelic content of KIR haplotypes in individuals of European ancestry using strategies to isolate haplotypes for DNA sequencing.
Results
KIR haplotype structures were examined in a panel of 76 unrelated individuals with European ancestry. Previously, gene amplification assays had been used to identify the presence or absence of KIR genes in these individuals and comprehensive DNA sequencing of essentially all exons had been applied to identify alleles at the 14 expressed KIR loci (32-39). Sixty eight individuals appeared to carry only the centromeric (A1, B1, B2) and telomeric (A1, B1) haplotype structures previously defined (18) (Tables 1 and 2). Eight individuals carried “other” haplotype organizations. Isolation of haplotype-specific DNA fragments and limited DNA sequencing were used to define the allelic linkages in these 76 individuals.
Table 2.
Haplotype structures in 68 individuals with European ancestry1
| Centromeric Haplotype2 | Percent (No. Individuals) | Telomeric Haplotype | Percent (No. Individuals) |
|---|---|---|---|
| Cen-A1/Cen-A1 | 48.5 (33) | Tel-A1/Tel-A1 | 61.8 (42) |
| Cen-A1/Cen-B1 | 16.2 (11) | Tel-A1/Tel-B1 | 35.3 (24) |
| Cen-A1/Cen-B2 | 26.5 (18) | Tel-B1/Tel-B1 | 2.9 (2) |
| Cen-B1/Cen-B1 | 0 | ||
| Cen-B1/Cen-B2 | 1.5 (1) | ||
| Cen-B2/Cen-B2 | 4.4 (3) |
Eight of the 76 individuals had at least one haplotype that did not conform to the five centromeric and telomeric organizations and are not included in these totals.
Two of 68 individuals carried either Cen-B1/Cen-B1 or Cen-B1/Cen-B2 and are not included in the centromeric tally. Both cells carried the same centromeric assignments: KIR3DL3*00301,*01402, KIR2DS2*00101, KIR2DL2*001, KIR2DL5B*0020101 (or *0020103), KIR2DS3*00103, KIR2DL1*00401.
The frequencies at which centromeric or telomeric common haplotypes were found in the 76 individuals studied are similar to that observed by Pyo and colleagues in a random Caucasian population (Figure 1). The frequencies of Cen-A1, Cen-B1, and Cen-B2 were 67.1%, 8.6%, and 17.8% in our study compared to 71.9%, 10.4% and 16.7% in the earlier study. The frequencies of Tel-A1 and Tel-B1 were 76.3% and 19.1% in our study versus 82.3% and 16.7% in the Pyo et al. study (18). The frequency of “other” haplotypes was higher in our study (eight of 76 individuals) compared to one in 48 individuals in the Pyo et al. study. This difference may be the result of the inability of the screening assays used in the earlier study to detect less common gene organizations. The distribution of haplotype structures in individuals carrying only common structures is given in Table 2.
Figure 1.
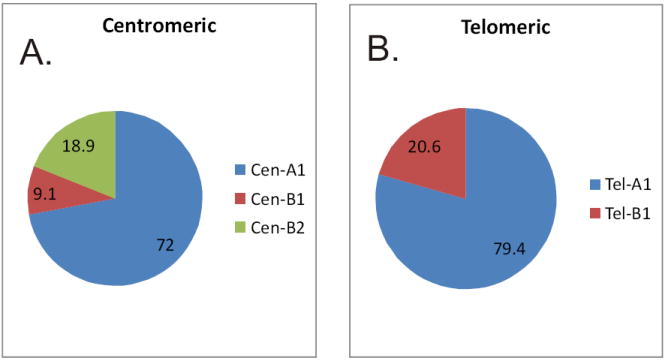
Frequencies of (A.) Cen-A1, Cen-B1, Cen-B2 and (B.) Tel-A1 and Tel-B1 haplotype structures in 76 unrelated individuals of European ancestry. The centromeric structures in two individuals could not be defined (ND). In addition to common haplotype structures, eight individuals carried extended or contracted haplotypes (labeled as “other”).
Although this study was not able to link specific centromeric and telomeric haplotypes, the combinations of centromeric and telomeric structures found were consistent with their frequencies in the population. The majority of individuals (31.8%, n=21 of 66 with known haplotypes) carried Cen-A1/Cen-A1 and Tel-A1/Tel-A1 haplotypes. Also frequent were Cen-A1/Cen-A1 with Tel-A1/Tel-B1 (16.7%) and Cen-A1/Cen-B2 with Tel-A1/Tel-A1 (21.2%). The associations between centromeric and telomeric KIR haplotype structures appear to be random (p=0.47).
Redefining the centromeric framework gene, KIR3DL3
Extensive diversity is found in the KIR3DL3 alleles associated with the centromeric haplotypes. Cen-A1 haplotypes alone were found with 26 different KIR3DL3 alleles. However, if the KIR3DL3 gene is divided into two segments based on a hotspot for recombination separating exons encoding the extracellular domains (exons 1-5) from exons encoding the intracellular segment (exons 7-9) (40), the extensive nucleotide variation is localized to exons 1-5. The nucleotide sequences of exons 7-9 of KIR3DL3 are highly conserved (Table 3). Only five consensus sequences for this 3’ region are found among all the known alleles of KIR3DL3 (n=75). These conserved sequences have specific associations with centromeric haplotype structures, Cen-A1 and Cen-B (18) and with specific consensus KIR haplotypes within those haplotype structures as described below.
Table 3.
Nucleotide sequence differences in exons 7-9 among KIR3DL3 alleles
| Position of Nucleotide Differences | |||||||
|---|---|---|---|---|---|---|---|
| Exons 7-91 | 961 | 969 | 971 | 1042 | 1074 | 1119 | Observed2 KIR3DL3 Alleles Included |
| 00101 | A | C | T | G | A | A | *00101, *00103,*00602, *01302, *01303, *01306, *026 |
| 00102 | - | - | - | - | G | - | *00102, *00601, *01305, *01307, *017 |
| 00201 | C | - | - | - | - | - | *00201, *00202, * 00206, *00207, *005, *00801, *00901, *010, *01102, *015, *030 |
| 00301 | T | - | C | - | - | - | *00301, *007, *01402, *01404, *01405, *016 |
| 009023 | C | T | - | - | - | - | *00902 |
| 025 | C | - | - | C | - | T | *025, *027, *031 |
| 0293 | C | - | A | C | - | T | *029 |
The numerical “allele” designation of exons 7-9 is based on the lowest numbered allele with the consensus sequence in exons 7-9.
Only alleles observed in this study are included in the table.
KIR3DL3*00902 and KIR3DL3*029 are microvariants of KIR3DL3*00201 and KIR3DL3*025 exon 7-9 sequences, respectively.
Exons 1-5 of KIR3DL3 found associated with the centromeric haplotype structures exhibit a similar level diversity as found when all known KIR3DL3 alleles are compared. Figure 2 shows a mismatch comparison of pairs of KIR3DL3 alleles based on the nucleotide sequences of exons 1-5. The plot compares the frequency distribution of pairwise comparisons of all KIR3DL3 alleles (n=75) to the 16 alleles found within a related cluster (consensus haplotype I as described below) of Cen-A1 haplotypes. Both comparisons exhibit a bimodal distribution suggesting that they are composed of recently related lineages with a small number of nucleotide variations and more distantly related lineages with greater sequence differences. So, the KIR3DL3 alleles associated with a consensus haplotype of Cen-A1 are as different from one another as is found if all the KIR alleles are examined without regard to shared haplotypes. This diversity in KIR lineages associated with a conserved haplotype has been previously noted (18).
Figure 2.
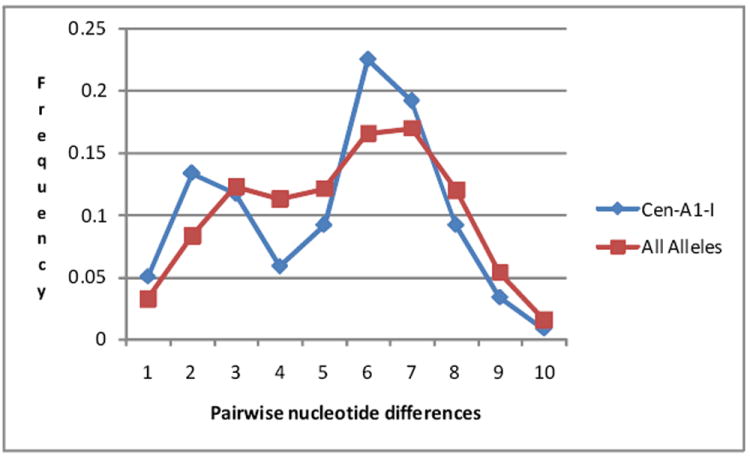
Mismatch distribution of pairwise comparisons of exon 1-5 nucleotide sequences among alleles at the KIR3DL3 locus. Blue is the distribution observed in a comparison of 16 KIR3DL3 alleles that were identified within the Cen-A1-I consensus group. Red is the distribution observed in a comparison of 75 KIR3DL3 alleles.
Cen-A1
Thirty two different haplotypes were observed from 1 to 17 times in the 102 haplotypes designated as Cen-A1 (Table 4). KIR3DL3*00901--KIR2DL3*001--KIR2DL1*00302 (17 copies) and KIR3DL3*00101--KIR2DL3*002--KIR2DL1*002 (15copies) were the two most common Cen-A1 haplotypes. The 32 Cen-A1 haplotypes can be clustered into three consensus haplotypes where haplotypes within each cluster share essentially all alleles at the included loci if exons 1-5 of the most centromeric locus, KIR3DL3, are excluded. The consensus haplotype is the most frequent haplotype within the consensus cluster. While haplotypes differ from the consensus sequence by ≤3 nucleotides In the coding regions of expressed alleles, the three consensus haplotypes differ from one another by seven to 11 nucleotides (Figure 3). The frequencies of the three centromeric consensus haplotype clusters are shown in Figure 4.
Table 4.
Cen-A1 haplotypes1
| Haplotype | 3DL3* | 3DL3 Exons 7-92 |
2DL3* | 2DL1* | No. Observed |
|---|---|---|---|---|---|
| I--Consensus | 00201 | 001 | 00302 | 57 | |
| 00901 | 00201 | 001 | 00302 | 173 | |
| 00901 | 00201 | 001 | 009 | 1 | |
| 00902 | 00201v4 | 001 | 00302 | 3 | |
| 00902 | 00201v | 003 | 00302 | 1 | |
| 00202 | 00201 | 001 | 00302 | 6 | |
| 00206 | 00201 | 001 | 00302 | 33 | |
| 030 | 00201 | 001 | 00302 | 2 | |
| 010 | 00201 | 001 | 00302 | 23 | |
| 00201 | 00201 | 001 | 00302 | 2 | |
| 00207 | 00201 | 001 | 00302 | 4 | |
| 00801 | 00201 | 001 | 00302 | 2 | |
| 005 | 00201 | 001 | 00302 | 1 | |
| 01102 | 00201 | 001 | 00302 | 1 | |
| 015 | 00201 | 001 | 00302 | 2 | |
| 00101 | 00101 | 001 | 00302 | 6 | |
| 01302 | 00101 | 001 | 00302 | 1 | |
| 01306 | 00101 | 001 | 00302 | 2 | |
| 01303 | 00101 | 001 | 00302 | 1 | |
| II--Consensus | 00101 | 002 | 002 | 40 | |
| 00101 | 00101 | 002 | 002 | 153 | |
| 00101 | 00101 | 002 | 008 | 1 | |
| 00103 | 00101 | 002 | 002 | 1 | |
| 026 | 00101 | 002 | 002 | 1 | |
| 01302 | 00101 | 002 | 002 | 3 | |
| 00102 | 00102 | 002 | 002 | 6 | |
| 00601 | 00102 | 002 | 002 | 3 | |
| 01305 | 00102 | 002 | 002 | 3 | |
| 01307 | 00102 | 002 | 002 | 1 | |
| 017 | 00102 | 002 | 002 | 6 | |
| III--Consensus | 025 | 005 | 001 | 5 | |
| 025 | 025 | 005 | 001 | 2 | |
| 031 | 025 | 005 | 001 | 1 | |
| 029 | 025v | 005 | 001 | 1 | |
| 027 | 025 | 005 | 001 | 1 |
Based on 74 individuals where the centromeric haplotypes could be defined. Two other individuals carrying expected haplotype structures were not included because it was not possible to determine whether they carried Cen-B1/-B1 or Cen-B1/-B2.
The designation is based on the lowest numbered allele with the consensus sequence in exons 7-9 (Table 3).
Haplotype previously reported (18).
Blue shading marks alleles that differ by one or two nucleotide changes from the consensus allele. Only the exon 7-9 segment of KIR3DL3 is included in this comparison. The 3’ nucleotide sequences of KIR3DL3*00902 (labeled 00201v) and KIR3DL3*029 (labeled 025v) are one nucleotide different from their consensus sequences (Table 3).
Figure 3.
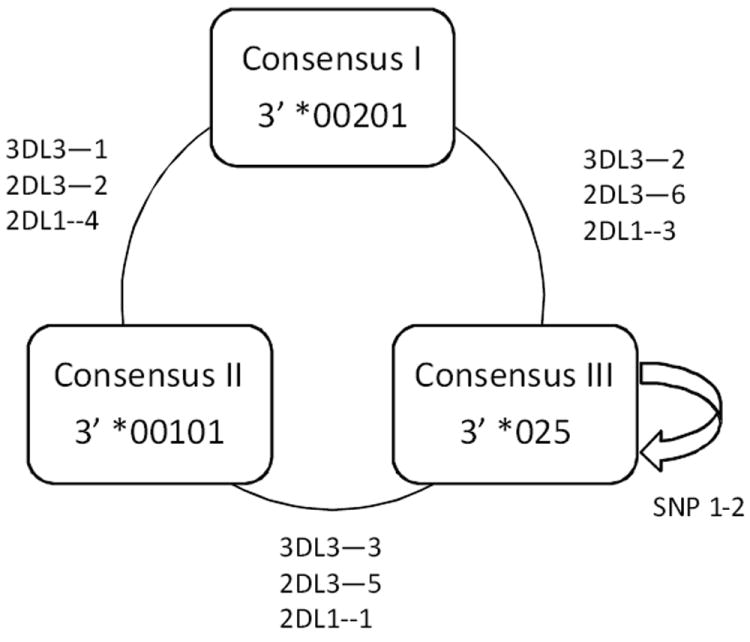
The 32 Cen-A1 haplotypes described in Table 4 were reduced to ten haplotypes based on the nucleotide sequences of KIR3DL3 exons7-9, KIR2DL3, and KIR2DL1. The evolutionary history among these ten haplotypes was then inferred using the Neighbor-Joining method. The alleles carried by each haplotype are indicated in the figure; for example, CenA2 00101-002-002 refers to the Cen-A1 haplotype in consensus group II that carries KIR3DL3*00101 (exons7-9)-KIR2DL3*002-KIR2DL1*002. Brackets indicate the haplotypes within the three consensus groups; arrows indicate the three consensus haplotypes defining those groups. Haplotypes within each consensus group differ from the consensus by ≤3 single nucleotide polymorphisms. The consensus haplotypes differ from one another by from seven to 11 single nucleotide polymorphisms. The nucleotide sequences of the alleles at each locus in each Cen-A1 haplotype were concatenated for this analysis. The bootstrap consensus tree was inferred from 1000 replicates. The percentage of replicate trees in which the associated haplotypes clustered together in the bootstrap test is shown next to the branches.
Figure 4.
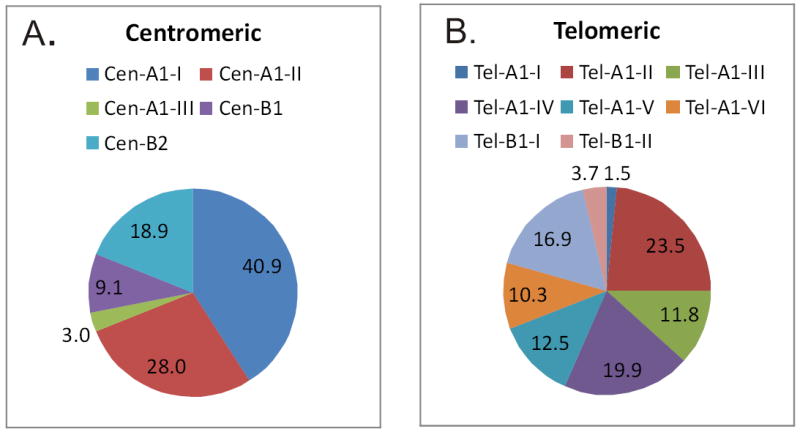
Frequencies of (A.) centromeric and (B.) telomeric consensus haplotypes in 76 individuals of European ancestry. The centromeric structures in two individuals were unclear (labeled “ND”). In addition to common haplotype structures, eight individuals carried extended or contracted haplotypes (labeled as “other”).
Eighteen distinct haplotypes share a consensus haplotype (designated Cen-A1-I for Cen-A1, consensus haplotype I) comprised of KIR3DL3*00201 (exons 7-9)--KIR2DL3*001—KIR2DL1*00302 (Table 4). The majority (37.5%) of the centromeric haplotypes carry this consensus (Figure 4). Minor differences from the Cen-A1-I consensus include a haplotype carrying KIR2DL3*003, an allele differing by two nucleotides from KIR2DL3*001, and a haplotype carrying KIR2DL1*009, an allele differing by two nucleotides from KIR2DL1*00302. Four haplotypes differ by a single nucleotide substitution in exons 7-9 creating KIR3DL3*00101. A single nucleotide substitution in the exon 7-9 sequence is also observed for the haplotypes associated with KIR3DL3*00902 (Table 3). Thus, a conserved haplotype defines this group with less common variant haplotypes differing only subtly from the conserved coding sequences of the KIR genes present.
The ten Cen-A1 consensus II (Cen-A1-II) haplotypes share KIR3DL3*00101 (exons 7-9)-- KIR2DL3*002--KIR2DL1*002. This consensus appears at a frequency of 26.3% in individuals with European ancestry. Microvariation generates haplotypes carrying KIR2DL1*008 or KIR3DL3*00102 (exons 7-9); both alleles differ by a single nucleotide from the consensus allele at the locus. KIR3DL3*025 (exons 7-9)--KIR2DL3*005--KIR2DL1*001 form the third consensus haplotype (Cen-A1-III) with a frequency of 3.3%. Four haplotypes were identified that share these alleles but differ for the 5’ regions of KIR3DL3; one differs by a single nucleotide substitution within exons 7-9 of the consensus KIR3DL3 sequence (Table 3).
Cen-B
Six Cen-B1 haplotypes were observed with KIR3DL3*00301--KIR2DS2*00101--KIR2DL2*001--KIR2DL5B*0020101 (or*0020103)--KIR2DS3*00103—KIR2DL1*00401 as the most common, found in six of the 13 Cen-B1 copies (Table 5)(Figure 4). Of the seven allele divergent Cen-B2 haplotypes observed, there were 15 copies of KIR3DL3*00301--KIR2DS2*00101--KIR2DL2*003 out of 27 total Cen-B2 copies (Table 6). Based on the alleles shared at KIR3DL3 exons 7-9, KIR2DS2, and KIR2DL2, it appears that Cen-B2 was generated from Cen-B1 by a gene deletion event (or Cen-B1 generated by a gene insertion event). Allelic variation in the three functional loci shared by the two haplotypes is restricted to a single nucleotide difference at KIR2DL2*001 versus KIR2DL2*003. The Cen-B1-I and Cen- B2-I consensus haplotypes are found at frequencies of 8.6% and 17.8%, respectively. Microvariation similar to that observed for Cen-A1 within a consensus was observed.
Table 5.
Cen-B1 haplotypes1
| Haplotype | 3DL3* | 3DL3 Exons 7-92 |
2DS2* | 2DL2* | 2DL5B* | 2DS3* | 2DL1* | No. Observed |
|---|---|---|---|---|---|---|---|---|
| I--Consensus | 00301 | 00101 | 001 | 00201013 | 00103 | 00401 | 13 | |
| 00301 | 00301 | 00101 | 001 | 0020101 | 00103 | 00401 | 64 | |
| 00301 | 00301 | 0025 | 001 | 00201026 | 00103 | 00401 | 3 | |
| 00301 | 00301 | 002 | 001 | 0020102 | 00103 | 00402 | 1 | |
| 00301 | 00301 | 002 | 001 | 009 | 00103 | 00401 | 1 | |
| 01402 | 00301 | 00101 | 005 | 0020101 | 00103 | 00401 | 1 | |
| 007 | 00301 | 00101 | 001 | 0020101 | 00103 | 00401 | 1 |
Based on 74 individuals where the centromeric haplotypes could be defined. Two other individuals carrying expected haplotype structures were not included because it was not possible to determine whether they carried Cen-B1/-B1 or Cen-B1/-B2.
The designation is based on the lowest numbered allele with the consensus sequence in exons 7-9 (Table 3).
Not distinguished from KIR2DL5B*0020103 in this study.
Haplotype previously observed (18).
Blue shading marks alleles that differ by a single nucleotide change from the consensus allele. Only the exon 7-9 segment of KIR3DL3 is included in this comparison.
KIR2DL5B*0020102 has the same nucleotide coding sequence as the consensus allele.
Table 6.
Cen-B2 haplotypes1
| Haplotype | 3DL3* | 3DL3 Exons 7-92 |
2DS2* | 2DL2* | 2DL13 | No. Observed |
|---|---|---|---|---|---|---|
| I--Consensus | 00301 | 00101 | 003 | neg | 27 | |
| 00301 | 00301 | 00101 | 003 | neg | 154 | |
| 01405 | 00301 | 00101 | 003 | neg | 2 | |
| 00301 | 00301 | 00101 | 0015 | neg | 2 | |
| 01404 | 00301 | 00101 | 003 | neg | 1 | |
| 01402 | 00301 | 00101 | 001 | neg | 14 | |
| 007 | 00301 | 00101 | 003 | neg | 5 | |
| 016 | 00301 | 00101 | 001 | neg | 1 |
Based on 74 individuals where the centromeric haplotypes could be defined. Two other individuals carrying expected haplotype structures were not included because it was not possible to determine whether they carried Cen-B1/-B1 or Cen-B1/-B2.
The designation is based on the lowest numbered allele with the consensus sequence in exons 7-9 (Table 3).
Neg, negative for the gene. Some of these haplotypes were tested for the presence of KIR2DP1 and found to be negative.
Haplotype observed previously (18).
Blue shading marks an allele that differs by two nucleotide changes from the consensus allele. Only the exon 7-9 segment of KIR3DL3 is included in this comparison.
Tel-A1
Twenty three allele content haplotypes were identified for Tel-A1 (Table 7). The most common were KIR2DL4*00102--KIR3DL1*002--KIR2DS4*00101--KIR3DL2*002 (20 copies) and KIR2DL4*0080101 (or*0080103)--KIR3DL1*00101--KIR2DS4*003--KIR3DL2*00101 (19 copies). The haplotypes fall into six consensus haplotypes (I-VI), distinguished by the lineage of the KIR3DL1 alleles (41) and by the extent of nucleotide differences between the combined coding regions. Consensus haplotypes I-III carry the KIR3DL1*015 lineage alleles. Clusters IV and V include the lineage KIR3DL1*005 alleles. Tel-A1-VI carries the interlineage recombinant KIR3DL1*00101. The consensus haplotypes differ from one another by from six (consensus IV versus VI) to 30 nucleotides (consensus I versus V) with an average of 18.2 nucleotides. For this comparison, the 22 bp deletion found in some of the KIR2DS4 alleles was counted as a single alteration. The relationships among the Tel-A1 consensus haplotypes are shown in a Neighbor-Joining tree (Figure 5). The frequencies of the telomeric A consensus haplotypes are shown in Figure 4. They vary from 23.0% for Tel-A1-I to 2.0% for Tel-A1-III.
Table 7.
Tel-A1 haplotypes1
| Haplotype | 2DL4* | 3DL1* | 2DS4* | 3DL2* | No. Observed |
|---|---|---|---|---|---|
| I-Consensus | 00102 | 002 | 00101 | 002 | 35 |
| 00102 | 002 | 00101 | 002 | 20 | |
| 00102 | 015023 | 00101 | 002 | 112 | |
| 00102 | 01702 | 00101 | 021 | 1 | |
| 00103 | 002 | 00101 | 002 | 3 | |
| II-Consensus | 00103 | 008 | 003 | 00901 | 16 |
| 00103 | 008 | 003 | 00901 | 112 | |
| 00103 | 008 | 003 | 019 | 3 | |
| 00103 | 020 | 001014 | 00902 | 2 | |
| III-Consensus | 00602 | 007 | 004 | 008 | 3 |
| 00602 | 007 | 004 | 008 | 32 | |
| IV--Consensus | 011 | 00501 | 007 | 00103 | 19 |
| 011 | 00501 | 007 | 00103 | 14 | |
| 011 | 00501 | 007 | 010 | 5 | |
| V--Consensus | 00802 | 00401 | 006 | 00301 | 16 |
| 00802 | 00401 | 006 | 00301 | 4 | |
| 00802 | 00402 | 006 | 00301 | 1 | |
| 00802 | 00401 | 006 | 005 | 52 | |
| 00802 | 019 | 006 | 005 | 2 | |
| 00201 | 00401 | 006 | 005 | 1 | |
| 00802 | 00401 | 006 | 011 | 2 | |
| 00802 | 00401 | 006 | 020 | 1 | |
| VI--Consensus | 00801015 | 00101 | 003 | 00101 | 27 |
| 0080101 | 00101 | 003 | 00101 | 192 | |
| 00801046 | 00101 | 003 | 00101 | 1 | |
| 0080101 | 00101 | 008 | 00101 | 1 | |
| 0080101 | 00101 | 003 | 011 | 4 | |
| 0080101 | 0097 | 003 | 011 | 1 | |
| 0080101 | 00101 | 003 | 00901 | 1 |
Based on 76 individuals where the telomeric haplotypes could be defined.
Allele level haplotype previously observed (18). Five of the haplotypes on this table (without KIR2DS4 typing) were deduced by Norman et al. in a European population as part of a haplotype with a duplication (25).
Blue shading marks alleles that differ by one or two nucleotide changes from the consensus allele.
KIR2DS4*003 differs from KIR2DS4*00101 by a 22 base pair deletion.
KIR2DL4*0080103 not excluded.
KIR2DL4*0080104 has the same coding sequence as the consensus allele.
KIR3DL1*009 is an interlineage recombinant, sharing the sequences of exons 1-3 with KIR3DS1 and exons 4-9 with KIR3DL1*00101 (41).
Figure 5.
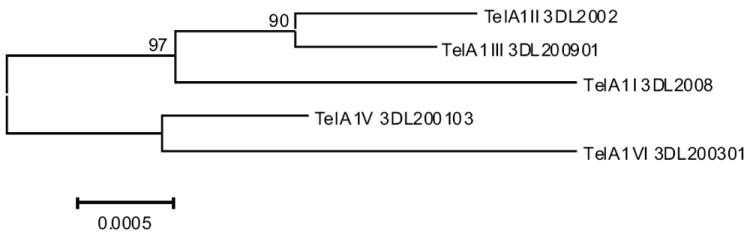
The evolutionary history among five consensus Tel-A1 haplotypes (I-V) was inferred using the Neighbor-Joining method. The nucleotide sequences of the alleles at each locus in each Tel-A1 consensus haplotype were concatenated for this analysis. The bootstrap consensus tree was inferred from 1000 replicates. The percentage of replicate trees in which the associated haplotypes clustered together in the bootstrap test is shown next to the branches. The sixth consensus haplotype IV was not included because of the presence of interlineage recombinants of KIR3DL1.
Within a consensus haplotype cluster, there is a frequent consensus haplotype with microvariation creating the others. For example, the consensus haplotype for Tel-A1-I is KIR2DL4*00102—KIR3DL1*002—KIR2DS4*00101—KIR3DL2*002 found 20 times within the 35 haplotypes in this group. The other three haplotypes in this group differ by one to two nucleotides from the consensus at individual loci forming microvariants KIR2DL4*00103, KIR3DL1*01502, KIR3DL1*01702, and KIR3DL2*021. The second most prevalent haplotype is KIR2DL4*00102—KIR3DL1*01502—KIR2DS4*00101—KIR3DL2*002 found 11 times. The other two haplotypes were observed one and three times in this population.
An exception to the microvariation is found within the consensus VI haplotype. The KIR3DL1*00101 interlineage recombinant fusing exons 1-3 of KIR3DL1*005 and exons 4-9 of KIR3DL1*015, is replaced by KIR3DL1*009. This allele is a second interlineage recombinant joining exons 1-3 of KIR3DS1*013 and exons 4-9 of KIR3DL1*00101 (41). Thus recombination may also cause variation within a consensus haplotype.
Tel-B1
Six Tel-B1 haplotypes were identified with KIR2DL4*00501--KIR3DS1*01301-- KIR2DL5A*0010101--KIR2DS5*00201--KIR2DS1*00201--KIR3DL2*00701 the most frequent at 21 copies (Table 8). Again, microvariation is noted. Furthermore, a single gene conversion event with a Cen-B1 haplotype may have created the second consensus haplotypes carrying KIR2DL5A*0050101— KIR2DS3*002. It has been noted previously that both KIR2DL5A*0050101 and KIR2DS3 are fusion genes (42) (18;43). KIR2DL5*0050101 shares the 5’ regulatory region and exon 1 with the consensus allele in the major Tel-B1 haplotype, KIR2DL5A*0010101 and the remainder of its sequence with KIR2DL5B*0020101 while KIR2DS3*002 shares its 5’ sequence with KIR2DS3*00103 and its 3’ sequence with KIR2DS5. Since the surrounding genes in Tel-B1 are shared, this suggests that a single gene conversion-like event created this haplotype from the consensus. The frequencies of the consensus haplotypes I and II are 15.8% and 3.3%, respectively.
Table 8.
Tel-B1 haplotypes1
| Haplotype | 2DL4* | 3DS1* | 2DL5A* | 2DS3* | 2DS5* | 2DS1* | 3DL2* | No. Observed |
|---|---|---|---|---|---|---|---|---|
| I--Consensus | 00501 | 01301 | 0010101 | 00201 | 00201 | 00701 | 24 | |
| 00501 | 01301 | 0010101 | 00201 | 00201 | 00701 | 212 | ||
| 00501 | 01301 | 0010101 | 00201 | 0063 | 00701 | 1 | ||
| 00501 | 01301 | 0010101 | 00201 | 00501 | 00701 | 1 | ||
| 00501 | 01301 | 0010101 | 00201 | 00201 | 018 | 12 | ||
| II-Consensus | 00501 | 01301 | 0050101 | 002 | 00201 | 00701 | 5 | |
| 00501 | 01301 | 0050101 | 002 | 00201 | 00701 | 42 | ||
| 00501 | 01301 | 00501024 | 002 | 00201 | 00701 | 1 |
Based on 76 individuals where the telomeric haplotypes could be defined.
Allele level haplotype previously reported (18).
Blue shading marks alleles that differ by one to three nucleotide changes from the common allele in the cluster.
KIR2DL5A*0050102 has the same coding region sequence as the consensus allele.
Gene extended/contracted haplotypes
Eight of the 76 individuals carried allele combinations that did not fit the expected common haplotype structures. Comparison to previously reported expanded or contracted haplotypes and to the centromeric and telomeric haplotype structures, and physical linkage of adjacent alleles were used to assign genes and alleles to haplotypes in each cell. The number of KIR2DL4 genes in each of the eight individuals was confirmed by a quantitative polymerase chain reaction.
Four of the eight individuals carried haplotypes with deletions of KIR genes resulting in hybrid genes fusing the 5’ region of a centromeric gene with the 3’ region of a telomeric gene at the recombination site. For example, RDP71 carried a haplotype with a large deletion and only four genes: KIR3DL3*00602, KIR2DS1*00502, KIR3DL2*017, and a KIR2DL3/2DP1 fusion gene (Table 9). This haplotype had been previously observed in a CEPH family (designated haplotype “j”)(23). KIR2DS1*00502 is a second hybrid allele in this haplotype linking the 5’ sequence of KIR2DL1 and the 3’ sequence of KIR2DS1. The alleles carried by the three loci of the CEPH haplotype are shared with cell RDP71 with the exception of a single nucleotide substitution in the KIR3DL2 allele (*007 in the CEPH family vs *017 in this study). Partial sequences of KIR2DL3/2DP1 covering exons 1-4 of KIR2DL3 and exons 6-9 of KIR2DP1 showed identity to the previously reported fusion gene sequence (GenBank accession numbers CU041340 and CU633846).
Table 9.
Gene contracted or expanded KIR haplotypes
| Sample ID |
Haplotype1 | Reference | |
|---|---|---|---|
| Contracted | Fusion Gene(s) | ||
| RDP27 | 3DL3*00301--2DS2*002--2DL2*001--2DL5B*0020102--2DS3*002--2DS1*00201--3DL2*007012 | (16) | KIR2DS3*002 (2DS3/2DS5) |
| RDP69 | 3DL3*00301--2DS2*00101--2DL2*001--2DL5B*008--2DS5*00201--2DS1*00201--3DL2*007012 | (21) | KIR2DL5B*008 (2DL5B/2DL5A) |
| RDP71 | 3DL3*00602--2DL3/2DP1--2DS1*00502--3DL2*0172 | (23) | KIR2DL3/KIR2DP1 KIR2DS1*005023 (2DL1/2DS1) |
| RDP76 | 3DL3*00301--2DS2*005—2DP1--2DL1*00401—3DP1--known telomeric region | (21) | KIR2DS2*005 (2DS2/2DS3) |
| Expanded | |||
| RDP10 | Known centromeric region--2DL4*00501--3DS1*01301--2DL5A/3DP1--2DL4*00102-- 3DL1*002--2DS4*00101--3DL2*002 |
(22;24;44) | KIR2DL5A/3DP1 |
| RDP64 | Known centromeric region-2DL4*00501--3DS1*01301--2DL5A/3DP1--2DL4*0080101/0080103-- 3DL1*00101--2DS4*003--3DL2*009variant4 |
||
| RDP81 | 3DL3*015--2DL3*001—[2DP15]--2DL1*00302—[3DP1]--2DL4*00501--3DS1*01301-- 2DL5A*0050101--2DS3*00103—[2DP1]--2DL1*00401—[3DP1]--2DL4*00501--3DS1*01301-- 2DL5A*0010101--2DS5*002--2DS1*00201--3DL2*00701 |
(16) | |
| RDP14 | 3DL3*015--2DL3*001—[2DP1]--2DL1*00302—[3DP1]--2DL4*00501--3DS1*01301 (or *058)-- 2DL5A*0050101--2DS3*00103—[2DP1]--2DL1*00401—[3DP1]--2DL4*00501--3DS1*058 (or *01301)--2DL5A*0010101--2DS5*002--2DS1*00201--3DL2*00701 |
||
The order of the genes based on physical linkage of alleles, by known allele-level haplotype observed in this study, and/or on previous literature.
These haplotypes do not appear to carry intact KIR2DP1 and KIR3DP1 genes.
KIR2DS1*00502 had previously been named as KIR2DS1*007.
Allele designation requested, not yet assigned.
Positions of KIR2DP1 and KIR3DP1 based on Ordonez et al. (16)
In the deletion haplotype found in RDP69, KIR2DL5B*008 is a fusion between KIR2DL5B*0020101 and KIR2DL5A*0010101 in which the recombination can be localized between nucleotide 16 in exon 1 and nucleotide 75 in exon 3. Likewise, in a third contracted haplotype in RDP76, KIR2DS2*005 is a fusion between KIR2DS2*0010101 and KIR2DS3*00103 with the recombination occurring between exon 5 and exon 7. The haplotypes observed in RDP69 and RDP76 were observed, at the gene presence/absence level, by Gourraud and colleagues (21). A final contracted haplotype links the centromeric KIR2DL5B*0020102 to telomeric KIR2DS3*002 in RDP27. KIR2DS3*002 has previously been noted to be a hybrid gene of KIR2DS3*00103 and KIR2DS5 (43). A haplotype apparently carrying the same deletion as RDP27 on the basis of genes present or absent was observed previously in a Spanish family (C43)(16).
Two (RDP10, RDP64) of the 76 individuals studied (2.6%) carried a haplotype with an insertion of several KIR genes, previously found in European populations at a frequency of 0.8-4.5% (22;24;44) (Table 9). The haplotype is marked by the presence of a chimeric gene carrying the 5’ regulatory and exon 1 sequence of KIR2DL5A fused to the 3’ sequence of KIR3DP1. Because the chimeric gene was identified by sequence-specific amplification, it is not known if its sequence is identical to the original report identifying the gene as KIR3DP1*004 (24). Comparison of the haplotypes in RDP10 and RDP64 suggests that they originated independently. RDP10 arose from a Tel-A1-I consensus haplotype based on the four telomeric genes while RDP64 arose from Tel-A1-VI (Table 7). Other allele level haplotypes carrying the KIR2DL5A/3DP1 fusion gene identified in families (22;24;25) carry other alleles in the telomeric segment suggesting that this insertion has arisen independently several times.
Two individuals (RDP81, RDP14) (2.6%) carried a haplotype with an extensive insertion, previously observed in a Northern Irish family (12/04) (16). The two individuals carried the same alleles at each locus with the exception that one (RDP14) carried KIR3DS1*058 at one of the two KIR3DS1 loci instead of KIR3DS1*01301. The two alleles differ by a single nucleotide substitution. The previously reported haplotype carries the same alleles at the ten loci that were typed as the cells identified here suggesting that these haplotype are ancestrally related. The haplotype carries 14 functional genes and is comprised of segments of known Cen-A1-I (genes 1-3 starting from the centromere), Tel-B1-II (genes 4- 6), Cen-B1-I (genes 7-8), Tel-B1-I (genes 9-14) allelic haplotypes. Partnering haplotypes in the eight individuals with extended or contracted haplotypes comprised the more common centromeric and telomeric structures.
Discussion
Using a panel of 76 European Americans with alleles previously identified for all functional KIR loci, we isolated haplotype-specific DNA fragments to identify linked alleles. These data complement and extend haplotype information obtained by the comprehensive DNA sequencing of overlapping genomic clones obtained from 27 KIR haplotypes derived from multiple ethnic groups (18;19;45). Our in-depth analysis of the structure and frequency of KIR allele level haplotypes in individuals of European ancestry suggests that conserved haplotypes predominant in the population and shows the extent to which microvariation has impacted those haplotypes.
The five conserved gene presence/absence haplotype structures (Cen-A1, Cen-B1, Cen-B2, Tel- A1, Tel-B1) were the only structures found in the majority of individuals (89.5%). Conserved structures were also found in single haplotypes from the eight individuals who also carried an extended or a contracted haplotype. The haplotypes characterized in this study using physical linkage carry essentially all of the two locus allele combinations previously noted in a linkage study of families from Northern Ireland (21). For example, the earlier study noted the association of KIR3DL1*01502 with KIR3DL2*002 with a 100% positive predictive value. This association was observed in our cohort as a variant of the Tel-A1 consensus I haplotype (Table 7). There were two exceptions from the many associations observed in the family linkage study which are explained by the lower resolution typing of the earlier study. The association of KIR3DL1*005 (and KIR2DL4*011) with KIR2DS4*003 was not observed in our study. KIR2DS4*003 differs by a single nucleotide from KIR2DS4*007, the allele we observed in association with KIR3DL1*005 (Table 7, Tel-A1-IV) and an allele not yet identified when the family samples were typed (20). Each of the consensus haplotypes described here has been previously observed (18) with a few exceptions. While the consensus haplotypes for Tel-A1-I and Tel-A1-V were not observed, one of the microvariant haplotypes in those clusters were characterized in the earlier study. Two consensus haplotypes, Cen-A1-III and Tel-A1-IV, were not found in the earlier study but this may be due to the limited number of cells investigated by fosmid sequencing. Six of the allele level haplotypes observed in our study were identified as extended haplotypes in a previous study of linkage disequilibrium (21). Thus, our independent dataset and analysis supports and extends the allele-level haplotype data previously reported in world populations.
The majority of the centromeric and telomeric consensus haplotypes observed in this cohort with European ancestry are found at frequencies between 10-20%. Three exceptions are haplotypes that appear at frequencies less than 5% (Cen-A1-III, Tel-A1-III, Tel-B1-II). These haplotypes may have originated more recently. Two other exceptions are the Cen-A1-I and Cen-A1-II haplotypes that are found at frequencies of 37.5% and 26.3%, respectively. These two haplotypes exhibit the most variation with 17 and 9 microvariant haplotypes, respectively. Many of the microvariant haplotypes are found in multiple individuals suggesting that these two consensus haplotypes may be more ancient or may be favored by selection.
At first glance, the diversity of KIR haplotypes appears extensive. Forty five different haplotypes were observed within the centromeric region with 32 classified as Cen-A1. However, the diversity within Cen-A1 was reduced to three consensus haplotypes by eliminating the KIR3DL3 sequences 5’ of a hotspot of recombination and by identifying variation at other loci as limited to ≤3 nucleotide substitutions. Likewise, the 29 telomeric haplotypes fell into 8 consensus haplotypes. The telomeric haplotypes are more diverse than the centromeric haplotypes as previously noted (18;21).
It has been noted that hot spots of recombination, as found in the intron dividing extracellular from intracellular coding segments of KIR3DL3 and in the region separating the centromeric and telomeric KIR haplotypes, are often flanked by regions with reduced crossing over called haplotype blocks (46;47). This is consistent with the conserved allele level KIR haplotype structures observed. This is especially true for the exon 7-9 region of human KIR3DL3 which is thought be very ancient (40). The conserved haplotypes noted here with modest variation within the alleles carried suggests that these extended KIR haplotypes comprise optimal allele combinations that have been preserved over evolutionary time.
But how do the diverse 5’ sequences of KIR3DL3 fit into the picture? The 5’ exon 1-5 sequences of KIR3DL3 appear to form a region distinct from the conserved centromeric and telomeric segments. There appears to have been extensive exchange among the various KIR haplotypes so that there is no association of this region with exons 7-9 nor with specific centromeric haplotype structures. The impact of a hotspot of recombination on the KIR3DL3 protein itself is not clear. KIR3DL3 lacks exon 6 which encodes the stem region of KIR (48) and cell surface expression has not been observed (49) suggesting that this locus might not be functional. However, comparison of the sequence of the extracellular region with that of higher primates shows extensive conservation and the impact of purifying selection (40). Solution to this conundrum may come when the function of KIR3DL3, if there is any, is understood.
Gene expanded or contracted haplotype appeared in 10.5% of individuals with European ancestry studied. The creation of these haplotypes appears to be facilitated by the unique features of the KIR gene complex in terms of the dense clustering of repeat elements in the KIR introns (23). Such domain shuffling as a mechanism for generating new receptors has been noted in a comparison among higher primates (50). These less common haplotypes are sometimes difficult to detect in routine testing but are often marked by unique fusion genes providing a marker for these “other” haplotypes. By comparison with previous reports of similar structures, it is clear that some of these allele level haplotypes, such as the three gene-content contracted haplotype first described by Traherne et al. (23) will be found multiple times in the population. In contrast, other haplotypes such as those bearing the KIR2DL5A/3DP1 fusion gene (22;24;25), vary in their allele content suggesting that the same gene organization has arisen independently. Regardless, it is possible to identify the allele level consensus centromeric and telomeric haplotypes that gave rise to these expanded and contracted haplotypes.
KIR alleles are inherited on conserved haplotypes and it is this association of alleles on a haplotype that generates an immune response profile (Table 10). For example, the Cen-A1-I haplotype carries two inhibitory KIR with HLA-C ligands. KIR2DL3*001, a receptor with lower ligand binding affinity for HLA-C groups 1 and 2 compared to KIR2DL2 (28) is paired with KIR2DL1*00302 which strongly binds HLA-C group 2 (51). Likewise, Tel-A1 includes haplotypes with strong (KIR3DL1*00501, KIR3DL1*00101, consensus IV, VI), intermediate (KIR3DL1*01502, KIR3DL1*020, consensus I, II), and weak (KIR3DL1*007, consensus III) HLA-Bw4 binding. The presence of Tel-B1-I with Cen-B2 in an individual who expresses only HLA-C group 2 may weaken the inhibitory signal of KIR2DL2 by providing a stimulatory signal through the KIR2DS1 receptor with its similar ligand specificity (52;53). Some haplotypes carry apparently nonfunctional alleles such as Cen-B1 with KIR2DL5B and KIR2DS3 (42;54) or Tel-A1-V with two secreted receptors (KIR2DL4*00802, KIR2DS4*006)(55;56) and one predominantly misfolded receptor (KIR3DL1*00401)(57). The impacts of haplotypes encoding KIR receptors with unknown ligands (eg KIR2DL5) or where our understanding of any functional differences among allelic products is rudimentary (e.g., alleles of KIR3DL2) are yet to be determined. Furthermore, licensing induced by HLA allelic products in NK cells expressing these haplotypes will also impact immune activity (58). However, since KIR haplotypes are conserved, it will be possible to define immune profiles for these consensus haplotypes and apply this knowledge to our understanding of the role of KIR in health and disease.
Table 10.
Immune response profile1 of KIR Haplotypes
| Centromeric | |||||||
|---|---|---|---|---|---|---|---|
| Gene | 3DL3 | 2DS2 | 2DL2 | 2DL3 | 2DL1 | 2DL5B | 2DS3 |
| Ligand | ? | ? | HLA-Cg1/HLA-Cg2 (affinity for Cg2 is weak) | HLA-Cg1/HLA-Cg2 (ligand affinity lower than 2DL2) | HLA-Cg2 | ? | ? |
| Cen-A1-I | ++2 | Absent | Absent | Weak (001)3 | Strong (00302) | Absent | Absent |
| Cen-A1-II | ++ | Absent | Absent | ++ (002) | Strong (002) | Absent | Absent |
| Cen-A1-III | ++ | Absent | Absent | ++ (005) | Strong (001) | Absent | Absent |
| Cen-B1 | ++ | ++ | Strong (001) | Absent | Weak (00401) | Not expressed (0020101) | Misfolded (00103) |
| Cen-B2 | ++ | ++ | Strong (003) | Absent | Absent | Absent | Absent |
| Telomeric | |||||||||
| Gene | 2DL4 | 3DS1 | 3DL1 | 2DS4 | 2DS1 | 3DL2 | 2DL5A | 2DS3 | 2DS5 |
| Ligand | HLA-G | ? | HLA-Bw4 | HLA-A*1102 C subset | HLA-Cg2 | HLA-A3,11, B27d; CpG | ? | ? | ? |
| Tel-A1-I | ++ (00102) | Absent | Intermediate (01502) | ++ (00101) | Absent | ++ | Absent | Absent | Absent |
| Tel-A1-II | ++ (00103) | Absent | Intermediate (020) | Secreted (003) | Absent | ++ | Absent | Absent | Absent |
| Tel-A1-III | ++ (00602) | Absent | Weak (007)a | Secreted (004) | Absent | ++ | Absent | Absent | Absent |
| Tel-A1-IV | Secreted (011) | Absent | Strong (00501) | Secreted (007) | Absent | ++ | Absent | Absent | Absent |
| Tel-A1-V | Secreted (00802) | Absent | Misfolded (00401) | Secreted (006) | Absent | ++ | Absent | Absent | Absent |
| Tel-A1-VI | Secreted (0080101) | Absent | Strong (00101) | Secreted (003) | Absent | ++ | Absent | Absent | Absent |
| Tel-B1-I | ++ (00501) | ++ | Absent | Absent | Weak | ++ | ++ (0010101) | Absent | ++ |
| Tel-B1-II | ++ (00501) | ++ | Absent | Absent | Weak | ++ | ++ (0050101) | Misfolded (002) | Absent |
The characteristics of KIR allelic products and their ligands have been described: KIR2DL1 (51;73), KIR2DL2/KIR2DL3 (28), KIR2DL4 (11;55), KIR2DL5 (42), KIR2DS1 (52), KIR2DS3 (54), KIR2DS4 (9;56;74), KIR3DL1 (29;57); KIR3DL2 (6;75;76).
++ indicates gene is present for loci in which the functional impact of allelic variation has not been characterized.
Alleles are given in parentheses.
Materials and Methods
KIR alleles carried by 76 unrelated individuals with European ancestry were identified by DNA sequencing of the coding regions of all functional KIR loci and are described in previous publications (32- 39). These individuals were either donors or recipients of hematopoietic stem cells. The characteristics of this cohort have been previously described (59); KIR genotyping was performed retrospectively. The cells were obtained from the National Marrow Donor Program Research Repository.
PyPop (Python for Population genetics, version 0.7.0 http://www.pypop.org) was used to carry out Hardy-Weinberg testing for three KIR loci found in all haplotypes, one in the centromeric gene cluster and two in the telomeric cluster (60;61). Allele frequencies were determined by gene counting and assumed that each individual carried two copies of each locus. Individuals (8) carrying gene expanded/contracted haplotypes were not included in the analysis. Allele frequencies at each locus were evaluated for deviations from Hardy-Weinberg equilibrium proportions using the exact test of Guo and Thompson (62). Chi-square tests were investigated for a pooled set of all heterozygotes and a pooled set of all homozygotes (63). The tests measured the goodness of fit of KIR2DL2/2DL3, KIR3DL1/3DS1, and KIR3DL2 genotypes to Hardy-Weinberg proportions. This test measures the degree to which observed genotype frequencies differ from those expected based on the allele frequencies for that population, assuming that the population is suitably large and experiences random mating. No overall deviation from Hardy-Weinberg proportions were noted (p-values=0.9165, 0.4587, 0.7527, respectively).
Centromeric and telomeric haplotype structures were predicted based on the structures of Cen- A1, Cen-B1, Cen-B2, Tel-A1 and Tel-B1 (18). Cells were selected for further haplotype characterization based on these predictions. Haplotype specific extraction (HSE) (Qiagen, Valencia, CA) was used to isolate a DNA fragment carrying a single allele at a locus. Supplemental Table 1 lists the HSE and amplification reagents used. Once isolated, the alleles carried by the DNA fragment were identified by DNA sequencing for expressed genes and sequence-specific amplification for the pseudogenes and some fusion genes. For example, haplotypes predicted as Cen-A1 were confirmed by physically linking KIR3DL3 to KIR2DL3 using haplotype specific extraction with KIR3DL3 group specific probes (3DL3-1018A or 3DL3-161A) followed by DNA sequencing of segments of KIR3DL3 and KIR2DL3 coding regions found on that fragment of DNA. A similar strategy was used for cells carrying unexpected haplotype structures. Exceptions included the following: An extended polymerase chain reaction (PCR) (32) was performed to link KIR2DL2 to KIR2DL5 in haplotypes carrying Cen-B1 with DNA sequencing to identify the presence of the alleles at the loci. An extended PCR and sequencing were also used to link KIR3DS1 and KIR2DL5 in Tel-B1 haplotypes. Alleles in the second KIR haplotype in a cell were defined as those alleles not assigned by physical linkage to the first haplotype. In some cells, KIR2DL3-KIR2DL1 Cen-A1 haplotypes were assumed based on common associations. Amplification reactions were used to identify the presence of KIR2DP1, KIR3DP1, a KIR2DL5A-3DP1 fusion gene (24;64-66) and a 2DL3/2DP1 fusion gene (23)(Supplemental Table 1).
TaqMan copy number assays were used to measure KIR2DL4 copy number variation (Applied Biosystems, Inc., Foster City, CA). The 20ul reaction volume contained 4 ul (5 ng/ul) DNA, 10 ul TaqMan 2X PCR Master Mix, 1 ul 2DL4 CNV assay solution (Sense Primer- TCAGGACAAGCCCTTCTG, Antisense Primer- ACCCCATCTTTCTTGTACAGTG, and KIR2DL4-probe- CTGTGGTGCCTCAAGGAGG-labeled with FAM dye as previously described (22)), 1 ul reference assay RNaseP solution, and 4ul water. The quantitative PCR was performed using a StepOnePlus Real-time PCR cycler (Applied Biosystems, Inc.) with the following cycling conditions: 95°C for 10 min hold and 40 cycles of 95°C for 15 sec and 60°C for 60 sec. The results were analyzed using CopyCaller v1.0 (relative quantitation) (Applied Biosystems).
A mismatch distribution for exons 1-5 of the KIR3DL3 alleles using a constant population size was calculated using DnaSP v5 (67-69). SNAP (http://www.hiv.lanl.gov) was used to identify the number of nucleotide differences among the pairs. A Neighbor-Joining method (70) was used to infer the evolutionary history of haplotypes. The bootstrap consensus tree was inferred from 1000 replicates (71) using MEGA4 (72) with complete deletion to adjust for gaps and missing data and using a nucleotide--maximum composite likelihood model with uniform rates among sites. Independence of the centromeric and telomeric haplotype structures was tested using the exact option for the chi-square test in StatXact (Cytel Software Corp., Cambridge, MA).
Supplementary Material
Acknowledgments
This research is supported by funding from the Office of Naval Research N00014-08-1-1078 and N00014- 10-1-0199. The views expressed in this article are those of the authors and do not reflect the official policy or position of the Department of the Navy, the Department of Defense, or the U.S. government. We would like to thank Rebecca Slack for her assistance.
Footnotes
Conflict of interest
The authors have no competing financial interests related to this work.
References
- 1.Bryceson YT, Long EO. Line of attack: NK cell specificity and integration of signals. Curr Opin Immunol. 2008;20(3):344–352. doi: 10.1016/j.coi.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–274. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- 3.Moretta A, Vitale M, Bottino C, Orengo AM, Morelli L, Augugliaro R, et al. P58 molecules as putative receptors for major histocompatibility complex (MHC) class I molecules in human natural killer (NK) cells. Anti-p58 antibodies reconstitute lysis of MHC class I-protected cells in NK clones displaying different specificities. J Exp Med. 1993;178(2):597–604. doi: 10.1084/jem.178.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colonna M, Borsellino G, Falco M, Ferrara GB, Strominger JL. HLA-C is the inhibitory ligand that determines dominant resistance to lysis by NK1- and NK2-specific natural killer cells. Proc Natl Acad Sci U S A. 1993;90(24):12000–12004. doi: 10.1073/pnas.90.24.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med. 1994;180(2):537–543. doi: 10.1084/jem.180.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kollnberger S, Chan A, Sun MY, Chen LY, Wright C, di Gleria K, et al. Interaction of HLA-B27 homodimers with KIR3DL1 and KIR3DL2, unlike HLA-B27 heterotrimers, is independent of the sequence of bound peptide. Eur J Immunol. 2007;37(5):1313–1322. doi: 10.1002/eji.200635997. [DOI] [PubMed] [Google Scholar]
- 7.Winter CC, Gumperz JE, Parham P, Long EO, Wagtmann N. Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol. 1998;161(2):571–577. [PubMed] [Google Scholar]
- 8.Biassoni R, Falco M, Cambiaggi A, Costa P, Verdiani S, Pende D, et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by “group 2” or “group 1” NK clones. J Exp Med. 1995;182(2):605–609. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graef T, Moesta AK, Norman PJ, Abi-Rached L, Vago L, Older Aguilar AM, et al. KIR2DS4 is a product of gene conversion with KIR3DL2 that introduced specificity for HLA-A*11 while diminishing avidity for HLA-C. J Exp Med. 2009;206(11):2557–2572. doi: 10.1084/jem.20091010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kikuchi-Maki A, Yusa S, Catina TL, Campbell KS. KIR2DL4 is an IL-2-regulated NK cell receptor that exhibits limited expression in humans but triggers strong IFN-gamma production. J Immunol. 2003;171(7):3415–3425. doi: 10.4049/jimmunol.171.7.3415. [DOI] [PubMed] [Google Scholar]
- 11.Rajagopalan S, Bryceson YT, Kuppusamy SP, Geraghty DE, van der MA, Joosten I, et al. Activation of NK cells by an endocytosed receptor for soluble HLA-G. PLoS Biol. 2006;4(1):e9. doi: 10.1371/journal.pbio.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- 13.Valiante NM, Uhrberg M, Shilling HG, Lienert-Weidenbach K, Arnett KL, D’Andrea A, et al. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity. 1997;7(6):739–751. doi: 10.1016/s1074-7613(00)80393-3. [DOI] [PubMed] [Google Scholar]
- 14.Norman PJ, Stephens HA, Verity DH, Chandanayingyong D, Vaughan RW. Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics. 2001;52(3-4):195–205. doi: 10.1007/s002510000281. [DOI] [PubMed] [Google Scholar]
- 15.Hsu KC, Chida S, Dupont B, Geraghty DE. The killer cell immunoglobulin-like receptor (KIR) genomic region: gene-order, haplotypes and allelic polymorphism. Immunological Reviews. 2002;190(1):40–52. doi: 10.1034/j.1600-065x.2002.19004.x. [DOI] [PubMed] [Google Scholar]
- 16.Ordonez D, Meenagh A, Gomez-Lozano N, Castano J, Middleton D, Vilches C. Duplication, mutation and recombination of the human orphan gene KIR2DS3 contribute to the diversity of KIR haplotypes. Genes Immun. 2008;9(5):431–437. doi: 10.1038/gene.2008.34. [DOI] [PubMed] [Google Scholar]
- 17.Gomez-Lozano N, Gardiner CM, Parham P, Vilches C. Some human KIR haplotypes contain two KIR2DL5 genes: KIR2DL5A and KIR2DL5B. Immunogenetics. 2002;54:314–319. doi: 10.1007/s00251-002-0476-2. [DOI] [PubMed] [Google Scholar]
- 18.Pyo CW, Guethlein LA, Vu Q, Wang R, Abi-Rached L, Norman PJ, et al. Different patterns of evolution in the centromeric and telomeric regions of group A and B haplotypes of the human killer cell Ig-like receptor locus. PLoS ONE. 2010;5(12):e15115. doi: 10.1371/journal.pone.0015115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, et al. Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci U S A. 2000;97(9):4778–4783. doi: 10.1073/pnas.080588597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Middleton D, Meenagh A, Gourraud PA. KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics. 2007;59(2):145–158. doi: 10.1007/s00251-006-0181-7. [DOI] [PubMed] [Google Scholar]
- 21.Gourraud PA, Meenagh A, Cambon-Thomsen A, Middleton D. Linkage disequilibrium organization of the human KIR superlocus: implications for KIR data analyses. Immunogenetics. 2010;62(11-12):729–740. doi: 10.1007/s00251-010-0478-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. 2003;171(5):2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- 23.Traherne JA, Martin M, Ward R, Ohashi M, Pellett F, Gladman D, et al. Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet. 2010;19(5):737–751. doi: 10.1093/hmg/ddp538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomez-Lozano N, Estefania E, Williams F, Halfpenny I, Middleton D, Solis R, et al. The silent KIR3DP1 gene (CD158c) is transcribed and might encode a secreted receptor in a minority of humans, in whom the KIR3DP1, KIR2DL4 and KIR3DL1/KIR3DS1 genes are duplicated. Eur J Immunol. 2005;35(1):16–24. doi: 10.1002/eji.200425493. [DOI] [PubMed] [Google Scholar]
- 25.Norman PJ, Abi-Rached L, Gendzekhadze K, Hammond JA, Moesta AK, Sharma D, et al. Meiotic recombination generates rich diversity in NK cell receptor genes, alleles, and haplotypes. Genome Res. 2009;19(5):757–769. doi: 10.1101/gr.085738.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson J, Mistry K, McWilliam H, Lopez R, Marsh SG. IPD--the Immuno Polymorphism Database. Nucleic Acids Res. 2010;38(Database issue):D863–D869. doi: 10.1093/nar/gkp879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.VandenBussche CJ, Dakshanamurthy S, Posch PE, Hurley CK. A single polymorphism disrupts the killer Ig-like receptor 2DL2/2DL3 D1 domain. J Immunol. 2006;177(8):5347–5357. doi: 10.4049/jimmunol.177.8.5347. [DOI] [PubMed] [Google Scholar]
- 28.Moesta AK, Norman PJ, Yawata M, Yawata N, Gleimer M, Parham P. Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol. 2008;180(6):3969–3979. doi: 10.4049/jimmunol.180.6.3969. [DOI] [PubMed] [Google Scholar]
- 29.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203(3):633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the bi-directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet. 2008;4(11):e1000254. doi: 10.1371/journal.pgen.1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shilling HG, Guethlein LA, Cheng NW, Gardiner CM, Rodriguez R, Tyan D, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168(5):2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 32.Hou L, Chen M, Jiang B, Wu D, Ng J, Hurley CK. Thirty allele-level haplotypes centered around KIR2DL5 define the diversity in an African American population. Immunogenetics. 2010;62:491–498. doi: 10.1007/s00251-010-0458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou L, Steiner NK, Chen M, Belle I, Kubit AL, Ng J, et al. Limited Allelic Diversity of Stimulatory Two Domain Killer Immunoglobulin-Like Receptors. Human Immunology. 2008;69:174–178. doi: 10.1016/j.humimm.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 34.Mulrooney TJ, Hou L, Steiner NK, Chen M, Belle I, Ng J, et al. Promoter variants of KIR2DL5 add to diversity and may impact gene expression. Immunogenetics. 2008;60(6):287–294. doi: 10.1007/s00251-008-0273-7. [DOI] [PubMed] [Google Scholar]
- 35.Belle I, Hou L, Chen M, Steiner NK, Ng J, Hurley CK. Investigation of killer cell immunoglobulinlike receptor gene diversity in KIR3DL1 and KIR3DS1 in a transplant population. Tissue Antigens. 2008;71(5):434–439. doi: 10.1111/j.1399-0039.2008.01017.x. [DOI] [PubMed] [Google Scholar]
- 36.Hou L, Chen M, Steiner NK, Belle I, Turino C, Ng J, et al. Seventeen novel alleles add to the already extensive KIR3DL3 diversity. Tissue Antigens. 2007;70(6):449–454. doi: 10.1111/j.1399-0039.2007.00930.x. [DOI] [PubMed] [Google Scholar]
- 37.Gedil MA, Steiner NK, Hurley CK. KIR3DL2: diversity in a hematopoietic stem cell transplant population. Tissue Antigens. 2007;70(3):228–232. doi: 10.1111/j.1399-0039.2007.00880.x. [DOI] [PubMed] [Google Scholar]
- 38.Hou LH, Steiner NK, Chen M, Belle I, Ng J, Hurley CK. KIR2DL1 allelic diversity: four new alleles characterized in a bone marrow transplant population and three families. Tissue Antigens. 2007;69(3):250–254. doi: 10.1111/j.1399-0039.2006.00793.x. [DOI] [PubMed] [Google Scholar]
- 39.Shulse C, Steiner NK, Hurley CK. Allelic diversity in KIR2DL4 in a bone marrow transplant population: description of three novel alleles. Tissue Antigens. 2007;70(2):157–159. doi: 10.1111/j.1399-0039.2007.00864.x. [DOI] [PubMed] [Google Scholar]
- 40.Jones DC, Hiby SE, Moffett A, Trowsdale J, Young NT. Nature of allelic sequence polymorphism at the KIR3DL3 locus. Immunogenetics. 2006;58(8):614–627. doi: 10.1007/s00251-006-0130-5. [DOI] [PubMed] [Google Scholar]
- 41.Norman PJ, Abi-Rached L, Gendzekhadze K, Korbel D, Gleimer M, Rowley D, et al. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat Genet. 2007;39(9):1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- 42.Gomez-Lozano N, Trompeter HI, de Pablo R, Estefania E, Uhrberg M, Vilches C. Epigenetic silencing of potentially functional KIR2DL5 alleles: Implications for the acquisition of KIR repertoires by NK cells. Eur J Immunol. 2007;37(7):1954–1965. doi: 10.1002/eji.200737277. [DOI] [PubMed] [Google Scholar]
- 43.Ordonez D, Gomez-Lozano N, Vilches C. The 5’ intergenic, promoter, pseudoexon 3 and complete coding sequences of the hybrid gene KIR2DS3*002. Tissue Antigens. 2008;72(5):504–505. doi: 10.1111/j.1399-0039.2008.01126.x. [DOI] [PubMed] [Google Scholar]
- 44.Williams F, Maxwell LD, Halfpenny IA, Meenagh A, Sleator C, Curran MD, et al. Multiple copies of KIR 3DL/S1 and KIR 2DL4 genes identified in a number of individuals. Hum Immunol. 2003;64(7):729–732. doi: 10.1016/s0198-8859(03)00089-2. [DOI] [PubMed] [Google Scholar]
- 45.Martin AM, Kulski JK, Gaudieri S, Witt CS, Freitas EM, Trowsdale J, et al. Comparative genomic analysis, diversity and evolution of two KIR haplotypes A and B. Gene. 2004;335:121–131. doi: 10.1016/j.gene.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 46.Wall JD, Pritchard JK. Haplotype blocks and linkage disequilibrium in the human genome. Nat Rev Genet. 2003;4(8):587–597. doi: 10.1038/nrg1123. [DOI] [PubMed] [Google Scholar]
- 47.Kauppi L, Sajantila A, Jeffreys AJ. Recombination hotspots rather than population history dominate linkage disequilibrium in the MHC class II region. Hum Mol Genet. 2003;12(1):33–40. doi: 10.1093/hmg/ddg008. [DOI] [PubMed] [Google Scholar]
- 48.Torkar M, Norgate Z, Colonna M, Trowsdale J, Wilson MJ. Isotypic variation of novel immunoglobulin-like transcript/killer cell inhibitory receptor loci in the leukocyte receptor complex. Eur J Immunol. 1998;28(12):3959–3967. doi: 10.1002/(SICI)1521-4141(199812)28:12<3959::AID-IMMU3959>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 49.Trundley AE, Hiby SE, Chang C, Sharkey AM, Santourlidis S, Uhrberg M, et al. Molecular characterization of KIR3DL3. Immunogenetics. 2006;57(12):904–916. doi: 10.1007/s00251-005-0060-7. [DOI] [PubMed] [Google Scholar]
- 50.Rajalingam R, Parham P, Abi-Rached L. Domain shuffling has been the main mechanism forming new hominoid killer cell Ig-like receptors. J Immunol. 2004;172(1):356–369. doi: 10.4049/jimmunol.172.1.356. [DOI] [PubMed] [Google Scholar]
- 51.Bari R, Bell T, Leung WH, Vong QP, Chan WK, Das GN, et al. Significant functional heterogeneity among KIR2DL1 alleles and a pivotal role of arginine 245. Blood. 2009;114(25):5182–5190. doi: 10.1182/blood-2009-07-231977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moesta AK, Graef T, Abi-Rached L, Older Aguilar AM, Guethlein LA, Parham P. Humans differ from other hominids in lacking an activating NK cell receptor that recognizes the C1 epitope of MHC class I. J Immunol. 2010;185(7):4233–4237. doi: 10.4049/jimmunol.1001951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sivori S, Carlomagno S, Falco M, Romeo E, Moretta L, Moretta A. Natural killer cells expressing the KIR2DS1 activating receptor efficiently kill T cell blasts and dendritic cells: implications in haploidentical HSCT. Blood. 2011;117(16):4284–4292. doi: 10.1182/blood-2010-10-316125. [DOI] [PubMed] [Google Scholar]
- 54.VandenBussche CJ, Mulrooney TJ, Frazier WR, Dakshanamurthy S, Hurley CK. Dramatically reduced surface expression of NK cell receptor KIR2DS3 is attributed to multiple residues throughout the molecule. Genes and Immunity. 2009;10:162–173. doi: 10.1038/gene.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goodridge JP, Lathbury LJ, Steiner NK, Shulse CN, Pullikotil P, Seidah NG, et al. Three common alleles of KIR2DL4 (CD158d) encode constitutively expressed, inducible and secreted receptors in NK cells. Eur J Immunol. 2007;37(1):199–211. doi: 10.1002/eji.200636316. [DOI] [PubMed] [Google Scholar]
- 56.Middleton D, Gonzalez A, Gilmore PM. Studies on the expression of the deleted KIR2DS4*003 gene product and distribution of KIR2DS4 deleted and nondeleted versions in different populations. Hum Immunol. 2007;68(2):128–134. doi: 10.1016/j.humimm.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 57.Taner SB, Pando MJ, Roberts A, Schellekens J, Marsh SG, Malmberg KJ, et al. Interactions of NK cell receptor KIR3DL1*004 with chaperones and conformation-specific antibody reveal a functional folded state as well as predominant intracellular retention. J Immunol. 2011;186(1):62–72. doi: 10.4049/jimmunol.0903657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jonsson AH, Yokoyama WM. Natural killer cell tolerance licensing and other mechanisms. Adv Immunol. 2009;101:27–79. doi: 10.1016/S0065-2776(08)01002-X. [DOI] [PubMed] [Google Scholar]
- 59.Shah R, Selby ST, Yokley B, Slack RS, Hurley CK, Posch PE. TNF, LTA and TGFB1 genotype distributions among acute graft-vs-host disease subsets after HLA-matched unrelated hematopoietic stem cell transplantation: a pilot study. Tissue Antigens. 2009;74(1):50–56. doi: 10.1111/j.1399-0039.2009.01257.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lancaster AK, Single RM, Solberg OD, Nelson MP, Thomson G. PyPop update - a software pipeline for large-scale multilocus population genomics. Tissue Antigens. 2007;69(Suppl 1):192–197. doi: 10.1111/j.1399-0039.2006.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lancaster AK, Nelson MP, Single RM, Meyer D, Thomson G. PyPop: a software framework for population genomics: analyzing large-scale multi-locus genotype data. In: Altman RB, Dunker K, Hunter L, Jung T, Klein T, editors. Pacific Symposium on Biocomputing. Vol. 8. Singapore: World Scientific; 2003. pp. 514–525. [PMC free article] [PubMed] [Google Scholar]
- 62.Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- 63.Meyer D, Single R, Mack SJ, Lancaster A, Nelson MP, Erlich HA, et al. 13th IHWS anthropology/human genetic diversity joint report. Single locus polymorphism of classical HLA genes. In: Hansen JA, editor. Immunobiology of the human MHC. Chapter 4. Seattle: International Histocompatibility Working Group Press; 2006. pp. 653–704. [Google Scholar]
- 64.Vilches C, Castano J, Gomez-Lozano N, Estefania E. Facilitation of KIR genotyping by a PCR-SSP method that amplifies short DNA fragments. Tissue Antigens. 2007;70(5):415–422. doi: 10.1111/j.1399-0039.2007.00923.x. [DOI] [PubMed] [Google Scholar]
- 65.Vilches C, Gardiner CM, Parham P. Gene structure and promoter variation of expressed and nonexpressed variants of the KIR2DL5 gene. J Immunol. 2000;165(11):6416–6421. doi: 10.4049/jimmunol.165.11.6416. [DOI] [PubMed] [Google Scholar]
- 66.Gomez-Lozano N, Vilches C. Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: An update. Tissue Antigens. 2002;59:184–193. doi: 10.1034/j.1399-0039.2002.590302.x. [DOI] [PubMed] [Google Scholar]
- 67.Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9(3):552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 68.Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129(2):555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 70.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 71.Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 72.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 73.Mandelboim O, Reyburn HT, Vales-Gomez M, Pazmany L, Colonna M, Borsellino G, et al. Protection from lysis by natural killer cells of group 1 and 2 specificity is mediated by residue 80 in human histocompatibility leukocyte antigen C alleles and also occurs with empty major histocompatibility complex molecules. J Exp Med. 1996;184(3):913–922. doi: 10.1084/jem.184.3.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Katz G, Markel G, Mizrahi S, Arnon TI, Mandelboim O. Recognition of HLA-Cw4 but not HLA-Cw6 by the NK cell receptor killer cell Ig-like receptor two-domain short tail number 4. J Immunol. 2001;166:7260–7267. doi: 10.4049/jimmunol.166.12.7260. [DOI] [PubMed] [Google Scholar]
- 75.Sivori S, Falco M, Carlomagno S, Romeo E, Soldani C, Bensussan A, et al. A novel KIR-associated function: evidence that CpG DNA uptake and shuttling to early endosomes is mediated by KIR3DL2. Blood. 2010;116(10):1637–1647. doi: 10.1182/blood-2009-12-256586. [DOI] [PubMed] [Google Scholar]
- 76.Hansasuta P, Dong T, Thananchai H, Weekes M, Willberg C, Aldemir H, et al. Recognition of HLAA3 and HLA-A11 by KIR3DL2 is peptide-specific. Eur J Immunol. 2004;34(6):1673–1679. doi: 10.1002/eji.200425089. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


