Abstract
Reconstructed microbial metabolic networks facilitate a mechanistic description of the genotype-phenotype relationship through the deployment of methods in constraint-based reconstruction and analysis (COBRA). Since reconstructed networks leverage genomic data for insight and phenotype prediction, the development of COBRA methods has accelerated, following the advent of whole-genome sequencing. Here, we describe a phylogeny of COBRA methods that has rapidly evolved from early methods, such as flux balance analysis and elementary flux mode analysis, into a repertoire of more than 100 methods. These methods have enabled genome-scale analysis of microbial metabolism for numerous basic and applied uses, including antibiotic discovery, metabolic engineering, and modeling of microbial community behavior.
Introduction
The genotype-phenotype relationship is fundamental to biology. For decades this relationship has been subject to mostly argument, speculation and qualitative analysis. However, our ability to fundamentally understand the genotype-phenotype relationship began changing in the mid-1990s, on completion of the first bacterial genome sequencing projects. Full genome sequences provide comprehensive, albeit not yet complete, information about the genetic elements that create an organism. The comprehensive understanding for some cellular processes, such as metabolism, has resulted in structured knowledge-bases that can be mathematically represented1–3. This mathematical representation enables the computation of phenotypic states4–7 based on genetic and environmental parameters. Remarkably, this provides a mechanistic representation of the microbial metabolic genotype-phenotype relationship.
Constraint-based models of genome-scale metabolic networks capture the genotype-phenotype relationship by simultaneously accounting for constraints on phenotype imposed by physicochemical laws and genetics. The realization that these quantitative genotype-phenotype relationships could be constructed from a genome has driven the emergence of this area of research, and the flood of increasingly rich high-throughput data has accelerated the evolution of constraint-based reconstruction and analysis (COBRA) methods from a set of basic tools for metabolic network analysis into a powerful analytical framework that is increasingly used. Here, we describe basic features of the COBRA framework, the ‘phylogeny’ of evolving COBRA methods, and the COBRA ‘ecology,’ i.e., how COBRA methods complement each other in answering larger questions in biology.
Constraint-based modeling defined
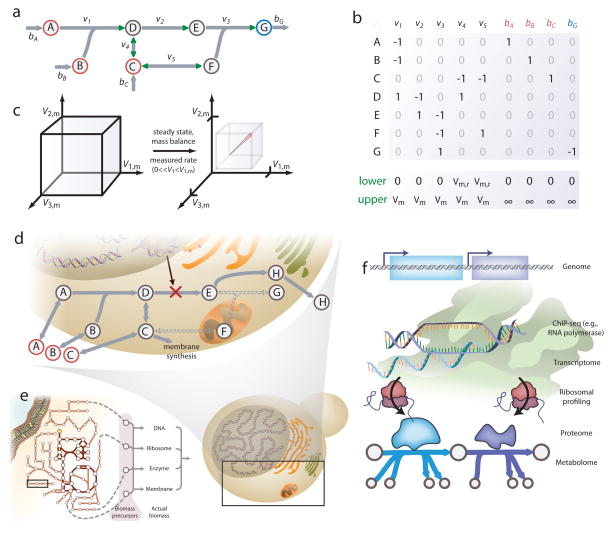
The COBRA approach is based on a few fundamental concepts. These concepts include the imposition of physicochemical constraints that limit computable phenotypes (Figure 1.a–d), the identification and mathematical description of evolutionary selective pressures (Figure 1.e), and a genome-scale perspective of cell metabolism that accounts of all metabolic gene products in a cell (Figure 1.d,f). These fundamental concepts are briefly described here.
Figure 1. Fundamentals of the genome-scale metabolic genotype-phenotype relationship.
The COBRA approach is based on three primary fundamental concepts: network constraints (a–d), objective functions (e), and the association of reactions with the genome. (a) A complex mixture of molecules (red) can react to yield end products (blue). (b) The stoichiometry of this reaction network is described mathematically in a stoichiometric matrix, with each column representing the stoichiometry of a reaction. Negative and positive values represent reactants and products, respectively. Reaction flux is limited by thermodynamics and catalytic capacities (Vm=Vmax), described by upper and lower bounds on flux for each reaction (green). (c) Reaction constraints result in a “solution space” that contains all feasible flux distributions. Additional constraints (e.g., mass balance, the steady-state assumption, and measured metabolite consumption rates) reduce the space of feasible flux distributions, as shown by the pink line. (d) In vivo biochemical networks involve additional complexity. Gene regulation can change the abundance of catalysts (e.g., the transformation of D to E). Often components are also localized in different organelles (e.g., E and F), thereby blocking reactions. (e) The biomass objective function describes an evolutionary pressure for microbial growth, and describes the metabolic demands to make basic metabolite building blocks for all cellular components (e.g., membranes, macromolecules, ATP, etc.). (f) The association of metabolism with the genome is done by mathematically linking the genome to transcripts, proteins, and chemical reactions. The gene-protein-reaction schema is used to describe gene association in the models, and provide an interface for the integration of high-throughput data.
Constraints on reaction networks
Metabolism is a complex network of biochemical reactions. The reaction occurrence is limited by three primary constraints: reaction substrate and enzyme availability, mass and charge conservation, and thermodynamics. For metabolism, reaction substrates must be present in a cell’s microenvironment or produced from other reactions, and enzymes must be available. Mass conservation further limits the possible reaction products and their stoichiometry, while thermodynamics constrain reaction directionality. For a given organism, this information can be obtained from careful biochemical and genetic studies or inferred from related organisms, and then catalogued in metabolic reconstruction knowledgebases1, 2.
In the COBRA framework, a metabolic reconstruction is converted into an in silico model by mathematically describing the reactions and adding network inputs and outputs (e.g., uptake and secretion products). Much like a cell has one genome and many transcriptional states, an organism has one metabolic reconstruction from which context-specific models can be derived, each representing cellular functions under different conditions.
Physicochemical constraints on the metabolic network are mathematically described by a matrix representing the stoichiometric coefficients of each reaction (Figure 1.a–b)8. Known upper and lower bounds on each reaction flux are imposed as additional constraints. Mathematically, these constraints define a multi-dimensional “solution space” of allowable reaction flux distributions, and the actual expressed flux state resides in this solution space. Additional constraints can further shrink the solution space to focus in on the actual flux state of the network (Figure 1.c). These additional constraints may include enzyme capacity, spatial localization, metabolite sequestration, and multiple levels of gene, transcript, and protein regulation (Figure 1.d).
Mathematical statement of cell objectives: a reflection of evolution
In non-biological chemical networks, the material flow through pathways can be predicted in a “cause and effect” manner, using mathematical models that describe the associated physical laws. This description can be achieved in a “time-invariant” manner, since reproducing the same physical conditions will drive flux through the same pathways. By contrast, causation in biology is “time-variant”. A plethora of chemical reactions may occur inside a cell, and many “pathways” can link a starting molecule to a given product. However, regulatory mechanisms have evolved to select when and where pathways will be used in an organism under a given condition. Thus, if the cellular objectives that drive evolution are understood or can be inferred, optimal flux states of biochemical reaction networks can be predicted. In the COBRA framework these cellular objectives are described mathematically and used for computation of phenotypic states.
Many cellular objectives can be defined in the context of metabolism. For example, as a proxy for growth, a biomass reaction9 can be defined that contains all necessary precursors for synthesizing the cell components for growth (e.g., with amounts of amino acids for proteins and the nucleic acids for RNA) (Figure 1.e). The biomass function and other objective functions can be used with optimization algorithms, such as linear programming10, 11 to predict metabolic pathway usage and cellular phenotypes11. Since these objective functions mathematically state cellular aims and can predict phenotypes, they capture pressures guiding evolution, and therefore represent a determinant of causation in biology. The objective function is thus an important part of the COBRA framework. It is not based on fundamental physical principles, but based on biological functions that are selected for over many generations.
A genome-wide basis for modeling metabolism
Constraint-based modeling has rapidly developed since the advent of whole-genome sequencing12, 13. A genome provides the genetic basis for an organism’s metabolic network, and genome annotation defines the relationships between genes, enzymes, and the reactions they catalyze (Figure 1.f)14. Annotated genomes and their associated biochemical and genetic data have facilitated the development of carefully curated metabolic network reconstructions containing thousands of reactions. When a reconstruction knowledgebase for an organism is converted into a genome-scale model (GEM), the mathematical representation provides constraints, and the objective function can be used to represent the optimal biological functions the organism strives to achieve. Thus, simulation of an organism’s phenotypes can be performed using its GEM.
The genome-scale view of metabolism of these models has two primary implications. First, in principle, they account for all known metabolic genes in a cell. Thus, when used in genome-scale dataset analysis (e.g., proteomics, metabolomics, etc.)15, they provide novel insight since they account for real chemical connections between components (Figure 1.f). Second, since metabolic genes are associated with the biochemical functions of their gene products, simulations of metabolite flow through the network can provide mechanistic predictions of how each gene product affects the metabolic network function. Thus, cell phenotypes can be computed and data can be interpreted with GEMs, thereby providing mechanistic insight into how the cell genotype may contribute to the cell phenotype.
A phylogeny of constraint-based methods
COBRA methods have ‘evolved’ and ‘diversified’ over the past decade, leading to more than 100 different methods (Supplementary Table 1 and http://sourceforge.net/apps/mediawiki/opencobra/), many of which have been implemented in available software packages (Supplementary Table 2). These developments may be likened to an evolutionary process, in which specific scientific questions have selected for algorithmic innovations, yielding a phylogenetic tree of COBRA methods (Figure 2). We classify these methods into major groups and describe examples that address the broader scientific questions.
Figure 2. The “phylogeny” of constraint-based modeling methods.
Over the past years, the constraint-based modeling community has rapidly expanded. Because of the versatility and scalability of these models, more than 100 methods have been developed for their modeling and analysis, all based on the analysis of the underlying metabolic network structure (i.e., the stoichiometric matrix). A phylogenetic tree is used to depict the similarities between application and use of the methods, and the underlying algorithms for many of the methods. See Supplementary Table 1 for a more complete list of methods and descriptions of methods.
Global characterization of solution spaces
Metabolic pathways are conceptual abstractions that group reactions. However, sometimes these “pathways” fail to reflect actual metabolic network usage16, since textbook pathways often reflect the order of enzyme discovery or pathway usage in one model organism. Fortunately, through computational analysis of metabolic networks, the required “pathways” for specific metabolic functions can be identified without biases from traditional pathway concepts. In constraint-based modeling, this is approached through unbiased and biased methods, represented by the two primary branches of the phylogenetic tree (Figure 2). Unbiased methods describe all steady-state flux distributions, including reaction sets that function together without belonging to the same traditional pathway concepts.
Two such unbiased approaches, elementary flux mode (EFM) analysis and extreme pathway (ExPa) analysis, globally characterize allowable phenotypes, and have been reviewed and compared previously17–19. These methods identify reaction sets (i.e., pathways) that achieve specific metabolic functions, and combinations of these reaction sets describe the entire solution space (i.e., all steady-state phenotypes). These methods have enjoyed many applications. For example, in studying E. coli metabolism, they have helped assess global pathway regulation20, facilitated the design of an ethanol-secreting strain21, identified synthetic lethal gene interactions22, and demonstrated the trade-off between reducing translation costs and rapidly responding to environmental changes23. These methods are usually applied to small models or portions of GEMs24, since their computational complexity scales exponentially25, 26. However, their use on larger models is becoming possible through simplifications that, for example, calculate a subset of potential pathways or find minimal pathways that accomplish a biological function27–30.
Alternative approaches can also describe the entire “solution space” in an unbiased fashion31, 32. For example, Markov-chain Monte Carlo sampling (MCMC) methods32 characterize all feasible steady-state reaction fluxes. This provides a probability distribution of feasible fluxes for each reaction under the user-provided growth conditions. These methods have provided insight into several biological properties, such as the high flux backbone of central metabolism in E. coli33, condition-specific regulation of yeast34, 35 and E. coli36 metabolism, and disease states in cardiac myocytes37, erythrocytes38, and the human brain39.
Finding the ‘optimal’ metabolic state with FBA methods
EFM, ExPa, and MCMC methods characterize all flux states a metabolic network can deploy. However, a cell does not use most possible flux states. Thus, biased COBRA methods include the optimization of an objective function to identify physiologically relevant flux distributions. Flux Balance Analysis (FBA) is the most basic and commonly used biased method for simulating genome-scale metabolism. In FBA, the cellular objective is defined, and metabolites in the media are supplied to the metabolic network. Using linear programming, an objective function is optimized (e.g., the biomass objective function) subject to the constraints imposed by the metabolic network and metabolite uptake rates10, 11, 40. This calculation finds one solution in the solution space that is believed to best represent the true cellular phenotype. The solution includes a prediction of the optimal objective magnitude (e.g., biomass yield or growth rate) and potential flux values for each reaction (Figure 3.a).
Figure 3. Flux balance analysis (FBA).
(a) In FBA, a cellular objective (e.g., biomass production) is optimized. This provides the predicted flux for each reaction in the network. (b) FBA solutions are typically not unique, i.e., there are alternate optimal solutions that use different pathways to achieve the same objective value (e.g., growth rate). (c) Additional constraints can be applied to reduce the solution space size, and may remove competing optimal solutions, or (d) change the optimal solution. If the optimal solution is moved, then the choice of the new optimal solution may depend on the solver and/or algorithm, as shown for the MOMA50 method. (e) The addition of constraints can enhance predictions. For example, when constraints on molecular crowding are added, the model-predicted order of substrate metabolism is consistent with experimental observation. Panel e reproduced from57, Copyright 2007, National Academy of Sciences, USA. NTPs, nucleotide triphosphates; AAs, amino acids; FVA, flux variability analysis; v, reaction flux; μmax, predicted maximum growth rate.
FBA successfully makes quantitative predictions using a few governing constraints on the model. For example, a pre-genome era application of FBA recapitulated the acetate overflow phenotype of E. coli41, in which acetate is excreted at high growth rates. Using GEMs, FBA has since predicted growth rates42, pathway usage43, 44, and the effect of gene expression noise on fitness45. It allowed the analysis complex phenotypes, such as metabolism in non-growing cells46, and numerous variations on FBA have been developed to assess alternative optimal solutions or to account for additional constraints on metabolic flux in cells (Figure 2).
Predicted flux values from FBA can vary due to alternate optimal solutions (i.e., the same objective value using different reactions) (Figure 3.b–d). Alternate optimal solutions are enumerated using mixed-integer linear programming (MILP)47 and the ranges spanned by alternate optima are found for each reaction using flux variability analysis (Figure 3.b)48, 49. The consideration of all alternate optima is crucial when interpreting an FBA solution, since the flux through a single reaction can vary considerably depending on which solution is found. For example, the COBRA method Minimization of Metabolic Adjustment (MOMA)50 predicts a new flux vector and objective value after a perturbation (e.g., gene deletion). To do this, MOMA computes one “wild type” FBA solution, and finds the nearest solution after perturbing the network (i.e., the minimum change to reaction fluxes from the FBA solution). Since the new predicted flux vector and growth rate can differ considerably depending on which alternate optimal solution is used (Figure 3.d), all possible results from alternate optima must be assessed.
To identify realistic microbial phenotypes in FBA predictions, additional biologically-relevant constraints have been proposed. These include constraints imposed by economy in enzyme usage43, 51–53, metabolite dilution54, and changes in transcript level55, 56. These FBA refinements further decrease the range of feasible reaction fluxes to obtain solutions closely resembling cellular physiology under certain growth conditions. For example, constraints from enzyme crowding have been applied to FBA solutions (FBAwMC)57, 58. In FBAwMC, reaction flux is constrained to reflect internal spatial limitations on enzyme abundance in the crowded cytoplasm. This method predicted that molecular crowding contributes to substrate preferences in E. coli57. In a medium with multiple carbon substrates, FBAwMC accurately predicted that glucose would be preferentially consumed, followed by mixed-substrate consumption and a late usage of glycerol and the excreted acetate (Figure 3.e), suggesting that molecular crowding may contribute to substrate preference. A similar variation on FBA accounts for cytoplasmic membrane crowding (FBAME) by limiting the flux through the glucose transporter and the three cytochromes in E. coli59. This constraint recapitulated the simultaneous use of respiratory and fermentative pathways and predicted the effect of glucose and oxygen availability on cytochrome oxidase expression. Thus, the imposition of crowding constraints on metabolic flux has provided additional insights into cell physiology57–59.
Modeling genetic perturbations
Since genome-scale metabolic networks capture the activities of hundreds of enzymes, mutant phenotypes can be assayed through in silico gene perturbation and simulation. On the first GEMs12, 13, such approaches demonstrated the predictive power of COBRA methods when metabolic genes were “knocked out” in the model by restricting flux through their associated reactions. When growth of mutant E. coli was simulated with FBA, 86% of the mutant phenotypes (i.e., growth or no growth) were accurately predicted13. This success rate exceeded any other phenotype-predicting algorithm at the time. Subsequent studies have identified growth conditions60 and genetic backgrounds61 for which genes in S. cerevisiae are conditionally essential. For example, combinations of gene knockouts were simulated and tested for essentiality. This demonstrated that 74% of yeast metabolic genes contribute to essential metabolic processes, and most of these are masked by isozymes and alternative pathways61. To address additional questions concerning gene deletion, new methods have been introduced such as MOMA50, Regulatory On/Off Minimization62, and Metabolite Essentiality Analysis (MEA)63(Figure 2).
Gene and reaction perturbation studies have aided health-related applications, such as predicting metabolic side-effects of off-target protein-drug interactions64 and predicting novel anti-microbial targets65. For example, MEA63 was applied to the Vibrio vulnificus GEM66 to identify potential antibiotic targets for this pathogenic relative to Vibrio cholerae. MEA was used since it identifies metabolites that, if removed, inhibit biomass production. These metabolites could possibly be blocked in vivo with analogues that bind or modify active sites on their associated enzymes. This analysis identified five metabolites as potential antibiotic targets. Thus, only 352 analogues had to be tested for antimicrobial properties, allowing for a smaller screen than commonly required for drug discovery. One of screened molecules with antimicrobial properties was subjected to additional study, and this candidate molecule considerably out-performed sulfamethoxazole, an existing therapeutic for V. vulnificus infection. Although additional drug safety assessment and optimization is required for this candidate drug, this study demonstrates how COBRA methods can guide antibiotic screens and provide immediate insight into their mode of action.
In silico design of production strains
Metabolic engineering approaches often perturb and screen cells for desired phenotypes. However, engineered strains can decrease product yield over time, since products drain cellular resources. Thus, several COBRA methods aim to predict perturbations (e.g., gene deletions or additions) that force the strain to couple product yield to a cellular objective. For example, the secretion of a product can be coupled to growth if its precursor provides an essential biomass component, and if pathways are removed that would metabolize the desired product. Thus, as cells grow exponentially, they can actually increase productivity67 (Figure 4.a).
Figure 4. Principles of model-guided strain design.
(a) Non-growth-coupled production strains witness a decrease in product yield over time, while growth-coupled strains can enhance product yield. (b) Growth-coupled strain designs are predicted to force product secretion while growing optimally. Several methods have been developed to predict growth-coupled production strains by modeling reaction deletion, gene deletion, or reaction addition. Different reaction deletion algorithms, such as OptKnock68, Objective tilting69, and RobustKnock70 can provide different optimal growth-coupled strain designs, due to algorithmic differences. (d) Many algorithms predict the set of reactions that must be blocked to obtain a desired product. However, methods like OptGene71 and GDLS72, provide a more realistic view by modeling genetic modifications, since some genes catalyze multiple reactions, and other reactions are spontaneous.
Most COBRA strain-design methods systematically identify reactions that, when perturbed, may couple a product to a selective pressure (Figure 2). For example, OptKnock68 employs MILP on a wild-type model (Figure 4.b.i) to find reaction deletions that force product secretion under optimal growth (Figure 4.b.ii). However, since OptKnock optimizes both the biomass objective function and product yield, strain designs occasionally have alternate optima with other secretion products (Figure 4.b.iii). To avoid this, the product can be added to the biomass function (Objective Tilting69) or MILP can be used (RobustKnock70) to find designs that provide the maximum lower bound on product yield while maximizing growth (Figure 4.b.iv).
For algorithmic simplicity, most strain design methods perturb reactions. However, strain designs based on reactions can require additional gene deletions (isozymes). Moreover, predictions are occasionally not feasible if they require the removal of one reaction catalyzed by a multi-specific enzyme (Figure 4.c). To avoid such predictions, heuristic approaches, such as OptGene71 and GDLS72, identify growth-coupled production strain designs that directly involve gene deletions. Thus, these strain designs are more realistic and easier to test in vivo.
Strain-design predictions are not limited to manipulations of the host cell’s metabolic pathways. The repertoire of products may be expanded in silico by adding genes from other organisms to confer novel metabolic functions. In silico methods have used graph theoretical approaches73, 74 or kinetic parameters75 to build novel biosynthetic pathways, which were subsequently tested or ranked using COBRA methods. Unfortunately, without accounting for the host metabolic network, these approaches cannot guarantee growth-coupled strain designs. Thus, without further engineering (e.g., with scaffolds that physically couple enzymes76) predicted biosynthetic pathways may not yield product in vivo. However, this concern has been addressed through a few approaches, such as by manually removing genes to growth-couple the new pathways75, or systematically following pathway prediction with OptKnock77. Optstrain goes further by conducting the novel pathway search within the host-cell metabolic network to optimize the balance between reaction addition and deletion78. Thus, COBRA approaches allow the coupling of non-native product synthesis to a cellular objective.
The concept of designing strains that couple a product to a defined selective pressure is not only intriguing, but a few COBRA-based in silico predictions have been implemented in vivo67, 77. It is anticipated that these tools will continue to aid metabolic engineering projects.
Refining representations of biological causation
Simulating cell phenotypes requires accurate representations of metabolic network stoichiometry and objective functions. Although metabolic reconstructions are usually carefully built and rigorously tested, they are often incomplete, and may contain a few errors in stoichiometry, thermodynamics, gene associations, or biomass composition, resulting from ambiguities in associated biochemical studies79 or genome annotation80. Moreover, biomass composition and cellular objectives can vary between environments81, 82, especially under nutrient limitation, stationary phase, or stress46, 83. To address these concerns, phenotypic screens have been analyzed with gap-filling COBRA methods (Figure 2) to predict missing pathways 84, 85, to identify incorrect reaction directionality or inclusion79, 86–88, and to suggest subcellular reaction localization in microorganisms with multiple organelles89. Complementary COBRA methods also improve the definition of cellular objectives by integrating data to systematically assess90–92, predict93, or modify objective functions79, 81, 87.
Recently, high-throughput genetic interaction screens have helped refine metabolic networks and the biomass objective function of yeast79, 94. For example, model-predicted epistasis in S. cerevisiae was compared with 176,821 experimentally measured genetic interaction pairs. Although the COBRA model predictions were enriched for high-confidence measured genetic interactions, it did not predict many epistatic interactions. The authors developed an algorithm that reconciled discrepancies between model-predicted and experimentally measured interactions. Several predicted model improvements were experimentally validated. For example, the authors found that quinolinate formation from aspartate was mistakenly included in the yeast reconstruction. In addition, the algorithm predicted that glycogen should be removed as an essential component in the biomass objective function, since it is not essential for growth. Thus, this study demonstrated that COBRA methods could be deployed to improve the yeast metabolic network and provide condition-specific updates to the biomass objective function.
Thermodynamics
COBRA methods provide quantitative predictions without detailed parameterization of each reaction, beyond declaring directionality to reflect reaction thermodynamics. Directionality is often determined from biochemical assays, but such assays may not recapitulate the conditions and metabolite concentrations inside the cell. Therefore, reaction directionality in vitro may be inconsistent with in vivo flux. In addition, unrealistic fluxes can be predicted in silico if a reaction is reversible in a model, but irreversible in vivo. Thus, methods are now applying more rigorous thermodynamic constraints (Figure 2) by removing thermodynamically infeasible pathway usage95–97 or constraining flux based on Gibbs free energy calculations51, 98, 99. Methods are also being used to infer thermodynamic parameters100.
Most COBRA models contain sets of reactions that can cycle metabolites amongst themselves (Figure 5.a). In these cases, FBA cannot predict flux values for these reactions, since their metabolites are cycled infinitely. Such “loops” are biologically unrealistic since no net thermodynamic driving force exists, akin to Kirchhoff’s second law for electric circuits. Thus the net flux around these loops should be zero95. Although these loops often do not affect non-loop reaction flux, their existence can upset some model predictions. Approaches to systematically remove loops have been proposed95–97. For example, loopless-COBRA96 improves FBA solutions by employing MILP to cancel out loop flux (Figure 5.b).
Figure 5. Refining thermodynamic constraints.
Thermodynamic constraints in COBRA models can be refined. (a) For example, when a metabolic network is not adequately constrained, metabolites can cycle infinitely in loops. Akin to Kirchhoff’s loop law for electrical circuits, this property is thermodynamically infeasible. (b) Thus, methods like ll-FVA, which uses the loopless-COBRA96 constraints on flux variability analysis, are able to systematically remove these loops by adding a constraint that limits flux to the solution space regions that are not involved in these loops.
Although loop-removal methods can be easily deployed without extra parameterization, detailed thermodynamic approaches may provide more biologically meaningful reaction flux predictions. Thermodynamic parameters for many metabolites are not known. Fortunately, recent advances in group contribution theory provide Gibbs free energy of formation estimates for metabolites in COBRA models101. With these predicted values, the standard Gibbs free energy change can be predicted for each reaction. These values can help determine reaction directionality51, 102, predict reasonable concentration levels98, and allow the use of metabolite concentrations103 and ranges on kinetic parameters99 as constraints. A recent study104 used estimated metabolite free energy with experimentally measured equilibrium constants to quantitatively assign reaction directionality. This approach also incorporated in vivo pH, temperature, and ionic strength to quantitatively assign reaction directionality to the E. coli metabolic network. When the authors compared the model-predicted and experimentally measured growth rates, they found that the quantitative assignment of directionality matched model predictions with experimental data, and only required qualitative directionality assignment for certain reactions (e.g., ABC and proton coupled transporters). Since thermodynamics represents one primary model constraint necessary for accurate COBRA predictions, it is expected that further developments in this area will be of great importance to the field.
Incorporating regulatory constraints and signaling
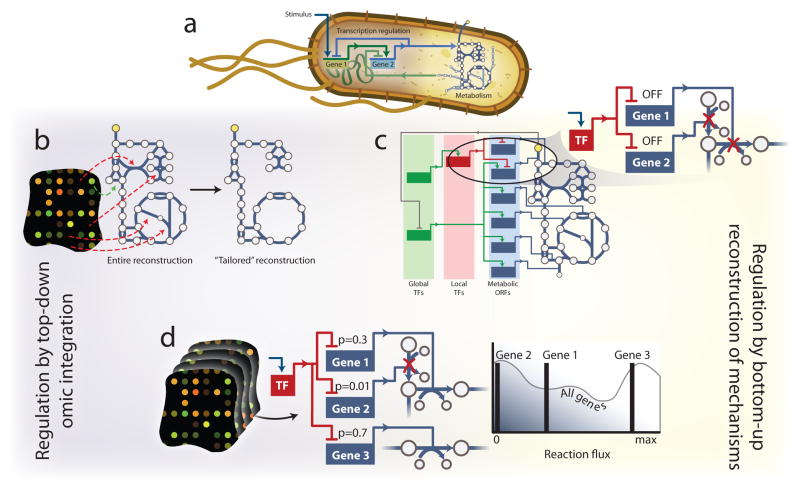
Transcriptional regulation and signaling networks interface extensively with metabolism to produce cellular phenotypes (Figure 6.a). By incorporating regulatory and signaling constraints into metabolic network models, interactions between the systems can be captured to enhance COBRA predictions. There are two primary paradigms on how regulatory constraints are implemented in constraint-based models (Figure 2). Either experimental data are used55, 56, 105–108 to constrain flux through specific reactions (Figure 6.b), or a mathematical representation of transcription regulation109, 110 or signaling111, 112 is interfaced directly with the metabolic network to aid in modeling (Figure 6.c).
Figure 6. Incorporating and inferring regulation.
(a) Signaling, transcription regulation, and metabolism are interlinked in the cell. Therefore integrating the networks may provide more holistic modeling of organisms. Two primary paradigms exist in COBRA modeling for integrating transcription regulation and metabolism. (b) Algorithms such as GIMME105 and MBA107 use high-throughput data and model simulations to identify which pathways are likely expressed and active in the cells when the data were sampled. This results in a tailored context-specific representation of the metabolic network. (c) Algorithms such as rFBA109, iFBA111, and SR-FBA110 incorporate detailed mathematical representations of the known molecular mechanisms of transcription regulation. These approaches contain binary regulatory logic that dictates, under a specific signal, which metabolic pathways are suppressed and cannot carry flux. (d) Hybrid methods, such as PROM116 are arising, in which transcriptomic data are used to infer the regulatory network. This allows for the elucidation of novel regulatory interactions and their immediate incorporation into model simulations. PROM also uses probabilistic measures to allow for a more continuous regulation of reaction flux. For example, Gene 2 is tightly regulated by a transcription factor (TF). Thus, when the TF is activated by a signal, reaction flux is more tightly constrained than Gene 1, which is only loosely regulated.
Not all pathways are active under all growth conditions. Thus, ‘omic data can be used to constrain models accordingly (Figure 6.b)55, 56, 105–108. Methods such as GIMME105, Shlomi-NBT-08106, and MBA107 each remove pathways lacking expression in ‘omic data to obtain functional models that are consistent with the cell gene or protein expression. These approaches have provided novel insights and discoveries in tissue-specific human metabolism39, 64, 113, 114. However, they were also recently used to model metabolic interactions between M. tuberculosis and a macrophage81.
To expand model predictions beyond metabolism, regulatory mechanisms are being integrated with metabolic models (Figure 6.c). Such integrated metabolic and regulatory models can improve phenotype predictions and even suggest novel regulatory interactions. This was done for the nutrient-controlled transcriptional regulatory network for S. cerevisiae115, which included Boolean regulatory interactions between 55 transcription factors and 750 metabolic genes. This integrated regulatory-metabolic network could simulate growth under different environmental and genetic perturbations using regulatory FBA (rFBA). The model predicted new transcriptional regulatory interactions, and elucidated regulatory cascades using chromatin immunoprecipitation data and transcription factor binding motifs. While integrated models of metabolism and transcription regulation provide improved phenotype predictions, this study showed they can also expand regulatory knowledge. It is anticipated that such models may further demonstrate metabolic pathway usage in conditions for which ‘omic data are not available.
Variations on rFBA have been suggested110, 111. Despite their success, rFBA and related methods have two primary weaknesses. First, they assume binary responses for all transcriptional regulatory interactions, when real biological systems exhibit a range of behavior in transcriptional regulation, from binary to continuous. Second, few organisms have been studied enough to provide adequate regulatory information for rFBA. However, a method called probabilistic regulation of metabolism (PROM) addresses these concerns116. When ample transcriptomic data are available, PROM can infer an organism’s transcriptional regulatory network and integrate it with the metabolic network, yielding an improved regulatory-metabolic network model. Moreover, PROM can apply intermediate responses (as opposed to binary), since it uses conditional probabilities for modeling transcription regulation instead of hard Boolean rules (Figure 6.d).
PROM was deployed to infer the regulatory network of M. tuberculosis and integrate it with metabolism116. Each transcription factor (TF) modulating metabolic gene expression was systematically deleted from the model and in silico growth phenotypes were compared with experimentally measured phenotypes. PROM correctly predicted 96% of the TF knockout phenotypes, including 5 of the 6 TFs that were essential for optimal growth. This suggests that this method may help predict antibiotic targets for both regulatory and metabolic genes. Furthermore, the connections between the inferred regulatory network and metabolism may represent novel regulatory targets for uncharacterized transcription factors.
An ecosystem of COBRA methods
Individual COBRA methods can answer numerous scientific questions. However, multiple methods can be deployed in parallel to obtain additional insights into a question of interest. Moreover, different models can be easily swapped or combined to test hypotheses relevant to different species. Thus by using a community of methods and several data types, deeper insights into larger questions may be attained. For example, COBRA methods have complemented each other and provided insight into microbial community interactions.
The community structure in an organism’s microenvironment can shape metabolic pathways usage. Organisms compete for scarce resources or depend on the metabolic capabilities of their cohabitants. Evolution often selects for cells that leverage this community structure117. COBRA methods are now characterizing metabolism’s role in microbial community structure118–120. These studies are providing insight into mutualism121, competition122, parasitism81, 123, and community evolution117, 124.
Mutalism
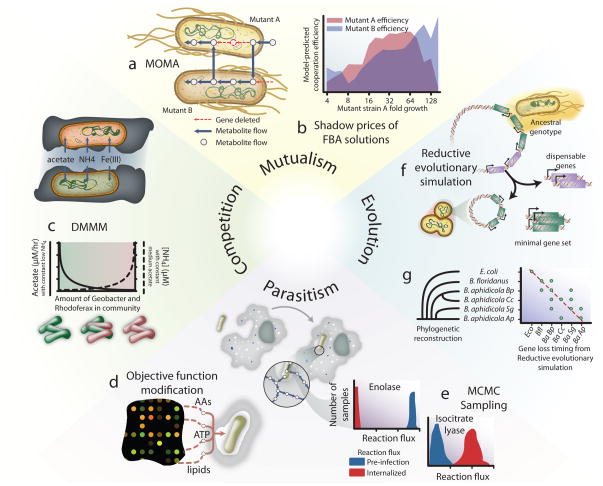
Synthetic mutualism between auxotrophic E. coli mutants was recently studied using COBRA methods121. The authors grew pairs of auxotrophic mutants and then modeled their coupled metabolism using MOMA to identify mutant pairs that exchange essential metabolites to improve growth (Figure 7.a). FBA shadow prices demonstrated the balance between the cost (from metabolite loss) and the benefit (from receiving missing essential metabolites) to each rescued auxotroph. The cooperative efficiency (i.e., the ratio of uptake benefit to production cost) recapitulated the observed growth of the co-cultures. Substantial increases in growth (Figure 7.b) were witnessed in co-cultures that exchanged beneficial, but less costly metabolites (i.e., higher cooperative efficiency). Although it is difficult to directly measure metabolite exchange between the auxotrophs, the computed cooperation efficiency provides an indirect quantitative assessment of the metabolite cross-feeding in this mutualistic system.
Figure 7. Integrating COBRA methods to study community interactions.
COBRA methods are providing insight into the metabolic interactions in various types of microbial communities. (a) To study the mutualistic behavior of co-dependent mutant E. coli, researchers used MOMA50 to simulate synergistic growth of pairs of auxotrophic E. coli. (b) Shadow prices from FBA simulations of these pairs were used to compute cooperation efficiencies between strains, which were subsequently compared with measured fitness improvements. (c) Competition in communities was modeled using DMMM122 to understand how communities of Geobacter and Rhodoferax compete for resources, and how the demographics vary under different nutrient ratios, thereby affecting the efficiency of bioremediation efforts. Host-pathogen interactions between M. tuberculosis and a human macrophage were studied using COBRA. (d) While transcriptomic data were employed to build host-pathogen models at different stages of infection, the cellular objective of internalized M. tuberculosis is not known, so refinements to the objective function were predicted from transcriptomic data to account for changes in required amounts of compounds like lipids and amino acids (AAs). (e) This information was used to compute flux states of internalized M. tuberculosis with MCMC sampling32. This demonstrated a suppression of central metabolism and activation of the glyoxylate shunt, represented here by enolase and isocitrate lyase, respectively. The role of communities in evolution has been studied using Reductive evolutionary simulation117. In particular, this method predicted the minimal set of genes needed to for Buchnera to grow in the rich innards of the aphid. The predicted minimial gene sets (f) and temporal order of gene loss (g) were consistent with the gene content and phylogenetic structure of several Buchnera species.
Competition
Metabolic competition for scarce nutrients has also been assessed with COBRA methods. Dynamic multi-species metabolic modeling (DMMM) characterized the competition for acetate, Fe(III), and ammonia between Geobacter sulfurreducens and Rhodoferax ferrireducens in subsurface anoxic environments (Figure 7.c)122. DMMM simulates the growth rate of both organisms and the rates of change of external metabolites, to dynamically predict population changes in the community. Using DMMM, the community composition was predicted under geochemically distinct conditions of low, medium, and high acetate flux. Under low acetate flux, DMMM predicts Rhodoferax dominates the community when sufficient ammonia is available, whereas Geobacter dominates under low ammonia and high acetate flux. This difference was attributed to the nitrogen fixation abilities of Geobacter, as well as its higher acetate uptake rate compared to Rhodoferax. Moreover, it was also predicted that under nitrogen fixing conditions, Geobacter increases its respiration at the expense of biomass production, thus showing how balancing community structure can impact the efficacy of uranium bioremediation in low ammonium zones.
Parasitism
Host-pathogen interactions have been studied with COBRA methods123. A recent study modeled the metabolic interactions between a human alveolar macrophage and M. tuberculosis81. Context-specific models of infection were built with GIMME105 and Shlomi-NBT-08106 using transcriptomic data from three types of M. tuberculosis infections. Next, the M. tuberculosis objective function was revised using infection-specific gene expression data to better represent the metabolic activity of the internalized pathogen (Figure 7.d). Gene deletion analysis was compared with in vivo gene essentiality data, and MCMC sampling was also used to demonstrate a substantial alteration in metabolic pathway usage in M. tuberculosis during macrophage infection, including a suppression of glycolysis and an increased dependency on glyoxylate metabolism (Figure 7.e). This constraint of central metabolism during M. tuberculosis infection was also suggested by DCP, another method related to FBA125. This suppression of certain metabolic pathways with an increased dependency on normally latent pathways may provide novel antibiotic targets.
Community evolution
In evolution, genetic drift and selective pressures cause organisms to optimize their cellular machinery for a particular niche126. This assumption of cellular optimization has made COBRA methods useful tools to investigate hypotheses concerning organismal evolution, as reviewed by Papp, et al.6 In nature, the optimization of microbial metabolism is a multi-species affair, as demonstrated by the aphid endosymbiant Buchnera aphidicola. This descendant of the Enterobacteriaceae family has suffered drastic loss of genomic material as it evolved in its host’s nutrient-rich innards. Since B. aphidicola is related to E. coli, reductive evolutionary simulation (a gene deletion analysis derivative)117 on the E. coli model provided minimal metabolic gene set predictions. These predicted minimal sets are highly consistent with the metabolic gene content of B. aphidicola (Figure 7.f). In addition, the predicted temporal order of gene loss was significantly consistent with the phylogenetically reconstructed gene loss timing among the genomes of five Buchnera species (Figure 7.g)124, thus suggesting that the bacterium optimized its pathway usage for its new rich habitat. Interestingly, metabolic pathways retained in the computed minimal gene sets highlight the bacterium’s role in symbiotic evolution. Retained pathways contained reactions needed for producing riboflavin and essential amino acids lacking from the aphid diet, thereby highlighting their role in the symbiotic relationship117. Thus, COBRA methods are helping to describe how the community shapes gene content in evolving symbiotic communities6.
Future directions
Constraint-based modeling has rapidly evolved over the past two decades and now forms a foundation for achieving a genome-scale science of microbial metabolism. Prior to 2004, studies in this field focused on its conceptualization and algorithmic development. Thus, the methods developed were largely conceptual and employed for studying fundamental properties of metabolic networks, such as robustness, alternate optima, and the functional consequences of metabolic network topology. After 2004, the field expanded to provide tools for addressing both basic and applied scientific questions focused on issues like strain design, gap-filling85, and evolution6. Despite limitations in constraint-based modeling, its scope and uses are growing. GEMs and their corresponding analytical methods are expanding in scope beyond microbial metabolism, facilitating ‘omic data analysis, and directing scientific inquiry.
COBRA methods have gained rapid acceptance since their focus on governing constraints facilitates genome-scale analysis. However, the simplifying assumptions can limit its scope. COBRA methods focus on steady state flux, so models do not address metabolite concentrations, changes in biochemistry from pH and SNPs, temporal metabolic changes, and spatial constraints. Initial efforts are addressing some limitations and providing insight into these properties of metabolism58, 103, 105, 122, 127, and additional efforts will further address these and other limitations.
Metabolism is involved in most cell processes and phenotypes. However, genome-scale models are extending beyond microbial metabolism to include transcription regulation109, 110, 116, protein and transcript synthesis128, 129, signaling112, plant and animal metabolism39, 58, 64, 113, 114, 130, 131, and host-pathogen interactions81, 123, 132. The advances beyond microbial metabolism, invite additional applications by providing additional targets for drug discovery and metabolic engineering133, and allowing studies on medicine and crop engineering. This expansion of models and applications is requiring further evolution of COBRA methods and theoretical breakthroughs to integrate non-stoichiometric networks (e.g., transcriptional regulation) with metabolism, and account for interactions with spatial constraints (e.g., multi-cell metabolism39, 81, 134).
The past decade has witnessed a deluge of high-throughput data ranging from phenotypic screens, sequencing data, proteomics, metabolomics, and so forth. Recent studies have demonstrated that novel insights can be gained when these data are analyzed in the context of GEMs34, 39, 64, 79, 113, 125, 135. As models expand, they will increasingly aid in data interpretation, since they provide a structured context for high-throughput data analysis. Moreover, the biochemical mechanisms in these models will leverage ‘omic analysis to inform experimental work.
Constraint-based modeling is already guiding discovery85 by identifying missing metabolic and regulatory functions84, 86, 94, 115, 116, 136, predicting enzyme localization89, suggesting novel drug targets65, 66, 114, and aiding in strain design for chemical production67, 77, 137–141 and biosensor development142. These studies are now increasingly directing experimental work. As models expand and are used to integrate ‘omic data, COBRA methods will increasingly be deployed to guide scientific inquiry.
Supplementary Material
Table S1. Brief details and categorization of 100+ constraint-based methods.
Table S2. Software packages implementing multiple constraint-based methods
Online summary.
Genotype-phenotype relationships have classically been qualitative, but recent advances are enabling us to overcome conceptual and technological barriers leading to quantitative relationships. Within the constraint-based modeling framework, generation of quantitative relationships is facilitated by the realization that cell phenotypes are limited by physical and genetic constraints.
Physical laws, such as mass conservation and thermodynamics, constrain the possible metabolic and biosynthetic transformations that can occur in nature, and genetics specify which sets of biochemical reactions have been selected through evolution. Genome sequencing and annotation have allowed the comprehensive reconstruction of microbial metabolic networks, and constraint-based modeling has emerged as a set of valuable tools that allow for detailed analyses of biochemical mechanisms underlying the metabolic genotype-phenotype relationship.
Network-based pathway analysis tools, such as elementary flux modes and extreme pathways analysis, delineate pathways that can perform a given metabolic function in an organism of interest. While these methods have been difficult to use in larger metabolic networks, simplifications are now beginning to allow their use on genome-scale models.
Since not all pathways are used in a cell at a given time, optimization algorithms are routinely used to identify pathway usage that best reflects the in vivo metabolic state. Flux balance analysis, which uses linear programming to optimize a mathematical description of the cellular objective, has been widely used to understand microbial physiology and the effects of environmental and genetic perturbations.
The ability to model genetic perturbations has allowed constraint-based modeling to be repeatedly deployed to help predict antimicrobial targets and aid in the design of production strains for chemical production.
Reconstructed metabolic networks are often incomplete and can have a small fraction of incorrect reactions therein. However, the integration of phenotypic screens with model simulations can provide a systematic approach to refine models and discover new metabolic functions in an organism.
COBRA methods are extending beyond metabolism, and approaches are beginning to incorporate transcription regulation implicitly by constraining models with multiple –omic datatypes or explicitly with detailed descriptions of regulatory mechanisms.
The diverse range of more than 100 constraint-based methods is being deployed to address many questions in microbiology. For example, several recent studies have begun to explore the roles of metabolism in community interactions, including symbiosis, competition, parasitism, and evolution.
Acknowledgments
The authors would like to thank Marc Abrams, Jonathan Monk, Douglas Taylor, Edward O’Brien, Adam Feist, Joshua Lerman, Daniel Zielinski, Roger Chang, Karsten Zengler, Aarash Bordbar, and Eytan Ruppin for thoughtful discussions and comments on this review, and numerous members of the constraint-based community for help with compiling methods. This work was funded by National Institutes of Health (R01GM057089) and the Office of Science (BER), U.S. Department of Energy (DE-SC0004485).
Glossary terms
- Metabolic reconstruction
A carefully curated and biochemically validated knowledgebase in which all known chemical reactions for an organism are detailed and catalogued
- Genome-scale model (GEM)
A condition-specific, mathematically-described, computable derivative of a metabolic reconstruction, containing comprehensive knowledge of metabolism
- Biomass function
A pseudo reaction formed to aid in predicting growth of a cell in COBRA models. Describes the rate and the accurate proportions at which all of the biomass precursors are made
- Flux distribution
A set of steady-state fluxes for all reactions in the metabolic network
- Linear programming (LP)
A mathematical optimization technique that determines a way to maximize a particular objective under a given set of conditions. Linear programming involves the optimization of a linear objective subject to linear equalities and inequalities as constraints. Typically used in FBA, where the objective is generally the biomass function (growth) and the constraints represent the growth conditions
- Mixed integer linear programming (MILP)
Similar to linear programming, but some of the constraints are integer values. Used for applications such as enumerating alternate optimal solutions, strain design, eliminating loops etc
- Solution space
The feasible region satisfying a set of constraints. In COBRA models, this represents the feasible flux values for all the reactions in the model
- Epistasis
The interaction between two genes where the phenotypic effects of one gene is masked by that of the other. Usually identified by the phenotype of the double mutant relative to the phenotype shown by the two single mutants
- Growth-coupled design
The situation where the production of a particular compound is positively correlated with the growth rate of the organism. Often preferred in strain design to increase product yield as the cell multiplies exponentially
- Shadow Prices
A mathematical term that refers to the dual of the linear programming problem. It represents the rate at which the objective value of the linear program (e.g. growth rate) changes as the supply of a particular resource (e.g. metabolite) increases
Biographies
Nathan E. Lewis is a Ph.D. candidate in bioengineering at the University of California, San Diego and has a degree in biochemistry from Brigham Young University. He is studying a range of means to integrate high-throughput data types with genome-scale metabolic networks, and novel means to analyze data with constraint-based modeling in an effort to drive biological discovery.
Harish Nagarajan is a Ph.D. candidate in Bioinformatics and Systems Biology at the University of California, San Diego and has a degree in Biotechnology from Indian Institute of Technology Madras. He is developing genome-scale metabolic networks of several bacteria and COBRA methods to characterize microbial community interactions and other metabolic engineering applications with the aid of several high-throughput data types.
Bernhard Ø. Palsson is the Galletti Professor of Bioengineering and an adjunct professor of medicine at the University of California-San Diego, USA. He received his Ph.D. in chemical engineering from the University of Wisconsin-Madison in 1984, after which he held a faculty position at the University of Michigan. His recent research focuses on the reconstruction and analysis of genome-scale models of metabolism, regulation and signaling and in deciphering microbial operon structure. He has developed undergraduate and graduate systems biology curricula, including two textbooks (Systems Biology: Properties of Reconstructed Networks (Cambridge University Press, 2006) and Systems Biology: Simulation of Dynamic Network States (Cambridge University Press, 2011)).
References
- 1.Feist AM, Herrgard MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microorganisms. Nat Rev Microbiol. 2009;7:129–143. doi: 10.1038/nrmicro1949. This review provides the detailed concepts of metabolic network reconstruction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiele I, Palsson BO. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protocols. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry CS, et al. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 4.Feist AM, Palsson BO. The growing scope of applications of genome-scale metabolic reconstructions using Escherichia coli. Nat Biotechnol. 2008;26:659–667. doi: 10.1038/nbt1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oberhardt MA, Palsson BO, Papin JA. Applications of genome-scale metabolic reconstructions. Mol Syst Biol. 2009;5:320. doi: 10.1038/msb.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Papp B, Notebaart RA, Pal C. Systems-biology approaches for predicting genomic evolution. Nat Rev Genet. 2011;12:591–602. doi: 10.1038/nrg3033. A thorough review of how COBRA methods aid in the study of evolution. [DOI] [PubMed] [Google Scholar]
- 7.Mahadevan R, Palsson BO, Lovley DR. In situ to in silico and back: elucidating the physiology and ecology of Geobacter spp. using genome-scale modelling. Nat Rev Microbiol. 2011;9:39–50. doi: 10.1038/nrmicro2456. [DOI] [PubMed] [Google Scholar]
- 8.Palsson B. Systems biology : properties of reconstructed networks. Cambridge University Press; Cambridge ; New York: 2006. p. 322. [Google Scholar]
- 9.Feist AM, Palsson BO. The biomass objective function. Curr Opin Microbiol. 2010;13:344–349. doi: 10.1016/j.mib.2010.03.003. A description of how the biomass objective function is formulated. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fell DA, Small JR. Fat synthesis in adipose tissue. An examination of stoichiometric constraints. Biochem J. 1986;238:781–786. doi: 10.1042/bj2380781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watson MR. Metabolic maps for the Apple II. Biochemical Society Transactions. 1984;12:1093. [Google Scholar]
- 12.Edwards JS, Palsson BO. Systems properties of the Haemophilus influenzae Rd metabolic genotype. J Biol Chem. 1999;274:17410–17416. doi: 10.1074/jbc.274.25.17410. [DOI] [PubMed] [Google Scholar]
- 13.Edwards JS, Palsson BO. The Escherichia coli MG1655 in silico metabolic genotype: its definition, characteristics, and capabilities. Proc Natl Acad Sci U S A. 2000;97:5528–5533. doi: 10.1073/pnas.97.10.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed JL, Famili I, Thiele I, Palsson BO. Towards multidimensional genome annotation. Nat Rev Genet. 2006;7:130–141. doi: 10.1038/nrg1769. [DOI] [PubMed] [Google Scholar]
- 15.Kim TY, Kim HU, Lee SY. Data integration and analysis of biological networks. Curr Opin Biotechnol. 2010;21:78–84. doi: 10.1016/j.copbio.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 16.Sauer U. Metabolic networks in motion: 13C-based flux analysis. Mol Syst Biol. 2006;2:62. doi: 10.1038/msb4100109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Papin JA, et al. Comparison of network-based pathway analysis methods. Trends Biotechnol. 2004;22:400–405. doi: 10.1016/j.tibtech.2004.06.010. An assessment of the differences between elementary flux modes and extreme pathway analysis. [DOI] [PubMed] [Google Scholar]
- 18.Trinh CT, Wlaschin A, Srienc F. Elementary mode analysis: a useful metabolic pathway analysis tool for characterizing cellular metabolism. Appl Microbiol Biotechnol. 2009;81:813–826. doi: 10.1007/s00253-008-1770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Llaneras F, Pico J. Which metabolic pathways generate and characterize the flux space? A comparison among elementary modes, extreme pathways and minimal generators. J Biomed Biotechnol. 2010;2010:753904. doi: 10.1155/2010/753904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stelling J, Klamt S, Bettenbrock K, Schuster S, Gilles ED. Metabolic network structure determines key aspects of functionality and regulation. Nature. 2002;420:190–193. doi: 10.1038/nature01166. [DOI] [PubMed] [Google Scholar]
- 21.Trinh CT, Unrean P, Srienc F. Minimal Escherichia coli cell for the most efficient production of ethanol from hexoses and pentoses. Appl Environ Microbiol. 2008;74:3634–3643. doi: 10.1128/AEM.02708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imielinski M, Belta C. Exploiting the pathway structure of metabolism to reveal high-order epistasis. BMC Syst Biol. 2008;2:40. doi: 10.1186/1752-0509-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wessely F, et al. Optimal regulatory strategies for metabolic pathways in Escherichia coli depending on protein costs. Mol Syst Biol. 2011;7:515. doi: 10.1038/msb.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schilling CH, Palsson BO. Assessment of the metabolic capabilities of Haemophilus influenzae Rd through a genome-scale pathway analysis. J Theor Biol. 2000;203:249–283. doi: 10.1006/jtbi.2000.1088. [DOI] [PubMed] [Google Scholar]
- 25.Yeung M, Thiele I, Palsson BO. Estimation of the number of extreme pathways for metabolic networks. BMC Bioinformatics. 2007;8:363. doi: 10.1186/1471-2105-8-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klamt S, Stelling J. Combinatorial complexity of pathway analysis in metabolic networks. Mol Biol Rep. 2002;29:233–236. doi: 10.1023/a:1020390132244. [DOI] [PubMed] [Google Scholar]
- 27.Kaleta C, de Figueiredo LF, Schuster S. Can the whole be less than the sum of its parts? Pathway analysis in genome-scale metabolic networks using elementary flux patterns. Genome Res. 2009;19:1872–1883. doi: 10.1101/gr.090639.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rezola A, et al. Exploring metabolic pathways in genome-scale networks via generating flux modes. Bioinformatics. 2011;27:534–540. doi: 10.1093/bioinformatics/btq681. [DOI] [PubMed] [Google Scholar]
- 29.Ip K, Colijn C, Lun DS. Analysis of complex metabolic behavior through pathway decomposition. BMC Syst Biol. 2011;5:91. doi: 10.1186/1752-0509-5-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan SH, Ji P. Decomposing flux distributions into elementary flux modes in genome-scale metabolic networks. Bioinformatics. 2011;27:2256–2262. doi: 10.1093/bioinformatics/btr367. [DOI] [PubMed] [Google Scholar]
- 31.Braunstein A, Mulet R, Pagnani A. Estimating the size of the solution space of metabolic networks. BMC Bioinformatics. 2008;9:240. doi: 10.1186/1471-2105-9-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schellenberger J, Palsson BO. Use of randomized sampling for analysis of metabolic networks. J Biol Chem. 2009;284:5457–5461. doi: 10.1074/jbc.R800048200. [DOI] [PubMed] [Google Scholar]
- 33.Almaas E, Kovacs B, Vicsek T, Oltvai ZN, Barabasi AL. Global organization of metabolic fluxes in the bacterium Escherichia coli. Nature. 2004;427:839–843. doi: 10.1038/nature02289. [DOI] [PubMed] [Google Scholar]
- 34.Bordel S, Agren R, Nielsen J. Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes. PLoS Comput Biol. 2010;6:e1000859. doi: 10.1371/journal.pcbi.1000859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mo ML, Palsson BO, Herrgard MJ. Connecting extracellular metabolomic measurements to intracellular flux states in yeast. BMC Syst Biol. 2009;3:37. doi: 10.1186/1752-0509-3-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrett CL, Herrgard MJ, Palsson B. Decomposing complex reaction networks using random sampling, principal component analysis and basis rotation. BMC Syst Biol. 2009;3:30. doi: 10.1186/1752-0509-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thiele I, Price ND, Vo TD, Palsson BO. Candidate metabolic network states in human mitochondria. Impact of diabetes, ischemia, and diet. J Biol Chem. 2005;280:11683–11695. doi: 10.1074/jbc.M409072200. [DOI] [PubMed] [Google Scholar]
- 38.Price ND, Schellenberger J, Palsson BO. Uniform sampling of steady-state flux spaces: means to design experiments and to interpret enzymopathies. Biophys J. 2004;87:2172–2186. doi: 10.1529/biophysj.104.043000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lewis NE, et al. Large-scale in silico modeling of metabolic interactions between cell types in the human brain. Nat Biotechnol. 2010;28:1279–1285. doi: 10.1038/nbt.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orth JD, Thiele I, Palsson BO. What is flux balance analysis? Nat Biotechnol. 2010;28:245–248. doi: 10.1038/nbt.1614. A primer to flux balance analysis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varma A, Palsson BO. Stoichiometric flux balance models quantitatively predict growth and metabolic by-product secretion in wild-type Escherichia coli W3110. Appl Environ Microbiol. 1994;60:3724–3731. doi: 10.1128/aem.60.10.3724-3731.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edwards JS, Ibarra RU, Palsson BO. In silico predictions of Escherichia coli metabolic capabilities are consistent with experimental data. Nat Biotechnol. 2001;19:125–130. doi: 10.1038/84379. [DOI] [PubMed] [Google Scholar]
- 43.Lewis NE, et al. Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol. 2010;6:390. doi: 10.1038/msb.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teusink B, Wiersma A, Jacobs L, Notebaart RA, Smid EJ. Understanding the adaptive growth strategy of Lactobacillus plantarum by in silico optimisation. PLoS Comput Biol. 2009;5:e1000410. doi: 10.1371/journal.pcbi.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Z, Zhang J. Impact of gene expression noise on organismal fitness and the efficacy of natural selection. Proc Natl Acad Sci U S A. 2011;108:E67–76. doi: 10.1073/pnas.1100059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goffin P, et al. Understanding the physiology of Lactobacillus plantarum at zero growth. Mol Syst Biol. 2010;6:413. doi: 10.1038/msb.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee S, Palakornkule C, Domach MM, Grossmann IE. Recursive MILP model for finding all the alternate optima in LP models for metabolic networks. Computers & Chemical Engineering. 2000;24:711–716. [Google Scholar]
- 48.Gudmundsson S, Thiele I. Computationally efficient flux variability analysis. BMC Bioinformatics. 2010;11:489. doi: 10.1186/1471-2105-11-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burgard AP, Vaidyaraman S, Maranas CD. Minimal reaction sets for Escherichia coli metabolism under different growth requirements and uptake environments. Biotechnol Prog. 2001;17:791–797. doi: 10.1021/bp0100880. [DOI] [PubMed] [Google Scholar]
- 50.Segre D, Vitkup D, Church GM. Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A. 2002;99:15112–15117. doi: 10.1073/pnas.232349399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Holzhutter HG. The principle of flux minimization and its application to estimate stationary fluxes in metabolic networks. Eur J Biochem. 2004;271:2905–2922. doi: 10.1111/j.1432-1033.2004.04213.x. [DOI] [PubMed] [Google Scholar]
- 52.Ponce de Leon M, Cancela H, Acerenza L. A strategy to calculate the patterns of nutrient consumption by microorganisms applying a two-level optimisation principle to reconstructed metabolic networks. J Biol Phys. 2008;34:73–90. doi: 10.1007/s10867-008-9067-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murabito E, Simeonidis E, Smallbone K, Swinton J. Capturing the essence of a metabolic network: a flux balance analysis approach. J Theor Biol. 2009;260:445–452. doi: 10.1016/j.jtbi.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 54.Benyamini T, Folger O, Ruppin E, Shlomi T. Flux balance analysis accounting for metabolite dilution. Genome Biol. 2010;11:R43. doi: 10.1186/gb-2010-11-4-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Colijn C, et al. Interpreting expression data with metabolic flux models: predicting Mycobacterium tuberculosis mycolic acid production. PLoS Comput Biol. 2009;5:e1000489. doi: 10.1371/journal.pcbi.1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Berlo RJP, et al. Predicting Metabolic Fluxes Using Gene Expression Differences As Constraints. Computational Biology and Bioinformatics, IEEE/ACM Transactions on. 2011;8:206–216. doi: 10.1109/TCBB.2009.55. [DOI] [PubMed] [Google Scholar]
- 57.Beg QK, et al. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc Natl Acad Sci U S A. 2007;104:12663–12668. doi: 10.1073/pnas.0609845104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vazquez A, Markert EK, Oltvai ZN. Serine biosynthesis with one carbon catabolism and the glycine cleavage system represents a novel pathway for ATP generation. PLoS One. 2011;6:e25881. doi: 10.1371/journal.pone.0025881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhuang K, Vemuri GN, Mahadevan R. Economics of membrane occupancy and respiro-fermentation. Mol Syst Biol. 2011;7:500. doi: 10.1038/msb.2011.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Papp B, Pal C, Hurst LD. Metabolic network analysis of the causes and evolution of enzyme dispensability in yeast. Nature. 2004;429:661–664. doi: 10.1038/nature02636. [DOI] [PubMed] [Google Scholar]
- 61.Deutscher D, Meilijson I, Kupiec M, Ruppin E. Multiple knockout analysis of genetic robustness in the yeast metabolic network. Nat Genet. 2006;38:993–998. doi: 10.1038/ng1856. [DOI] [PubMed] [Google Scholar]
- 62.Shlomi T, Berkman O, Ruppin E. Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proc Natl Acad Sci U S A. 2005;102:7695–7700. doi: 10.1073/pnas.0406346102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim PJ, et al. Metabolite essentiality elucidates robustness of Escherichia coli metabolism. Proc Natl Acad Sci U S A. 2007;104:13638–13642. doi: 10.1073/pnas.0703262104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chang RL, Xie L, Xie L, Bourne PE, Palsson BO. Drug off-target effects predicted using structural analysis in the context of a metabolic network model. PLoS Comput Biol. 2010;6:e1000938. doi: 10.1371/journal.pcbi.1000938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen Y, et al. Blueprint for antimicrobial hit discovery targeting metabolic networks. Proc Natl Acad Sci U S A. 2010;107:1082–1087. doi: 10.1073/pnas.0909181107. Metabolic networks are used to search for antimicrobials. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim HU, et al. Integrative genome-scale metabolic analysis of Vibrio vulnificus for drug targeting and discovery. Mol Syst Biol. 2011;7:460. doi: 10.1038/msb.2010.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fong SS, et al. In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol Bioeng. 2005;91:643–648. doi: 10.1002/bit.20542. [DOI] [PubMed] [Google Scholar]
- 68.Burgard AP, Pharkya P, Maranas CD. Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng. 2003;84:647–657. doi: 10.1002/bit.10803. [DOI] [PubMed] [Google Scholar]
- 69.Feist AM, et al. Model-driven evaluation of the production potential for growth-coupled products of Escherichia coli. Metab Eng. 2009 doi: 10.1016/j.ymben.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tepper N, Shlomi T. Predicting metabolic engineering knockout strategies for chemical production: accounting for competing pathways. Bioinformatics. 2010;26:536–543. doi: 10.1093/bioinformatics/btp704. [DOI] [PubMed] [Google Scholar]
- 71.Patil KR, Rocha I, Forster J, Nielsen J. Evolutionary programming as a platform for in silico metabolic engineering. BMC Bioinformatics. 2005;6:308. doi: 10.1186/1471-2105-6-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lun DS, et al. Large-scale identification of genetic design strategies using local search. Mol Syst Biol. 2009;5:296. doi: 10.1038/msb.2009.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yousofshahi M, Lee K, Hassoun S. Probabilistic pathway construction. Metab Eng. 2011;13:435–444. doi: 10.1016/j.ymben.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 74.Rodrigo G, Carrera J, Prather KJ, Jaramillo A. DESHARKY: automatic design of metabolic pathways for optimal cell growth. Bioinformatics. 2008;24:2554–2556. doi: 10.1093/bioinformatics/btn471. [DOI] [PubMed] [Google Scholar]
- 75.Bar-Even A, Noor E, Lewis NE, Milo R. Design and analysis of synthetic carbon fixation pathways. Proc Natl Acad Sci U S A. 2010;107:8889–8894. doi: 10.1073/pnas.0907176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 77.Yim H, et al. Metabolic engineering of Escherichia coli for direct production of 1,4-butanediol. Nat Chem Biol. 2011;7:445–452. doi: 10.1038/nchembio.580. A detailed study in which a microbe is engineered to synthesize 1,4-butanediol using several computational and experimental technologies. [DOI] [PubMed] [Google Scholar]
- 78.Pharkya P, Burgard AP, Maranas CD. OptStrain: a computational framework for redesign of microbial production systems. Genome Res. 2004;14:2367–2376. doi: 10.1101/gr.2872004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Szappanos B, et al. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat Genet. 2011;43:656–662. doi: 10.1038/ng.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hsiao TL, Revelles O, Chen L, Sauer U, Vitkup D. Automatic policing of biochemical annotations using genomic correlations. Nat Chem Biol. 2010;6:34–40. doi: 10.1038/nchembio.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bordbar A, Lewis NE, Schellenberger J, Palsson BO, Jamshidi N. Insight into human alveolar macrophage and M. tuberculosis interactions via metabolic reconstructions. Mol Syst Biol. 2010;6:422. doi: 10.1038/msb.2010.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schuster S, Pfeiffer T, Fell DA. Is maximization of molar yield in metabolic networks favoured by evolution? J Theor Biol. 2008;252:497–504. doi: 10.1016/j.jtbi.2007.12.008. A critical assessment of assumptions in flux balance analysis. [DOI] [PubMed] [Google Scholar]
- 83.Milne CB, et al. Metabolic network reconstruction and genome-scale model of butanol-producing strain Clostridium beijerinckii NCIMB 8052. BMC Syst Biol. 2011;5:130. doi: 10.1186/1752-0509-5-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Reed JL, et al. Systems approach to refining genome annotation. Proc Natl Acad Sci U S A. 2006;103:17480–17484. doi: 10.1073/pnas.0603364103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Orth JD, Palsson BO. Systematizing the generation of missing metabolic knowledge. Biotechnol Bioeng. 2010;107:403–412. doi: 10.1002/bit.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Satish Kumar V, Dasika MS, Maranas CD. Optimization based automated curation of metabolic reconstructions. BMC Bioinformatics. 2007;8:212. doi: 10.1186/1471-2105-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumar VS, Maranas CD. GrowMatch: an automated method for reconciling in silico/in vivo growth predictions. PLoS Comput Biol. 2009;5:e1000308. doi: 10.1371/journal.pcbi.1000308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Suthers PF, Zomorrodi A, Maranas CD. Genome-scale gene/reaction essentiality and synthetic lethality analysis. Mol Syst Biol. 2009;5:301. doi: 10.1038/msb.2009.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mintz-Oron S, Aharoni A, Ruppin E, Shlomi T. Network-based prediction of metabolic enzymes’ subcellular localization. Bioinformatics. 2009;25:i247–52. doi: 10.1093/bioinformatics/btp209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Burgard AP, Maranas CD. Optimization-based framework for inferring and testing hypothesized metabolic objective functions. Biotechnol Bioeng. 2003;82:670–677. doi: 10.1002/bit.10617. [DOI] [PubMed] [Google Scholar]
- 91.Knorr AL, Jain R, Srivastava R. Bayesian-based selection of metabolic objective functions. Bioinformatics. 2007;23:351–357. doi: 10.1093/bioinformatics/btl619. [DOI] [PubMed] [Google Scholar]
- 92.Schuetz R, Kuepfer L, Sauer U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol Syst Biol. 2007;3:119. doi: 10.1038/msb4100162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gianchandani EP, Oberhardt MA, Burgard AP, Maranas CD, Papin JA. Predicting biological system objectives de novo from internal state measurements. BMC Bioinformatics. 2008;9:43. doi: 10.1186/1471-2105-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zomorrodi AR, Maranas CD. Improving the iMM904 S. cerevisiae metabolic model using essentiality and synthetic lethality data. BMC Syst Biol. 2010;4:178. doi: 10.1186/1752-0509-4-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Beard DA, Liang SD, Qian H. Energy balance for analysis of complex metabolic networks. Biophys J. 2002;83:79–86. doi: 10.1016/S0006-3495(02)75150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Schellenberger J, Lewis NE, Palsson BO. Elimination of thermodynamically infeasible loops in steady-state metabolic models. Biophys J. 2011;100:544–553. doi: 10.1016/j.bpj.2010.12.3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Price ND, Thiele I, Palsson BO. Candidate states of Helicobacter pylori’s genome-scale metabolic network upon application of “loop law” thermodynamic constraints. Biophys J. 2006;90:3919–3928. doi: 10.1529/biophysj.105.072645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Henry CS, Broadbelt LJ, Hatzimanikatis V. Thermodynamics-based metabolic flux analysis. Biophys J. 2007;92:1792–1805. doi: 10.1529/biophysj.106.093138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fleming RM, Thiele I, Provan G, Nasheuer HP. Integrated stoichiometric, thermodynamic and kinetic modelling of steady state metabolism. J Theor Biol. 2010;264:683–692. doi: 10.1016/j.jtbi.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kummel A, Panke S, Heinemann M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol Syst Biol. 2006;2:2006.0034. doi: 10.1038/msb4100074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Jankowski MD, Henry CS, Broadbelt LJ, Hatzimanikatis V. Group contribution method for thermodynamic analysis of complex metabolic networks. Biophys J. 2008;95:1487–1499. doi: 10.1529/biophysj.107.124784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Henry CS, Jankowski MD, Broadbelt LJ, Hatzimanikatis V. Genome-scale thermodynamic analysis of Escherichia coli metabolism. Biophys J. 2006;90:1453–1461. doi: 10.1529/biophysj.105.071720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hoppe A, Hoffmann S, Holzhutter HG. Including metabolite concentrations into flux balance analysis: thermodynamic realizability as a constraint on flux distributions in metabolic networks. BMC Syst Biol. 2007;1:23. doi: 10.1186/1752-0509-1-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fleming RM, Thiele I, Nasheuer HP. Quantitative assignment of reaction directionality in constraint-based models of metabolism: application to Escherichia coli. Biophys Chem. 2009;145:47–56. doi: 10.1016/j.bpc.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Becker SA, Palsson BO. Context-specific metabolic networks are consistent with experiments. PLoS Comput Biol. 2008;4:e1000082. doi: 10.1371/journal.pcbi.1000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shlomi T, Cabili MN, Herrgard MJ, Palsson BO, Ruppin E. Network-based prediction of human tissue-specific metabolism. Nat Biotechnol. 2008;26:1003–1010. doi: 10.1038/nbt.1487. [DOI] [PubMed] [Google Scholar]
- 107.Jerby L, Shlomi T, Ruppin E. Computational reconstruction of tissue-specific metabolic models: application to human liver metabolism. Mol Syst Biol. 2010;6:401. doi: 10.1038/msb.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jensen PA, Papin JA. Functional integration of a metabolic network model and expression data without arbitrary thresholding. Bioinformatics. 2011;27:541–547. doi: 10.1093/bioinformatics/btq702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Covert MW, Knight EM, Reed JL, Herrgard MJ, Palsson BO. Integrating high-throughput and computational data elucidates bacterial networks. Nature. 2004;429:92–96. doi: 10.1038/nature02456. [DOI] [PubMed] [Google Scholar]
- 110.Shlomi T, Eisenberg Y, Sharan R, Ruppin E. A genome-scale computational study of the interplay between transcriptional regulation and metabolism. Mol Syst Biol. 2007;3:101. doi: 10.1038/msb4100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Covert MW, Xiao N, Chen TJ, Karr JR. Integrating metabolic, transcriptional regulatory and signal transduction models in Escherichia coli. Bioinformatics. 2008;24:2044–2050. doi: 10.1093/bioinformatics/btn352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lee JM, Gianchandani EP, Eddy JA, Papin JA. Dynamic analysis of integrated signaling, metabolic, and regulatory networks. PLoS Comput Biol. 2008;4:e1000086. doi: 10.1371/journal.pcbi.1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Folger O, et al. Predicting selective drug targets in cancer through metabolic networks. Mol Syst Biol. 2011;7:501. doi: 10.1038/msb.2011.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Frezza C, et al. Haem oxygenase is synthetically lethal with the tumour suppressor fumarate hydratase. Nature. 2011 doi: 10.1038/nature10363. [DOI] [PubMed] [Google Scholar]
- 115.Herrgard MJ, Lee BS, Portnoy V, Palsson BO. Integrated analysis of regulatory and metabolic networks reveals novel regulatory mechanisms in Saccharomyces cerevisiae. Genome Res. 2006;16:627–635. doi: 10.1101/gr.4083206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chandrasekaran S, Price ND. Probabilistic integrative modeling of genome-scale metabolic and regulatory networks in Escherichia coli and Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2010;107:17845–17850. doi: 10.1073/pnas.1005139107. An approach to integrate inferred regulatory networks with metabolic modeling. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pal C, et al. Chance and necessity in the evolution of minimal metabolic networks. Nature. 2006;440:667–670. doi: 10.1038/nature04568. [DOI] [PubMed] [Google Scholar]
- 118.Stolyar S, et al. Metabolic modeling of a mutualistic microbial community. Mol Syst Biol. 2007;3:92. doi: 10.1038/msb4100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Taffs R, et al. In silico approaches to study mass and energy flows in microbial consortia: a syntrophic case study. BMC Syst Biol. 2009;3:114. doi: 10.1186/1752-0509-3-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Klitgord N, Segre D. Environments that induce synthetic microbial ecosystems. PLoS Comput Biol. 2010;6:e1001002. doi: 10.1371/journal.pcbi.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Mol Syst Biol. 2010;6:407. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhuang K, et al. Genome-scale dynamic modeling of the competition between Rhodoferax and Geobacter in anoxic subsurface environments. ISME J. 2011;5:305–316. doi: 10.1038/ismej.2010.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huthmacher C, Hoppe A, Bulik S, Holzhutter HG. Antimalarial drug targets in Plasmodium falciparum predicted by stage-specific metabolic network analysis. BMC Syst Biol. 2010;4:120. doi: 10.1186/1752-0509-4-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yizhak K, Tuller T, Papp B, Ruppin E. Metabolic modeling of endosymbiont genome reduction on a temporal scale. Mol Syst Biol. 2011;7:479. doi: 10.1038/msb.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bonde BK, Beste DJ, Laing E, Kierzek AM, McFadden J. Differential Producibility Analysis (DPA) of Transcriptomic Data with Metabolic Networks: Deconstructing the Metabolic Response of M. tuberculosis. PLoS Comput Biol. 2011;7:e1002060. doi: 10.1371/journal.pcbi.1002060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nam H, Conrad TM, Lewis NE. The role of cellular objectives and selective pressures in metabolic pathway evolution. Curr Opin Biotechnol. 2011 doi: 10.1016/j.copbio.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Srinivasan K, Mahadevan R. Characterization of proton production and consumption associated with microbial metabolism. BMC Biotechnol. 2010;10:2. doi: 10.1186/1472-6750-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Thiele I, Jamshidi N, Fleming RM, Palsson BO. Genome-scale reconstruction of Escherichia coli’s transcriptional and translational machinery: a knowledge base, its mathematical formulation, and its functional characterization. PLoS Comput Biol. 2009;5:e1000312. doi: 10.1371/journal.pcbi.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Thiele I, Fleming RM, Bordbar A, Schellenberger J, Palsson BO. Functional characterization of alternate optimal solutions of Escherichia coli’s transcriptional and translational machinery. Biophys J. 2010;98:2072–2081. doi: 10.1016/j.bpj.2010.01.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sigurdsson MI, Jamshidi N, Steingrimsson E, Thiele I, Palsson BO. A detailed genome-wide reconstruction of mouse metabolism based on human Recon 1. BMC Syst Biol. 2010;4:140. doi: 10.1186/1752-0509-4-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sweetlove LJ, Ratcliffe RG. Flux-balance modelling of plant metabolism. Frontiers in Plant Science. 2011;2 doi: 10.3389/fpls.2011.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gille C, et al. HepatoNet1: a comprehensive metabolic reconstruction of the human hepatocyte for the analysis of liver physiology. Mol Syst Biol. 2010;6:411. doi: 10.1038/msb.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim J, Reed JL. OptORF: Optimal metabolic and regulatory perturbations for metabolic engineering of microbial strains. BMC Syst Biol. 2010;4:53. doi: 10.1186/1752-0509-4-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bordbar A, et al. A multi-tissue type genome-scale metabolic network for analysis of whole-body systems physiology. BMC Syst Biol. 2011;5:180. doi: 10.1186/1752-0509-5-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhang Y, et al. Three-dimensional structural view of the central metabolic network of Thermotoga maritima. Science. 2009;325:1544–1549. doi: 10.1126/science.1174671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Barua D, Kim J, Reed JL. An automated phenotype-driven approach (GeneForce) for refining metabolic and regulatory models. PLoS Comput Biol. 2010;6:e1000970. doi: 10.1371/journal.pcbi.1000970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lee KH, Park JH, Kim TY, Kim HU, Lee SY. Systems metabolic engineering of Escherichia coli for L-threonine production. Mol Syst Biol. 2007;3:149. doi: 10.1038/msb4100196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Alper H, Jin YS, Moxley JF, Stephanopoulos G. Identifying gene targets for the metabolic engineering of lycopene biosynthesis in Escherichia coli. Metab Eng. 2005;7:155–164. doi: 10.1016/j.ymben.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 139.Kennedy CJ, Boyle PM, Waks Z, Silver PA. Systems-level engineering of nonfermentative metabolism in yeast. Genetics. 2009;183:385–397. doi: 10.1534/genetics.109.105254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xu P, Ranganathan S, Fowler ZL, Maranas CD, Koffas MA. Genome-scale metabolic network modeling results in minimal interventions that cooperatively force carbon flux towards malonyl-CoA. Metab Eng. 2011;13:578–587. doi: 10.1016/j.ymben.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 141.Chemler JA, Fowler ZL, McHugh KP, Koffas MA. Improving NADPH availability for natural product biosynthesis in Escherichia coli by metabolic engineering. Metab Eng. 2010;12:96–104. doi: 10.1016/j.ymben.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 142.Tepper N, Shlomi T. Computational design of auxotrophy-dependent microbial biosensors for combinatorial metabolic engineering experiments. PLoS One. 2011;6:e16274. doi: 10.1371/journal.pone.0016274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Brief details and categorization of 100+ constraint-based methods.
Table S2. Software packages implementing multiple constraint-based methods