Abstract
Escherichia coli isolates (n = 300) collected from six sites in subtropical Brisbane, Australia, prior to and after storm events were tested for the presence of 11 virulence genes (VGs) specific to diarrheagenic pathotypes. The presence of eaeA, stx1, stx2, and ehxA genes specific for the enterohemorrhagic E. coli (EHEC) pathotype was detected in 56%, 6%, 10%, and 13% of isolates, respectively. The VGs astA (69%) and aggR (29%), carried by enteroaggregative (EAEC) pathotypes, were frequently detected in E. coli isolates. The enteropathogenic E. coli (EPEC) gene bfp was detected in 24% of isolates. In addition, enteroinvasive E. coli (EIEC) VG ipaH was also detected in 14% of isolates. During dry periods, isolates belonging to the EAEC pathotype were most commonly detected (23%), followed by EHEC (11%) and EPEC (11%). Conversely, a more uniform prevalence of pathotypes, EPEC (14%), EAEC (12%), EIEC (10%), EHEC (7%), and ETEC (7%), was observed after the storm events. The results of this study highlight the widespread occurrence of potentially diarrheagenic pathotypes in the urban aquatic ecosystems. While the presence of VGs in E. coli isolates alone is insufficient to determine pathogenicity, the presence of diarrheagenic E. coli pathotypes in high frequency after the storm events could lead to increased health risks if untreated storm water were to be used for nonpotable purposes and recreational activities.
INTRODUCTION
Storm events can result in mobilization and transport of fecal contaminants from point sources, such as urban wastewater treatment plants, and nonpoint sources, in particular animal fecal material, to receiving water bodies. The presence of fecal contamination in rivers, lakes, and creeks can lead to the degradation of water quality and subsequently result in the water becoming unfit for potable/nonpotable uses, aquaculture, and recreational activities such as swimming and fishing (1–4).
Escherichia coli and Enterococcus spp. commonly found in mammalian feces have been traditionally used as indicators of fecal pollution in fresh and marine waters (5, 6). After storm events, a severalfold increase in the fecal indicator bacteria (FIB) numbers occur in the surface waters (7–9). There may be several sources of E. coli that contribute to sudden increases in numbers of this bacterium in waterways, including sewage overflows, farm animals, pets, and birds. The elevated microbial contaminants in storm runoff (7, 10) and subsequently in receiving water bodies may pose a serious public health risk. Disease outbreaks related to exposure to contaminated freshwater are well documented (11–14). Exposure to recreational water has been linked to high numbers (21 out of 31) of reported E. coli O157:H7 disease outbreaks in the United States from 1982 to 2002 (15). Despite the significant disease burden linked to contaminated water exposure, the prevalence of E. coli pathotypes in the urban aquatic environment is not well characterized.
The majority of E. coli strains are commensal; however, some strains have acquired specific virulence attributes that allow them to cause a wide spectrum of intestinal and extraintestinal infections, such as diarrhea, urinary tract infection, meningitis, and septicemia (16, 17). Diarrheagenic E. coli has been classified into five well-described groups: enterotoxigenic E. coli (ETEC) strains, which are associated with traveler's diarrhea and porcine and bovine diarrhea; enteropathogenic E. coli (EPEC) strains, which cause diarrhea in children; enterohemorrhagic E. coli (EHEC) strains, which are associated with hemorrhagic colitis and hemolytic-uremic syndrome in humans; enteroaggregative E. coli (EAEC) strains, which are associated with persistent diarrhea in humans; and enteroinvasive E. coli (EIEC) strains, which are involved in invasive intestinal infections, watery diarrhea, and dysentery in humans and animals (16, 17).
Despite increasing evidence that E. coli strains originating from human and animal feces contain several VGs (2, 16, 18), only a few studies have determined whether E. coli strains isolated from fresh and marine water contain VGs and are potentially pathogenic (3, 19–21). The presence of E. coli strains with VG profiles similar to EHEC (21), EPEC (3, 19), and ETEC (21) have been reported previously in the fresh and estuarine waters. However, the distribution of diarrheagenic E. coli pathotypes in freshwater, especially after storm events, still remains relatively unknown. In a recent study, an increase in E. coli VGs after enrichment of water samples collected after rainfall have been reported (22); however, in the absence of actual isolates, no conclusions could be drawn regarding the relative prevalence of pathotypes carrying VGs or prevalence of potentially pathogenic E. coli.
The main aim of this study was to determine the frequency of occurrence of diarrheagenic E. coli pathotypes in surface waters and the impact of storm runoff on their prevalence and distribution. The specific objectives of the study were to (i) determine the frequency of occurrence of potentially pathogenic E. coli strains in the surface water in subtropical Brisbane, Australia; (ii) characterize the VG profile of E. coli isolates to determine the most common pathotypes; and (iii) assess the influence of storm events and runoff on the distribution of potentially pathogenic E. coli strains. This was done to determine the extent of potential human health risks posed by the prevalence of diarrheagenic E. coli pathotypes in surface waters used for potable, nonpotable, and recreational purposes.
MATERIALS AND METHODS
Water sampling and enumeration of E. coli.
One-liter grab samples were collected in sterile Nalgene containers from six sites in Brisbane prior to and after storm events, representing diverse fecal pollution sources, ranging from high-density urban areas to a relatively unpolluted site. A brief site description along with GPS coordinates are provided in Table 1. Samples were collected 1 m from the shore and at a depth of about 0.5 m below water surface with a telescopic water sampler following a dry period (no rainfall in 48 h prior to sampling) and 10 to 12 h after a storm event (>20 mm rainfall). Collected samples were transported to the laboratory on ice and processed within 6 to 8 h of collection.
Table 1.
Sampling site description and location in Brisbane, Australia
| Site name | Site description | GPS coordinates |
|---|---|---|
| Fitzgibbon Drain | Receives runoff from low-density urban areas, some animal input, such as cattle and horses | 27°20′08.7′′S; 153°.01′14.5′′E |
| Cabbage Tree Creek | Medium-density residential and industrial developments | 27°20′59.7′′S; 153°02′06.6′′E |
| Brisbane River | Storm water drain outlets from urban area, dilution effect due to large vol and tidal influence | 27°28′50.05′′S; 152°59′53.84′′E |
| Pine River | Rural area with large block size, animal input (cattle, horses, and sheep) | 27°22′37.45′′S; 152°.59′54.1′′E |
| Oxley Creek | Tributary of the Brisbane River, industrial area, animal input (cattle, horses, and sheep), medium-density population, and close by wastewater treatment plant | 27°32′07.8′′S; 152°59′31.4′′E |
| Enoggera Creek | Moderately populated area, some animal inputs, such as cattle and horses | 27°26′41.96′′S; 152°57′16.90′′E |
The standard membrane filtration method was used for the quantification of E. coli from the collected water samples (8). Briefly, 1- and 10-ml samples were filtered through 0.45-μm nitrocellulose (Millipore) filters (47 mm) and placed on Chromocult coliform agar (Merck). Plates were incubated at 37°C overnight, and typical E. coli colonies were counted to determine the average number of CFU per 100 ml.
E. coli isolation and extraction of DNA.
Individual well-isolated typical E. coli colonies were picked from the Chromocult coliform agar plates and streaked on fresh Chromocult agar plates. During the dry period, from six sampling sites a total of 90 E. coli colonies were isolated during two sampling events. The aim was to collect around 10 E. coli isolates each time from each site, with no more than 2 to 3 colonies isolated from each plate. Similarly, during two wet-period sampling events from the same six sites, around 15 isolates per site were collected, resulting in 210 E. coli isolates. After purification (twice), single colonies were picked from agar plates and inoculated into 2-ml centrifuge tubes containing 1.5 ml nutrient broth (Oxoid). Inoculated tubes were incubated overnight at 37°C in the shaking platform incubator at 100 rpm. All E. coli isolates were stored at −80°C in nutrient broth and 15% (vol/vol) glycerol. At the time of DNA extraction, E. coli isolates were grown in 5 ml of nutrient broth at 37°C overnight. One milliliter of overnight cell culture from each isolate was centrifuged at 6,000 × g for 3 min. The resulting supernatants were removed, and the cell pellets were resuspended in 200 μl of sterile water by vortexing. DNA was extracted from the cell pellets by using the InstaGene matrix according to the manufacturer's instructions (Bio-Rad Laboratories). Presumptive E. coli isolates were confirmed by PCR amplification of the uidA gene as described previously (23).
PCR-positive controls.
E. coli ATCC 9637 was used as a positive control (uidA gene) in PCR assays to confirm presumptive E. coli. E. coli O157:H7 (ATCC 35150) was used as a positive control for the eaeA, stx1, and stx2 genes. Shigella sonnei (ATCC 29930) was used as a positive control for the ipaH gene. The E. coli strain belonging to serotype O138 of porcine origin was used as a positive control for heat-stable (ST) and heat-labile (LT) toxin genes. For the remaining target VGs, pure cultures of clinical E. coli isolates containing target genes were used as positive controls.
PCR detection of E. coli toxin and other target genes.
The list of VGs and the corresponding pathotypes tested in this study are shown in Table 2. Each isolate was screened for a number of adhesion, invasion, and toxin genes to correctly identify them under five main pathotypes. PCR-confirmed E. coli isolates (n = 300) were screened for the presence of 11 diarrheagenic E. coli VGs by using previously published primer sets, stx1 and stx2 (24, 39), eaeA (25), ehxA (18), LT and bfp (26), ST (27), aggR (28), ipaH (29), astA (30), cdtB (31), and thermal cycling conditions. PCRs were performed on a Bio-Rad iQ5 thermocycler system (Bio-Rad Laboratories, California), using iQ supermix (Bio-Rad). Each 25-μl PCR mixture contained 12.5 μl of supermix, 120 to 200 nM each primer, and 3 μl of template DNA. For each PCR experiment, corresponding positive (i.e., target positive-control DNA) and negative (sterile water) controls were included. A melt curve analysis was performed after each PCR run to differentiate between actual products and primer dimers and to eliminate the possibility of false-positive results. The melt curve was generated using 80 cycles of 10 s each starting at 55°C and increasing in 0.5°C intervals to a final temperature of 95°C. The midpoint temperature (Tm) for each amplicon was determined using the iQ5 software (Bio-Rad).
Table 2.
Escherichia coli pathotypes and associated virulence genes tested in this study
| Pathotype | Adhesion/invasion gene | Function | Toxin gene | Function |
|---|---|---|---|---|
| EHEC | eaeAa | Intimin | stx1 | Shiga toxin I |
| stx2 | Shiga toxin II | |||
| ehxAb | Enterohemolysin | |||
| ETEC | LT1 | Heat-labile toxin 1 | ||
| ST1 | Heat-stable toxin 1 | |||
| EPEC | eaeAa | Intimin | ||
| bfp | Type IV bundle-forming pili | |||
| cdtBc | Cytolethal distending toxin | |||
| EAEC | aggR | Transcriptional regulator for chromosomal gene | EAST1 (astA) | EaggEC heat-stable enterotoxin |
| EIEC | ipaH | Invasion plasmid antigen |
Gene carried by both EHEC and EPEC.
Gene also carried by EPEC.
Gene also carried by ExEPC. Typical EHEC strains carry the eaeA gene along with stx1, stx2, or both; typical EPEC strains carry eaeA and bfp genes; typical EAEC strains carry astA and aggR.
Statistical analysis.
The difference in VG distribution among the six sites and existence of variation among VG patterns was determined by analysis of variance (ANOVA) on the pooled E. coli data from the dry- and wet-weather isolates, with significance defined as P values of <0.05. All data on E. coli numbers from all sites were log10 transformed prior to statistical analysis. The Student t test was performed to compare the significance of difference between E. coli numbers across sites and during the dry and wet periods. The critical P value for the t test was set at 0.05, and all tests were considered significant if the P value was <0.05. A linear regression analysis was applied to investigate existence of any correlation between E. coli numbers and VGs during the dry and wet weather.
RESULTS
Prevalence of E. coli during dry and wet periods.
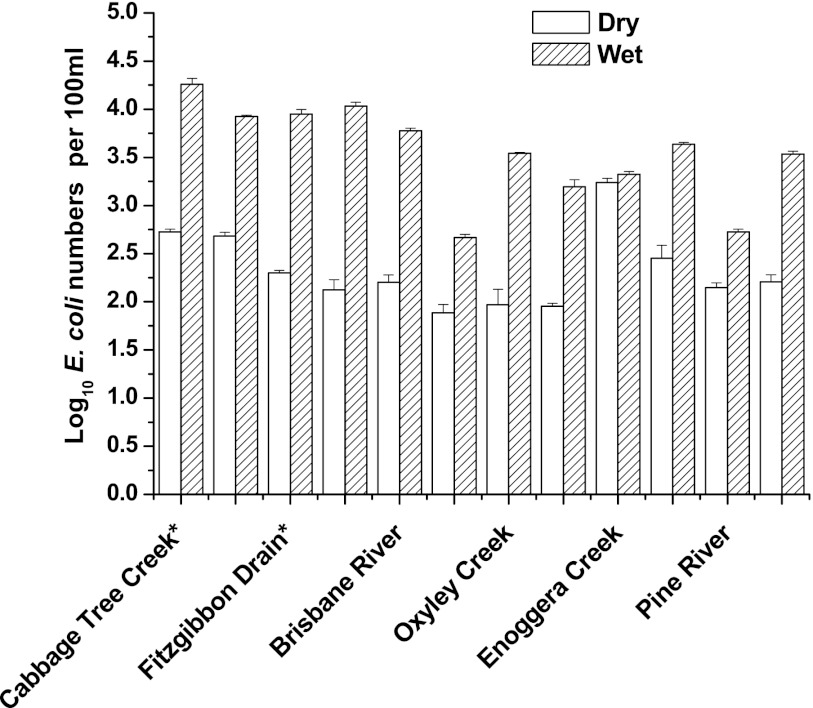
The log10-transformed results for the detection of E. coli are shown in Fig. 1. In general, E. coli numbers in water samples from all sites varied between 1.8 to 4.2 log10 per 100 ml between dry and wet periods. The mean E. coli numbers after the storm events were significantly higher (P < 0.05) than the dry period. Samples collected from Cabbage Tree Creek and the Fitzgibbon Drain sites had significantly higher (P < 0.05) E. coli counts after the storm events than the other four sites tested.
Fig 1.
Comparative E. coli numbers during the dry and wet weather conditions at the six sampling sites during two wet- and dry-period sampling events. *, E. coli counts significantly higher (P < 0.05) from other sites after storm events.
Prevalence of VGs among E. coli isolates.
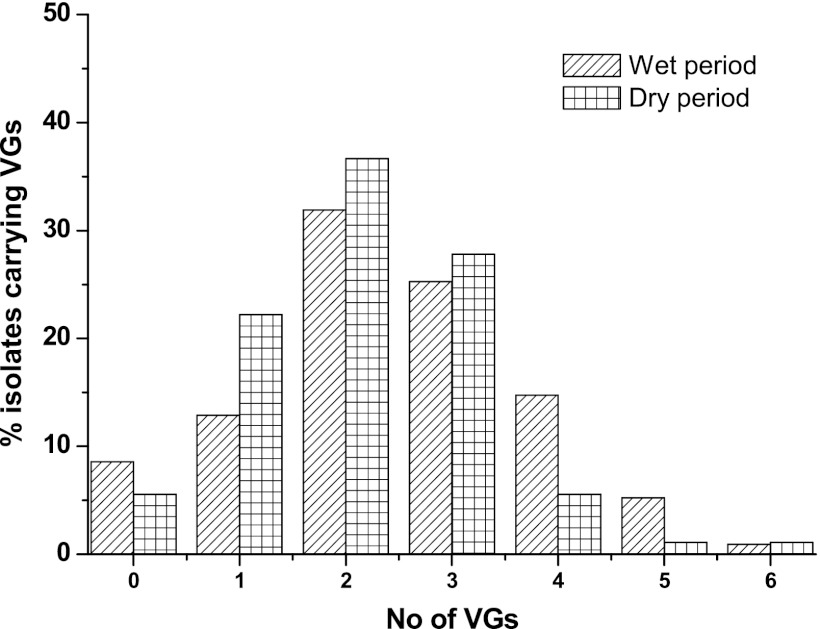
Among the 300 E. coli isolates tested, 256 (85%) carried at least one VG, and 44 isolates (15%) carried no VGs. During the dry period, 72 out of 90 isolates (80%) carried 1 to 3 VGs, and only 7 isolates (8%) carried >4 VGs (Fig. 2). However, after the storm, 145 isolates (69%) were found to harbor 1 to 3 VGs, and a further 32 (15%) carried >4 VGs. The prevalence of multiple VGs in the E. coli isolates was higher after the storm events than during dry periods. However, no correlation was observed between the total number of E. coli isolates and E. coli isolates that were carrying VGs in water samples during the dry or wet period.
Fig 2.
Comparative distribution of the numbers of virulence genes (VGs) carried by individual E. coli isolates.
Among the adhesion and invasion VGs, eaeA, which codes for intimin protein in both EHEC and EPEC pathotypes, was the single most prevalent gene (56%) (Table 3). Among 300 E. coli isolates, only nine (3%) carried only the eaeA gene, and these isolates were classified as atypical EPEC. The EAEC transcriptional regulator gene aggR was detected in 29% of the isolates, which was followed by the EPEC bundle-forming pili gene bfp (24%) and the EIEC invasion plasmid antigen gene ipaH (14%). The eaeA gene was detected more frequently (61%) in E. coli isolates after the storm events than during the dry periods (42%). Similarly, the bfp and ipaH genes were detected more frequently, 27% and 18%, respectively, after the storm events than during dry periods, where their prevalence was 17% and 6%, respectively. In contrast, the aggR gene had a higher prevalence (36%) during the dry periods than during the wet periods (26%).
Table 3.
Occurrence of VGs in Escherichia coli isolated from surface water samples across six sampling sites in Brisbane, Australia
| Period (no. of isolates) | No of E. coli strains carrying virulence gene and distribution (%) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| eaeAa | stx1 | stx2 | LT1 | ST | bfp | cdtBa | ipaH | aggR | astA | ehxAa | |
| Dry period (90) | 38 (42) | 4 (5) | 12 (13) | 2 (2) | 0 (0) | 15 (17) | 8 (9) | 5 (6) | 32 (36) | 67 (75) | 9 (10) |
| Wet period (210) | 129 (61) | 15 (7) | 18 (9) | 4 (2) | 12 (6) | 56 (27) | 17 (8) | 37 (18) | 55 (26) | 139 (66) | 29 (14) |
| Total (300) | 167 (56) | 19 (6) | 30 (10) | 6 (2) | 12 (4) | 71 (24) | 25 (8) | 42 (14) | 87 (29) | 206 (69) | 38 (13) |
These genes are shared by more than one E. coli pathotype.
Among the toxin genes, the enteroaggregative heat-stable enterotoxin 1 (EAST1)-encoding astA gene carried primarily by EAEC but also by EHEC was the single most prevalent gene (69%). The ehxA gene primarily carried by EHEC was the second most commonly detected toxin gene (13%), with a slight increase in detection from 10 to 14% from the dry to wet periods. The cdtB toxin gene was detected in 9% of dry-period isolates and 8% in wet-period isolates. The stx2 EHEC toxin gene (10%) was more frequently detected than stx1 (6%), with the stx2 gene more prevalent in dry-period isolates (13%) than in wet-period isolates (9%). The heat-stable toxin gene (ST gene), generally carried by ETEC, was detected only in E. coli isolates during the wet period (6%). In contrast to all the other toxin genes, the heat-labile toxin gene (LT gene) was detected in a small number of isolates (2%) during both the dry and wet periods (Table 3).
In order to further explore the distribution of the 11 VGs among all six sites, an ANOVA was performed on the pooled data of wet and dry periods. There was a highly significant difference (P < 0.001) between the occurrence of eaeA and ST and LT genes. Similarly a highly significant difference (P < 0.001) was observed in the occurrence of the astA gene compared to that of the stx1, ST, LT, and cdtB genes. A highly significant difference (P < 0.001) was also observed between the occurrence of the aggR gene and the ST and LT genes. The difference between the occurrence of ipaH and astA, ST and bfp, stx2 and astA, and eaeA and stx1 genes was significant (P < 0.01).
Comparative prevalence of E. coli pathotypes.
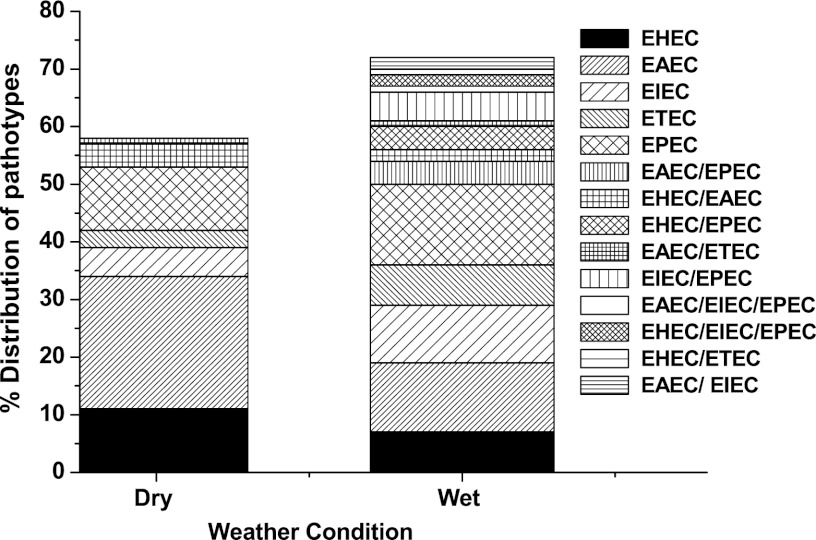
The percentage of E. coli isolates with defined pathotypes from the six sampling locations during the dry and wet periods are shown in Fig. 3. On the basis of combinations of VGs, approximately 53% of the E. coli isolates could be placed into five main pathotypes (EHEC, EAEC, EIEC, ETEC, and EPEC) during the dry periods, and a further 4% of isolates were observed to have combinations of genes from both EHEC and EAEC pathotypes. During the dry periods, isolates belonging to the EAEC pathotype were the most commonly detected (23%), followed by EHEC (11%) and EPEC (11%). Nearly 40% of the dry-period isolates carrying VGs could not be placed under defined categories due to random distribution of single or multiple genes from different defined pathotypes.
Fig 3.
Comparative distribution of E. coli pathotypes during dry and wet weather conditions from all six sites in Brisbane, Australia.
Approximately 50% of the E. coli isolates collected after storm events could be placed under five main pathotypes; however, the distribution of pathotypes was more uniform compared to that during the dry periods. The pathotypes EPEC (14%), EAEC (12%), and EIEC (10%) were more commonly detected than EHEC (7%) and ETEC (7%). In addition, due to a more common occurrence of multiple VGs in the E. coli isolates collected after the storm events, nearly 20% of E. coli isolates could be placed under more than one pathotype. Approximately 9% of the isolates carried a combination of EPEC, EIEC, and EAEC VGs. The remaining 30% of the isolates carrying single or multiple VGs could not be classified into known pathotypes due to a random distribution of genes from more than one pathotype.
Comparison of E. coli VG profiles from six sites.
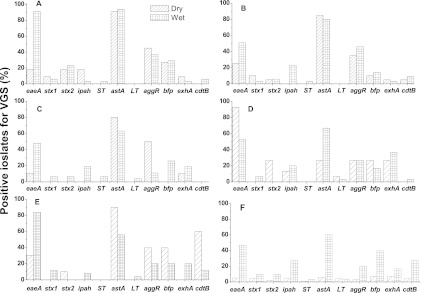
A comparative analysis of the distribution of VGs across all six sites during the dry periods and after the storm events is presented in Fig. 4. In general, the frequency of occurrence of VGs in E. coli isolates collected after the storm event conditions was higher than that during the dry period. However, at the Oxley Creek site, apart from the eaeA gene, all other genes were detected in higher frequency in isolates from the dry periods (Fig. 4E). The Enoggera Creek site has the lowest overall frequency of occurrence of VGs. In contrast, the Fitzgibbon Drain has the highest occurrence of VGs. The VGs astA and eaeA were most frequently detected across all the sites, whereas the LT and ST genes were infrequently detected (Fig. 4). Among the toxin genes screened in this study, ehxA and stx2 genes were the two most commonly detected genes across all sites after the astA gene, which was highly prevalent (>60%). Out of the six sites, the toxin gene cdtB was detected only in more than 10% of isolates from the Oxley Creek and Enoggera Creek sites.
Fig 4.
Comparative distribution of VGs in E. coli isolates at the six sites during the dry periods and after storm events. (A) Fitzgibbon Drain; (B) Cabbage Tree Creek; (C) Brisbane River; (D) Pine River; (E) Oxley Creek; and (F) Enoggera Creek.
A comparison between sites was made (ANOVA) to determine if the sites were similar or different on the basis of occurrence of VGs. The Enoggera Creek site was significantly different (P < 0.05) from the Brisbane River site, whereas differences between the other sites was found to be statistically nonsignificant. In the E. coli isolates from Brisbane River, the stx1, stx2, ST, and LT toxin genes and ipah and bfp adhesion genes were detected only after the storm events. In contrast, at the Enoggera Creek site, all these genes were detected during both dry and wet periods, with a slight increase in frequency after rain fall.
DISCUSSION
Storm runoff may lead to an increased prevalence of microbial pathogens, including diarrheagenic E. coli pathotypes in the surface water bodies due to transport of fecal contamination from land (8). This study compared the distribution and frequency of occurrence of the potentially diarrheagenic E. coli pathotypes in surface water prior to and after storm events to assess if storm runoff could lead to elevated health risks.
The results show a significant increase (P < 0.05) in the E. coli numbers after rainfall at all sites. A potential cause could be fresh human sewage input from sewage leakage and overflow; other likely sources include input from animal sources (32) and mobilization of E. coli surviving in the soil (33), sediments (34), and aquatic environment (35). These findings are in agreement with the previously reported observation of severalfold increase in FIB numbers in the surface water bodies after storm events (7–9). In this study, an increase in the VG distribution among E. coli isolates was observed after the storm events; however, no correlation could be found between the E. coli numbers and the occurrence of VGs during the dry period or storm events. This demonstrates a potential risk of infection from E. coli carrying VGs in storm water even when E. coli numbers in water are not high.
The presence of a single or multiple VGs in an E. coli strain does not necessarily indicate that a strain is pathogenic unless that strain has the appropriate combination of VGs to cause disease in the host (36). The pathogenic E. coli strains use a complex multistep mechanism of pathogenesis involving a number of virulence factors depending upon the pathotype, which consists of attachment, host cell surface modification, invasin, a variety of toxins, and secretion systems which eventually lead toxins to the target host cells (16). Thus, VGs are ideal targets for determining the pathogenic potential of a given E. coli isolate (37).
Enteroaggregative E. coli (EAEC) strains cause persistent diarrhea in children and adults and are defined by the presence of heat-stable enterotoxin 1 (EAST1) along with aggR (16). In this study, the astA gene, encoding E. coli heat-stable enterotoxin 1 (EAST1), was found to be widely distributed in E. coli isolates from all sites (Table 3). The prevalence of the astA gene was statistically higher (ANOVA, P < 0.001) than that of the stx1, ST, LT, and cdtB genes. The high prevalence of E. coli strains carrying the astA gene in fresh and estuarine water has also been reported previously (22) and could potentially be due to its reported presence in many commensal E. coli isolates (38). The results of this study showing the presence of the astA gene in diarrheagenic E. coli pathotypes EAEC, EHEC, and EPEC are in agreement with the previously reported wide distribution of this gene among diarrheagenic E. coli isolates from humans and animals (30, 39, 40). The high prevalence of the toxin gene astA in the E. coli isolates from the storm water is a cause of concern, as E. coli strains carrying astA toxin gene alone have been shown to cause diarrhea in studies from Japan and Spain (41, 42). Since the astA gene is also reported to be carried by many commensal E. coli strains, the implications of the wide prevalence of this gene in surface water remains an open question (39). All E. coli isolates were screened for the presence of the aggR gene to determine if they belong to the EAEC pathotype, and E. coli strains carrying both the astA and aggR genes were classified as typical EAEC. The aggR gene was detected in isolates from the dry period (36%) at a slightly higher frequency than in those from the wet periods (26%), suggesting a relatively high overall prevalence of this pathotype.
The eaeA gene, which codes for intimin protein, was the second most prevalent gene (56%) in the E. coli isolates from storm water. This gene is necessary for intimate attachment to host epithelial cells in both the EHEC and EPEC pathotypes. The frequency of occurrence of this gene was statistically higher at all times (ANOVA, P < 0.001) that that of ST, LT, and other VGs, which were infrequently detected and had a noticeably higher prevalence in the isolates collected after the storm event (61%) than during dry periods (42%). This observation is in agreement with a previously reported finding of significantly higher prevalence of the eaeA gene (up to 96%) in surface water (12, 22). EHEC causes hemorrhagic colitis and hemolytic uremic syndrome in humans, and key virulence factors include intimin (eaeA gene) and Shiga toxins (stx1 and stx2 genes) (36). The relatively high occurrence of the stx2 gene (10%) compared to stx1 (6%) in the storm water E. coli isolates suggests that E. coli carrying a combination of the eaeA and stx2 genes is more common than the combination of eaeA and stx1 genes. This observation is of concern, as the former combination of genes is known to cause more severe diarrhea in humans (18, 36). Typical EPEC strains carry the LEE pathogenicity island, which encodes for several virulence factors, including intimin (eaeA) and the plasmid-encoded bundle-forming pilus (bfp), which mediates adhesion to intestinal epithelial cells (3, 16). Therefore, all isolates were further tested for the presence of the bfp gene to determine if they belong to the EPEC pathotype. In this study, a noticeably higher prevalence of the bfp gene in isolates from storm water (27%) than from dry-period (17%) isolates suggests that a higher prevalence of the EPEC pathotype could be expected in the surface water bodies after storm events.
In addition to the presence of the eaeA gene in both EHEC and EPEC pathotypes, it was also detected in isolates which lacked other typical genes from both groups, and 3% of isolates carried only the eaeA gene. This suggests that there is a wide prevalence of this gene in E. coli found in aquatic ecosystems. Similarly, high prevalence of the eaeA gene in surface water has been reported in other studies (12, 22). This is a cause of concern, as an atypical EPEC pathotype which lacks the bfp gene but carries the eaeA gene has been found to be a major cause of gastroenteritis worldwide (43), in patients suffering from community-acquired gastroenteritis in Melbourne, Australia (44), and from children with diarrhea in Germany (45).
The ehxA toxin gene, which is carried by EHEC and non-Shiga-toxin-producing E. coli pathotypes (2, 16, 18, 46), was the second most commonly detected toxin gene (13%), with a slight increase in the detection frequency from 11 to 16% from dry to wet periods. Enteroinvasive E. coli (EIEC) strains carry the invasion plasmid antigen H (ipaH), which has been used for identification of isolates belonging to this pathotype (27). The frequency of occurrence of this gene was noticeably higher during the wet periods (18%) than during the dry periods (6%), which suggests noticeable movement of this pathotype into surface water from storm runoff.
The results of this study show that diarrheagenic E. coli pathotypes occur at each of the sampling sites during both dry and wet periods (Fig. 3). During the dry periods, a high percentage (53%) of isolates could be grouped under five main diarrheagenic E. coli pathotypes. EAEC strains, which cause persistent diarrhea in children and adults, were the single most common pathotype (23%). This was expected, as the heat-stable enterotoxin 1 (EAST1) gene (also known as astA) and the aggR gene which define this pathotype (16) were two of the most frequently detected genes. This is in agreement with the previously reported high prevalence (up to 80%) of the EAEC pathotype in fresh and estuarine samples (22). The high prevalence of this pathotype is of concern, as EAEC strains are the second-most-common agent of traveler's diarrhea after ETEC, with food and water being the most likely means of transmission (47, 48). EHEC and EPEC were the second- and third-most-common pathotypes detected in this study, with each group represented by 11% of isolates. This suggests that the three pathotypes EAEC, EHEC, and EPEC occur widely in the surface water at all sites.
A more uniform distribution of E. coli pathotypes was observed in E. coli isolates after the storm events, with EPEC (14%), EAEC (12%), and EIEC (10%) strains being the three most commonly detected pathotypes, followed by EHEC (7%) and ETEC (7%). Furthermore, the frequency of occurrence of EAEC pathotypes declined noticeably from 23 to 12% between the dry and wet periods. The observed decline in EAEC pathotypes and more uniform distribution of E. coli pathotypes after rainfall could possibly be due to mobilization of E. coli from point sources, such as wastewater treatment plant discharge, and nonpoint sources, such as animal sources (17, 18, 49, 50) and E. coli surviving in the soil (33), sediments (34), and aquatic environment (35). This could also be a possible explanation of the observed increase (5 to 20%) in the frequency of occurrence of isolates which could be placed under more than one pathotype. The occurrence of unusual combinations of VGs in E. coli isolates observed in this study could be explained on the basis of horizontal gene transfer between cells, which enables the exchange of genetic material located on mobile elements (transposons, integrons, or plasmids) among related or unrelated bacterial species (51). Further screening of the E. coli isolates with these unusual VG patterns in tissue culture or animal models would be required to demonstrate their pathogenicity.
In this study, we collected water samples from areas with diverse human population density and land use to determine if these factors influence the distribution of VGs (Fig. 4). The results of this study did not show any clear pattern of occurrence of VGs across the sites apart from a noticeable difference of occurrence of the cdtB gene (>10% isolates) at Oxley Creek and Enoggera Creek. This suggests that, overall, the contamination sources (point and nonpoint) were potentially similar across sites. There was a difference in the overall occurrence of VGs, with the Fitzgibbon Drain having a high occurrence and the Enoggera Creek site with one of the lowest occurrences of VGs. However, the difference in occurrences was statistically significant (ANOVA, P < 0.05) only between the Enoggera Creek and Brisbane River sites, with the latter site showing prevalence of stx1, stx2, ST, LT, ipah, and bfp genes only after rainfall, unlike the former site, which had a prevalence of these genes during both the dry period and after the storm events.
A better understanding of the prevalence and distribution of E. coli pathotypes in water sources used for potable, nonpotable, or recreation purposes could be an important tool in the development of public health risk mitigation strategies. Pathotyping of E. coli isolates may also provide useful information to identify potential sources of pollution, as the principal reservoirs of EAEC, EIEC, and EPEC pathotypes are humans, whereas the bovine intestinal tract is the main source of the EHEC pathotype (16, 50). The lower prevalence of the EHEC pathotype than other pathotypes suggests that human fecal contamination of the waterways is the main source of diarrheagenic E. coli pathotypes in the surface water as opposed to contamination from animals. This underscores the importance of managing municipal wastewater sources, such as sewage leaks and overflows and wastewater treatment plant discharge, in aquatic environments. Although the frequency of occurrence of certain VGs and pathotypes clearly increased after the rainfall, the presence of these genes could not be attributed to storm water runoff alone. The prevalence of VGs in the E. coli isolates collected during the dry periods suggests that there is always the presence of pathogenic E. coli in the surface water. The results demonstrate that the risk of contracting infection, however, may increase after the storm event.
Since this study was focused on the detection of E. coli pathotypes carrying VGs, it is plausible to assume that actual distribution of these VGs in surface water could be higher. While the ability of E. coli isolates described in this study to cause human diarrheal diseases was not demonstrated, a high proportion of isolates carried a full set of VGs linked to known pathotypes. Further screening for other VGs along with serotype testing and other assays may provide further information on pathogenicity of these isolates.
In conclusion, we found E. coli bacteria with defined pathotypes which originate mainly from human sources, as opposed to contamination from animals, in surface water samples. This underscores the importance of controlling sources of human fecal pollution, such as managing municipal wastewater sources to reduce potential risks to human health. This highlights the need for some degree of treatment of captured storm water prior to its reuse for potable and nonpotable purposes for public health risk mitigation. This study clearly indicates that there is a need to develop a better understanding of public health implications of occurrence of E. coli carrying VGs in water sources used for potable, nonpotable, and recreational purposes.
ACKNOWLEDGMENTS
This research was undertaken and funded as part of the Urban Water Security Research Alliance, a scientific collaboration in South East Queensland, Australia, between the Queensland government, CSIRO Water for a Healthy Country Flagship Program, The University of Queensland, and Griffith University.
Thanks to Nicole Masters for kindly providing clinical isolates of E. coli.
Footnotes
Published ahead of print 2 November 2012
REFERENCES
- 1. Marsalek J, Rochfort Q. 2004. Urban wet-weather flows: sources of fecal contamination impacting on recreational waters and threatening drinking-water sources. J. Toxicol. Environ. Health. Part A 67:1765–1777 [DOI] [PubMed] [Google Scholar]
- 2. Ishii S, Meyer KP, Sadowsky MJ. 2007. Relationship between phylogenetic groups, genotypic clusters, and virulence gene profiles of Escherichia coli strains from diverse human and animal sources. Appl. Environ. Microbiol. 73:5703–5710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hamilton MJ, Hadi AZ, Griffith JF, Ishii S, Sadowsky MJ. 2010. Large scale analysis of virulence genes in Escherichia coli strains isolated from Avalon Bay, CA. Water Res. 44:5463–5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Noble RT, Griffith JF, Blackwood AD, Fuhrman JA, Gregory JB, Hernandez X, Liang X, Bera AA, Schiff K. 2006. Multitiered approach using quantitative PCR to track sources of fecal pollution affecting Santa Monica Bay, California. Appl. Environ. Microbiol. 72:1604–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. NHMRC 2008. The guidelines for managing risks in recreational water. NHMRC Publications, Canberra, Australia: www.nhmrc.gov.au/_files_nhmrc/publications/attachments/eh38.pdf [Google Scholar]
- 6. USEPA 1986. Ambient water quality criteria for bacteria—1986. United States Environmental Protection Agency, Washington, DC [Google Scholar]
- 7. Parker JK, McIntyre D, Noble RT. 2010. Characterizing fecal contamination in stormwater runoff in coastal North Carolina, U.S.A. Water Res. 44:4186–4196 [DOI] [PubMed] [Google Scholar]
- 8. Sidhu JP, Hodgers L, Ahmed W, Chong MN, Toze S. 2012. Prevalence of human pathogens and indicators in stormwater runoff in Brisbane, Australia. Water Res. [Epub ahead of print.] doi:10.1016/j.watres.2012.03.012 [DOI] [PubMed] [Google Scholar]
- 9. Brownell MJ, Harwood VJ, Kurz RC, McQuaig SM, Lukasik J, Scott TM. 2007. Confirmation of putative stormwater impact on water quality at a Florida beach by microbial source tracking methods and structure of indicator organism populations. Water Res. 41:3747–3757 [DOI] [PubMed] [Google Scholar]
- 10. Sauer EP, Vandewalle JL, Bootsma MJ, McLellan SL. 2011. Detection of the human specific Bacteroides genetic marker provides evidence of widespread sewage contamination of stormwater in the urban environment. Water Res. 45:4081–4091 [DOI] [PubMed] [Google Scholar]
- 11. Ackman D, Marks S, Mack P, Caldwell M, Root T, Birkhead G. 1997. Swimming-associated haemorrhagic colitis due to Escherichia coli O157:H7 infection: evidence of prolonged contamination of a fresh water lake. Epidemiol. Infect. 119:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shelton DR, Karns JS, Higgins JA, Van Kessel JA, Perdue ML, Belt KT, Russell-Anelli J, Debroy C. 2006. Impact of microbial diversity on rapid detection of enterohemorrhagic Escherichia coli in surface waters. FEMS Microbiol. Lett. 261:95–101 [DOI] [PubMed] [Google Scholar]
- 13. Chalmers RM, Aird H, Bolton FJ. 2000. Waterborne Escherichia coli O157. Symp. Ser. Soc. Appl. Microbiol. 29:124S–132S [DOI] [PubMed] [Google Scholar]
- 14. Olsen SJ, Miller G, Breuer T, Kennedy M, Higgins C, Walford J, McKee G, Fox K, Bibb W, Mead P. 2002. A waterborne outbreak of Escherichia coli O157: H7 infections and hemolytic uremic syndrome: implications for rural water systems. Emerg. Infect. Dis. 8:370–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rangel JM, Sparling PH, Crowe C, Griffin PM, Swerdlow DL. 2005. Epidemiology of Escherichia coli O157: H7 outbreaks, United States, 1982–2002. Emerg. Infect. Dis. 11:603–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2:123–140 [DOI] [PubMed] [Google Scholar]
- 17. Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Paton AW, Paton JC. 1998. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J. Clin. Microbiol. 36:598–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lauber CL, Glatzer L, Sinsabaugh RL. 2003. Prevalence of pathogenic Escherichia coli in recreational waters. J. Great Lakes Res. 29:301–306 [Google Scholar]
- 20. Hamelin K, Bruant G, El-Shaarawi A, Hill S, Edge TA, Bekal S, Fairbrother JM, Harel J, Maynard C, Masson L, Brousseau R. 2006. A virulence and antimicrobial resistance DNA microarray detects a high frequency of virulence genes in Escherichia coli isolates from Great Lakes recreational waters. Appl. Environ. Microbiol. 72:4200–4206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chern EC, Tsai YL, Olson BH. 2004. Occurrence of genes associated with enterotoxigenic and enterohemorrhagic Escherichia coli in agricultural waste lagoons. Appl. Environ. Microbiol. 70:356–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Masters N, Wiegand A, Ahmed W, Katouli M. 2011. Escherichia coli virulence genes profile of surface waters as an indicator of water quality. Water Res. 45:6321–6333 [DOI] [PubMed] [Google Scholar]
- 23. Frahm E, Obst U. 2003. Application of the fluorogenic probe technique (TaqMan PCR) to the detection of Enterococcus spp. and Escherichia coli in water samples. J. Microbiol. Methods 52:123–131 [DOI] [PubMed] [Google Scholar]
- 24. Lopez-Saucedo C, Cerna JF, Villegas-Sepulveda N, Thompson R, Velazquez FR, Torres J, Tarr PI, Estrada-Garcia T. 2003. Single multiplex polymerase chain reaction to detect diverse loci associated with diarrheagenic Escherichia coli. Emerg. Infect. Dis. 9:127–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang G, Clark CG, Rodgers FG. 2002. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 serotype, and components of the type 2 Shiga toxin family by multiplex PCR. J. Clin. Microbiol. 40:3613–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vidal R, Vidal M, Lagos R, Levine M, Prado V. 2004. Multiplex PCR for diagnosis of enteric infections associated with diarrheagenic Escherichia coli. J. Clin. Microbiol. 42:1787–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Guion CE, Ochoa TJ, Walker CM, Barletta F, Cleary TG. 2008. Detection of diarrheagenic Escherichia coli by use of melting-curve analysis and real-time multiplex PCR. J. Clin. Microbiol. 46:1752–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Toma C, Lu Y, Higa N, Nakasone N, Chinen I, Baschkier A, Rivas M, Iwanaga M. 2003. Multiplex PCR assay for identification of human diarrheagenic Escherichia coli. J. Clin. Microbiol. 41:2669–2671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sethabutr O, Venkatesan M, Murphy GS, Eampokalap B, Hoge CW, Echeverria P. 1993. Detection of Shigellae and enteroinvasive Escherichia coli by amplification of the invasion plasmid antigen H DNA sequence in patients with dysentery. J. Infect. Dis. 167:458–461 [DOI] [PubMed] [Google Scholar]
- 30. Yamamoto T, Nakazawa M. 1997. Detection and sequences of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene in enterotoxigenic E. coli strains isolated from piglets and calves with diarrhea. J. Clin. Microbiol. 35:223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnson JR, Stell AL. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261–272 [DOI] [PubMed] [Google Scholar]
- 32. Davies CM, Bavor HJ. 2000. The fate of stormwater-associated bacteria in constructed wetland and water pollution control pond systems. J. Appl. Microbiol. 89:349–360 [DOI] [PubMed] [Google Scholar]
- 33. Brennan FP, O'Flaherty V, Kramers G, Grant J, Richards KG. 2010. Long-term persistence and leaching of Escherichia coli in temperate maritime soils. Appl. Environ. Microbiol. 76:1449–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Czajkowska D, Witkowska-Gwiazdowska A, Sikorska I, Boszczyk-Maleszak H, Horoch M. 2005. Survival of Escherichia coli serotype O157:H7. Pol. J. Environ. Stud. 14:423–430 [Google Scholar]
- 35. Lothigius A, Sjoling A, Svennerholm AM, Bolin I. 2010. Survival and gene expression of enterotoxigenic Escherichia coli during long-term incubation in sea water and freshwater. J. Appl. Microbiol. 108:1441–1449 [DOI] [PubMed] [Google Scholar]
- 36. Boerlin P, McEwen SA, Boerlin-Petzold F, Wilson JB, Johnson RP, Gyles CL. 1999. Associations between virulence factors of Shiga toxin-producing Escherichia coli and disease in humans. J. Clin. Microbiol. 37:497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuhnert P, Boerlin P, Frey J. 2000. Target genes for virulence assessment of Escherichia coli isolates from water, food and the environment. FEMS Microbiol. Rev. 24:107–117 [DOI] [PubMed] [Google Scholar]
- 38. Ménard LP, Dubreuil JD. 2002. Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): a new toxin with an old twist. Crit. Rev. Microbiol. 28:43–60 [DOI] [PubMed] [Google Scholar]
- 39. Savarino SJ, McVeigh A, Watson J, Cravioto A, Molina J, Echeverria P, Bhan MK, Levine MM, Fasano A. 1996. Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative E. coli. J. Infect. Dis. 173:1019–1022 [DOI] [PubMed] [Google Scholar]
- 40. Yatsuyanagi J, Saito S, Miyajima Y, Amano KI, Enomoto K. 2003. Characterization of atypical enteropathogenic Escherichia coli strains harboring the astA gene that were associated with a waterborne outbreak of diarrhea in Japan. J. Clin. Microbiol. 41:2033–2039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Itoh Y, Nagano I, Kunishima M, Ezaki T. 1997. Laboratory investigation of enteroaggregative Escherichia coli O untypeable:H10 associated with a massive outbreak of gastrointestinal illness. J. Clin. Microbiol. 35:2546–2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Viljanen MK, Peltola T, Junnila SY, Olkkonen L, Jarvinen H, Kuistila M, Huovinen P. 1990. Outbreak of diarrhoea due to Escherichia coli O111:B4 in schoolchildren and adults: association of Vi antigen-like reactivity. Lancet 336:831–834 [DOI] [PubMed] [Google Scholar]
- 43. Hernandes RT, Elias WP, Vieira MA, Gomes TA. 2009. An overview of atypical enteropathogenic Escherichia coli. FEMS Microbiol. Lett. 297:137–149 [DOI] [PubMed] [Google Scholar]
- 44. Robins-Browne RM, Bordun AM, Tauschek M, Bennett-Wood VR, Russell J, Oppedisano F, Lister NA, Bettelheim KA, Fairley CK, Sinclair MI, Hellard ME. 2004. Escherichia coli and community-acquired gastroenteritis, Melbourne, Australia. Emerg. Infect. Dis. 10:1797–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kozub-Witkowski E, Krause G, Frankel G, Kramer D, Appel B, Beutin L. 2008. Serotypes and virutypes of enteropathogenic and enterohaemorrhagic Escherichia coli strains from stool samples of children with diarrhoea in Germany. J. Appl. Microbiol. 104:403–410 [DOI] [PubMed] [Google Scholar]
- 46. Paton AW, Paton JC. 2002. Direct detection and characterization of Shiga toxigenic Escherichia coli by multiplex PCR for stx1, stx2, eae, ehxA, and saa. J. Clin. Microbiol. 40:271–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang DB, Mohanty A, DuPont HL, Okhuysen PC, Chiang T. 2006. A review of an emerging enteric pathogen: enteroaggregative Escherichia coli. J. Med. Microbiol. 55:1303–1311 [DOI] [PubMed] [Google Scholar]
- 48. Mohamed JA, Huang DB, Jiang ZD, DuPont HL, Nataro JP, Belkind-Gerson J, Okhuysen PC. 2007. Association of putative enteroaggregative Escherichia coli virulence genes and biofilm production in isolates from travelers to developing countries. J. Clin. Microbiol. 45:121–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lyautey E, Lu Z, Lapen DR, Wilkes G, Scott A, Berkers T, Edge TA, Topp E. 2010. Distribution and diversity of Escherichia coli populations in the South Nation River drainage basin, eastern Ontario, Canada. Appl. Environ. Microbiol. 76:1486–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Levine MM. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive, enterohemorrhagic, and enteroadherent. J. Infect. Dis. 155:377–389 [DOI] [PubMed] [Google Scholar]
- 51. Davison J. 1999. Genetic exchange between bacteria in the environment. Plasmid 42:73–91 [DOI] [PubMed] [Google Scholar]