Abstract
The type VI secretion system (T6SS) is a versatile secretion machine dedicated to various functions in Gram-negative bacteria, including virulence toward eukaryotic cells and antibacterial activity. Activity of T6SS might be followed in vitro by the release of two proteins, Hcp and VgrG, in the culture supernatant. Citrobacter rodentium, a rodent pathogen, harbors two T6SS gene clusters, cts1 and cts2. Reporter fusion and Hcp release assays suggested that the CTS1 T6SS was not produced or not active. The cts1 locus is composed of two divergent operons. We therefore developed a new vector allowing us to swap the two divergent endogenous promoters by Ptac and PBAD using the λ red recombination technology. Artificial induction of both promoters demonstrated that the CTS1 T6SS is functional as shown by the Hcp release assay and confers on C. rodentium a growth advantage in antibacterial competition experiments with Escherichia coli.
INTRODUCTION
Citrobacter rodentium is an extracellular Gram-negative rodent pathogen that causes transmissible colonic hyperplasia in mice (1, 2). It colonizes the lumen of the mouse gut mucosa, causing attaching and effacing (A/E) lesions on the surface of the enterocytes. Its virulence strategy is similar to that of the clinical human pathovar enteropathogenic Escherichia coli (EPEC) and enterohemorrhagic E. coli (EHEC) (3). For this reason, C. rodentium is a valuable model to study the mechanisms underlying A/E lesions under physiological conditions, with the ability to manipulate both the pathogen and the host (4). The A/E lesions are mediated by the locus of enterocyte effacement (LEE) pathogenicity island (5), which encodes the well-characterized type III secretion system (T3SS) and cognate effector proteins (6, 7). The C. rodentium ICC168 genome has been recently sequenced (8). Aside from the T3SS, several other secretion machines have been identified, such as two type VI secretion systems (T6SSs) that were named T6SS clusters 1 (CTS1) and 2 (CTS2) (8).
The T6SS is a macromolecular transenvelope machine composed of at least 13 subunits, called core components, required for proper assembly and function of the secretion apparatus (9, 10). Some of these proteins are structurally analogous to that assembling the tail of contractile bacteriophages (11). The secreted T6SS hemolysin-coregulated protein (Hcp), postulated to form a tube-like structure (12), is a structural analogue of gp19, the phage tail tube protein of bacteriophage T4 (13, 14). The valine-glycine repeat (VgrG) protein, localized to the tip of the Hcp tube (15), may function similarly to the structurally analogous gp5/gp27 complex of bacteriophage T4, forming the spike required for puncturing the targeted cells (13). A recent work revealed that the TssB (VipA) and TssC (VipB) proteins assemble a dynamic structure in the cytoplasm of Vibrio cholerae resembling the bacteriophage sheath. Based on this observation, it has been proposed that cycles of extension and contraction of the T6SS sheath propel the Hcp tube toward adjacent target cells prior to effector translocation (16). This inverted tail-like structure is supposed to be anchored to the cell envelope by a membrane complex composed of the TssM, TssL, and TssJ proteins (17–21).
Gene clusters encoding T6SS are widely distributed among proteobacteria, and it is not rare that they are found in several copies on the chromosome (9, 10, 22). A few examples of T6SSs that have been studied are directly responsible for pathogenesis by the injection of toxins into eukaryotic cells that interfere with the cytoskeleton (15, 23–26). In contrast, a growing number of T6SSs are shown to kill target bacterial cells by translocating antibacterial effectors directly into the periplasm or the cytoplasm of the prey upon cell-cell contact (27–33).
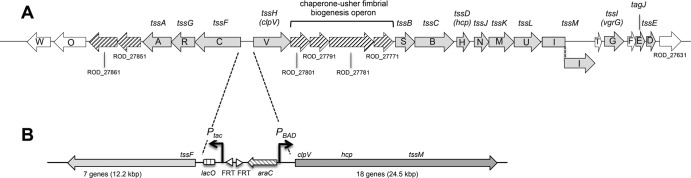
In this study, we focused our work on the Citrobacter rodentium CTS1 T6SS. CTS1 is composed of two operons organized in opposite orientations, which bear all the genes encoding the T6SS core components (Fig. 1A). These T6SS genes were originally named cts followed by a letter (e.g., ctsV, ctsB, and ctsG) (8). However, we adhere to the general nomenclature proposed by Shalom et al. (34). The longest operon contains tssH (clpV), tssB, tssC, tssD (hcp), tssJ, tssK, tssL, tssM, tssI (vgrG), and tssE. Interestingly, four genes encoding chaperone-usher fimbriae are embedded within this operon, suggesting a probable coproduction of the T6SS and fimbriae (Fig. 1A) (8). The second operon carries the core component genes tssF, tssG, and tssA (Fig. 1A). To assemble a functional T6SS, it is therefore essential that both operons are coexpressed. However, the CTS1 tssM gene, formerly named cts1I (8), is disrupted by a frameshift. TssM is an inner-membrane protein with a large periplasmic domain. The C terminus of the periplasmic domain interacts with the outer-membrane lipoprotein TssJ. This interaction is critical for T6SS function in enteroaggregative Escherichia coli (20). In C. rodentium CTS1, the frameshift mutation leads to a shorter TssM protein which lacks the C-terminal domain. To test whether the cts1 gene cluster encodes a functional T6SS, we searched for Hcp1 in the culture supernatant. Hcp release assay suggested that the CTS1 T6SS was not produced or not functional. To investigate the functionality of CTS1, we created a genetic tool allowing the simultaneous swapping of the two divergent chromosomal promoters by Ptac and PBAD using λ red recombination. This tool was validated and further demonstrated that CTS1 assemble a functional T6SS that confers a bacterial fitness benefit on C. rodentium.
Fig 1.
Organization of the C. rodentium CTS1 locus in the WT and engineered strains. (A) Organization of the CTS1 locus in strain ICC168. Genes are indicated using the original nomenclature (cts) (8) and the general T6SS gene nomenclature (tss) (34). Arrowheads show gene orientations. Gray arrows indicate T6SS core and accessory genes found in other T6SS gene clusters. Striped arrows indicate genes encoding putative fimbriae. Open arrows designate genes that are not conserved among T6SS gene clusters. (B) Schematic organization of the engineered CTS1 locus in which the divergent promoter region has been swapped with inducible Ptac and PBAD promoters (strain RLC55). The lacO operator and araC genes are indicated.
MATERIALS AND METHODS
Bacteria and growth conditions.
Bacterial strains used in this study are listed in Table S1 in the supplemental material. Strains were routinely grown in lysogeny broth (LB) or on LB agar plates (35), supplemented with ampicillin (Amp, 100 μg · ml−1), kanamycin (Kan, 50 μg · ml−1), chloramphenicol (Cm, 30 μg · ml−1), or nalidixic acid (Nal, 20 μg · ml−1) as required. The C. rodentium DBS100 isolate used in this work originates from the same stock as the sequenced strain ICC168 (4, 8) and was generously provided by H. Le Moual (McGill University, Montreal, Canada). A spontaneous mutant of C. rodentium DBS100 resistant to nalidixic acid (20 μg · ml−1) was obtained by growing DBS100 at 37°C in LB medium supplemented with Nal at 5 μg · ml−1 for 7 h before spreading the liquid culture onto LB agar plates supplemented with Nal at 20 μg · ml−1.
Recombinant DNA methods.
Colony PCR was performed with Taq DNA polymerase with standard Taq buffer (New England BioLabs). PCR fragments used for plasmid construction and λ red technology were amplified with Phusion high-fidelity DNA polymerase (Thermo Scientific). Phusion-generated fragments and final constructs were verified by DNA sequencing. Restriction enzymes were purchased from New England BioLabs. Custom oligonucleotides were synthesized by Sigma-Aldrich and are listed in Table S2 in the supplemental material.
Plasmids construction.
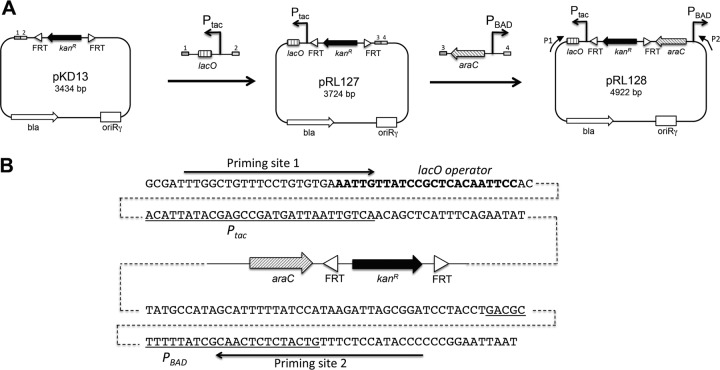
The pKD13-derived pRL128 plasmid was constructed in two steps by restriction-free (RF) cloning (36, 37) using oligonucleotides with a 3′ target-specific sequence and a 5′ extension complementary to the desired insertion site in the pKD13 destination plasmid (38). First, the Ptac-lacO DNA region of the pMAL-p2 plasmid was amplified by PCR with the pKD13-ptac#1 and pKD13-ptac#2 primers (see Table S2 in the supplemental material). The PCR product was then used as oligonucleotides for a second PCR using pKD13 as a template (Fig. 2A). The resulting plasmid, carrying the Ptac-lacO DNA fragment downstream of the Flp recognition target (FRT) site and Kanr-encoding gene, was named pRL127 (Fig. 2A). Then, the araC-PBAD DNA region of pBAD18 was amplified by PCR with the pKD13-pBAD#1 and pKD13-pBAD#2 oligonucleotides (see Table S2). The PCR product was used as oligonucleotides for a second PCR using pRL127 as a template. In the resulting plasmid, designated pRL128, the Ptac and PBAD promoters are in divergent orientations and flank the two FRT sites (Fig. 2). pRL127 and pRL128 were verified by restriction to check the presence of the inserts at the desired positions and orientations. Inserts were sequenced. pRL128 was made available with the Addgene plasmid library (deposited under accession number 40180).
Fig 2.
Construction and features of pRL128. (A) Different cloning steps during construction of the pRL128 plasmid template. The parental pKD13 vector is a template plasmid used in the λ red technology (38). A Ptac-lacO DNA fragment was amplified with primers carrying 5′ extremities corresponding to DNA sequences of pKD13 symbolized by rectangles 1 and 2. This Ptac-lacO fragment was inserted into pKD13 by RF cloning (see Materials and Methods). Then, an araC-PBAD DNA fragment was inserted into pRL127. For pRL128, P1 and P2 arrows indicate the orientation and location of the primers carrying the priming site sequence P1 and P2 (see Table S2 in the supplemental material). (B) Sequences of the pRL128 regions around the Ptac and PBAD promoters. Promoter sequences are underlined. The lacO sequence is indicated in bold letters. Priming sites 1 and 2 are indicated by arrows.
Construction of the complementation vector expressing the tssM1 gene (pRL39) is described in the supplemental material.
Strain construction.
The construction of the C. rodentium strain in which the CTS1 promoter region is swapped with the Ptac and PBAD promoters in divergent orientations was achieved by following the PCR-based method of Datsenko and Wanner (38) using pRL128 as the template, yielding strain RLC55 (see Fig. S1 in the supplemental material). The ΔtssM1 and ΔlacYZ isogenic mutant strains were generated using pKD13 (38). The chromosomal LacZ translational fusions were generated using plasmid pFUSE (39). Details regarding these constructions can be found in the supplemental material.
Isolation of extracellular proteins (Hcp release assay).
C. rodentium was grown to an optical density at 600 nm (OD600) of 0.8 in LB medium at 37°C. A total of 2 × 109 cells were harvested by centrifugation at 2,000 × g for 5 min and resuspended in denaturing loading buffer. When required, gene expression was induced by addition of isopropyl-β-d-thiogalactopyranoside (IPTG; 500 μM) and/or arabinose (0.2%) for 30 min. The supernatant was subjected to centrifugation at 20,000 × g for 15 min, and extracellular proteins were precipitated by addition of trichloroacetic acid (TCA; 15% final concentration) for at least 2 h on ice. Precipitated proteins were recovered by centrifugation at 20,000 × g at 4°C for 45 min. Pellets were washed with 1 ml acetone, air dried, and resuspended in denaturing loading buffer. Proteins were separated by electrophoresis on denaturing 15% acrylamide gels and visualized by Coomassie blue staining. The bands from the supernatant fraction of the RLC55 C. rodentium strain were excised and subjected to in-gel trypsin digestion followed by matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (Proteomic Facility, CNRS, Marseille, France).
Antibacterial competition assay.
The wild-type (WT) E. coli strain W3110 bearing the pUA66-rrnB plasmid (Kanr [40]) was used as prey in the competition assay. The pUA66-rrnB plasmid provides a strong constitutive green fluorescent (GFP+) phenotype to E. coli. Cells were grown in LB at 37°C to an OD600 of 1, normalized to an OD600 of 0.5, and mixed to a 4:1 ratio (predator to prey). Then, 25 μl of the mixture was spotted in triplicate onto a prewarmed dry agar plate supplemented or not with 2% arabinose and 500 μM IPTG (this high concentration of arabinose was used to circumvent the catabolism of this sugar by C. rodentium and E. coli during the overnight [14-h] incubation, which could limit the full induction of the T6SS genes in the RLC55 C. rodentium strain). Agar plates were incubated overnight at 30°C. Fluorescent images of the competition assays were taken with a LI-COR Odyssey imager. The bacterial spots were cut off, and cells were resuspended in 1 ml of LB. Triplicates of 150 μl were transferred into wells of a black 96-well plate (Greiner), and the absorbance at 600 nm and fluorescence (excitation, 485 nm; emission, 530 nm) were measured with a Tecan Infinite M200 microplate reader (9 measures per mixture per experiment). The relative fluorescence was expressed as the intensity of fluorescence divided by the absorbance at 600 nm, after subtracting the values of a blank sample. These results are given in arbitrary units (AU) because the intensity of fluorescence is acquired with a variable gain and hence varies from one experiment to the other. The experiments were done in triplicate, with identical results, and we report here the results of a representative experiment. For enumeration of viable cells, bacterial suspensions recovered from the spots were serially diluted and spotted onto selective LB agar plates supplemented with kanamycin (for E. coli) or nalidixic acid (for C. rodentium).
RESULTS AND DISCUSSION
The C. rodentium CTS1 T6SS is inactive under laboratory conditions.
The release of the Hcp protein in the culture supernatant is a hallmark feature of functional T6SSs. Hcp is an ∼20-kDa protein assembling the extracellular portion of the T6SS apparatus. Hcp is usually abundantly produced and accumulates in the medium when T6SS expression is activated. To test whether the C. rodentium CTS1 T6SS is functional under laboratory conditions, we attempted to detect the presence of the CTS1 Hcp (Hcp1) protein in the supernatant. Whereas samples were harvested in exponential and stationary phases of cells grown under various conditions, including rich (LB) and minimal (M9 and M63) media, we did not detect Hcp1 in the culture supernatant (data not shown and Fig. 3A, lanes 1 and 2). We hypothesize that (i) the CTS1 T6SS gene operons were not expressed under the various conditions tested or (ii) the CTS1 T6SS is not functional because of the frameshift mutation in the tssM1 gene previously reported (8). Translational fusions in which the promoters of tssF1 (cts1C) and clpV1 (cts1V) control the expression of the reporter lacZ gene were constructed. Comparison of β-galactosidase activities of the RLC11 (ΔlacZY), RLC15 (tssF1-lacZ), and RLC16 (clpV1-lacZ) strains demonstrated that these promoters were silent (data not shown). It is noteworthy that the production of T6SS is often repressed under standard laboratory conditions in many bacterial species (41, 42). Many regulatory mechanisms have been described so far, and it will be interesting to identify the proteins controlling the expression of the cts1 gene cluster using transposon mutagenesis and the lacZ reporter translational fusions, as done previously for the Sci-1 T6SS of enteroaggregative E. coli (43). To discriminate between the two hypotheses raised above, we decided to artificially induce the expression of the chromosomally encoded T6SS gene operons. However, to circumvent the difficulty of the C. rodentium CTS1 T6SS being encoded on two divergent operons, we constructed a plasmid template allowing us to engineer a strain in which the endogenous intergenic region has been swapped for two divergent and inducible promoters.
Fig 3.
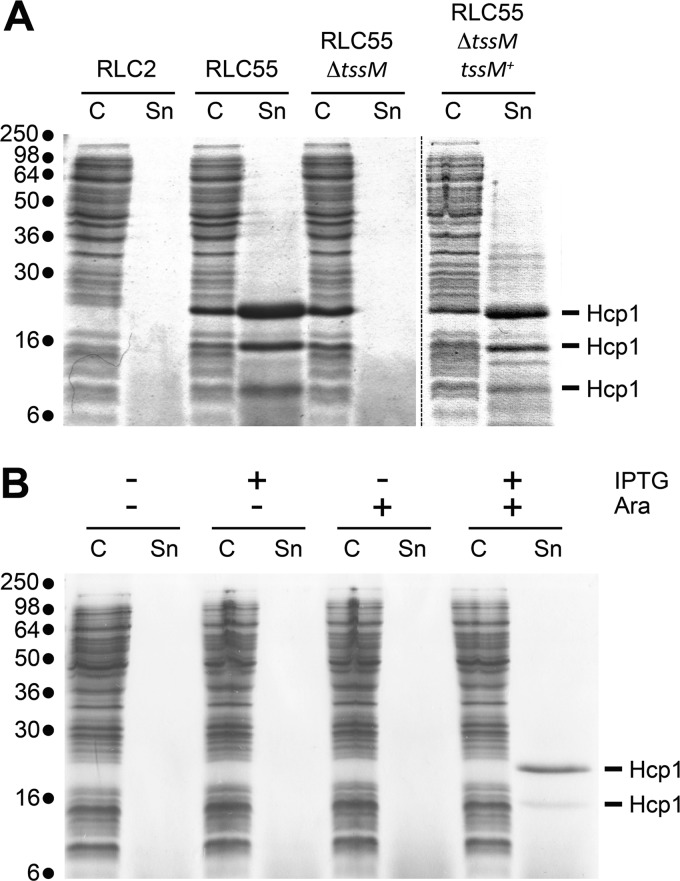
Hcp release assay. (A) Hcp release assay using the WT strain RLC2, the engineered RLC55 strain (in which the endogenous promoters have been exchanged with the inducible Ptac and PBAD promoters), its ΔtssM1 derivative in the presence of IPTG (500 μM) and arabinose (0.2%), and the ΔtssM1 derivative complemented with the tssM1 gene (tssM1+). The extracellular proteins were isolated by separating whole cells (C) and the supernatant fraction (Sn). Proteins were visualized by Coomassie blue staining after 15% SDS-PAGE. Proteins detected in the RLC55 Sn fraction were identified by mass spectrometry and correspond to the CTS1 Hcp protein (see text for details). Molecular mass markers are indicated on the left. (B) Hcp release assay using the engineered RLC55 strain after 30 min of gene expression induction in the presence of the indicated inducers (IPTG [500 μM] and arabinose [0.2%]). Molecular mass markers are indicated on the left.
Construction and validation of pRL128, a new pKD13 derivative vector for divergent promoter swapping.
Several studies reported chromosomal promoter swapping with inducible promoters. Promoter replacement has been shown to be a successful approach to characterize the function of genes, including those for which the expression is modulated by a feedback mechanism (44, 45), or to control the expression of a given gene in order to change the metabolic flux and thus optimize the production of a biological compound (46). Recently, a promoter replacement tool based on a SacB suicide plasmid carrying Plac was described and employed to modulate the expression of the pil locus gene, encoding a type IVb pilus in Pseudomonas aeruginosa (47). However, this method requires cloning steps and selection of double recombination events with the sacB counterselection marker, which can be time-consuming. However, none of the template plasmids or techniques allowed the quick exchange of two divergent chromosomal promoters for inducible promoters in opposite orientations (reviewed in reference 48). Our promoter swapping strategy was based on the one-step gene modification technology allowing short template recombination of electroporated PCR products with the chromosome by the λ red proteins. We constructed a pKD13-derived plasmid, named pRL128, allowing swapping of chromosomal endogenous divergent promoters by Ptac and PBAD promoters (see Materials and Methods). pRL128 bears on either side of the Kanr cassette flanked by the FLP recognition target sites the Ptac and PBAD promoters oriented in opposite directions (Fig. 2). The pRL128 plasmid was constructed from pRL127, a pKD13 derivative carrying the Ptac promoter and the lacO operator, adjacent to the Kanr cassette and an FRT sequence. pRL127 is very similar to pKE2, a pKD4-derived vector designed in a promoter swapping strategy in order to analyze the function of the PhoU phosphate regulatory protein in E. coli (45). pRL128 has the advantage of carrying both promoters Ptac and PBAD flanking the Kanr cassette and the FRT sequences. Depending on the primers designed for the PCR, pRL128 can thus be used as a template to insert either Ptac or PBAD or, if required, both promoters in opposite directions, anywhere into the chromosome.
Using pRL128 as the template, we constructed the RLC55 strain, in which the expression of the genes of the longest operon, including tssH1 (cts1V), hcp1, and tssM1 (cts1I), is under the control of the PBAD promoter. The shortest operon, including tssF1 (cts1C), is under the control of the Ptac promoter (Fig. 1B).
The inducible C. rodentium CTS1 gene operons encode a functional T6SS.
To test whether the CTS1 gene operons encode a functional T6SS, we assayed for Hcp secretion under inducible conditions. Since gene expressions from Ptac and PBAD are, respectively, activated in the presence of IPTG and arabinose, RLC55 was grown in LB supplemented with 0.2% (wt/vol) arabinose and 500 μM IPTG. These concentrations of arabinose and IPTG were used to maximize the level of expression of the T6SS genes. The wild-type, nonengineered strain RLC2 and a RLC55 derivative carrying a deletion of the tssM1 gene (i.e., a gene previously shown to be critical for T6SS function [21, 49, 50]) were used as controls. Figure 3A shows that while no protein is found in the culture supernatant of the wild-type RLC2 strain, three proteins with apparent molecular masses of 20, 16, and 10 kDa are released from RLC55. MALDI-TOF mass spectrometry analyses demonstrated that the three bands corresponded to the Hcp1 protein (ROD_27741; accession number NC_013716). While peptides corresponding to the C terminus of Hcp1 were not identified in the protein of the lower band, mass spectrometry analysis did not detect any truncation of Hcp that could explain the difference of migration that is observed in the SDS-PAGE gel for the two upper bands. However, an internal peptide was not recovered in the upper band, suggesting that a posttranslational modification occurred on this fragment. Identification of the putative posttranslational modification is under investigation. These bands were not observed in the culture supernatant of the ΔtssM1 strain (Fig. 3A), suggesting that Hcp1 is released in a CTS1-dependent manner. The presence of Hcp1 confirmed that the CTS1 T6SS is functional despite the frameshift mutation in the tssM1 gene (8). Using PCR amplification of the tssM1 gene and sequencing, we confirmed that it encodes a truncated form (data not shown). However, the tssM1 nucleotide sequence has a stretch of 11 adenosines upstream of its stop codon. Because polyadenosine tracks may influence transcription by RNA polymerase slippage (51), it will be interesting to determine whether the truncated form of TssM is functional or whether transcriptional frameshifting occurs in vivo.
To test whether CTS1 function requires both operons, the Hcp release assay was repeated in the absence of arabinose (Fig. 3B, lanes 3 and 4) or of IPTG (Fig. 3B, lanes 5 and 6). As shown in Fig. 3B, both inductions with IPTG and arabinose are required for efficient Hcp1 release, suggesting that proteins encoded by the two operons are necessary to assemble a functional T6SS.
CTS1 confers a growth advantage on C. rodentium.
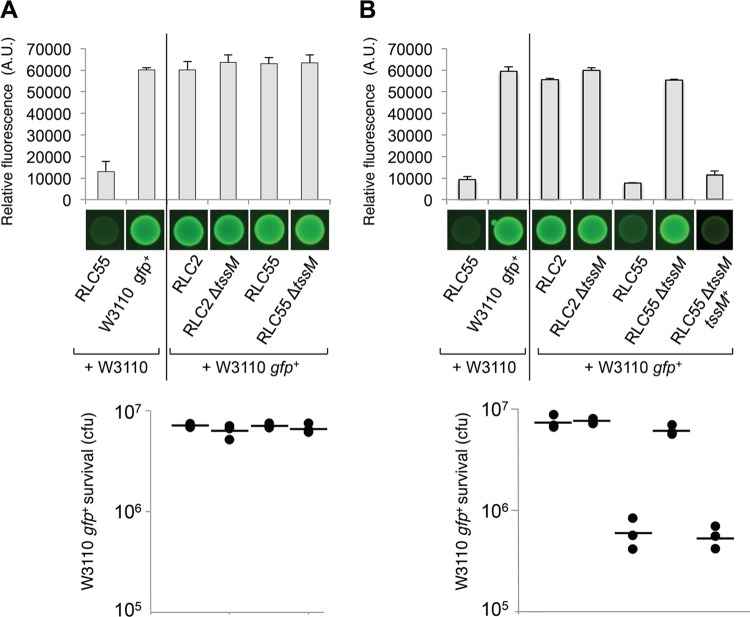
Since Hcp is released from RLC55 cells, we hypothesized that the CTS1 T6SS is functional and that it could achieve its function, so far undetermined. The T6SS has been shown to be versatile in function and suggested to be adapted to the specific needs of each bacterium. So far, while a few T6SSs are required for full virulence toward eukaryotic cells (52–54), most T6SSs display an antibacterial activity able to kill or prevent the growth of competitor bacteria (32). We therefore performed several assays. Using the worm Caenorhabditis elegans, we first observed that the CTS1 T6SS did not confer reduced or increased virulence on C. rodentium (data not shown). We therefore tested whether CTS1 confer a growth advantage in an interspecies competition assay. The growth competition experiments were conducted between C. rodentium strains as predators and a fluorescent E. coli K-12 strain as prey. Control experiments showed that RLC2, RLC55, its ΔtssM1 derivative, the ΔtssM1 complemented C. rodentium, and the E. coli strains used here have similar growth behaviors and similar generation times. The predator and the prey were mixed and incubated onto an LB agar plate overnight at 30°C. The outcome was then determined by measuring the level of fluorescence and the number of surviving E. coli cells (Fig. 4). C. rodentium cells grown with a nonfluorescent E. coli strain displayed a basal, low fluorescence level at about 12,000 AU (Fig. 4A, top). In the absence of inducers, fluorescence levels of the mixes were comparable to those in a control experiment in which the fluorescent E. coli strain was mixed with the nonfluorescent E. coli strain (∼60,000 AU). This indicated that WT C. rodentium does not prevent the growth of E. coli. This was confirmed by CFU counts to measure the survival of the E. coli strain (Fig. 4A, bottom). The number of fluorescent E. coli cells surviving after overnight incubation with C. rodentium was similar to that mixed with the nonfluorescent E. coli. From these data, we concluded that the CTS1 T6SS does not harbor an antibacterial activity or that the expression of the CTS1 gene operons is not induced by direct contact with prey cells. The experiment was then repeated in the presence of both arabinose and IPTG (Fig. 4B). Under these conditions, a 7-fold decrease of the fluorescence level and a 12-fold decrease of survival were observed for the E. coli cells cocultivated with the Ptac/PBAD RLC55 strain. This decrease was due to the controlled expression of a functional CTS1 T6SS, since the RLC55 ΔtssM1 derivative did not confer a growth advantage on C. rodentium. This result indicates the CTS1 T6SS confers a growth advantage on C. rodentium. The role of T6SS for competition toward neighboring bacterial cells has already been made evident in several Gram-negative bacteria, including P. aeruginosa, V. cholerae, and Serratia marcescens (27–29). This result also suggests that the CTS1 T6SS delivers effector proteins that carry antibacterial activities to the prey. A few T6SS effectors have been described so far. In P. aeruginosa, two enzymes, Tse1 and Tse3, are translocated to the periplasm of the recipient cell and target the peptidoglycan (27, 30, 55), while a third effector, Tse2, has been described, but its activity and its molecular target are still unknown (27). It is noteworthy that a superfamily of T6SS effectors with amidase activity was recently identified in various organisms using a heuristic approach (31). None of these effectors are encoded on the C. rodentium genome, suggesting that CTS1 translocates an effector(s) with a novel activity. Further experiments will be performed to identify proteins delivered by the CTS1 T6SS.
Fig 4.
The CTS1 T6SS confers a growth advantage on C. rodentium. A growth competition assay was conducted using a fluorescent E. coli strain as prey (W3110 gfp+) and different strains of C. rodentium (RLC2, the engineered RLC55, its ΔtssM1 derivative, and the complemented ΔtssM1 strain [tssM1+]) as predators. C. rodentium cells were mixed with GFP+ E. coli W3110 cells, spotted onto LB agar supplemented (B) or not (A) with IPTG (500 μM) and arabinose (2%), and incubated for 14 h at 30°C. As controls, W3110 gfp+ and RLC55 cells were mixed with nonfluorescent W3110 E. coli cells. The relative fluorescence (upper graph) is expressed in arbitrary units (A.U.) and is the mean of fluorescence levels obtained from three independent experiments (each measured in triplicate). Fluorescent images of the competition assays (obtained with a LI-COR Odyssey imager) are shown below the upper graph. The number of recovered E. coli cells (survival expressed in CFU) is indicated in the lower graph (the closed circles indicate values from three independent assays, and the average is indicated by the bar).
Supplementary Material
ACKNOWLEDGMENTS
We thank Hervé Le Moual (McGill University, Montreal) for providing the wild-type C. rodentium DBS100 strain and the pFUSE plasmid; the members of the Cascales, Lloubès, Bouveret, and Sturgis research groups for discussion; Emmanuelle Bouveret for the Tecan microplate reader; Régine Lebrun and Remy Puppo (Proteomic Facility, CNRS, Marseille, France) for mass spectrometry analyses; Isabelle Bringer, Annick Brun, and Olivier Uderso for technical assistance; and Marcel Éloem for encouragement.
Work in the laboratory of E.C. is supported by the CNRS and funded by a grant from the Agence National de la Recherche (ANR-10-JCJC-1303-03). E.G. was supported by a postdoctoral grant from the Fondation pour la Recherche Medicale (SPF-2009-12-17571).
Footnotes
Published ahead of print 12 October 2012
We heartily dedicate this work to Guy Duval-Valentin (LMGM, Toulouse, France).
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02504-12.
REFERENCES
- 1. Barthold SW, Coleman GL, Bhatt PN, Osbaldiston GW, Jonas AM. 1976. The etiology of transmissible murine colonic hyperplasia. Lab. Anim. Sci. 26:889–894 [PubMed] [Google Scholar]
- 2. Barthold SW, Coleman GL, Jacoby RO, Livestone EM, Jonas AM. 1978. Transmissible murine colonic hyperplasia. Vet. Pathol. 15:223–236 [DOI] [PubMed] [Google Scholar]
- 3. Luperchio SA, Schauer DB. 2001. Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect. 3:333–340 [DOI] [PubMed] [Google Scholar]
- 4. Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. 2005. Citrobacter rodentium of mice and man. Cell. Microbiol. 7:1697–1706 [DOI] [PubMed] [Google Scholar]
- 5. McDaniel TK, Jarvis KG, Donnenberg MS, Kaper JB. 1995. A genetic locus of enterocyte effacement conserved among diverse enterobacterial pathogens. Proc. Natl. Acad. Sci. U. S. A. 92:1664–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jarvis KG, Giron JA, Jerse AE, McDaniel TK, Donnenberg MS, Kaper JB. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. U. S. A. 92:7996–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schmidt MA. 2010. LEEways: tales of EPEC, ATEC and EHEC. Cell. Microbiol. 12:1544–1552 [DOI] [PubMed] [Google Scholar]
- 8. Petty NK, Bulgin R, Crepin VF, Cerdeno-Tarraga AM, Schroeder GN, Quail MA, Lennard N, Corton C, Barron A, Clark L, Toribio AL, Parkhill J, Dougan G, Frankel G, Thomson NR. 2010. The Citrobacter rodentium genome sequence reveals convergent evolution with human pathogenic Escherichia coli. J. Bacteriol. 192:525–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boyer F, Fichant G, Berthod J, Vandenbrouck Y, Attree I. 2009. Dissecting the bacterial type VI secretion system by a genome wide in silico analysis: what can be learned from available microbial genomic resources? BMC Genomics 10:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cascales E. 2008. The type VI secretion toolkit. EMBO Rep. 9:735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cascales E, Cambillau C. 2012. Structural biology of type VI secretion systems. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367:1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ballister ER, Lai AH, Zuckermann RN, Cheng Y, Mougous JD. 2008. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl. Acad. Sci. U. S. A. 105:3733–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, Burley SK, Almo SC, Mekalanos JJ. 2009. Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U. S. A. 106:4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pell LG, Kanelis V, Donaldson LW, Howell PL, Davidson AR. 2009. The phage lambda major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U. S. A. 106:4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. 2007. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U. S. A. 104:15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. 2012. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483:182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aschtgen MS, Bernard CS, De Bentzmann S, Lloubes R, Cascales E. 2008. SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 190:7523–7531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Aschtgen MS, Gavioli M, Dessen A, Lloubes R, Cascales E. 2010. The SciZ protein anchors the enteroaggregative Escherichia coli type VI secretion system to the cell wall. Mol. Microbiol. 75:886–899 [DOI] [PubMed] [Google Scholar]
- 19. Durand E, Zoued A, Spinelli S, Watson PJ, Aschtgen MS, Journet L, Cambillau C, Cascales E. 2012. Structural characterization and oligomerization of the TssL protein, a component shared by bacterial type VI and type IVb secretion systems. J. Biol. Chem. 287:14157–14168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Felisberto-Rodrigues C, Durand E, Aschtgen MS, Blangy S, Ortiz-Lombardia M, Douzi B, Cambillau C, Cascales E. 2011. Towards a structural comprehension of bacterial type VI secretion systems: characterization of the TssJ-TssM complex of an Escherichia coli pathovar. PLoS Pathog. 7:e1002386 doi:10.1371/journal.ppat.1002386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma LS, Narberhaus F, Lai EM. 2012. IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J. Biol. Chem. 287:15610–15621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bingle LE, Bailey CM, Pallen MJ. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11:3–8 [DOI] [PubMed] [Google Scholar]
- 23. Burtnick MN, DeShazer D, Nair V, Gherardini FC, Brett PJ. 2010. Burkholderia mallei cluster 1 type VI secretion mutants exhibit growth and actin polymerization defects in RAW 264.7 murine macrophages. Infect. Immun. 78:88–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Durand E, Derrez E, Audoly G, Spinelli S, Ortiz-Lombardia M, Raoult D, Cascales E, Cambillau C. 16 August 2012. Crystal structure of the VgrG1 actin cross-linking domain of the Vibrio cholerae type VI secretion system. J. Biol. Chem. [Epub ahead of print.] doi:10.1074/jbc.M112.390153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rosales-Reyes R, Skeldon AM, Aubert DF, Valvano MA. 2012. The type VI secretion system of Burkholderia cenocepacia affects multiple Rho family GTPases disrupting the actin cytoskeleton and the assembly of NADPH oxidase complex in macrophages. Cell. Microbiol. 14:255–273 [DOI] [PubMed] [Google Scholar]
- 26. Suarez G, Sierra JC, Erova TE, Sha J, Horneman AJ, Chopra AK. 2010. A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192:155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD. 2010. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7:25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. MacIntyre DL, Miyata ST, Kitaoka M, Pukatzki S. 2010. The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc. Natl. Acad. Sci. U. S. A. 107:19520–19524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murdoch SL, Trunk K, English G, Fritsch MJ, Pourkarimi E, Coulthurst SJ. 2011. The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J. Bacteriol. 193:6057–6069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD. 2011. Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475:343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Russell AB, Singh P, Brittnacher M, Bui NK, Hood RD, Carl MA, Agnello DM, Schwarz S, Goodlett DR, Vollmer W, Mougous JD. 2012. A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11:538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwarz S, Hood RD, Mougous JD. 2010. What is type VI secretion doing in all those bugs? Trends Microbiol. 18:531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Schwarz S, West TE, Boyer F, Chiang WC, Carl MA, Hood RD, Rohmer L, Tolker-Nielsen T, Skerrett SJ, Mougous JD. 2010. Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6:e1001068 doi:10.1371/journal.ppat.1001068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shalom G, Shaw JG, Thomas MS. 2007. In vivo expression technology identifies a type VI secretion system locus in Burkholderia pseudomallei that is induced upon invasion of macrophages. Microbiology 153:2689–2699 [DOI] [PubMed] [Google Scholar]
- 35. Miller JH. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 36. Unger T, Jacobovitch Y, Dantes A, Bernheim R, Peleg Y. 2010. Applications of the Restriction Free (RF) cloning procedure for molecular manipulations and protein expression. J. Struct. Biol. 172:34–44 [DOI] [PubMed] [Google Scholar]
- 37. van den Ent F, Lowe J. 2006. RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 67:67–74 [DOI] [PubMed] [Google Scholar]
- 38. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bäumler AJ, Tsolis RM, van der Velden AW, Stojiljkovic I, Anic S, Heffron F. 1996. Identification of a new iron regulated locus of Salmonella typhi. Gene 183:207–213 [DOI] [PubMed] [Google Scholar]
- 40. Zaslaver A, Bren A, Ronen M, Itzkovitz S, Kikoin I, Shavit S, Liebermeister W, Surette MG, Alon U. 2006. A comprehensive library of fluorescent transcriptional reporters for Escherichia coli. Nat. Methods 3:623–628 [DOI] [PubMed] [Google Scholar]
- 41. Bernard CS, Brunet YR, Gueguen E, Cascales E. 2010. Nooks and crannies in type VI secretion regulation. J. Bacteriol. 192:3850–3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Leung KY, Siame BA, Snowball H, Mok YK. 2011. Type VI secretion regulation: crosstalk and intracellular communication. Curr. Opin. Microbiol. 14:9–15 [DOI] [PubMed] [Google Scholar]
- 43. Brunet YR, Bernard CS, Gavioli M, Lloubes R, Cascales E. 2011. An epigenetic switch involving overlapping Fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet. 7:e1002205 doi:10.1371/journal.pgen.1002205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Horstman NK, Darwin AJ. 2012. Phage shock proteins B and C prevent lethal cytoplasmic membrane permeability in Yersinia enterocolitica. Mol. Microbiol. 85:445–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rice CD, Pollard JE, Lewis ZT, McCleary WR. 2009. Employment of a promoter-swapping technique shows that PhoU modulates the activity of the PstSCAB2 ABC transporter in Escherichia coli. Appl. Environ. Microbiol. 75:573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee KH, Park JH, Kim TY, Kim HU, Lee SY. 2007. Systems metabolic engineering of Escherichia coli for L-threonine production. Mol. Syst. Biol. 3:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Spagnolo J, Bigot S, Denis Y, Bordi C, de Bentzmann S. 2012. Development of a genetic tool for activating chromosomal expression of cryptic or tightly regulated loci in Pseudomonas aeruginosa. Plasmid 67:245–251 [DOI] [PubMed] [Google Scholar]
- 48. McCleary WR. 2009. Application of promoter swapping techniques to control expression of chromosomal genes. Appl. Microbiol. Biotechnol. 84:641–648 [DOI] [PubMed] [Google Scholar]
- 49. Ma LS, Lin JS, Lai EM. 2009. An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its Walker A motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J. Bacteriol. 191:4316–4329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zheng J, Leung KY. 2007. Dissection of a type VI secretion system in Edwardsiella tarda. Mol. Microbiol. 66:1192–1206 [DOI] [PubMed] [Google Scholar]
- 51. Wagner LA, Weiss RB, Driscoll R, Dunn DS, Gesteland RF. 1990. Transcriptional slippage occurs during elongation at runs of adenine or thymine in Escherichia coli. Nucleic Acids Res. 18:3529–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ma AT, McAuley S, Pukatzki S, Mekalanos JJ. 2009. Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5:234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pukatzki S, Ma AT, Sturtevant D, Krastins B, Sarracino D, Nelson WC, Heidelberg JF, Mekalanos JJ. 2006. Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U. S. A. 103:1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Suarez G, Sierra JC, Sha J, Wang S, Erova TE, Fadl AA, Foltz SM, Horneman AJ, Chopra AK. 2008. Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb. Pathog. 44:344–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chou S, Bui NK, Russell AB, Lexa KW, Gardiner TE, Leroux M, Vollmer W, Mougous JD. 2012. Structure of a peptidoglycan amidase effector targeted to Gram-negative bacteria by the type VI secretion system. Cell Rep. 1:656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.