Abstract
Fresh vegetables have been recurrently associated with salmonellosis outbreaks, and Salmonella contamination of retail produce has been correlated positively with the presence of soft rot disease. We observed that population sizes of Salmonella enterica serovar Typhimurium SL1344 increased 56-fold when inoculated alone onto cilantro leaves, versus 2,884-fold when coinoculated with Dickeya dadantii, a prevalent pathogen that macerates plant tissue. A similar trend in S. enterica populations was observed for soft-rotted lettuce leaves. Transcriptome analysis of S. enterica cells that colonized D. dadantii-infected lettuce and cilantro leaves revealed a clear shift toward anaerobic metabolism and catabolism of substrates that are available due to the degradation of plant cells by the pectinolytic pathogen. Twenty-nine percent of the genes that were upregulated in cilantro macerates were also previously observed to have increased expression levels in the chicken intestine. Furthermore, multiple genes induced in soft rot lesions are also involved in the colonization of mouse, pig, and bovine models of host infection. Among those genes, the operons for ethanolamine and propanediol utilization as well as for the synthesis of cobalamin, a cofactor in these pathways, were the most highly upregulated genes in lettuce and cilantro lesions. In S. Typhimurium strain LT2, population sizes of mutants deficient in propanediol utilization or cobalamin synthesis were 10- and 3-fold lower, respectively, than those of the wild-type strain in macerated cilantro (P < 0.0002); in strain SL1344, such mutants behaved similarly to the parental strain. Anaerobic conditions and the utilization of nutrients in macerated plant tissue that are also present in the animal intestine indicate a niche overlap that may explain the high level of adaptation of S. enterica to soft rot lesions, a common postharvest plant disease.
INTRODUCTION
The association of food-borne illness with contaminated produce has prompted numerous investigations into the ability of human enteric pathogens to attach to and survive on fresh fruits and vegetables. Since enteric pathogens are unlikely to land on plants at high densities, it still remains unclear how they achieve the population sizes required to cause human illness at infectious doses in that habitat. Therefore, the factors that drive the growth of enteric pathogens on plants still need to be explored. We have previously demonstrated that Salmonella enterica and Escherichia coli serovar O157:H7 can multiply in the cilantro and lettuce phyllosphere under optimal conditions of warm temperatures and free water on the leaves (1, 2). Based on our observations that enteric pathogens appear less fit in the phyllosphere than plant-associated bacterial species, even under optimal growth conditions (2), and that their growth on middle-aged lettuce leaves is limited by nitrogen availability (1), we hypothesized that these human pathogens have not evolved to utilize the full range of nutrients present on leaf surfaces (3). The occurrence of plant lesions may enhance the abundance and range of nutrients in the phyllosphere. Mechanically damaged plant tissue supports larger population sizes of E. coli O157:H7 on lettuce (4, 5), presumably because of the leaching of plant cell contents that may provide substrates for bacterial growth. Indeed, our E. coli O157:H7 transcriptome analysis of lettuce lysates, which modeled the chemical environment of leaf lesions, revealed the upregulation of multiple carbohydrate transport systems involved in the utilization of substrates that are prevalent in plant cells (6).
Lesions produced by plant pathogens may also promote the growth of human pathogens on fruits and vegetables. Wells and Butterfield previously reported increased incidences of presumptive Salmonella on retail produce affected by fungal decay (7) and by soft rot (8), a common postharvest plant disease caused by pectinolytic bacterial pathogens. The coinoculation of Dickeya dadantii (formerly Erwinia chrysanthemi) and E. coli O157:H7 onto lettuce leaves (5, 9) and the coinoculation of Pectobacterium pectovorum (formerly Erwinia carotovora) and S. enterica onto tomato fruit (10) significantly increased the growth rates and population sizes of the human pathogens in tissue macerated by these soft rot pathogens. Spoilage also had a positive effect on the colonization of endive leaves by Listeria monocytogenes (11), whereas the pathogen was inhibited strongly in potato tuber slices infected with Pseudomonas fluorescens and Pseudomonas viridiflava, possibly due to iron competition (12). Furthermore, it was reported previously that some plant-pathogenic fungi are more likely to increase the proliferation of human pathogens in infected fruit tissue by neutralizing the pH in the lesions and thereby enabling bacterial growth (13–15).
While increased substrate abundance due to the degradation of plant cells likely underlies the positive effect of D. dadantii and P. pectovorum on enteric pathogens, a detailed investigation of the precise mechanisms by which this growth promotion occurs has not been carried out. Since various combinations of enteric pathogens and soft rot pathogens have diverse outcomes (12), more specific conditions than overall enhanced nutrient availability can be hypothesized to be at play as the driving force of the observed bacterial proliferation in macerated tissue.
Here, we present the results from a global transcriptome analysis of S. enterica cells that co-colonized cilantro leaves with D. dadantii. Our study reveals the physiological state of the human pathogen and the specific biochemical pathways that are involved in its growth in macerated cilantro leaf tissue. We conclude that S. enterica in soft rot dedicates a significant part of its transcriptome to the assimilation of substrates that are important for anaerobic growth and pathogenesis in the host intestinal environment and which are uniquely provided in our system via the degradation of the plant cell wall by the soft rot pathogen.
MATERIALS AND METHODS
Strains and culture conditions.
All strains and plasmids are described in Table 1. Salmonella enterica serovar Typhimurium strain SL1344 and Dickeya dadantii strain 3937 were used for most studies. SL1344 mutants defective in propanediol (strain SS371) and ethanolamine (strain SS377) utilization and in cobalamin synthesis (strain SS374) were generated by using the λ Red recombinase system (19), by replacing pduD, eutK, and cobS, respectively, with a kanamycin resistance cassette. A second set of SL1344 mutants was generated with the same approach to fully delete the propanediol utilization (PDU) operon (strain MB784) or the ethanolamine utilization (EUT) operon (strain MB789) with the kanamycin resistance cassette. The propanediol- and ethanolamine-minus double deletion strain of SL1344 was constructed in the propanediol utilization-minus mutant strain MB784 background by replacing the ethanolamine utilization operon with the chloramphenicol resistance cassette, to give strain MB792. Primers used for mutagenesis are listed in Table S1 in the supplemental material. Rifampin-resistant derivatives of D. dadantii 3937 and of S. Typhimurium SL1344 and some of its mutants were isolated on Luria-Bertani (LB) agar containing rifampin (100 μg/ml), as described previously (2).
Table 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Strains | ||
| S. enterica serovar Typhimurium SL1344 | ||
| MB282 | Wild type; Strepr | 16 |
| MB427 | Rifr derivative of wild-type strain SL1344 | This study |
| SS371 | MB427 ΔpduD::kan; Strepr Kanr Rifr | This study |
| SS374 | MB427 ΔcobS::kan; Strepr Kanr Rifr | This study |
| SS377 | MB427 ΔeutK::kan; Strepr Kanr Rifr | This study |
| MB713 | SS371 with pMBpduD-X | This study |
| MB717 | SS377 with pMBeutRK | This study |
| MB784 | SL1344 ΔpduA-X::kan; Strepr Kanr | This study |
| MB789 | SL1344 ΔeutK-S::kan; Strepr Kanr | This study |
| MB792 | SL1344 ΔpduA-X::kan ΔeutK-S::cat; Strepr Kanr Cmr | This study |
| S. enterica serovar Typhimurium LT2 | ||
| TT22362 | Wild type | 17 |
| MB580 | Rifr derivative of wild-type strain TT22362 | This study |
| TT22344 | TT22362 pdu-12::MudA; Ampr | 17 |
| TT22341 | TT22362 ΔeutS-K; Tetr | 17 |
| TT20945 | TT22362 cbiA::MudJ ΔcbiB-cobT; Kanr | Gift of J. R. Roth |
| Dickeya dadantii | ||
| 3937 | Wild type | 18 |
| MB392 | Rifr derivative of strain 3937 | This study |
| Plasmids | ||
| pMBpduD-X | pBBR1-mcs5 with pduD-X from SL1344 cloned into the SacI site, driven by PlacZ; Gentr | This study |
| pMBeutRK | pBBR1-mcs5 with eutRK from SL1344 cloned into the SacI site, driven by PlacZ; Gentr | This study |
pduA-X consists of pduA, pduX, and all intergenic material; eutK-S is eutK, eutS, and all genetic material in between; and pduD-X contains pduD, pduX, and all intergenic material.
Complementation plasmids were constructed by PCR cloning of pduD-X (i.e., pduD to pduX, inclusively) and eutKR and ligation into the single SacI site on pBBR1-mcs5 (20) in the same orientation as the transcription of lacZ, using primers for the amplification of each clone that harbored a SacI site at their 5′ end. Primers used for the construction of complementation plasmids are listed in Table S1 in the supplemental material. The presence and orientation of the clones on the complementation plasmids were verified by PCR and by sequencing.
Propanediol and ethanolamine utilization in various strains was assessed by streaking cells onto MacConkey acid indicator agar containing 150 nM cyanocobalamin and 1% 1,2-propanediol or 1% ethanolamine; after 48 h of incubation on this medium at 28°C, red colonies were positive for the utilization of these substrates, whereas those that were white were defective in these pathways (21, 22).
All strains were cultured to the early stationary phase of growth on a rotary shaker at 28°C in half-salt (0.5% NaCl) LB broth, and the following antibiotics were added, as appropriate: streptomycin (30 μg/ml), rifampin (100 μg/ml), kanamycin (50 μg/ml), and gentamicin (15 μg/ml). For the preparation of the inoculum, the cultures were washed twice by centrifugation in potassium phosphate (KP) buffer (1 mM, pH 7) before being resuspended in KP buffer to the desired concentration based on the optical density at 600 nm (OD600).
Plant material.
Cilantro (Coriandrum sativum cv. Leisure) and romaine lettuce (Lactuca sativa cv. Parris Island) were grown to maturity in a greenhouse without pesticide application or overhead watering. The cilantro plants were used shortly before the emergence of flower shoots, and the lettuce plants were used when a full closed head had formed. For the inoculation of lettuce, the outer damaged leaves were discarded, and the remaining leaves on the head were used. For cilantro, fully expanded leaves from mature plants at the growth stage just prior to bolting were used. Only leaves that were free of visible mechanical and disease lesions were picked for all the experiments. The cilantro and lettuce leaves were cut into approximately 0.25-cm2 pieces and 1-cm-wide strips, respectively, before inoculation, in order to ensure rapid colonization by D. dadantii and, thus, more homogenous soft-rotted tissue over time.
Plant inoculation.
In order to measure bacterial population sizes over time, D. dadantii 3937 and S. enterica serovar Typhimurium were each inoculated at a starting concentration of ca. 105 CFU/g leaf by the addition of 4 ml or 15 ml of a mixed-inoculum suspension in 1 mM KP buffer to 3 g and 100 g of cut cilantro or lettuce, respectively, in a bag. For fitness competition experiments, a suspension containing a ratio of 1:1:1 of wild-type (WT) S. enterica, its mutant, and D. dadantii, each at 105 CFU/g, was prepared and used for the inoculum, as described above. For microarray hybridization experiments, the inoculum was resuspended in 1 mM KP buffer containing rifampin (100 μg/ml), and the rifampin-resistant derivatives of D. dadantii 3937 and of wild-type strain SL1344 were used at a starting concentration of 106 CFU/g leaf tissue; each replicate bag contained 15 g or 100 g cut cilantro or lettuce leaf tissue, respectively. The bags were shaken by hand to distribute the inoculum, opened ajar, and placed into a chamber under high humidity for 18 h at 28°C. Three replicate bags were prepared per inoculum mixture per experiment. Each plant inoculation experiment to carry out microarray analysis or to assess the population sizes of SL1344 and its mutants, as well as that of D. dadantii, was repeated at least three times.
Recovery of bacteria from leaf tissue.
To each bag of cilantro or lettuce, 100 ml or 650 ml of 10 mM KP buffer, respectively, was added. Bags with cilantro were then placed into a Stomacher 400 instrument (Seward, West Sussex, United Kingdom) on high for 1 min, followed by sonication in an Astramax Generator sonicator bath (Misonix Inc., Farmingdale, NY) at 250 W for 75 s. Bags with lettuce were massaged by hand, due to the large volume of buffer added to the cut leaves, before and after sonication, as described above. To assess bacterial population sizes, two aliquots of 1 ml of the leaf washings per bag were diluted separately before being plated with an Autoplate 4000 automated plater (Spiral Biotech Inc., Norwood, MA) onto LB agar containing antibiotics, as appropriate. The plate counts from each separate dilution event per replicate bag were averaged to obtain the bacterial population size on the plant tissue in each replicate sample. S. enterica colonies were counted on LB agar with streptomycin, whereas D. dadantii colonies were counted on LB agar with rifampin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), which also enabled us to count D. dadantii colonies (blue colonies due to β-galactosidase activity) and S. enterica colonies (white) separately on the same plate when rifampin-resistant strains were used for inoculation in microarray experiments. For competitive fitness experiments between wild-type and mutant S. enterica strains, the total S. enterica population and the mutant population were estimated by counts on LB agar with streptomycin or with the antibiotic that enabled the selection of the particular mutant tested, respectively, and the S. enterica wild-type population was assessed as the difference between total and mutant counts.
RNA extraction procedures.
To obtain bacterial RNA from the plant samples, the leaf wash samples described above were vacuum filtered through a 20-μm-pore-size nylon filter (Millipore) in 25-ml aliquots, before continuing with RNA extraction procedures. An ice-cold phenol-ethanol (5%–95%) solution was immediately added to filtered leaf wash samples or to cultures in LB medium with rifampin, and the mixture was incubated on ice for 30 min. The cells were then centrifuged, and the pellet was stored at −80°C. RNA extraction was performed with the Promega SV Total RNA kit according to the manufacturer's instructions, except that bacterial pellets were first treated with 50 mg/ml of lysozyme (Fisherbrand) and 1 U/μl of anti-RNase (Ambion). Total RNA was quantified with a Nanodrop ND 1000 spectrophotometer (Thermo Scientific), examined for quality on an Agilent Bioanalyzer, and stored at −80°C until used for microarray analysis or quantitative reverse transcription-PCR (qRT-PCR).
qRT-PCR.
Total bacterial RNA was treated with Turbo DNase I (Ambion), and the absence of DNA was confirmed by qRT-PCR in the absence of reverse transcriptase by using primers for gyrB. All qRT-PCRs were performed with the Brilliant II SYBR green qRT-PCR 1-Step kit (Stratagene) on an MxPro 3000P cycler (Stratagene). For each gene, the ratio of expression of S. enterica on rotten cilantro to that in LB cultures was normalized to the expression of gyrB based on an equation reported previously by Pfaffl (23). Lysates were tested for the presence of cilantro RNA by using primers directed to the C. sativum 26S ribosomal subunit. Only insignificant amounts of cilantro RNA were detected; these were estimated to have minimal effects on the calculated ratios of S. enterica gene expression. All primers used for qRT-PCR are listed in Table S1 in the supplemental material.
Microarray procedures.
Microarray procedures for gene expression profiling were based on previously described methods (6, 24, 25), using previously described S. enterica in-house microarrays (25). Microarray analysis was based on competitive hybridization between Cy5-DNA and Cy3-cDNA. Twenty micrograms of total RNA from each experimental condition and each biological replicate was used for three replicate arrays per microarray experiment. When the bacterial RNA sample originated from leaves coinoculated with S. enterica and D. dadantii, 40 μg of total RNA was used for three replicate arrays. In this way, the amount of S. enterica cDNA in the hybridizations was equivalent for the control (LB medium) and target (macerated leaves) environments. The population sizes of the two strains on leaves were very similar throughout the incubation period, and only bacterial pellets coming from samples that contained similar population sizes of both strains, as determined by the plating of the leaf wash specimens, were processed for microarray analysis. Since rifampin was added to the leaves at the time of inoculation, few bacteria other than the inoculated species multiplied during the incubation of the plant tissue.
Statistical analysis of microarray data.
Three biological replicates were prepared for each environment, and each replicate cDNA-DNA solution was hybridized onto three separate arrays on different slides (technical replicates). Technical replicates were analyzed for outliers with the Dixon “Q” test (26), using a critical value of 0.1, and values lying outside the cutoff were discarded. All remaining technical replicate values were averaged to obtain the value for each biological replicate, and the data from all three biological replicates were further tested by an unpaired t test with unequal variance using Genespring, version 10.0, software (Agilent/Stratagene). Genes showing a >2-fold upregulation or downregulation and a Benjamini-Hochberg false discovery rate (FDR)-adjusted P value of ≤0.05, were considered to be differentially regulated in D. dadantii-infected leaves compared with LB medium. The expression of a subset of genes of interest was confirmed by qRT-PCR. Two independent experiments were performed. Due to the variation in the magnitude of the transcriptional ratios across experiments, averages from the two replicate experiments were not computed, and our tables present data from a single experiment for each plant species. However, the final transcriptional data sets from these single experiments (one each for cilantro and lettuce) were obtained by comparing the lists of genes in both independent experiments, and only genes that were also regulated significantly differentially at least 2-fold in the replicate experiment are shown in the final lists.
RESULTS
Effect of soft rot on growth of S. enterica.
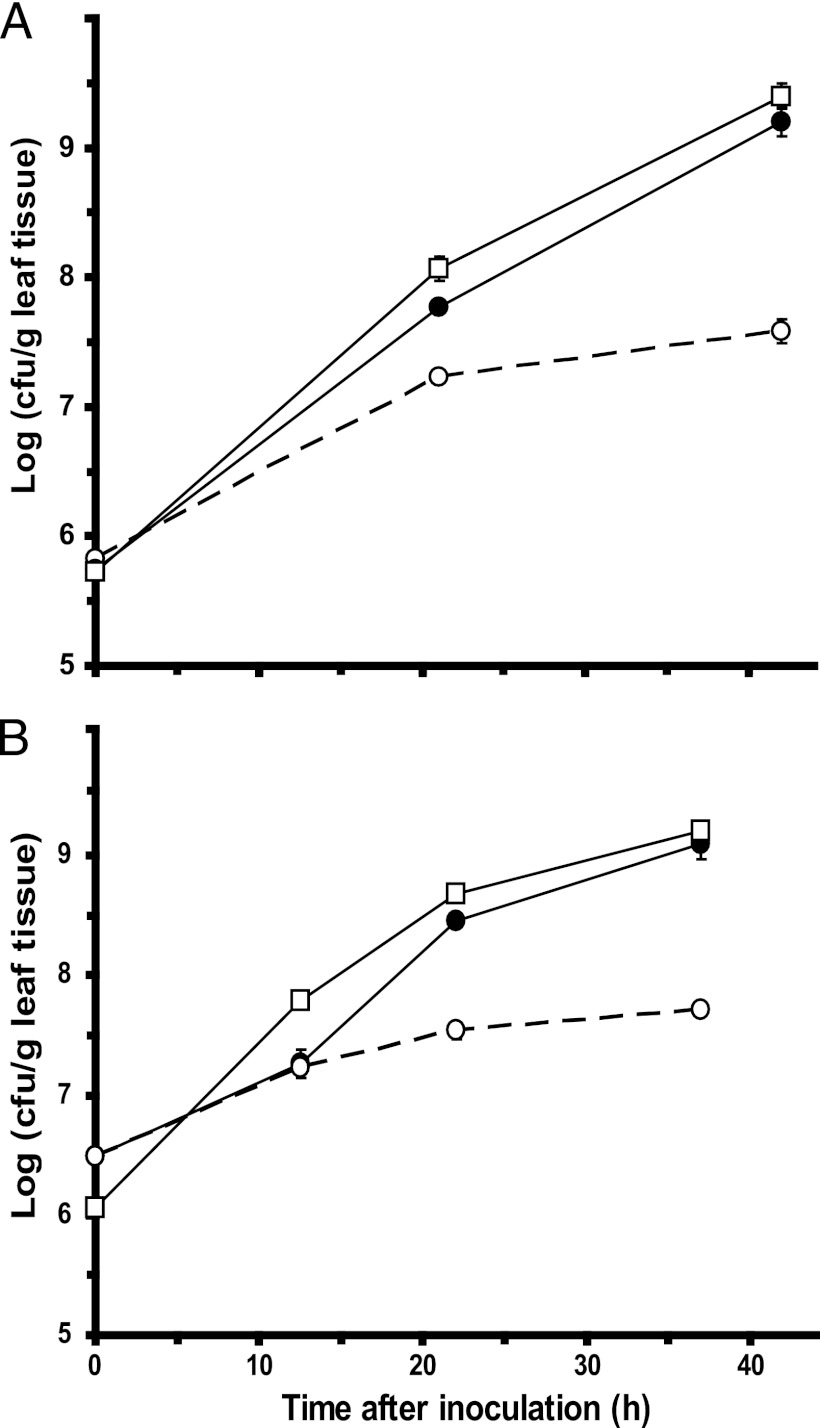
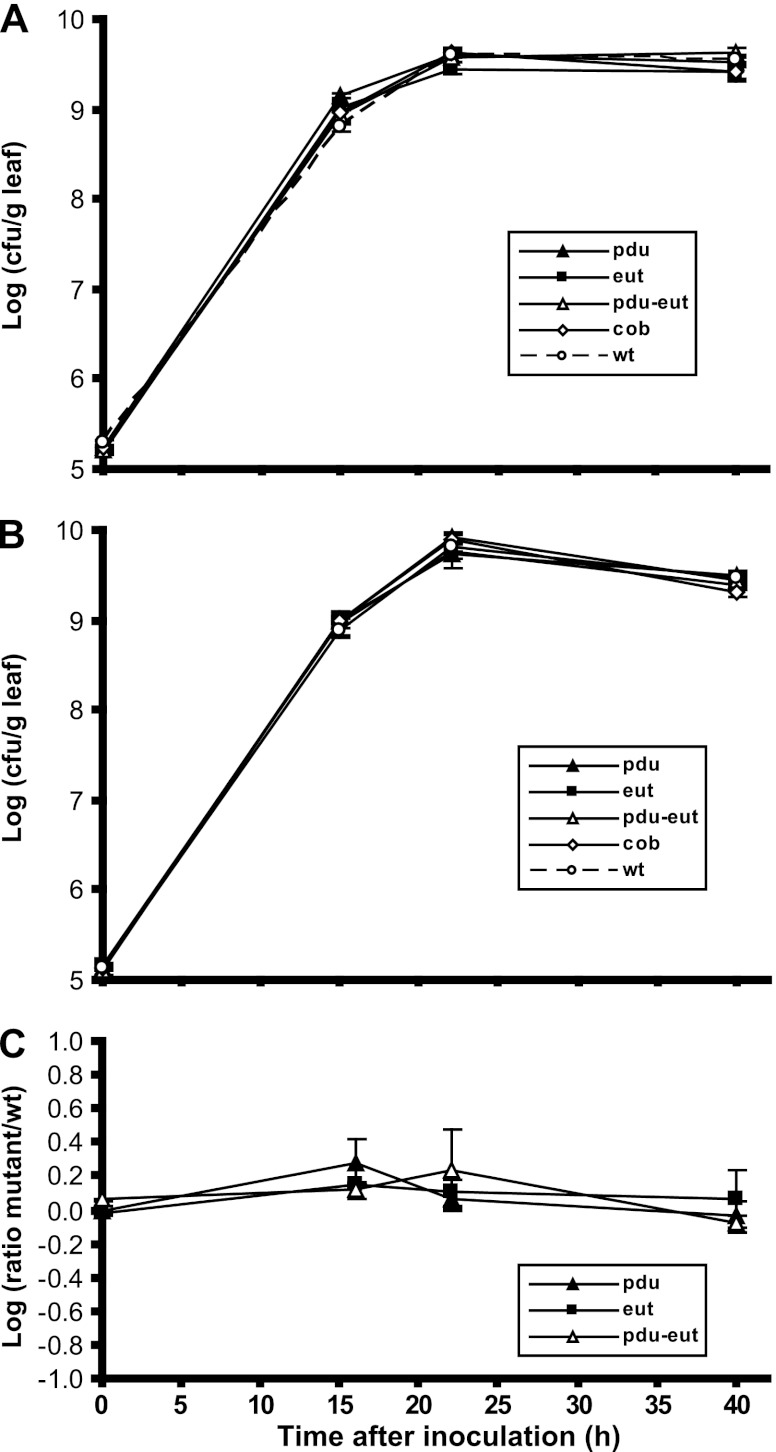
S. enterica population sizes increased 56-fold when inoculated alone onto cut cilantro leaves, versus 2,884-fold when coinoculated with D. dadantii, which macerated the leaf tissue (Fig. 1A). On cut lettuce leaves, S. enterica populations increased 17-fold on healthy tissue, versus 407-fold on D. dadantii-macerated tissue (Fig. 1B). The maximum population sizes reached by S. enterica on healthy cilantro and lettuce leaves were ca. 107 CFU/g, suggesting that both plant species had similar carrying capacities for the pathogen. Therefore, its lower overall increase in population size on healthy lettuce than on cilantro may be due to a slightly higher starting inoculation concentration on the lettuce leaves rather than to a factor related to the plant species per se. Not only was the multiplication of S. enterica faster in the presence of plant tissue infected and degraded by D. dadantii, but the maximum densities achieved by this human pathogen were also considerably higher. It is noteworthy that the growth of the plant pathogen was highly correlated with that of the human pathogen on leaves, achieving population sizes very similar to those of S. enterica throughout the different stages of tissue maceration (Fig. 1A and B). Because the leaves were cut into small pieces, maceration by D. dadantii progressed rapidly. Soft rot symptoms had increased far beyond the margins of the cut leaf blade by 22 h, and the plant tissue was fully macerated by 40 h.
Fig 1.
Effect of soft rot disease caused by D. dadantii on colonization of cilantro and lettuce leaves by S. enterica. Cut cilantro (A) and lettuce (B) leaves were inoculated with both S. enterica (●) and D. dadantii (■) or with S. enterica alone (○). Each datum point represents the mean of the log value of the population size and standard error of the mean for three replicate samples.
Transcriptome of S. enterica in soft rot lesions. (i) Global trends.
Global analysis of the transcriptome of S. enterica in D. dadantii-infected cilantro leaves showed that 374 and 448 genes were upregulated and downregulated, respectively, at least 2-fold in macerated cilantro leaf tissue compared with LB broth (see Table S2 in the supplemental material). In macerated lettuce leaf tissue, 256 and 462 genes were up- and downregulated, respectively (see Table S3 in the supplemental material). LB culture broth rather than a minimal culture medium was used as the control environment in our gene expression analysis, because the chemical environment in soft-rotted tissue was hypothesized to be overall complex. This allowed the identification of pathways that are more uniquely differentially regulated in soft rot lesions and distinct from housekeeping metabolism and comparisons of our data with those of a large number of S. enterica transcriptomic studies. Additionally, this control environment was used successfully in a previous study by Okinaka et al. (27) to identify D. dadantii genes that are differentially expressed during plant infection. It is noteworthy that 121 out of 374 genes and 175 out of 448 genes that were upregulated and downregulated, respectively, in S. enterica in cilantro soft rot compared with LB medium changed similarly in romaine lettuce soft-rotted tissue (see Table S4 in the supplemental material).
Since the cDNA used for microarray hybridization originated equally from S. enterica and D. dadantii, we tested the extent of hybridization of D. dadantii genes to our S. enterica array. Of the 4,360 S. enterica open reading frames (ORFs) spotted onto the array, 297 had sufficient homology to D. dadantii genes to produce a Cy3/Cy5 signal ratio greater than 2.0 in microarrays of competitive hybridization of Cy3 genomic D. dadantii and Cy5 genomic S. enterica DNAs. These D. dadantii genes are listed in Table S5 in the supplemental material.
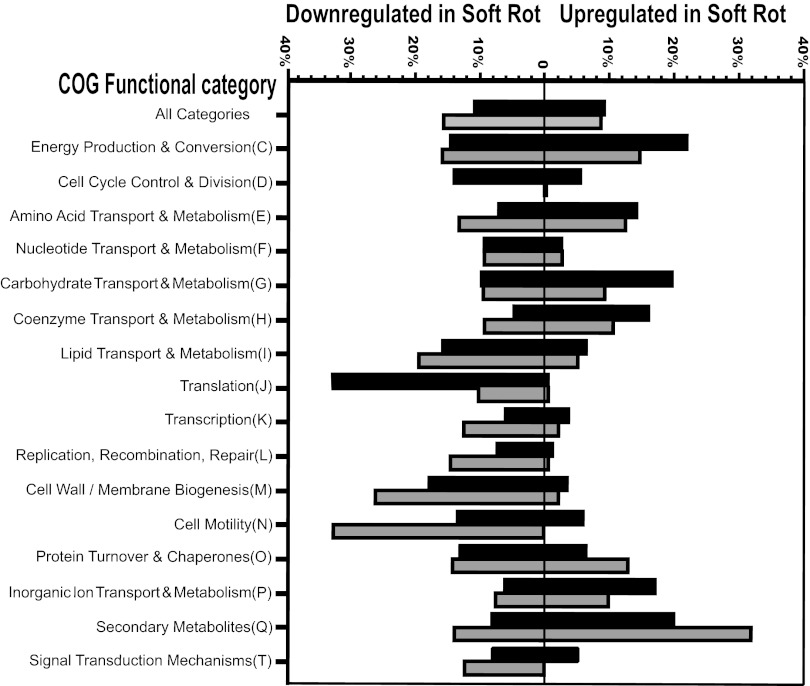
Among the S. enterica genes that have been assigned to a category of orthologous genes (COG), those that were overrepresented in upregulation in macerated cilantro leaf tissue belonged to the following functional categories: energy production and conversion; transport and metabolism of amino acids, carbohydrates, coenzymes, and inorganic ions; and production of secondary metabolites (Fig. 2). Genes that were overrepresented in downregulation in soft rot lesions belonged to functional categories such as cell cycle control and division, translation, cell wall/membrane biogenesis, and protein turnover and chaperones (Fig. 2). The broad downregulation of operons encoding the 30S (rps) and 50S (rpl and rpm) ribosomal subunit proteins, as evidenced in the cilantro microarray data (see Table S2 in the supplemental material), corroborates this slower growth than in LB broth. Also notable is the higher percentage of genes that are downregulated than upregulated and that are involved in motility, possibly due to the large amount of nutrients available in the lesions and, hence, the lack of necessary movement toward new substrates.
Fig 2.
Differential expression of genes within categories of orthologous genes (COGs) in S. enterica cells colonizing cilantro (black bars) and lettuce (gray bars) soft rot lesions caused by D. dadantii relative to that in LB broth culture, as revealed by microarray analysis. Bars represent the percentages of genes with a change in transcription of at least 2-fold within a given category.
Overall, similar trends in COG patterns were observed for S. enterica in macerated lettuce, with the most important difference concerning metabolism (Fig. 2). More specifically, upregulated genes involved in energy production and conversion and in carbohydrate transport and metabolism were represented at a higher percentage in cilantro than lettuce soft rot, whereas upregulated genes involved in secondary metabolite pathways were represented at a higher percentage in lettuce than cilantro soft rot. Thus, S. enterica cells may have been in a more advanced state of secondary metabolism in macerated lettuce than cilantro despite similar stages of soft rot disease at the 18-h-postinoculation sampling time used for microarray analysis.
(ii) General metabolism.
Microarray transcriptional analysis provided considerable information about the physiological state and metabolism of S. enterica cells in macerated leaf tissue. A close examination of the detailed gene lists revealed that soft rot conditions in both cilantro and lettuce induced numerous genes involved in growth under oxygen-limiting conditions. These genes included, as listed in Table 2 (see also Tables S2 and S3 in the supplemental material), the hydrogenase operons hya, hyb, and hyp and hydN; most genes of the nar and nap operons and the nrf operon, which function in nitrate and nitrite reduction, respectively; ccmC, ccmE, and ccmG, which encode proteins for the biogenesis of c-type cytochromes that function as the terminal electron acceptor in the anaerobic electron transport chain during growth in the presence of nitrate or nitrite (28); and nrfA and hmpA, which have major and minor roles, respectively, in protecting cells from the toxic effect of nitric oxide (NO) species under anoxic conditions (29, 30). Furthermore, the downregulation of the cytochrome o oxidase genes by 9- to 10-fold in cilantro lesions (cyoABC) and by 4- to 12-fold in lettuce lesions (cyoABCD) corroborates the above-described observations, since these genes are repressed in E. coli when oxygen levels are low (31). Although the precise function and regulation of yqjF are unknown, it is noteworthy that this very highly expressed gene is predicted to encode a quinol oxidase and thus may be involved in the electron transfer chain under microaerophilic or anaerobic conditions in macerated leaves. yhaK, a gene encoding a predicted pirin-related protein, was one of the most highly upregulated genes, with an increase in the expression level in macerated cilantro of 111-fold (12-fold in lettuce). This observation may be relevant to the low-oxygen conditions in soft rot lesions, since a pirin ortholog in Serratia marcescens was reported previously to divert central carbohydrate metabolism toward fermentation (32). Additional evidence for low oxygen availability in soft rot lesions is provided by the increased transcriptional activities of several operons that drive anaerobic catabolic pathways, as described below.
Table 2.
List of select genes in S. enterica that are indicative of its metabolism in macerated leaf tissue due to their upregulation in cilantro and lettuce soft rot lesionsa
| Gene category and name(s) | Fold increase in expressionb | Function |
|---|---|---|
| Anaerobic metabolism | ||
| hmpA | 7.9 | Nitric oxide dioxygenase |
| ccmC, ccmE, ccmG | 7.3–19.5 | Cytochrome biogenesis/function |
| hyaA-F | 3.1–7.3 | Hydrogenase 1 |
| hybA-G | 2.4 15.0 | Hydrogenase 2 |
| hydN | 3.8 | Formate dehydrogenase H |
| hypB, hypE, hypO | 2.5–6.9 | Hydrogenase formation protein |
| narH-J, narQ | 3.6–11.8 | Nitrate reductase 1 |
| napC, napB, napH | 3.4–4.8 | Nitrate reductase; electron transfer |
| nrfA-D, nrfG | 2.5–8.6 | Nitrite/nitrate reductase; nitric oxide detoxification |
| yhaK | 110.6 | Pirin-related protein |
| yqjF | 141.5 | Predicted quinol oxidase subunit |
| Substrate utilization | ||
| caiB, caiD-F, fixCc | 2.5–3.0 | l-Carnitine metabolism |
| citC-Gb | 29.5–131.7 | Citrate degradation |
| citC2, citE2, citF2, citX2c | 4.3–11.0 | Citrate degradation |
| cobT-U, cbiP-Bb | 5.4–37.6 | Cobalamin synthesis; cofactor in PDU/EUT pathways |
| dpiBA | 2.5–3.8 | Regulation of cit/citrate transport |
| dgoD, dgoK, dgoT | 2.5–3.5 | d-Galactonate transport and metabolism |
| eutS-Rc | 9.7–115.2 | EUT |
| fruB, fruK; STM3255 | 3.4–3.8 | d-Fructose metabolism |
| fucAO | 2.5–2.8 | l-Fucose degradation; feeds into PDU |
| gntU, kdgT, kduD | 2.2–3.3 | d-Gluconate/derivative transport and metabolism |
| idnK-T | 2.2–12.5 | l-Idonate transport and metabolism |
| pduA-W, pduFc | 8.9–176.6 | 1,2-l-Propanediol utilization |
| prpC, prpB | 2.7–3.5 | Propionate catabolism via propionyl-CoA; from PDU |
| rhaB-D, rhaT | 2.6–6.6 | l-Rhamnose transport and degradation; feeds into PDU |
| yjfR-sgaT-Uc | 13.6–44.5 | l-Ascorbate transport and utilization |
| Fe and Zn acquisition | ||
| cirA | 5.1 | Siderophore complex transporter |
| feoABc | 3.0–4.1 | Iron transporter |
| fhuDd | 5.8 | Iron transporter |
| iroB | 3.0 | Enterobactin transferase |
| znuC | 2.9 | High-affinity zinc uptake system |
The increase in the gene expression level is given for the pathogen in cilantro soft rot lesions compared with that in LB culture; most genes in the list were also upregulated in lettuce soft rot lesions, which is indicated by gene names in boldface type. Gene names with only the root in boldface type indicate that at least one of the genes in the operon was also upregulated in lettuce. See Table S5 in the supplemental material for the list of all genes that were upregulated in both cilantro and lettuce soft rot lesions.
When several genes are listed with a given function, the lowest and highest values are provided for fold increases among all the genes listed.
Involved in anaerobic metabolism.
fhuD was not upregulated in lettuce soft rot lesions, but the fhuB expression level increased 2.7-fold.
(iii) Nutrient acquisition.
Whereas the transcription levels of S. enterica genes involved in general growth functions in cilantro and lettuce soft rot appeared lower than those in LB broth (Fig. 2), multiple pathways for the utilization of growth substrates were very highly and broadly upregulated in both cilantro and lettuce soft rot lesions. Increased transcription levels of fruB and fruK and the STM3255 gene, encoding fructose-specific component IIB of a putative phosphotransferase system (PTS) (increased levels in cilantro but not in lettuce), indicated the utilization of fructose, a predominant sugar in plant cells (Table 2). Several glycoside-pentoside-hexuronide (GPH), PTS, major facilitator superfamily (MFS), and ATP-binding cassette (ABC) systems for the transport of a variety of carbohydrates were activated. Among these, uhpT codes for a hexose phosphate transport protein of the MFS family. uhpT is a member of the same operon as uhpB, which was also upregulated. Both genes are transcriptionally activated by uhpA, a LuxR family regulator homologous to psoR, which is expressed in response to plant macerates in Pseudomonas fluorescens (33).
Several genes were upregulated in operons involved in the transport and metabolism of d-galactonate (dgo), d-gluconate (gntU, kdgT, and kduD), and l-idonate (idn) (Table 2). Okinaka et al. also reported the increased expression level of a dgoT homolog in D. dadantii during plant infection (27). Expression levels of genes from the cai operon and fixC, which have a role in l-carnitine metabolism, increased in both plants. Carnitine stimulates anaerobic growth in Salmonella (34). It is widely distributed in nature (35) and is associated with fatty acid transfer in plant mitochondria and plastids (36) and therefore may have been released during leaf maceration by D. dadantii. Additionally, six out of the seven genes for transport and anaerobic utilization of ascorbic acid (yjfR to sgaE), a vitamin that is abundant in many plant species, showed increased transcription levels in macerated cilantro, with most genes being upregulated at least 20-fold (Table 2), and in macerated lettuce albeit at lower levels (see Table S3 in the supplemental material). Another carbon source ubiquitously present in plants is citrate, for which the fermentation operons (cit) and regulator/transport genes dpiBA (37, 38) were highly activated in S. enterica in the cilantro and lettuce macerates (Table 2; see also Tables S2 and S3 in the supplemental material). The expression level of the citrate transporter gene citB increased over 2-fold in lettuce only. It is unclear whether the pathogen obtained citrate mostly from the plant tissue or tapped into an ample supply of its precursor acetyl coenzyme A (CoA) provided by the high level of activity of EutE (see below), an enzyme that belongs to the ethanolamine utilization pathway and converts acetaldehyde to acetyl-CoA. Evidence that the pathogen switched its metabolism to alternate nutrient acquisition pathways is further provided by the 4-fold upregulation of rspA, a gene for a putative starvation-sensing protein.
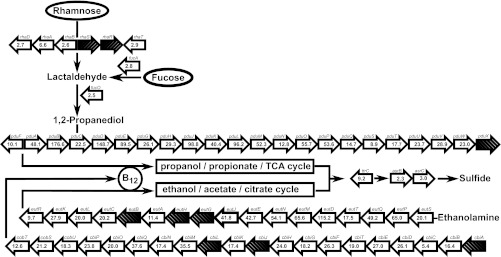
Our microarray data also clearly revealed that S. enterica appeared to devote a major part of its transcriptional activity in cilantro and lettuce soft rot lesions to the expression of operons for the utilization of 1,2-propanediol (pduA-W, i.e., pduA, pduW, and all genes in between) and ethanolamine (eutS-R). With only a few exceptions, these genes were among the most highly upregulated genes of the entire transcriptome (Fig. 3; see also Tables S2 and S3 in the supplemental material). For example, the expression levels of pduB and pduD increased 177- and 149-fold, respectively, and the eutD transcription level increased 115-fold (Fig. 3). These very large operons enable S. enterica to catabolize propanediol and ethanolamine under anaerobic conditions with cobalamin (vitamin B12) as a cofactor (39). The significant upregulation of most genes in the equally large operon that codes for cobalamin synthesis (cbi and cob) provides further evidence that the pathways for propanediol utilization (PDU) and ethanolamine utilization (EUT) are highly activated (Fig. 3).
Fig 3.
Schematic diagram of the S. enterica genes that are involved in propanediol (pdu) and ethanolamine (eut) utilization, the biosynthesis of their essential cofactor cobalamin (vitamin B12) (cob and cbi), and the catabolism of rhamnose (rha) and fucose (fuc), two precursors of the propanediol pathway. Numbers in block arrows represent the fold increases in gene expression levels in cilantro soft rot lesions compared with levels in LB culture. Cross-hatched arrows illustrate that the gene was not differentially regulated more than 2-fold. The orientation of block arrows indicates the orientation of transcription. Pathways are based on diagrams described previously by Price-Carter et al. (17). TCA, tricarboxylic acid.
The precursor of ethanolamine is phosphatidylethanolamine, a phospholipid constituent of plant membranes (40). On the other hand, propanediol is synthesized from l-rhamnose and l-fucose in S. enterica (41). l-Rhamnose in our soft rot system may have resulted from the degradation of rhamnogalacturonans in the pectin portion of the plant cell wall by D. dadantii (42). l-Fucose is also present in the plant cell wall (43) and may similarly have been released during maceration. The increased expression levels of the rha and fuc genes involved in propanediol synthesis support this hypothesis (Fig. 3). Propanediol utilization proceeds via propionate and propionyl-CoA, which is channeled to pyruvate, partly with functions encoded by the prpBCDE operon (44). Additional evidence for the high level of activity of the PDU pathway in S. enterica in soft rot came from the 3.5- and 2.7-fold upregulations of prpB and prpC, respectively. It is also notable in relation to the high-level activation of the PDU, EUT, and cobalamin pathways that S. enterica asr genes had increased expression levels in soft rot (Fig. 3; see also Tables S2 and S3 in the supplemental material). Asr proteins are involved in the pathway for the reduction of tetrathionate (45), which was proposed to be an electron acceptor for propanediol and ethanolamine degradation under conditions of anaerobiosis (17). Although it did not make the final list of genes upregulated at least 2-fold due to high levels of variation in expression across replicate samples, the expression level of ttrC, which has a role in tetrathionate reduction upstream of the asr genes (Fig. 3), increased 9.2-fold in cilantro soft rot, as revealed by qRT-PCR (see Fig. 5 and below). Taken together, the transcriptional units that are involved in PDU and EUT encompass a considerable portion of the S. enterica genome, and most of these had increased expression levels in soft rot.
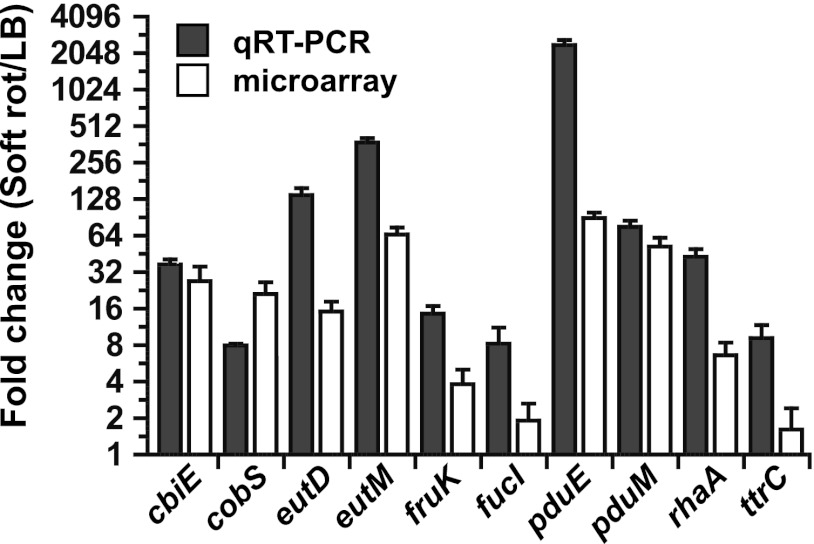
Fig 5.
Confirmation by qRT-PCR (gray bars) of the differential transcription of select S. enterica genes in cilantro soft rot relative to levels in LB broth culture, as assessed by microarray analysis (white bars). Thin bars represent standard errors of the means.
(iv) Iron and zinc acquisition.
Increased expression levels of cirA (siderophore transporter), iroB (enterobactin transferase), and fhuD and feoAB (iron transporters) revealed the activation of iron acquisition systems in S. enterica in cilantro macerates (see Table S2 in the supplemental material). Only the iron transporter fhuB was upregulated in lettuce (see Table S3 in the supplemental material), suggesting a potential difference in iron availability in the macerates of the two plant species. Homologs of fhuA and feoB as well as several iron acquisition genes homologous to those in S. enterica genes were also induced in D. dadantii in African violet soft rot (27, 46). The feo operon is anaerobically induced (47), and thus, its upregulation in S. enterica in our study provides further evidence that low-oxygen conditions prevail in D. dadantii-infected leaf tissue. znuC is part of an operon coding for a high-affinity zinc transporter required for Salmonella to overcome inflammation-induced zinc sequestration and colonize the host intestine (48). Its increased expression levels in soft rot lesions suggest that the human pathogen is limited in zinc in addition to iron in macerated leaf tissue.
(v) Response to inhibitory compounds.
Among the genes that showed the highest increases in transcriptional levels in soft rot compared with LB medium were several genes that have a role in the degradation of inhibitory compounds (see Tables S2 and S3 in the supplemental material). For example, yhhW, a homolog of the human pirin gene, which in E. coli is involved in the degradation of the plant flavonoid quercetin (49), was upregulated 93- and 23-fold in cilantro and lettuce soft rot, respectively. It is noteworthy that a gene encoding a pirin-related protein was similarly induced in D. dadantii during soft rot disease (27). azoR, which codes for a flavin mononucleotide (FMN)-dependent NADH-azoreductase that cleaves aromatic azo compounds, showed 144- and 101-fold increases in cilantro and lettuce, respectively. The expression level of ygiD, a predicted dioxygenase gene, increased 105-fold. Dioxygenases are involved in the catabolism of aromatic compounds (50) and are also induced in D. dadantii during plant disease (46), but it is unclear if YgiD would have sufficient available oxygen to function in this role under the conditions of our system.
Besides aromatic compounds, reactive oxygen species also are known to be released from diseased or injured plant cells. yqjG encodes a putative glutathione S-transferase and is highly upregulated in both cilantro (75-fold) and lettuce (37-fold); glutathione S-transferases protect bacterial cells against oxidative stress and other antimicrobials (51). Also transcribed at high levels in cilantro but not in lettuce were hcr, hcp, hmp, ygbA, and yftE (32-, 54-, 8-, 6-, and 4-fold, respectively), which are involved in protection from nitrosative stress (52). hcr was similarly greatly induced in D. dadantii during soft rot disease (27). The superoxide dismutase gene sodA was downregulated in both lettuce and cilantro. However, the transcription level of ahpF, which has a role in the detoxification of hydroperoxides, increased in lettuce, as was also reported for D. dadantii during soft rot of African violet (27), whereas smvA, which confers resistance to methyl viologen, a strong generator of superoxide radicals (53), showed increased expression levels in macerated cilantro. Also of note is the increased expression level of ycfR, which codes for a regulator involved in multiple-stress resistance, including oxidative stress (54), and which was upregulated in E. coli O157:H7 in shredded lettuce, as reported in our previous study (6). Finally, mdtH and yhcQ, which code for a drug efflux protein and a multidrug resistance protein, respectively, were upregulated. Along with YgjT, a putative resistance protein, the above-described genes may be required for resistance to antimicrobial compounds released from plant cells.
Comparison of transcriptomes of S. Typhimurium in cilantro soft rot and S. Enteritidis in the chicken intestine.
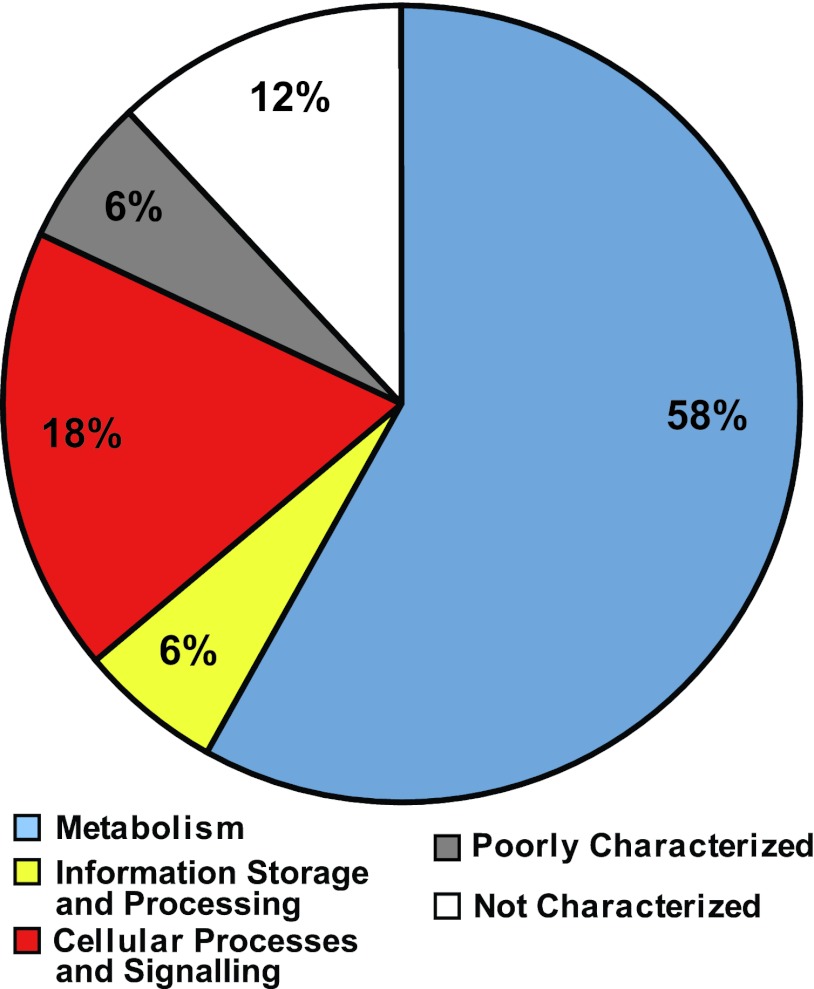
A comparison of the genes differentially regulated in S. Typhimurium in cilantro soft rot (this study) and in S. enterica serovar Enteritidis in the chicken intestine (55), both relative to those in LB broth, revealed striking commonalities in their physiologies. Table S6 in the supplemental material provides the lists of S. enterica genes that were upregulated (108 genes) or downregulated (127 genes) in both environments. Numerous genes whose expression levels increased in leaf macerates and in the chicken intestine play a role in the anaerobic respiratory chain, including the reduction of nitrate and nitrite; in transport systems; in iron and zinc acquisition; and in the utilization of nutrients such as carnitine, galactonate, gluconate, citrate, fucose, ascorbate, ethanolamine, and propanediol. The classification of the common upregulated genes in both study systems into categories of orthologous genes (COG) showed that 76% of these genes were related to metabolism (58%) and cellular processes (18%) (Fig. 4). Putative antimicrobial resistance genes (ycfR, ygjT, and yhcQ) and invasion genes (invG, invI, and prgH) also had increased transcription levels in both environments (see Table S6 in the supplemental material).
Fig 4.
Distribution of common genes upregulated in S. Typhimurium in cilantro soft rot (this study) and in S. Enteritidis in the chicken intestine (55), relative to levels in LB broth, into broad classes of categories of orthologous genes (COG). The percentages of genes that belong to categories that relate to metabolism (COG categories C, G, E, F, H, I, and Q), information storage and processing (COG categories J, K, and L), and cellular processes and signaling (COG categories D, O, M, N, P, and T) and those that are poorly characterized (COG categories R and S) and not characterized are illustrated. General gene functions of each COG by letter appellation are as shown in Fig. 2.
Confirmation of microarray data by qRT-PCR.
qRT-PCR was performed on select genes with the same bacterial RNA as that used for microarray hybridization. This approach confirmed the increased transcriptional activity observed by microarray analysis of eutD, eutM, fucI, rhaA, pduE, pudM, cbiE, and cobS, which are required for EUT and PDU (Fig. 5). The expression level of ttrC, which is involved in the reduction of tetrathionate, a sulfur compound acting as an electron acceptor during EUT and PDU under anaerobic conditions, was shown to increase 9-fold in macerated cilantro by qRT-PCR (Fig. 5). For most of the genes described above, the magnitude of upregulation was greater when assessed by qRT-PCR than when assessed by microarray analysis.
Behavior of mutants in PDU, EUT, and cobalamin pathways. (i) Phenotypes on indicator plates.
In light of the broad upregulation of the propanediol and ethanolamine utilization pathways and that of the biosynthesis of their cofactor, cobalamin, we generated deletion mutants of S. enterica SL1344 in these operons. Mutants that have a deletion in pduD (strain SS371), eutK (strain SS374), or cobS (strain SS377) were constructed. Complementation plasmids for these mutants were constructed by the cloning of pduD-X, eutRK, or cobTS into pBBR-MCS5 (20).
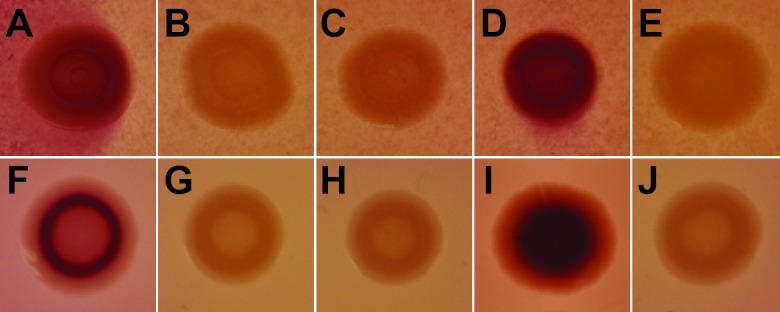
The transformation of the appropriate mutants with these plasmids resulted in strains MB713, MB715, and MB717, complemented in trans with pduD-X, eutRK, and cobTS, respectively. Due to the failure of the single-gene deletion mutants to show a defective colonization phenotype in cilantro and lettuce soft rot lesions (see below), we constructed individual mutants with a full pdu operon deletion (ΔpduA-X) (strain MB784), a full eut operon deletion (ΔeutK-S) (strain MB789), or a full deletion of both the pdu and eut operons (ΔpduA-X ΔeutK-S) (strain MB791). All deletion mutations were confirmed by PCR. Figure 6 shows the phenotypes of the parental strain and various eut and pdu mutants, as well as the complemented single-gene deletion mutants, on MacConkey agar containing ethanolamine or propanediol and vitamin B12. After incubation under aerobic conditions, colonies of cells that were able to utilize propanediol or ethanolamine and thus produce acid on these indicator plates appeared red, whereas colonies of cells defective in these pathways remained white (21, 22). Figure 6 clearly indicates that parental S. enterica strain SL1344 and the complemented PDU- and EUT-defective mutants had the red-colony phenotype but that the single-gene deletion and full-operon deletion mutants as well as the PDU-EUT double mutant produced white colonies. Therefore, all strains showed the expected phenotype on the indicator plates.
Fig 6.
Single colonies of WT S. enterica SL1344 and mutant strains on MacConkey agar containing cobalamin and 1,2-propanediol (top) or ethanolamine (bottom). The accumulation of deep red pigment in the colonies indicates the utilization of propanediol or ethanolamine. The WT strain (A and F) and PDU-minus and EUT-minus complemented mutant strains MB713 (D) and MB717 (I) produced distinct red colonies. None of the following mutant strains produced red colonies: strains MB784 and MB789, with a full PDU (B) or EUT (G) operon deletion; strains SS371 and SS377, with a deletion of pduD (C) or eutK (H); and the double PDU- and EUT-minus strain MB792 (E and J).
(ii) Phenotypes in planta.
S. enterica wild-type strain SL1344 or its pduD, eutK, or cobS deletion mutant was coinoculated with D. dadantii onto cut cilantro leaves, and their population sizes were measured over time. None of the above-described mutants showed a growth defect in soft-rotted tissue compared with the wild-type strain (data not shown). Coinoculations of the wild type and any one of the above-mentioned mutants at a ratio of 1:1 also failed to reveal any difference in their growths in D. dadantii-infected cilantro and, thus, in their competitive fitness. These experiments were repeated with the PDU and EUT full-operon deletion mutants and with the mutant that has a deletion of both operons. Figure 7A demonstrates that these mutants also behaved very similarly to the wild-type strain when coinoculated singly with D. dadantii. The soft rot pathogen itself was unaffected in growth during infection of cilantro by the presence of either the wild-type or the mutant strains, and its populations sizes were very similar to those of the human pathogen (Fig. 7B), as was also observed for cilantro and lettuce in Fig. 1. Competitive fitness experiments by the coinoculation of the mutants with the wild-type strain and D. dadantii at a ratio of 1:1:1 demonstrated that the initial proportion of the S. enterica strains in each pair did not change significantly over the course of maceration (Fig. 7C). Similar results were obtained with cut lettuce leaves (data not shown).
Fig 7.
Colonization of cilantro leaves by WT S. enterica SL1344 and mutant strains after their coinoculation with the soft rot pathogen D. dadantii. (A) Means of the log values of the population sizes of WT S. enterica, PDU-minus strain MB784, EUT-minus strain MB789, and the double PDU- and EUT-minus strain MB792 when inoculated individually with D. dadantii. (B) Means of the log values of the population sizes of D. dadantii when inoculated individually with one of the above-mentioned S. enterica strains. Symbols correspond to coinoculated S. enterica strains, as shown in panel A. (C) Means of the log values of the ratio of the population size of the S. enterica mutant strain to that of the WT strain when coinoculated with D. dadantii at a ratio of 1:1:1. Symbols correspond to S. enterica mutant strains MB784, MB789, and MB792 coinoculated with the WT strain. Bars represent standard errors of the means for three (A and B) and four (C) replicate leaf samples.
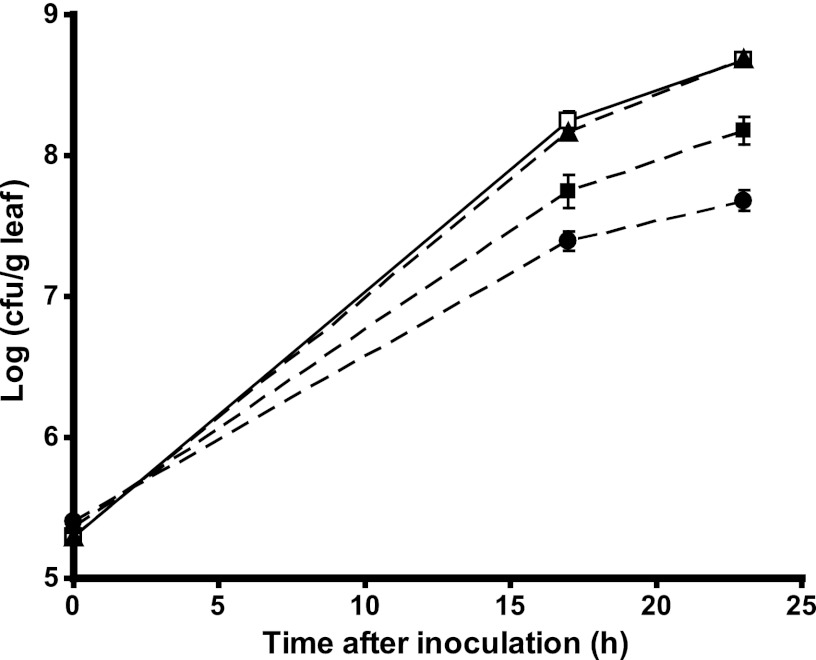
The above-described results diverge from those obtained with another S. enterica serovar Typhimurium strain, LT2. Mutants in this background that are impaired in propanediol utilization (strain TT22344) and in cobalamin synthesis (TT20945), kindly provided by J. Roth, had attenuated growth and achieved population sizes in macerated cilantro that were 9.8- and 3.1-fold lower than those of the wild-type strain (P < 0.0001 and P < 0.0002, respectively, by Student's t test) (Fig. 8). The transformation of the LT2 PDU-minus mutant with pMBpduD-X failed to complement its attenuated growth in soft rot lesions (data not shown). As in the SL1344 background, the LT2 ethanolamine utilization-minus mutant behaved like the wild-type strain.
Fig 8.
Colonization of cilantro leaves by WT S. enterica LT2 and mutant strains when coinoculated individually with the soft rot pathogen D. dadantii. □, WT strain MB580; ●, PDU-minus strain TT22344; ▲, EUT-minus strain TT22341; ■, cobalamin-minus strain TT20945. Datum points and bars represent the means of the log values for the population size in three replicate leaf samples and standard errors of the means.
DISCUSSION
For human enteric pathogens, the phyllosphere is a habitat with considerably lower nutrient availability than that of the intestine of their hosts. Nevertheless, numerous outbreaks of food-borne illness have occurred due to the contamination of leafy vegetables with S. enterica and E. coli O157:H7. Hence, on plants, these pathogens probably encounter conditions that allow them to multiply to the threshold population densities necessary to infect humans. We have previously provided evidence that the population sizes of E. coli O157:H7 on lettuce leaves were significantly larger in the presence of soft rot disease after the leaves were infected with the soft rot pathogen D. dadantii than when the human pathogen was inoculated alone (5). Yamazaki et al. (9) previously reported a similar effect of D. dadantii infection and provided evidence that this proliferation of E. coli O157:H7 is due primarily to the pectinolytic activity of the plant pathogen. The prevailing dogma has been that plant cell degradation during tissue maceration releases nutrients that enteric pathogens may utilize for growth, but the exact nature of these substrates remains unknown.
The results of our present study clearly indicate that like E. coli O157:H7, S. enterica benefits from increased nutrient availability during plant soft rot disease caused by D. dadantii. Cilantro and lettuce leaves appeared to have very similar carrying capacities for S. enterica, since its population sizes on both plants species were comparable; this was observed for healthy leaves as well as for infected leaves. Additionally, the population sizes of the human pathogen were very highly correlated with those of D. dadantii after their coinoculation onto leaves and during disease progression. This finding suggested either that each bacterial species competed equally for the same resources and that the availability of these resources in the macerated tissue was sufficient to promote the multiplication of both strains equally or that there was a lack of competition for nutrients between the two bacterial species due to resource partitioning, as shown previously by Wilson and Lindow (56) for epiphytic bacteria in the bean phyllosphere. The global gene expression profile of S. enterica in soft-rotted tissue, as we describe below, provides evidence that resource partitioning may be the predominant scenario driving the high levels of correlated growth of the human and plant pathogens in this environment.
Our transcriptome analysis indeed revealed much about the metabolism of S. enterica in the cilantro and lettuce leaf macerates produced by D. dadantii. The increased expression levels of numerous genes that function in anaerobiosis, including nitrate respiration, the synthesis of cytochromes and electron acceptors, and others that are involved in the acquisition or catabolism of compounds that serve as nutrients for anaerobic growth, indicate that low oxygen tension prevails in the lesions. Soft rot pathogens have also been shown to experience low oxygen levels in the plant tissue that they degrade, a condition that promotes disease via the induction of genes coding for pectinolytic enzymes (57). In particular, anaerobic nitrate respiration is induced in Erwinia carotovora subsp. atroseptica in soft-rotting potato tuber tissue (58), and a gene homolog with a role in nitrate/nitrite respiration and another functioning in iron transport under conditions of anaerobiosis (feoB) were both upregulated in D. dadantii during maceration of African violet leaves (27). Although it is likely that oxygen depletion in soft rot results at least partly from rapid microbial growth in the degraded tissue, its cause has not been investigated.
In addition to anaerobic/microaerobic metabolism, other commonalities in behavior were observed for the human pathogen and the soft rot pathogen in infected leaf tissue. For example, S. enterica appeared to respond to inhibitory compounds (reactive oxygen species, flavonoids, and other aromatic compounds), some of which may have been produced as part of the plant defense response to invasion by D. dadantii (59). Indeed, several of these genes had homology or were related in function to those upregulated in D. dadantii during infection of African violet (27, 46). Also, based on the increased gene expression levels in both systems, both the plant and the human pathogens appeared to metabolize substrates made available in macerated leaf tissue, such as d-galactonate (dgo) (27), d-gluconate (gntU, kdgT, and kduD) (60), and l-idonate, which is tied to gluconate metabolism in Erwinia spp. (61). Notably, gluconate metabolism is essential for the virulence of the soft rot pathogen Pectobacterium carotovorum in potato and in Arabidopsis thaliana leaves (60).
It is evident, however, that S. enterica in soft-rotted leaves devoted an extensive part of its transcriptional machinery to the catabolism of propanediol and ethanolamine and the synthesis of the required coenzyme cobalamin. With few exceptions, all genes in these three large operons had increased expression levels in macerated tissue, with certain genes being upregulated more than 100-fold. Furthermore, the expression levels of most genes for pathways that feed into and out of propanediol and ethanolamine catabolism to provide cellular energy during anaerobiosis (rha and fuc genes for rhamnose and fucose fermentation, prp genes for the production of pyruvate, and ttr and asr genes for the synthesis of tetrathionate as the alternate electron acceptor and its reduction to sulfide) increased significantly (Fig. 3). In total, all of the above-described transcriptional units represent approximately 1.4% of the S. Typhimurium genome, a sizeable portion of its genetic capability.
The production of propanediol and its catabolism in S. enterica were likely induced by the fermentation of the plant cell wall carbohydrates fucose and rhamnose, which are released during maceration by the soft rot pathogen and can be converted by the human pathogen into lactaldehyde, the precursor of propanediol (22). This is supported by the increased expression levels of the genes for the degradation of rhamnose (rha) and fucose (fuc) in S. enterica in infected leaves. Ethanolamine is a major constituent of lipids in eukaryotic cells (62), including plants (63), and therefore, it may be available to the human pathogen through the breakdown of plant cells by D. dadantii. Plants do not produce cobalamin (39), and the genome sequence of D. dadantii 3937 indicates that it does not have the capability to synthesize the coenzyme (64). This may explain the high level of induction of the cobalamin biosynthetic genes in S. enterica in soft rot and the lack of differential regulation of the btu operon, which is required for cobalamin uptake into this pathogen (39). The requirement for anaerobic conditions for the synthesis of cobalamin in S. enterica (65) corroborates our observation that the human pathogen is exposed to low oxygen tension in the plant macerates. D. dadantii can obtain energy from oligogalacturonides released from the plant cell wall through its pectinolytic activity (57) but does not have the genetic determinants necessary to metabolize ethanolamine and propanediol (64). Therefore, the dominant activity of these pathways may spare S. enterica extensive competition with the plant pathogen for nutrients in lettuce and cilantro soft-rotted tissue and, hence, may effect the highly correlated growth of the two bacterial species in that habitat via the segregation of nutritional resources.
Whereas a deficiency in propanediol utilization and cobalamin synthesis significantly decreased the fitness of S. Typhimurium strain LT2 in cilantro soft rot, a deficiency in ethanolamine had no detectable effect at the population level. Additionally, mutations resulting in the loss of cobalamin synthesis, of propanediol or ethanolamine catabolism, or of the degradation of both compounds did not affect the colonization of macerated cilantro or lettuce leaves by strain SL1344. This observation is surprising in light of the high-level and broad upregulation of these operons in strain SL1344 in soft rot. Although an increased gene expression level is not de facto indicative of function, the metabolic cost of the transcription of such large operons makes their high-level expression without subsequent enzymatic activity unlikely. Rather, the discrepancy may be explained by the remarkable metabolic robustness of S. enterica, which enables its adaptation to a diverse range of nutritional conditions (66). Its transcriptional profile in soft rot is evidence of its ability to utilize numerous other complex carbon sources available in that environment, such as citrate and ascorbate. Thus, the PDU- and EUT-minus mutants of strain SL1344 may have used alternate nutrient acquisition pathways in order to compensate for the loss of the latter operons. The difference in the behaviors of the PDU- and cobalamin-minus mutants in the strain LT2 and strain SL1344 backgrounds in macerated plant tissue may stem from low levels of the starvation-induced sigma factor RpoS in LT2 (67). The cobalamin and PDU pathways are under the control of several regulators, including ArcA and Crp (68) and CsrA (69), but are not known to be regulated by RpoS. It is possible that a weak ability to respond to starvation via RpoS combined with the deficiency in the PDU and cobalamin pathways attenuated the adaptation to alternate substrate assimilation in soft rot in LT2 to a greater extent than in SL1344.
Importantly, the transcriptional signature of S. enterica in leaf macerates uncovered considerable commonality with its physiological profile during the colonization of its hosts. A comparison of its transcriptome in cilantro soft rot with that in the chicken intestine further corroborates this observation by revealing that one-third of the genes that were upregulated in cilantro macerates were regulated similarly in the chicken intestine. A great portion of these genes had a role in anaerobic metabolism as well as substrate transport and fermentation. It appears that the anaerobic conditions in the leaf macerates allowed S. enterica to utilize plant-derived substrates that are likely also encountered in the anaerobic intestinal environment of its animal hosts due to dietary intake. Other carbon sources such as fucose and ethanolamine are abundant not only as plant derivatives but also as intestinal mucin glycoconjugates (70) and as phospholipids in intestinal epithelial cell membranes (62, 71), respectively.
Increased expression levels of the pdu, eut and cob/cbi genes in S. enterica were reported not only in the chicken intestine (55) but also in the chicken lumen (72) and in the blood of bacteremic patients (73). Ethanolamine utilization confers a competitive advantage to S. Typhimurium in the lumen of the inflamed intestine in the mouse colitis model (74) and to E. coli O157:H7 in bovine intestinal contents (75). Furthermore, propanediol utilization and cobalamin synthesis in S. Typhimurium are required for replication in macrophages (76) and are involved in the colonization of the chicken lumen (72). The high levels of activity of these pathways in soft rot lesions, combined with the enhanced transcription of genes involved in the degradation of other carbon sources that are also present in the animal intestine (carnitine, gluconate, and ascorbate) as well as of genes implicated in anaerobic metabolism, in the response to nitrosative and oxidative stress, and in iron uptake, some of which have a known role in host colonization or virulence (52, 77–79), suggests a niche overlap that allows the human pathogen to readily adapt to this habitat outside its primary host. The fitness of S. enterica and its ability to reach high population densities in macerated leaf tissue thus point to soft rot in produce as a potential risk to public health.
Supplementary Material
ACKNOWLEDGMENTS
We thank Aileen Haxo and Steven Huynh for technical assistance and John Roth for the gift of S. Typhimurium LT2 strains.
This research was supported by United States-Israel Binational Agricultural Research and Development Fund (BARD) award no. US-3949-06 to M.T.B. and S.S. and by U.S. Department of Agriculture Agricultural Research Service CRIS projects 5325-42000-046 and 5325-42000-047.
Footnotes
Published ahead of print 26 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02290-12.
REFERENCES
- 1. Brandl MT, Amundson R. 2008. Leaf age as a risk factor in contamination of lettuce with Escherichia coli O157:H7 and Salmonella enterica. Appl. Environ. Microbiol. 74:2298–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brandl MT, Mandrell RE. 2002. Fitness of Salmonella enterica serovar Thompson in the cilantro phyllosphere. Appl. Environ. Microbiol. 68:3614–3621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brandl MT. 2006. Fitness of human enteric pathogens on plants and implications for food safety. Annu. Rev. Phytopathol. 44:367–392 [DOI] [PubMed] [Google Scholar]
- 4. Aruscavage D, Miller SA, Ivey ML, Lee K, LeJeune JT. 2008. Survival and dissemination of Escherichia coli O157:H7 on physiologically and biologically damaged lettuce plants. J. Food Prot. 71:2384–2388 [DOI] [PubMed] [Google Scholar]
- 5. Brandl MT. 2008. Plant lesions promote the rapid multiplication of Escherichia coli O157:H7 on postharvest lettuce. Appl. Environ. Microbiol. 74:5285–5289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kyle JL, Parker CT, Goudeau D, Brandl MT. 2010. Transcriptome analysis of Escherichia coli O157:H7 exposed to lysates of lettuce leaves. Appl. Environ. Microbiol. 76:1375–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wells JM, Butterfield JE. 1999. Incidence of Salmonella on fresh fruits and vegetables affected by fungal rots and physical injury. Plant Dis. 83:722–726 [DOI] [PubMed] [Google Scholar]
- 8. Wells JM, Butterfield JE. 1997. Salmonella contamination associated with bacterial soft-rot of fresh fruits and vegetables in the marketplace. Plant Dis. 81:867–872 [DOI] [PubMed] [Google Scholar]
- 9. Yamazaki A, Li J, Hutchins WC, Wang L, Ma J, Ibekwe AM, Yang CH. 2011. Commensal effect of pectate lyases secreted from Dickeya dadantii on proliferation of Escherichia coli O157:H7 EDL933 on lettuce leaves. Appl. Environ. Microbiol. 77:156–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Noel JT, Joy J, Smith JN, Fatica M, Schneider KR, Ahmer BM, Teplitski M. 2010. Salmonella SdiA recognizes N-acyl homoserine lactone signals from Pectobacterium carotovorum in vitro, but not in a bacterial soft rot. Mol. Plant Microbe Interact. 23:273–282 [DOI] [PubMed] [Google Scholar]
- 11. Carlin F, Nguyen-the C, Da Silva AA. 1995. Factors affecting the growth of Listeria monocytogenes on minimally processed fresh endive. J. Appl. Microbiol. 78:636–646 [DOI] [PubMed] [Google Scholar]
- 12. Liao CH, Sapers GM. 1999. Influence of soft rot bacteria on growth of Listeria monocytogenes on potato tuber slices. J. Food Prot. 62:343–348 [DOI] [PubMed] [Google Scholar]
- 13. Conway WS, Leverentz B, Saftner RA, Janisiewicz WJ, Sams CE, Leblanc E. 2000. Survival and growth of Listeria monocytogenes on fresh-cut apple slices and its interaction with Glomerella cingulata and Penicillium expansum. Plant Dis. 84:177–181 [DOI] [PubMed] [Google Scholar]
- 14. Riordan DC, Sapers GM, Annous BA. 2000. The survival of Escherichia coli O157:H7 in the presence of Penicillium expansum and Glomerella cingulata in wounds on apple surfaces. J. Food Prot. 63:1637–1642 [DOI] [PubMed] [Google Scholar]
- 15. Wade WN, Beuchat LR. 2003. Metabiosis of proteolytic moulds and Salmonella in raw, ripe tomatoes. J. Appl. Microbiol. 95:437–450 [DOI] [PubMed] [Google Scholar]
- 16. Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239 [DOI] [PubMed] [Google Scholar]
- 17. Price-Carter M, Tingey J, Bobik TA, Roth JR. 2001. The alternative electron acceptor tetrathionate supports B12-dependent anaerobic growth of Salmonella enterica serovar Typhimurium on ethanolamine or 1,2-propanediol. J. Bacteriol. 183:2463–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lemattre M, Narcy JP. 1972. A new bacterial infection of Saintpaulia ionantha by Erwinia chrysanthemi. C. R. Acad. Agric. Fr. 58:227–231 [Google Scholar]
- 19. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, II, Peterson KM. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175–176 [DOI] [PubMed] [Google Scholar]
- 21. Kofoid E, Rappleye C, Stojiljkovic I, Roth J. 1999. The 17-gene ethanolamine (eut) operon of Salmonella typhimurium encodes five homologues of carboxysome shell proteins. J. Bacteriol. 181:5317–5329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Walter D, Ailion M, Roth J. 1997. Genetic characterization of the pdu operon: use of 1,2-propanediol in Salmonella typhimurium. J. Bacteriol. 179:1013–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 doi:10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 25. Rehfuss MYM, Parker CT, Brandl MT. 2011. Salmonella transcriptional signature in Tetrahymena phagosomes and role of acid tolerance in passage through the protist. ISME J. 5:262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dixon WJ. 1950. Analysis of extreme values. Ann. Math. Stat. 21:488–506 [Google Scholar]
- 27. Okinaka Y, Yang CH, Perna NT, Keen NT. 2002. Microarray profiling of Erwinia chrysanthemi 3937 genes that are regulated during plant infection. Mol. Plant Microbe Interact. 15:619–629 [DOI] [PubMed] [Google Scholar]
- 28. Thony-Meyer L, Fischer F, Kunzler P, Ritz D, Hennecke H. 1995. Escherichia coli genes required for cytochrome c maturation. J. Bacteriol. 177:4321–4326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gilberthorpe NJ, Poole RK. 2008. Nitric oxide homeostasis in Salmonella Typhimurium. J. Biol. Chem. 283:11146–11154 [DOI] [PubMed] [Google Scholar]
- 30. Mills PC, Rowley G, Spiro S, Hinton JCD, Richardson DJ. 2008. A combination of cytochrome c nitrite reductase (NrfA) and flavorubredoxin (NorV) protects Salmonella enterica serovar Typhimurium against killing by NO in anoxic environments. Microbiology 154:1218–1228 [DOI] [PubMed] [Google Scholar]
- 31. Tseng CP, Albrecht J, Gunsalus RP. 1996. Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J. Bacteriol. 178:1094–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Soo P-C, Horng Y-T, Lai M-J, Wei J-R, Hsieh S-C, Chang Y-L, Tsai Y-H, Lai H-C. 2007. Pirin regulates pyruvate catabolism by interacting with the pyruvate dehydrogenase E1 subunit and modulating pyruvate dehydrogenase activity. J. Bacteriol. 189:109–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Subramoni S, Gonzalez JF, Johnson A, Pechy-Tarr M, Rochat L, Paulsen I, Loper JE, Keel C, Venturi V. 2011. Bacterial subfamily of LuxR regulators that respond to plant compounds. Appl. Environ. Microbiol. 77:4579–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seim H, Löster H, Claus R, Kleber HP, Strack E. 1982. Stimulation of the anaerobic growth of Salmonella typhimurium by reduction of L-carnitine, carnitine derivatives and structure-related trimethylammonium compounds. Arch. Microbiol. 132:91–95 [DOI] [PubMed] [Google Scholar]
- 35. Fraenkel G. 1953. Studies on the distribution of vitamin Bt (carnitine). Biol. Bull. 104:359–371 [Google Scholar]
- 36. Bourdin B, Adenier H, Perrin Y. 2007. Carnitine is associated with fatty acid metabolism in plants. Plant Physiol. Biochem. 45:926–931 [DOI] [PubMed] [Google Scholar]
- 37. Bott M. 1997. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch. Microbiol. 167:78–88 [PubMed] [Google Scholar]
- 38. Chen Y-T, Liao T-L, Wu K-M, Lauderdale T-L, Yan J-J, Huang IW, Lu M-C, Lai Y-C, Liu Y-M, Shu H-Y, Wang J-T, Su I-J, Tsai S-F. 2009. Genomic diversity of citrate fermentation in Klebsiella pneumoniae. BMC Microbiol. 9:168 doi:10.1186/1471-2180-9-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roth JR, Lawrence JG, Bobik TA. 1996. Cobalamin (coenzyme B12): synthesis and biological significance. Annu. Rev. Microbiol. 50:137–181 [DOI] [PubMed] [Google Scholar]
- 40. Chapman KD. 2004. Occurrence, metabolism, and prospective functions of N-acylethanolamines in plants. Prog. Lipid Res. 43:302–327 [DOI] [PubMed] [Google Scholar]
- 41. Badia J, Ros J, Aguilar J. 1985. Fermentation mechanism of fucose and rhamnose in Salmonella typhimurium and Klebsiella pneumoniae. J. Bacteriol. 161:435–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Laatu M, Condemine G. 2003. Rhamnogalacturonate lyase RhiE is secreted by the Out system in Erwinia chrysanthemi. J. Bacteriol. 185:1642–1649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Reiter W-D. 2002. Biosynthesis and properties of the plant cell wall. Curr. Opin. Plant Biol. 5:536–542 [DOI] [PubMed] [Google Scholar]
- 44. Palacios S, Starai VJ, Escalante-Semerena JC. 2003. Propionyl coenzyme A is a common intermediate in the 1,2-propanediol and propionate catabolic pathways needed for expression of the prpBCDE operon during growth of Salmonella enterica on 1,2-propanediol. J. Bacteriol. 185:2802–2810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC. 1999. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol. Microbiol. 32:275–287 [DOI] [PubMed] [Google Scholar]
- 46. Yang S, Perna NT, Cooksey DA, Okinaka Y, Lindow SE, Ibekwe AM, Keen NT, Yang C-H. 2004. Genome-wide identification of plant-upregulated genes of Erwinia chrysanthemi 3937 using a GFP-based IVET leaf array. Mol. Plant Microbe Interact. 17:999–1008 [DOI] [PubMed] [Google Scholar]
- 47. Cartron ML, Maddocks S, Gillingham P, Craven CJ, Andrews SC. 2006. Feo—transport of ferrous iron into bacteria. Biometals 19:143–157 [DOI] [PubMed] [Google Scholar]
- 48. Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, Pasquali P, Lane TE, Chazin WJ, Vogl T, Roth J, Skaar EP, Raffatellu M. 2012. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11:227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adams M, Jia Z. 2005. Structural and biochemical analysis reveal pirins to possess quercetinase activity. J. Biol. Chem. 280:28675–28682 [DOI] [PubMed] [Google Scholar]
- 50. Fuchs G, Boll M, Heider J. 2011. Microbial degradation of aromatic compounds—from one strategy to four. Nat. Rev. Microbiol. 9:803–816 [DOI] [PubMed] [Google Scholar]
- 51. Allocati N, Federici L, Masulli M, Di Ilio C. 2009. Glutathione transferases in bacteria. FEBS J. 276:58–75 [DOI] [PubMed] [Google Scholar]
- 52. Gilberthorpe NJ, Lee ME, Stevanin TM, Read RC, Poole RK. 2007. NsrR: a key regulator circumventing Salmonella enterica serovar Typhimurium oxidative and nitrosative stress in vitro and in IFN-gamma-stimulated J774.2 macrophages. Microbiology 153:1756–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Etsuko H, Mitsuoki M, Kazuei M, Isamu M, Hiroko Hideo H-IT, Sachiko I, Yutaka N. 1994. The methyl viologen-resistance-encoding gene smvA of Salmonella Typhimurium. Gene 148:173–174 [DOI] [PubMed] [Google Scholar]
- 54. Deng K, Wang S, Rui X, Zhang W, Tortorello ML. 2011. Functional analysis of ycfR and ycfQ in Escherichia coli O157:H7 linked to outbreaks of illness associated with fresh produce. Appl. Environ. Microbiol. 77:3952–3959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dhawi AA, Elazomi A, Jones MA, Lovell MA, Li H, Emes RD, Barrow PA. 2011. Adaptation to the chicken intestine in Salmonella Enteritidis PT4 studied by transcriptional analysis. Vet. Microbiol. 153:198–204 [DOI] [PubMed] [Google Scholar]
- 56. Wilson M, Lindow SE. 1994. Coexistence among epiphytic bacterial populations mediated through nutritional resource partitioning. Appl. Environ. Microbiol. 60:4468–4477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. 1996. Regulation of pectinolysis in Erwinia chrysanthemi. Annu. Rev. Microbiol. 50:213–257 [DOI] [PubMed] [Google Scholar]
- 58. Smid EJ, Jansen AHJ, Tuijn CJ. 1993. Anaerobic nitrate respiration by Erwinia carotovora subsp. atroseptica during potato tuber invasion. Appl. Environ. Microbiol. 59:3648–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fagard M, Dellagi A, Roux C, Perino C, Rigault M, Boucher V, Shevchik VE, Expert D. 2007. Arabidopsis thaliana expresses multiple lines of defense to counterattack Erwinia chrysanthemi. Mol. Plant Microbe Interact. 20:794–805 [DOI] [PubMed] [Google Scholar]
- 60. Mole B, Habibi S, Dangl JL, Grant SR. 2010. Gluconate metabolism is required for virulence of the soft-rot pathogen Pectobacterium carotovorum. Mol. Plant Microbe Interact. 23:1335–1344 [DOI] [PubMed] [Google Scholar]
- 61. Truesdell SJ, Sims JC, Boerman PA, Seymour JL, Lazarus RA. 1991. Pathways for metabolism of ketoaldonic acids in an Erwinia sp. J. Bacteriol. 173:6651–6656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bakovic M, Fullerton MD, Michel V. 2007. Metabolic and molecular aspects of ethanolamine phospholipid biosynthesis: the role of CTP:phosphoethanolamine cytidylyltransferase (Pcyt2). Biochem. Cell Biol. 85:283–300 [DOI] [PubMed] [Google Scholar]
- 63. Miedema E, Richardson KE. 1966. Ethanolamine metabolism in plant tissues. Plant Physiol. 41:1026–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Glasner JD, Yang Reverchon C-HS, Hugouvieux-Cotte-Pattat N, Condemine G, Bohin JP, Van Gijsegem F, Yang S, Franza T, Expert D, Plunkett G, San Francisco MJ, Charkowski AO, Py B, Bell K, Rauscher L, Rodriguez-Palenzuela P, Toussaint A, Holeva MC, He SY, Douet V, Boccara M, Blanco C, Toth I, Anderson BD, Biehl BS, Mau B, Flynn SM, Barras F, Lindeberg M, Birch PRJ, Tsuyumu S, Shi X, Hibbing M, Yap MN, Carpentier M, Dassa E, Umehara M, Kim JF, Rusch M, Soni P, Mayhew GF, Fouts DE, Gill SR, Blattner FR, Keen NT, Perna NT. 2011. Genome sequence of the plant-pathogenic bacterium Dickeya dadantii 3937. J. Bacteriol. 193:2076–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jeter RM, Olivera BM, Roth JR. 1984. Salmonella typhimurium synthesizes cobalamin (vitamin B12) de novo under anaerobic growth conditions. J. Bacteriol. 159:206–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Becker D, Selbach M, Rollenhagen C, Ballmaier M, Meyer TF, Mann M, Bumann D. 2006. Robust Salmonella metabolism limits possibilities for new antimicrobials. Nature 440:303–307 [DOI] [PubMed] [Google Scholar]
- 67. Jarvik T, Smillie C, Groisman EA, Ochman H. 2010. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J. Bacteriol. 192:560–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Chen P, Ailion M, Bobik T, Stormo G, Roth J. 1995. Five promoters integrate control of the cob/pdu regulon in Salmonella typhimurium. J. Bacteriol. 177:5401–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lawhon SD, Frye JG, Suyemoto M, Porwollik S, McClelland M, Altier C. 2003. Global regulation by CsrA in Salmonella typhimurium. Mol. Microbiol. 48:1633–1645 [DOI] [PubMed] [Google Scholar]
- 70. Wesley A, Mantle M, Man D, Qureshi R, Forstner G, Forstner J. 1985. Neutral and acidic species of human intestinal mucin. Evidence for different core peptides. J. Biol. Chem. 260:7955–7959 [PubMed] [Google Scholar]
- 71. Kawai K, Fujita M, Nakao M. 1974. Lipid components of two different regions of an intestinal epithelial cell membrane of mouse. Biochim. Biophys. Acta 369:222–233 [PubMed] [Google Scholar]
- 72. Harvey PC, Watson M, Hulme S, Jones MA, Lovell M, Berchieri A, Young J, Bumstead N, Barrow P. 2011. Salmonella enterica serovar Typhimurium colonizing the lumen of the chicken intestine grows slowly and upregulates a unique set of virulence and metabolism genes. Infect. Immun. 79:4105–4121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Sheikh A, Charles RC, Rollins SM, Harris JB, Bhuiyan MS, Khanam F, Bukka A, Kalsy A, Porwollik S, Brooks WA, LaRocque RC, Hohmann EL, Cravioto A, Logvinenko T, Calderwood SB, McClelland M, Graham JE, Qadri F, Ryan ET. 2010. Analysis of Salmonella enterica serotype Paratyphi A gene expression in the blood of bacteremic patients in Bangladesh. PLoS Negl. Trop. Dis. 4:e908 doi:10.1371/journal.pntd.0000908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. 2011. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U. S. A. 108:17480–17485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bertin Y, Girardeau JP, Chaucheyras-Durand F, Lyan B, Pujos-Guillot E, Harel J, Martin C. 2011. Enterohaemorrhagic Escherichia coli gains a competitive advantage by using ethanolamine as a nitrogen source in the bovine intestinal content. Environ. Microbiol. 13:365–377 [DOI] [PubMed] [Google Scholar]
- 76. Klumpp J, Fuchs TM. 2007. Identification of novel genes in genomic islands that contribute to Salmonella typhimurium replication in macrophages. Microbiology 153:1207–1220 [DOI] [PubMed] [Google Scholar]
- 77. Maier RJ, Olczak A, Maier S, Soni S, Gunn J. 2004. Respiratory hydrogen use by Salmonella enterica serovar Typhimurium is essential for virulence. Infect. Immun. 72:6294–6299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sweeney NJ, Laux DC, Cohen PS. 1996. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect. Immun. 64:3504–3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Tsolis RM, Baumler AJ, Heffron F, Stojiljkovic I. 1996. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect. Immun. 64:4549–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.