Abstract
A strictly aerobic, halotolerant, rod-shaped bacterium, designated strain TG408, was isolated from a laboratory culture of the marine diatom Skeletonema costatum (CCAP1077/1C) by enrichment with polycyclic aromatic hydrocarbons (PAHs) as the sole carbon source. 16S rRNA gene sequence analysis placed this organism within the order Xanthomonadales of the class Gammaproteobacteria. Its closest relatives included representatives of the Hydrocarboniphaga-Nevskia-Sinobacter clade (<92% sequence similarity) in the family Sinobacteraceae. The strain exhibited a narrow nutritional spectrum, preferring to utilize aliphatic and aromatic hydrocarbon compounds and small organic acids. Notably, it displayed versatility in degrading two- and three-ring PAHs. Moreover, catechol 2,3-dioxygenase activity was detected in lysates, indicating that this strain utilizes the meta-cleavage pathway for aromatic compound degradation. Cells produced surface blebs and contained a single polar flagellum. The predominant isoprenoid quinone of strain TG408 was Q-8, and the dominant fatty acids were C16:0, C16:1 ω7c, and C18:1 ω7c. The G+C content of the isolate's DNA was 64.3 mol% ± 0.34 mol%. On the basis of distinct phenotypic and genotypic characteristics, strain TG408 represents a novel genus and species in the class Gammaproteobacteria for which the name Polycyclovorans algicola gen. nov., sp. nov., is proposed. Quantitative PCR primers targeting the 16S rRNA gene of this strain were developed and used to show that this organism is found associated with other species of marine phytoplankton. Phytoplankton may be a natural biotope in the ocean where new species of hydrocarbon-degrading bacteria await discovery and which contribute significantly to natural remediation processes.
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are organic molecules composed of fused benzene rings and are considered to be among the most widespread organic pollutants. On the basis of their poor water solubility, toxicity, persistence, and potential to bioaccumulate, PAHs are recognized as high-priority pollutants by the U.S. and European environmental protection agencies and are of significant concern to human health (1). PAHs can enter the marine environment through various sources, such as coastal surface runoff, fluvial inputs, industrial discharge, atmospheric deposition, and natural oil seeps. They occur naturally in fossil fuels, constituting as much as 25 to 35% of crude oil (2). Oil seeps and spills from the oil and gas industry are a significant source of PAH input into marine waters and sediment. The Deepwater Horizon oil spill, for example, released an estimated 4.4 million barrels (0.7 million tons) of crude oil into the Gulf of Mexico over a period of 84 days (3). Once released, PAHs can travel into the food web, persist for many years in the environment, and have detrimental effects on biological systems.
Despite their enormous and continual inputs into the sea, the mere fact that PAH compounds do not accumulate is due largely to the activities of particular types of marine bacteria with a nutritional preference for the utilization of hydrocarbons as sole sources of carbon and energy. These organisms include members of the Alcanivorax, Marinobacter, Thalassolituus, and Cycloclasticus genera, which are strongly selected for in oil-impacted environments (4, 5), where they play a critical role in oxidizing and mineralizing PAHs and other hydrocarbons completely to CO2. In recent years, improvements in cultivation-based techniques and molecular biological methods have helped efforts to identify many new species of PAH-degrading bacteria in environmental samples. However, we know very little about the ecology of these organisms in their natural environment, how they interact with other micro- and macrobiota, and the niches that they occupy.
A number of studies have reported on the potential of marine phytoplankton to adsorb and concentrate PAH molecules from the surrounding water column (6, 7). In a study by Binark et al. (8), the cell surfaces of several marine microalgae were shown to adsorb up to 14 different types of PAHs to relatively high concentrations. In a field study, Kowalewska (9) found that phytoplankton significantly contributed to the transport of PAHs by sedimentation from the upper layers of the southern Baltic ecosystem to the sea floor. These findings have potentially profound implications for the natural purging of the water column and the overall health of the marine ecosystem. Given the association of PAHs with marine phytoplankton, the potential exists for PAH-degrading bacteria to be associated with the phytoplankton as well. Recently, we isolated several PAH-degrading bacteria from different species of marine phytoplankton. In the present study, we characterize one of those isolates, designated strain TG408, which was found to be affiliated with the class Gammaproteobacteria. On the basis of polyphasic analysis, we propose that this strain represents a novel genus and species of “specialist” hydrocarbon-degrading marine bacterium associated with a marine eukaryotic phytoplankton. By using a new set of quantitative PCR primers that target the 16S rRNA gene of strain TG408, this organism was identified in several laboratory cultures of phytoplankton comprising different species of dinoflagellates and diatoms—algal lineages representing major groups in the marine ecosystem. This discovery poses important questions about the relationship of these types of bacteria with phytoplankton and their role in the natural purging of the marine water column of hydrocarbon contamination.
MATERIALS AND METHODS
Enrichment experiments, isolation, and algal strains.
Strain TG408 was isolated from a nonaxenic laboratory culture of the marine diatom Skeletonema costatum CCAP1077/1C (origin, North Sea). Isolation was performed by inoculating 2 ml of a growing culture of the diatom into 48 ml of ZM/10 marine broth (10-fold dilution of 2216 marine medium) containing a mixture of the PAHs (final concentrations, mg/liter) phenanthrene (500), anthracene (50), fluorene (50), and dibenzothiophene (50). After an incubation period of 3 weeks with shaking in the dark (120 rpm; 28°C), samples of the culture were spread onto plates of ZM/10 and ONR7a (10) agar and sprayed with a thin film of phenanthrene, anthracene, fluorene, or dibenzothiophene as described by Kiyohara et al. (11). Strain TG408 was isolated from an ONR7a plate that had been sprayed with phenanthrene on the basis of its ability to form a clearing zone. The strain was purified by several transfers to ONR7a agar plates and then stored in 20% (vol/vol) glycerol at −80°C.
The following nonaxenic laboratory cultures of marine microalgae (origin of isolation shown in parentheses) were obtained from the Culture Collection of Algae and Protozoa (CCAP; Oban, United Kingdom) and from the National Center for Marine Algae and Microbiota (East Boothbay, ME): Lingulodinium polyedrum CCAP1121/2 (Loch Creran, Argyll, United Kingdom); Isochrysis sp. strain CCMP1324 (Mataiva, Tahiti, South Pacific Ocean); Heterosigma akashiwo CCMP1870 (Long Beach, CA); Pseudo-nitzschia CCAP1061/25 (Loch Creran, Argyll, United Kingdom); Skeletonema costatum CCAP1077/1C (North Sea); Thalassiosira weissflogii CCMP1587 (Jakarta Harbor, Indonesia). These dinoflagellate and diatom strains were maintained on f/2 and f/2+Si media (12), respectively, in temperature-controlled illuminated incubators. These six phytoplankton strains were independently enriched with pyruvate and phenanthrene for subsequent analysis of strain TG408 by quantitative PCR. Enrichments were conducted as described previously (13), by using 50-ml Erlenmeyer flasks containing 10 ml of algal medium used to maintain the strains. Pyruvate enrichments contained a 0.3% (wt/vol) final concentration of substrate; phenanthrene enrichments contained a 0.001% (wt/vol) final concentration. Each flask was inoculated with 1 ml of an exponentially growing phytoplankton strain. After 6 days of incubation in the dark with gentle shaking (100 rpm) at 21°C, biomass was collected from each flask and stored at −20°C. For nonenriched controls, biomass of all six phytoplankton strains was set aside at the time these enrichments were prepared and stored at −20°C. Whole DNA extractions (14) from all biomass samples were performed for subsequent analysis for DNA and 16S rRNA gene quantification of the target bacterial isolate.
Isolate characterization.
Phenotypic characterization typically used cell suspensions generated from colonies that were grown on ONR7a agar medium amended with pyruvate. ONR7a medium was found to be optimal because it accommodates the specific nutritional requirements of this fastidious strain. All of the compounds used in this study were of reagent quality or better. To determine whether different substrates could be used as sole sources of carbon, representative aliphatic and aromatic hydrocarbons, carbohydrates, and common metabolic intermediates were added to ONR7a liquid medium at concentrations between 0.1 and 0.2% (wt/vol for solid substrates, vol/vol for liquid substrates). Because of their relative toxicity, some substrates were added at lower concentrations (<0.1%), or in the case of volatile substrates, they were added to the lids of plates. The carbohydrate substrates tested included glucose, fructose, mannitol, mannose, sucrose, xylose, and arabinose. Organic intermediates/acids were tested as sodium salts. These included pyruvate, lactate, succinate, acetate, propionate, glutamate, benzoate, salicylate, and phthalate. The hydrocarbons tested included hexane, pentane, decane, pristane, phytane, n-hexadecane, phenanthrene, anthracene, fluorene, pyrene, fluoranthene, and dibenzothiophene. The volatile substrates (i.e., benzene, toluene, p-xylene, biphenyl, naphthalene, and phenol) were supplied via the vapor phase as previously described (15). The cultures were incubated at 30°C under aerobic conditions, and growth was monitored spectrophotometrically over 2 to 3 weeks. Uninoculated medium and medium without an added carbon source were used as controls. All incubations were performed at least in triplicate. Growth on methane was tested by inoculating ONR7a agar plates and placing them in a desiccator, after which 30% of the air atmosphere was replaced with methane. Anaerobic growth was tested by using ONR7a agar plates that were incubated in a desiccator that was rendered oxygen free.
The ability of strain TG408 to mineralize 14C-labeled compounds was tested by using sterile 40-ml amber glass EPA vials each containing a 14C-labeled test compound (to 40,000 dpm), 2.5 μg of the respective unlabeled test compound, and 4.5 ml of ONR7a medium. For the inoculum, cells were grown in 50 ml of ONR7a medium amended with pyruvate (0.1%, wt/vol). The cells were washed twice with filtered (0.2 μm) natural seawater and resuspended at the initial cell density. Then 0.5-ml volumes of the washed cell suspension were used to inoculate the vials. For the killed controls, this medium was amended with 85% phosphoric acid to lower the pH to <1 prior to inoculation. All treatments were conducted in triplicate. A sterile glass test tube (12 by 75 mm) containing a piece of filter paper saturated with 60 μl of 2 M KOH was inserted into each vial. The vials were sealed with foil-covered, Teflon-lined caps and incubated with shaking (100 rpm) at 21°C. The filter paper was removed from each vial every 3 days, and the 14C captured from any 14CO2 respired was counted on a Packard (Meriden, CT) Tri-Carb liquid scintillation analyzer (model 1900 TR). The KOH-saturated filter paper from each vial was replaced at each sampling point for the duration of the experiment. The percentage of 14C mineralized for each compound was calculated by subtracting the triplicate values of the acidified controls from those of the experimental samples and then dividing by the total number of dpm of 14C added.
The cell morphology and motility of the isolate were observed by transmission and scanning electron microscopy of cells that had been grown in ONR7a liquid medium amended with pyruvate as the sole carbon and energy source. For negative staining, bacterial suspensions were stained with 3% ammonium molybdate (pH 7.0) (16) prior to viewing with a LEO EM910 transmission electron microscope operating at 80 kV. Digital images were acquired with a Gatan Orius SC1000 charge-coupled device digital camera and Digital Micrograph 3.11.0 (Gatan, Inc., Pleasanton, CA). To prepare thin sections, glutaraldehyde fixation was followed by postfixation with 1% OsO4 before the samples were dehydrated gradually and embedded in Polybed 812. Sectioned samples (∼100 nm) were then stained with uranyl acetate and lead citrate prior to visualization with a JEOL 100CX transmission electron microscope. For scanning electron microscopy, glutaraldehyde fixation was followed by postfixation with 2% OsO4. After critical-point drying, the cells were sputter coated with gold-palladium and observed with a Hitachi 4700 FEG scanning electron microscope.
Motility was also determined by inoculating a loopful of cells into a test tube containing ONR7a agar amended with pyruvate and triphenyl tetrazolium chloride as previously described (17). The Gram reaction, oxidase, catalase, lipase (Tween 80), and gelatinase tests were performed as previously described (18). A 1% gelatin concentration was used in the gelatinase test. Total phosphatase activity was assayed by the sodium hydroxide method (17). Oxidative-fermentation tests were performed in ONR7a amended with bromocresol purple as an indicator and with glucose, mannitol, fructose, xylose, or arabinose at a 0.5% (wt/vol) final concentration. To determine nitrate reduction, the organism was grown in ONR7a medium amended with 0.1% KNO3, 0.1% Bacto agar, and pyruvate as the sole carbon source, and the result was observed after 1 week of incubation. Spectral and microscopic analyses (with Nile Blue) were used to determine whether the cells accumulate poly-β-hydroxybutyrate (PHB) granules (18). The Schaeffer-Fulton staining method was used for the determination of endospore formation. Growth on high-nutrient medium was evaluated by streaking the cells onto ZM/10 and ZM/1 agar plates. To determine the organism's requirement and tolerance of NaCl, it was inoculated into ONR7a liquid medium amended with pyruvate and containing increasing concentrations of NaCl, i.e., 0.0, 1.0, 3.0, 6.0, 9.0, 12.0, 15.0, 20.0, and 25.0% (wt/vol). Cultures were incubated with mild shaking (100 rpm) at room temperature unless otherwise specified. Tolerance of KCl was determined in the same way but in the absence of NaCl addition to ONR7a medium. To determine the pH range for growth, ONR7a amended with pyruvate was prepared with the following buffers: 25 mM 2-(N-morpholino)ethanesulfonic acid (MES), pH 5.5 to 6.5; 25 mM 3-[N-Tris(hydroxymethyl)methylamino]-2-hydroxypropanesulfonic acid (TAPSO), pH 7.5; and 25 mM Tris base/Tris HCl, pH 8.5 to 9.5. To determine the temperature range for growth, cultures in ONR7a amended with pyruvate were incubated at 4, 10, 15, 20, 30, 37, and 40°C. The organism's susceptibility to antibiotics was tested in ONR7a amended with pyruvate and increasing concentrations of chloramphenicol, ampicillin, kanamycin, and streptomycin.
Whole-cell fatty acid profiles were obtained following extraction of biomass of strain TG408 and methylation of the total lipid fraction (19). The cells used had been grown to exponential phase under aerobic conditions in ONR7a liquid medium amended with pyruvate for 4 days at 30°C. Isoprenoid quinones were extracted from lyophilized cells, and the samples were purified and analyzed by the DSMZ Identification Service (20, 21). Polar lipids were analyzed by using an Iatroscan MK V TH10 thin-layer chromatography–flame ionization detector analyzer (Iatron Laboratories, Tokyo, Japan) as previously described (22). The G+C content of strain TG408 DNA was determined by high-pressure liquid chromatography (23).
16S rRNA sequencing and phylogenetic analysis.
Total genomic DNA was recovered by using a Wizard genomic DNA purification kit (Promega, Madison, WI) according to the manufacturer's instructions. The 16S rRNA gene of strain TG408 was amplified by PCR with primers 27f (24) and 1492r (25). The resulting product was then cloned into plasmid pCR4-TOPO by using the TOPO-TA cloning kit for sequencing (Invitrogen, Carlsbad, CA). The insert was sequenced with primers M13f and M13r, primers 338f and 338r (26), and primers 907f (27) and 907r (24) at the University of North Carolina Genome Analysis Facility. Sequences were assembled by using the program Sequencher 4.8 (Gene Codes Corp., Ann Arbor, MI). The consensus sequence was submitted to GenBank and checked for close relatives and phylogenetic affiliation by using the BLAST search program and RDP-II (28). The search results were used as a guide for tree construction. Additional related 16S rRNA sequences identified from the BLASTN and RDP-II search were retrieved from GenBank. The CLUSTAL_X program (29) was used to align the sequences and construct neighbor-joining trees with TREEVIEW (WIN32) version 1.5.2 (30). The trees were bootstrapped 1,000 times, and gaps in the alignment were ignored. To evaluate dendrogram topology and confirm phylogenetic affiliation, sequences were imported and automatically aligned in the ARB-SILVA SSURef 94 database (31) and manually refined while taking into account the secondary structural information of the rRNA molecule. Tree reconstruction was performed by using the neighbor-joining, maximum-parsimony, and maximum-likelihood methods and applying various filters.
Quantitative PCR primer design.
To quantify the novel TG408 isolate, 16S rRNA-targeted primers for quantitative real-time PCR (qPCR) were developed. The Probe Design and Probe Match tools of the ARB software (31) were used to design primers specific for the 16S rRNA gene sequence of this strain. The primers were named Pcy_120F (5′-TAC ATA GGA ATC TGC CCG A-3′) and Pcy_223R (5′-AGA CAT AGG CTC CTC CAA-3′). Primer specificity was confirmed with the Probe Match tool of RDP-II. The Pcy_120F primer was highly specific, as it was complementary to no other bacterial sequence. The Pcy_223R primer was complementary to 8 sequences of Singularimonas and 17 sequences of unclassified Sinobacteraceae—members of which phylogenetically cluster with strain TG408. The optimal annealing temperature of the primer set (57°C) was determined by using an Eppendorf (Hauppauge, NY) Mastercycler gradient thermal cycler. The template used for the construction of a standard curve for quantitative PCR was a plasmid containing a representative sequence that had been linearized by using NcoI (New England BioLabs, Ipswich, MA) and purified by using the QIAquick nucleotide removal kit (Qiagen, Valencia, CA). The limit of quantification of the target strain with these primers was 3 gene copies per reaction. When threshold cycle (CT) values beyond the highest value in the linear range of the standard curve (CT range, 12.0 to 40.7) were measured, the gene was considered to have been detected but at a level below the quantification limit of the assay. The amplification efficiency of the primers (32) was determined to be 1.74. Further support for the specificity of the newly designed primers for strain TG408 was evaluated by performing qPCRs with DNA from nontarget organisms, including Escherichia coli, Pseudomonas putida, and other marine strains isolated from some of the phytoplankton species examined in this study. The CT values measured from these reactions (>40.7) were outside the limit of detection.
Quantitative detection of strain TG408 in phytoplankton cultures.
DNA from the six nonaxenic laboratory cultures of phytoplankton (enriched and nonenriched) was quantified by using a NanoDrop ND-3300 fluorospectrometer (Thermo, Waltham, MA) and the Quant-iT Picogreen double-stranded DNA kit (Invitrogen, Carlsbad, CA). Measurements of triplicate DNA extracts were performed. The qPCR primer set developed in this study to target the 16S rRNA gene of strain TG408 was used to identify this organism in these extracts by qPCR as described previously (33).
Enzyme assays.
Strain TG408 was cultured in 0.5 liter of ZM/10 medium amended with naphthalene (0.1%, vol/vol). After 5 to 7 days, cells were harvested at the mid- to late-exponential phase, washed twice with 0.1 M phosphate buffer saline (pH 7.0), and resuspended in the same buffer (total volume, 5 ml). The concentrated cell suspension was then disrupted by sonication with an MSE Soniprep 150 [MSE (UK) Ltd., London, United Kingdom] at 5°C. The lysed extract was then centrifuged (13,000 × g) to remove undisrupted cells and particulate matter, and the resultant cell-free lysate was assayed spectrophotometrically for catechol 1,2-dioxygenase by monitoring the formation of cis,cis-muconic acid from catechol at 260 nm as previously described (34). The activity of catechol 2,3-dioxygenase was spectrophotometrically assayed by measuring the absorbance increase for the meta-cleavage product at 375 nm as previously described (35). All assays were performed at room temperature. Total protein concentrations were determined by using the BCA protein assay kit (Sigma, St. Louis, MO) with bovine serum albumin as the standard.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of strain TG408 was deposited with GenBank under accession number FJ176554.
TAXONOMY
Description of Polycyclovorans gen. nov.
Polycyclovorans gen. nov.: pol.y.cy.clo.vo′rans.; Gr. adj. polys, numerous; Gr. n. kyklos, circle or ring; L. masc. adj. vorus, devouring; M. L. masc. n. polycyclovorans degrading/devouring multiple-ring compounds.
Cells stain Gram negative and are motile rods (1.0 to 1.2 by 0.5 μm) with a single polar flagellum. Oxidase and catalase positive. The cells are strictly aerobic and grow very poorly or not at all on complex media. They exhibit a narrow substrate spectrum, utilizing various hydrocarbon compounds, including decane, pristane, n-hexadecane, benzene, toluene, p-xylene, biphenyl, naphthalene, anthracene, and phenanthrene, as sole or principal sources of carbon and energy. The predominant isoprenoid quinone is Q-8, and the major fatty acids are C16:0, C16:1 ω7c, and C18:1 ω7c. The G+C content of the DNA of the type strain of the type species is 64.3 mol%.
Description of Polycyclovorans algicola sp. nov.
Polycyclovorans algicola sp. nov.: al.gi′co.la. M. L. n. alga alga or seaweed; L. suff. -cola from L. n. incola an inhabitant or dweller; N. L. masc. n. algicola alga dweller.
The following properties are also observed in addition to the genus description. Colonies less than 3 weeks old are flat, smooth, and translucent with irregular edges and diameters of 3 to 4 mm. After 3 weeks, colonies gradually become larger (5 to 6 mm), off-white, and slightly umbonate. Surface blebs are exuded from cell surfaces. Lipase (Tween 80) and phosphatase activities are positive. Gelatinase is negative. Reduction of nitrate to nitrite is negative. No spores are formed, and no accumulation of PHB is observed. Does not require Na+ or other special nutrition for growth. Growth occurs at 10 to 30°C (optimum, 30°C) and at pH 6.5 to 8.5 (optimum, 8.3). Growth is detected at 0 to 9% NaCl, although better growth is seen at 0 to 6% NaCl. Substitution of Na+ for K+ inhibits growth. Sugars, such as glucose, are not fermented. Methane and methanol are not utilized as sole carbon sources. Growth on pyruvate, succinate, acetate, and propionate is observed. Poor growth occurs on phenol and glutamate. No growth occurs on mannitol, fructose, glucose, mannose, xylose, arabinose, lactate, dextran, benzoate, fluorene, pyrene, fluoranthene, or dibenzothiophene. Mineralization of salicylate but not phthalate. Susceptible to chloramphenicol, kanamycin, and streptomycin. Resistant to ampicillin. The type and only species is Polycyclovorans algicola. The type strain is TG408 (= ATCC BAA-2242 = KCTC 23940).
RESULTS AND DISCUSSION
Strain isolation and colony morphology.
PAH-sprayed agar plates that were streaked from enrichments grown on a PAH mixture produced several colonies displaying clearing zones. Two colonial morphotypes were picked and passaged to obtain pure cultures. One of these isolates, designated strain TG408, developed a peculiar colony morphology and was selected for further study. On ONR7a agar plates amended with pyruvate as the sole carbon and energy source, young (less than 3 weeks old) colonies of strain TG408 were flat, smooth, and translucent with irregular edges and diameters of 3 to 4 mm. After 3 weeks, the colonies gradually became larger (5 to 6 mm), off-white, and slightly umbonate. The cells did not show any pigmentation on solid medium or in liquid medium, and no diffusible or fluorescent pigments were observed. Growth was markedly suppressed in diluted Zobell's medium (ZM/10), compared to growth in ONR7a with pyruvate or PAH, and not observed at all in full-strength Zobell's medium (ZM/1).
Phenotypic and biochemical characterization.
The strain displayed a narrow nutritional spectrum. Growth was observed on pyruvate, succinate, acetate, and propionate but not on mannitol, fructose, glucose, mannose, sucrose, xylose, arabinose, lactate, dextran, phthalate, benzoate, methane, or methanol. Growth on glutamate was weak. Experiments with 14C-labeled glucose as the sole carbon source were negative, confirming that glucose was not metabolized. The strain was able to grow on and/or mineralize various hydrocarbon substrates as sole sources of carbon and energy. As shown in Table 1, the following hydrocarbons supported growth: n-hexadecane, benzene, toluene, p-xylene, biphenyl, naphthalene, phenanthrene, and anthracene. Growth on pristane, phenol, fluorene, and dibenzothiophene was weak. No growth on hexane, pentane, decane, phytane, pyrene, or fluoranthene was observed. However, a change in the color of the culture was observed with fluoranthene, which is indicative that this compound was transformed without growth. On 14C-labeled substrates, the following were mineralized: decane, n-hexadecane, naphthalene, phenanthrene, anthracene, and salicylate. The strain was not able to mineralize pyrene, fluoranthene, or phthalate. On the basis of its preference for hydrocarbon substrates, in particular, aromatic compounds, this novel organism appears to represent a new marine hydrocarbon-degrading specialist.
Table 1.
Utilization of various aliphatic, monoaromatic hydrocarbons and PAHs, including some key aromatic metabolites, by strain TG408 in ONR7a liquid medium
| Substrate | Growtha,f | Mineralizationb,f |
|---|---|---|
| Hexane | − | ND |
| Pentane | − | ND |
| Decane | − | + |
| Pristane | W | ND |
| Phytane | − | ND |
| n-Hexadecane | + | + |
| Benzene | + | ND |
| Toluene | + | ND |
| p-Xylene | + | ND |
| Phenol | Wc | ND |
| Biphenyl | + | ND |
| Naphthalene | + | + |
| Phenanthrene | + | + |
| Anthracene | + | + |
| Fluorene | Wc | ND |
| Pyrene | − | − |
| Fluoranthene | −d | −d |
| Dibenzothiophene | We | ND |
| Salicylate | ND | + |
| Phthalate | − | − |
Growth was measured spectrophotometrically (optical density at 600 nm) or by measuring the disappearance of the substrate relative to that in uninoculated controls, as described in Materials and Methods.
Mineralization was measured as the release of 14CO2 from 14C-labeled substrates.
Spectrophotometric analysis of solvent-extracted samples confirmed that degradation of phenol and fluorene had occurred since significant decreases in their concentrations were measured.
Although growth and mineralization were not observed, the color of the culture became somewhat orange, indicating that transformation of fluoranthene had occurred.
Growth on dibenzothiophene on solid medium was better than in liquid medium.
+, positive for growth; −, no growth; W, weak growth; ND, not determined.
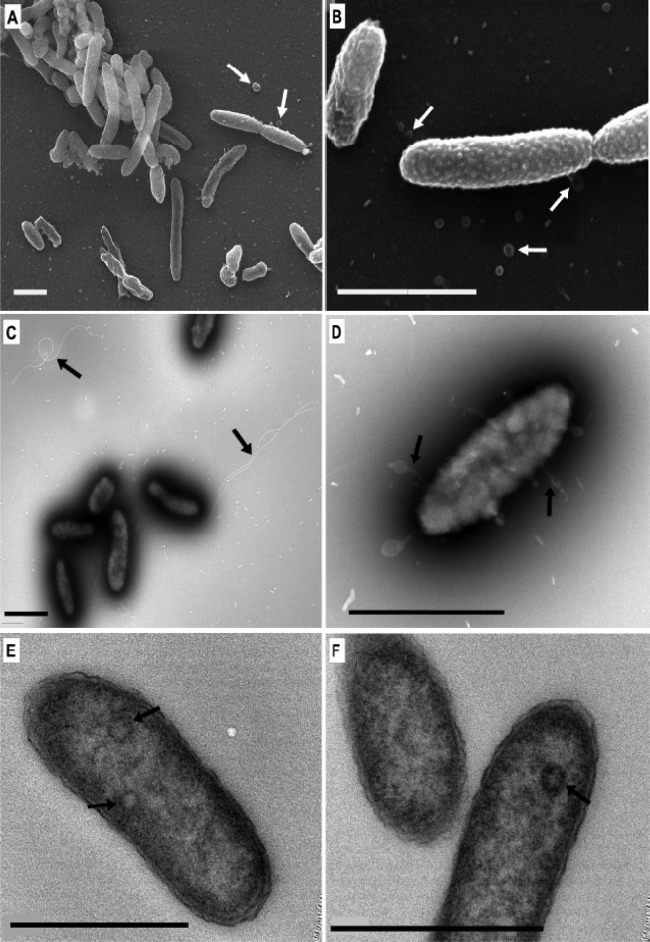
Cells of strain TG408 are small, non-spore-forming rods, 1.0 to 1.2 by 0.5 μm in average size, that occur singly or in pairs when grown on pyruvate (Fig. 1). They stain Gram negative and are motile by means of a single polar flagellum (Fig. 1C). Transmission and scanning electron micrographs showed numerous blebs at the cell surface and in the surrounding medium (Fig. 1D). Thin sections revealed that the strain has a cell envelope structure that is typical of Gram-negative bacteria (Fig. 1E and F). Intracellular inclusion bodies were observed in some cells. Although staining with Nile Blue revealed that some cells contained intracellular fluorescent bodies, these were unlikely to be PHB granules because a spectral analysis for PHB was negative. Strain TG408 is aerobic and catalase and oxidase positive. The strain grew at temperatures ranging from 10 to 30°C (optimal temperature, 30°C) and at pH values ranging from 6.5 to 8.5 (optimal pH, 8.3). The strain was positive for lipase (Tween 80) and phosphatase activities. It was negative for gelatinase activity and the fermentation of glucose and various other sugars. The strain exhibited slight halotolerance, since it grew well in medium containing NaCl concentrations ranging between 0 and 6% (optimum, 3%). Growth was partially inhibited in the presence of 9% NaCl and completely inhibited at higher concentrations. Strain TG408 is therefore a slightly halotolerant bacterium (41). The strain exhibited good growth in medium without Na+ added, although addition of Na+ stimulated growth slightly. Substitution of Na+ for K+ in the growth medium inhibited the growth of strain TG408. Growth was slightly inhibited at KCl concentrations of up to 2% and completely inhibited at concentrations of >3%. The strain was resistant to ampicillin at up to 41.5 μg/ml and susceptible to chloramphenicol, kanamycin, and streptomycin.
Fig 1.
Scanning (A, B) and transmission (negative staining, C, D; thin sectioning, E, F) electron micrographs of strain TG408T. (A, B, D) Arrows indicate cell surface and extracellularly released blebs. (C) Arrows indicate attached and detached flagella. (E, F) Arrows indicate inclusion bodies. Bars, 1 μm.
Table 2 shows the fatty acid (derived from the total extractable lipid) profiles of strain TG408 compared to those of reference strains that were grown according to their specific requirements. The fatty acid profile of strain TG408 was dominated by C16:0 (38.2%), C16:1 ω7c (31.3%), and C18:1 ω7c (17.4%). The fatty acids detected as minor components were C14:0 (1.4%), C12:0 2-OH (1.7%), C12:0 3-OH (2.4%), and C18:0 (2.6%). The predominant isoprenoid quinone for strain TG408 was Q-8. Strain TG408 contained phosphatidyl glycerol as the major phospholipid class (28%), followed by phosphatidic acid (25%) and phosphatidyl ethanolamine (19%), with an unidentified phospholipid also present (27%). The G+C content of strain TG408 DNA was found to be 64.3 mol%.
Table 2.
Cellular fatty acid compositions of strain TG408 and four reference strains
| Fatty acid | % of total fatty acidsa |
|||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| C12:0 | 4.3 | 16.3 | 3.2 | 4.6 | 4.8 | |
| Iso-C12:0 | 2.8 | |||||
| C12:0 2-OH | 1.7 | 1.7 | 2.9 | |||
| C12:0 3-OH | 2.4 | 3.4 | 1.1 | |||
| C13:0 | 1.3 | |||||
| C13:0 3-OH | 1.3 | 4.3 | ||||
| C14:0 | 1.4 | 1.2 | 2.2 | 20.5 | 5.9 | 5.1 |
| Iso-C14:0 | 2.7 | |||||
| Iso-C14:0 3-OH | 2.8 | |||||
| C14:0 2-OH | 2.1 | |||||
| C14:0 3-OH | 6.6 | 2.8 | 7.1 | 9.2 | ||
| C16:0 | 38.2 | 30.1 | 18.9 | 42.2 | 12.3 | 11.8 |
| Iso-C16:0 | 10.5 | |||||
| C16:0 2-OH | 1.6 | |||||
| C16:0 3-OH | 1.3 | |||||
| C16:1 ω5c | 5.6 | 6.6 | ||||
| C16:1 ω7c | 31.3 | 3.8 | 16.4 | 16.1 | 3.9b | 3.7b |
| C16:1 ω11c | 1.4 | |||||
| C17:0 | 4.3 | |||||
| C17:0 cyclo | 2.4 | |||||
| C17:1 ω6c | 1.1 | 3.2 | ||||
| C17:1 ω8c | 1.5 | |||||
| C18:0 | 2.6 | 14.9 | 2.1 | 1.1 | ||
| C18:1 ω7c | 17.4 | 15.8 | 37.6 | 17.9 | 21.3 | 26.3 |
| C18:1 ω9c | 2.3 | |||||
| C19:0 cycloc | 15.3 | |||||
| C19:0 cyclo ω8c | 3.3 | 4.3 | ||||
Strains: 1, TG408; 2, Alkanibacter difficilis MN154.3T (36); 3, Hydrocarboniphaga effusa ATCC BAA332T (36); 4, Nevskia ramosa DSM 11499T (36); 5, Sinobacter flavus CW-KD4T (37); 6, Solimonas soli DCY12T (37). Lipid extraction of biomass, followed by methylation of the total extractable lipids, was used for strain 1, with whole-cell methylation protocols for strains 2 to 6. The latter also recovers lipopolysaccharide-derived OH fatty acids. Components that represented <1% in all strains are not included. The genus Sinobacter was recently reclassified as synonymous with Solimonas (37).
May also contain C16:1 ω6c, as reported in the original publications.
C19:0 cyclopropyl fatty acids; cyclo11-12 for strain 2 (36).
Phylogenetic analysis and taxonomy.
We determined 1,494 bp of the 16S rRNA gene sequence. Phylogenetic analysis associated the strain with the class Gammaproteobacteria. From a BLAST analysis, the highest levels (≥95%) of sequence similarity for TG408 were to 13 environmental clones (Table 3). The highest levels of sequence similarity of TG408 to type strains occurred with Alkanibacter difficilis MN154.3T and Hydrocarboniphaga daqingensis B2-9T (91.7%), Nevskia soli GR15-1T (91.5%), Hydrocarboniphaga effusa AP103T (91.4%), Sinobacter flavus CW-KD4T and Solimonas variicoloris MN28T (91.3%), Nevskia ramosa Soe1T (90.8%), and Solimonas soli DCY12T (90.7%). These and other related sequences were used to construct a neighbor-joining tree (Fig. 2). The most closely related clones and type strains originated from contaminated soil, human skin, fresh and marine waters, or polluted environments.
Table 3.
Sources and characteristics of 16S rRNA gene sequences with the highest sequence similarity to strain TG408
| Source | Designationa | GenBank accession no. | Sequence length (bases) | % similarity | Reference |
|---|---|---|---|---|---|
| Zhongyuan oil field, China | BP76 | HQ190544 | 1,501 | 99 | Unpublished |
| Zhongyuan oil field, China | BP24 | HQ190510 | 1,501 | 99 | Unpublished |
| Oil-contaminated soil | B194 | EU328045 | 1,491 | 99 | Unpublished |
| NDb | DSW25-19 | HQ263242 | 1,473 | 99 | Unpublished |
| Rancho La Brea tar pits | 101-95 | EF157240 | 1,453 | 99 | 38 |
| Los Angeles, CA, contaminated soil | TERI-KL22 | JN217160 | 1,510 | 99 | Unpublished |
| Terrestrial mud volcano | SYNH02-ew01B-031 | JQ245626 | 1,388 | 99 | Unpublished |
| Human skin | nbw777e05c1 | GQ009517 | 1,351 | 99 | 39 |
| Human skin | nbw779c06c1 | GQ009776 | 1,351 | 99 | 39 |
| Human skin | ncd1111g03c1 | HM337987 | 1,351 | 99 | 40 |
| ND | ch-xj4 | JQ327935 | 1,521 | 98 | Unpublished |
| Contaminated soil | TERI-KL45 | JN217179 | 1,510 | 97 | Unpublished |
| Yellow Sea water | D13W-30 | HM057752 | 1,498 | 95 | Unpublished |
Sequence designation from the BLASTN database.
ND, no data available.
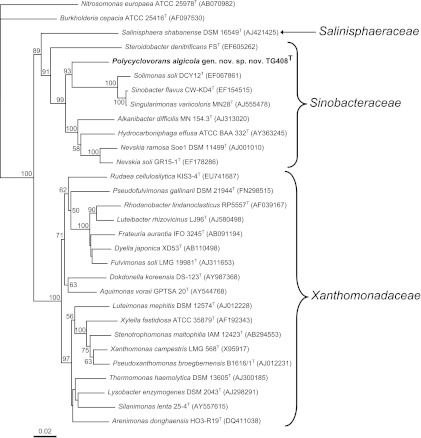
Fig 2.
Neighbor-joining phylogenetic tree, based on 16S rRNA gene sequences (>1,200 bp), showing the relationships between strain TG408T and representative strains within the order Xanthomonadales. Nitrosomonas europaea and Burkholderia cepacia in the class Betaproteobacteria were selected as the outgroup. Bootstrap values (expressed as percentages of 1,000 replications) of >50% are shown at each node. GenBank accession numbers are shown in parentheses. Bar, 0.02 substitution per nucleotide position. The maximum-parsimony and maximum-likelihood trees showed essentially the same topology (data not shown). The genus Sinobacter was recently reclassified as synonymous with Solimonas (37).
Although the phylogenetic position of strain TG408 clustered most closely with Solimonas and Sinobacter (bootstrap value of 85%), its highest level of sequence similarity was to A. difficilis MN154.3T. All of the related clones and type strains form a deeply rooted lineage—the so-called Hydrocarboniphaga-Nevskia-Sinobacter (HNS) clade—that is separated from the family Xanthomonadaceae (42). However, they have been allocated to this family exclusively on the basis of 16S rRNA gene sequence data (43, 44). In the systematic description of S. flavus CW-KD4T, Zhou et al. (42) proposed a new family—Sinobacteraceae—in order to distinguish this strain and members of the HNS clade from the family Xanthomonadaceae on the basis of distinct phenotypic characteristics. Analysis by Classifier in RDP-II (45) indicated that TG408 is affiliated with the family Sinobacteraceae. This was further supported by the strain's DNA G+C content of 64.3 mol%, which is similar to that of most members of the Sinobacteraceae family (60 to 65 mol%).
A number of phenotypic and other characteristics distinguish TG408 from closely related reference strains and from members of the Sinobacteraceae, Salinisphaeraceae, and Xanthomonadaceae families. Strains of Nevskia have a wide nutritional spectrum and are able to utilize sugars as growth substrates (46). In contrast, strain TG408 exhibits a narrow nutritional spectrum as it is unable to utilize sugars and prefers hydrocarbons over other carbon sources as growth substrates. A. difficilis MN154.3T, the only current representative of the genus Alkanibacter, also exhibits a narrow nutritional spectrum, but unlike strain TG408, it can grow on succinate (36)—a distinguishing feature that has been used to differentiate Nevskia from its closest relatives, such as Alkanibacter. The absence of diphosphatidylglycerol in the polar lipid profile of strain TG408 also differentiates it from A. difficilis MN154.3T. Strain TG408 grows at temperatures as low as 10°C, while higher growth temperatures are required by members of the family Xanthomonadaceae (≥20°C). Strain TG408 exhibits a higher pH optimum than members of the family Xanthomonadaceae. Strain TG408 was isolated from a marine habitat and tolerates NaCl concentrations as high as 9%. With a few exceptions, members of the families Xanthomonadaceae and Sinobacteraceae are from nonsaline environments and exhibit lower levels of salt tolerance. Only members of the family Salinisphaeraceae exhibit higher salt tolerances (up to 28%, wt/vol, NaCl), but they cannot grow in the absence of NaCl, which distinguishes them from TG408. Similarly to members of the families Sinobacteraceae and Salinisphaeraceae, strain TG408 possesses mostly monounsaturated fatty acids, while branched-chain or hydroxylated fatty acids are typically abundant in the xanthomonads.
On the basis of phylogeny, fatty acid composition, and phenotypic characteristics, strain TG408 represents a novel species of a novel genus, for which the name Polycyclovorans algicola is proposed.
Quantitative detection of strain TG408 in phytoplankton cultures.
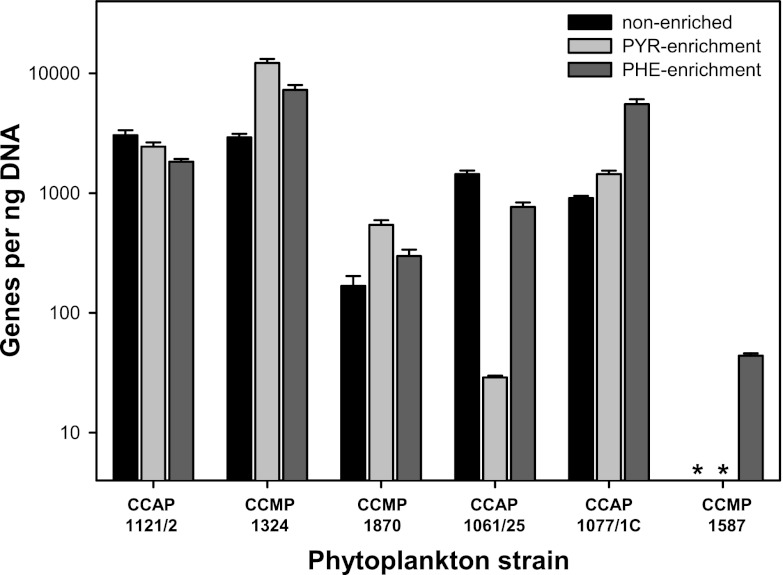
By using qPCR primers developed in this study to target the 16S rRNA gene of strain TG408, we showed that this novel organism, and possibly other Polycyclovorans-related bacteria, was present in all six laboratory cultures of phytoplankton examined (Fig. 3). As expected, strain TG408 was abundant in S. costatum CCAP1077/1C—the laboratory phytoplankton strain from which this novel bacterium was isolated. Although strain TG408 was present at levels below the quantification limit in CCMP1587 for cultures incubated with and without pyruvate, it was detected in the culture with phenanthrene. This result suggests that the levels of this bacterium associated with CCMP1587 are very low. A longer time of incubation with pyruvate might have yielded numbers of gene copies quantifiable by our qPCR assay. Except for strains CCAP1121/2 and CCAP1061/25, the 16S rRNA gene copy number of TG408 was higher for the other four phytoplankton strains after they had been enriched with pyruvate or phenanthrene. It is likely that these substrates, which serve as sole sources of carbon and energy for strain TG408, had stimulated its growth and thereby yielded higher cell numbers in these incubations. For CCAP1121/2, the abundance of TG408 genes was similar for cultures incubated with and without pyruvate or phenanthrene, whereas gene abundance was markedly lower for CCAP1061/25 incubated on pyruvate than for cultures incubated in the presence or absence of phenanthrene. Differences in gene copy number between the different treatments could have arisen because other microbiota associated with these phytoplankton cultures influenced the growth of strain TG408.
Fig 3.
Abundance of strain TG408 16S rRNA genes in phytoplankton species enriched with pyruvate (PYR-enriched) or phenanthrene (PHE-enriched) or with no added carbon source (non-enriched). Bars show the average results and standard deviations of duplicate qPCRs measuring the abundance of TG408-specific 16S rRNA genes per ng DNA. Phytoplankton strains: Lingulodinium polyedrum CCAP1121/2, Isochrysis sp. strain CCMP1324, Heterosigma akashiwo CCMP1870, Pseudo-nitzschia CCAP1061/25, Skeletonema costatum CCAP1077/1C, and Thalassiosira weissflogii CCMP1587. Asterisks represent values below the quantification limit of the assay (<3 gene copies per reaction).
As discussed in an earlier report describing the identification of a novel PAH-degrading bacterium in various species of marine phytoplankton (13), the association of hydrocarbon-degrading bacteria with phytoplankton cannot be a derivative of laboratory enrichment since hydrocarbons are not used in the maintenance of algal cultures. The bacterial community associated with any nonaxenic strain of laboratory phytoplankton is likely to closely represent the community that existed at the time when the strain was originally isolated from the field (47). Hence, strain TG408 is likely to be a member of the microbiota associated with the six phytoplankton species at the time when they were first collected in the field and brought into laboratory cultivation.
Ring cleavage pathway in the breakdown of naphthalene.
Catechol derivatives (dihydrodiols) are formed as key metabolic intermediates in the breakdown of aromatic hydrocarbons. Generally, these intermediates are formed during the first two steps of the breakdown pathway and act as precursors to subsequent cleavage of the aromatic ring either between (ortho cleavage) or adjacent to (meta cleavage) the vicinal diols. To discern the type of catechol dioxygenase produced by strain TG408, cell extracts were prepared following growth on naphthalene and assayed for catechol 1,2-dioxygenase or catechol 2,3-dioxygenase activity. Only catechol 2,3-dioxygenase activity was detected, indicating that catabolism proceeds via the meta-cleavage pathway. The specific enzyme activity from at least triplicate measurements was 3.33 ± 0.1 U/mg protein. In addition, a pale yellow coloration was observed during growth on naphthalene. A full-wavelength scan of the cell-free culture liquid revealed a maximal absorption peak at ca. 375 nm (result not shown), indicative of the formation of a muconic semialdehyde intermediate via the meta-cleavage pathway (48).
Physiology and ecology.
To our knowledge, the marine environment is the only place where bacteria that preferentially utilize hydrocarbons as sole sources of carbon and energy are found. Through our work, we have revealed novel genera (49 and this study) and species (13) of aromatic hydrocarbon “specialists” associated with laboratory cultures of eukaryotic phytoplankton. By using DNA-based stable-isotope probing, Gutierrez et al. (50) identified uncultivated species of PAH degraders associated with a phytoplankton bloom. Although further research is needed to understand why these and other species of PAH-degrading bacteria might be found living with phytoplankton, current evidence correlating the influence of phytoplankton, particularly during bloom periods, with the removal of hydrocarbons from the marine water column suggests that this is a widespread lifestyle (8, 9, 51–53). It has been inferred that the surface of phytoplankton cells can sequester PAHs from the surrounding seawater, subsequently transporting these compounds down to the sea floor by sedimentation (8, 9). More recently, research conducted on the formation of oil aggregates during the Deepwater Horizon oil spill in the Gulf of Mexico suggests that exopolymeric substances produced by bacteria and microalgae alike might act as a molecular trap for sequestering hydrophobic compounds such as PAHs (54). The possession of an inherent capacity to adsorb PAHs suggests that phytoplankton, or more specifically their cell surfaces, might act as an enrichment zone for PAH-degrading bacteria. Hence, the occurrence of strain TG408 and other aromatic-degrading bacteria living associated with marine phytoplankton may be attributed to the capacity of phytoplankton cells to adsorb PAHs on their surface and provide a favorable niche for bacteria with the ability to utilize these compounds.
In addition to the adsorbent qualities of phytoplankton cells for PAH molecules, there is evidence that phytoplankton may produce these chemicals. Studies conducted throughout the 1960s and 1970s provided the first evidence that microalgae are capable of synthesizing PAHs (6, 55) and incorporating them into their cell walls (55–58). More recently, Repeta et al. (59) discovered a suite of chlorinated aromatic compounds at two open-ocean sites that, on the basis of the global inventory and isomer distribution of this class of compounds, they suggested were likely to have been produced in situ by marine microbes such as phytoplankton. The association of PAH-degrading bacteria with eukaryotic phytoplankton may be viewed as a coevolutionary adaptation based on the ability of the phytoplankton to sequester and/or synthesize PAHs. Although photosynthetically enhanced biodegradation of toxic aromatic pollutants has been demonstrated by using artificial combinations of algal-bacterial consortia (60–63), little is known about this process in natural algal-bacterial assemblages. The association of these organisms may have potentially profound implications for the natural purging of the water column of PAH contaminants and help contribute to the overall health of the marine ecosystem.
ACKNOWLEDGMENTS
This research was supported by a Marie Curie International Outgoing Fellowship (PIOF-GA-2008-220129) within the 7th European Community Framework Programme. Partial support was provided through the U.S. National Institute of Environmental Health Sciences, grant 5 P42ES005948.
We thank Wallace Ambrose (University of North Carolina Analytical and Nanofabrication Laboratory) and Victoria Madden (University of North Carolina Microscopy Services Laboratory), who provided valuable assistance with the preparation of samples for electron microscopy and in the capture of electron micrographs. We also thank Danny Holdsworth, who managed the CSIRO gas chromatography and gas chromatography-mass spectrometry facility; Peter Mansour, who kindly assisted with the provision of phospholipid standards and the development of methods; CCAP (Oban, United Kingdom) for providing S. costatum; and two anonymous reviewers for their careful and concise criticism of the manuscript.
Footnotes
Published ahead of print 19 October 2012
REFERENCES
- 1. Schoeny R, Poirier K. 1993. Provisional guidance for quantitative risk assessment of polycyclic aromatic hydrocarbons. US Environmental Protection Agency, Office of Research and Development, Washington, DC [Google Scholar]
- 2. Head IM, Martin Jones D, Roling WFM. 2006. Marine microorganisms make a meal of oil. Nat. Rev. Microbiol. 4:173–182 [DOI] [PubMed] [Google Scholar]
- 3. Crone TJ, Tolstoy M. 2010. Magnitude of the 2010 Gulf of Mexico Oil Leak. Science 330:634. [DOI] [PubMed] [Google Scholar]
- 4. Kasai Y, Kishira H, Harayama S. 2002. Bacteria belonging to the genus Cycloclasticus play a primary role in the degradation of aromatic hydrocarbons released in a marine environment. Appl. Environ. Microbiol. 68:5625–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yakimov MM, Timmis KN, Golyshin PN. 2007. Obligate oil-degrading marine bacteria. Curr. Opin. Biotechnol. 18:257–266 [DOI] [PubMed] [Google Scholar]
- 6. Andelman JB, Suess MJ. 1970. Polynuclear aromatic hydrocarbons in the water environment. Bull. World Health Organ. 43:479–508 [PMC free article] [PubMed] [Google Scholar]
- 7. Mallet L, Sardou J. 1964. Investigation on the presence of the BP-type PH in the plankton environment of the region of the Bay of Villefranche (Alpes-Maritimes region). C. R. Hebd. Seances Acad. Sci. 258:5264–5267 [Google Scholar]
- 8. Binark N, Guven KC, Gezgin T, Unlu S. 2000. Oil pollution of marine algae. Bull. Environ. Contam. Toxicol. 64:866–872 [DOI] [PubMed] [Google Scholar]
- 9. Kowalewska G. 1999. Phytoplankton—the main factor responsible for transport of polynuclear aromatic hydrocarbons from water to sediments in the Southern Baltic ecosystem. ICES J. Mar. Sci. 56:219–222 [Google Scholar]
- 10. Dyksterhouse SE, Gray JP, Herwig RP, Cano Lara J, Staley JT. 1995. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int. J. Syst. Bacteriol. 45:116–123 [DOI] [PubMed] [Google Scholar]
- 11. Kiyohara H, Nagao K, Yana K. 1982. Rapid screen for bacteria degrading water-insoluble, solid hydrocarbons on agar plates. Appl. Environ. Microbiol. 43:454–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guillard RRL. 1975. Culture of phytoplankton for feeding marine invertebrates, p 29–60 In Smith WL, Chanley MH. (ed), Culture of marine invertebrate animals. Plenum Press, New York, NY [Google Scholar]
- 13. Gutierrez T, Nichols PD, Whitman WB, Aitken MD. 2012. Porticoccus hydrocarbonoclasticus sp. nov., an aromatic hydrocarbon-degrading bacterium identified in laboratory cultures of marine phytoplankton. Appl. Environ. Microbiol. 78:628–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tillet D, Neilan BA. 2000. Xanthogenate nucleic acid isolation from cultured and environmental cyanobacteria. J. Phycol. 36:251–258 [Google Scholar]
- 15. Paje ML, Neilan B, Couperwhite I. 1997. A Rhodococcus species that thrives on medium saturated with liquid benzene. Microbiology 143:2975–2981 [DOI] [PubMed] [Google Scholar]
- 16. Hayat MA, Miller SE. 1990. Negative staining. McGraw-Hill Publishing Co., New York, NY [Google Scholar]
- 17. MacFaddin JF. 2000. Biochemical tests for identification of medical bacteria, 3rd ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 18. Gerhardt P, Murray RGE, Wood WA, Krieg NR. 1994. Methods for general and molecular bacteriology. American Society for Microbiology, Washington, DC [Google Scholar]
- 19. Miller MR, Bridle AR, Nichols PD, Carter CG. 2008. Increased elongase and desaturase gene expression with stearidonic acid enriched diet does not enhance long-chain (n-3) content of seawater Atlantic salmon (Salmo salar L.). J. Nutr. 138:2179–2185 [DOI] [PubMed] [Google Scholar]
- 20. Tindall BJ. 1990. A comparative study of the lipid composition of Halobacterium saccharovorum from various sources. Syst. Appl. Microbiol. 13:128–130 [Google Scholar]
- 21. Tindall BJ. 1990. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol. Lett. 66:199–202 [Google Scholar]
- 22. Volkman JK, Nichols PD. 1991. Applications of thin layer chromatography-flame ionization detection to the analysis of lipids and pollutants in marine and environmental samples. J. Planar Chromatogr. 4:19–26 [Google Scholar]
- 23. Mesbah M, Premachandran U, Whitman WB. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39:159–167 [Google Scholar]
- 24. Wilmotte A, Van der Auwera G, De Wachter R. 1993. Structure of the 16S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (“Mastigocladus laminosus HTF”) strain PCC7518, and phylogenetic analysis. FEMS Microbiol. Lett. 317:96–100 [DOI] [PubMed] [Google Scholar]
- 25. Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–175 In Stackebrandt E, Goodfellow M. (ed), Nucleic acid sequencing techniques in bacterial systematics. John Wiley & Sons, New York, NY [Google Scholar]
- 26. Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lane DJ, Pace B, Olsen GJ, Stahl DA, Sogin ML, Pace NR. 1985. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. U. S. A. 82:6955–6959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Maidak BL, Cole, Parker CT, Jr, Garrity GM, Larsen N, Li B, Lilburn TG, McCaughey MJ, Olsen GJ, Overbeek R, Pramanik S, Schmidt TM, Tiedje JM, Woese CR. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL_X: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Page RDM. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357–358 [DOI] [PubMed] [Google Scholar]
- 31. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 doi:10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Singleton DR, Sangaiah R, Gold A, Ball LM, Aitken MD. 2006. Identification and quantification of uncultivated proteobacteria associated with pyrene degradation in a bioreactor treating PAH-contaminated soil. Environ. Microbiol. 8:1736–1745 [DOI] [PubMed] [Google Scholar]
- 34. Hegeman GD. 1966. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. J. Bacteriol. 91:1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bayly RC, Dagley S, Gibson DT. 1966. The metabolism of cresols by species of Pseudomonas. Biochem. J. 101:293–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Friedrich MM, Lipski A. 2008. Alkanibacter difficilis gen. nov., sp. nov. and Singularimonas variicoloris gen. nov., sp. nov., hexane-degrading bacteria isolated from a hexane-treated biofilter. Int. J. Syst. Evol. Microbiol. 58:2324–2329 [DOI] [PubMed] [Google Scholar]
- 37. Sheu S-Y, Cho N-T, Arun AB, Chen W-M. 2011. Proposal of Solimonas aquatic sp. nov., reclassification of Sinobacter flavus Zhou et al. 2008 as Solimonas flava comb. nov. and Singularimonas variicoloris Friedrich and Lipski 2008 as Solimonas variicoloris comb. nov. and emended descriptions of the genus Solimonas and its type species Solimonas soli. Int. J. Syst. Evol. Microbiol. 61:2284–2291 [DOI] [PubMed] [Google Scholar]
- 38. Kim J-S, Crowley DE. 2007. Microbial diversity in natural asphalts of the Rancho La Brea Tar Pits. Appl. Environ. Microbiol. 73:4579–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC; NISC Comparative Sequencing Program, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, Nomicos E, Polley EC, Komarow HD; NISC Comparative Sequence Program, Murray PR, Turner ML, Segre JA. 2012. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 22:850–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Larsen H. 1986. Halophilic and halotolerant microorganisms—an overview and historical perspective. FEMS Microbiol. Rev. 39:3–7 [Google Scholar]
- 42. Zhou Y, Zhang Y-Q, Zhi X-Y, Wang X, Dong J, Chen Y, Lai R, Li W-J. 2008. Description of Sinobacter flavus gen. nov., sp. nov., and proposal of Sinobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 58:184–189 [DOI] [PubMed] [Google Scholar]
- 43. Palleroni NJ, Port AM, Chang H-K, Zylstra GJ. 2004. Hydrocarboniphaga effusa gen. nov., sp. nov., a novel member of the γ-Proteobacteria active in alkane and aromatic hydrocarbon degradation. Int. J. Syst. Evol. Microbiol. 54:1203–1207 [DOI] [PubMed] [Google Scholar]
- 44. Saddler GS, Bradbury JF. 2005. Family I. Xanthomonadaceae fam. nov. The Proteobacteria, part B, p 63 In Brenner DJ, Krieg NR, Staley JT, Garrity GM. (ed), Bergey's manual of systematic bacteriology, 2nd ed, vol 2 Springer, New York, NY [Google Scholar]
- 45. Wang Q, Garrity GM, Tiedje JM, Cole JR. 2007. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Stürmeyer H, Overmann J, Babenzien H-D, Cypionka H. 1998. Ecophysiological and phylogenetic studies of Nevskia ramosa in pure culture. Appl. Environ. Microbiol. 64:1890–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jasti S, Sieracki ME, Poulton NJ, Giewat MW, Rooney-Varga JN. 2005. Phylogenetic diversity and specificity of bacteria closely associated with Alexandrium spp. and other phytoplankton. Appl. Environ. Microbiol. 71:3483–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cuskey SM, Olsen RH. 1988. Catabolism of aromatic biogenic amines by Pseudomonas aeruginosa PAO1 via meta cleavage of homoprotocatechuic acid. J. Bacteriol. 170:393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gutierrez T, Green DH, Whitman WB, Nichols PD, Semple KT, Aitken MD. 6 January 2012. Algiphilus aromaticivorans gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium isolated from a culture of the marine dinoflagellate Lingulodinium polyedrum, and proposal of Algiphilaceae fam. nov. Int. J. Syst. Evol. Microbiol. (Epub ahead of print.) doi:10.1099/ijs.0.033324-0 [DOI] [PubMed] [Google Scholar]
- 50. Gutierrez T, Singleton DR, Aitken MD, Semple KT. 2011. Stable isotope probing of an algal bloom to identify uncultivated members of the Rhodobacteraceae associated with low-molecular-weight polycyclic aromatic hydrocarbon degradation. Appl. Environ. Microbiol. 77:7856–7860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Buesseler KO. 1998. The decoupling of production and particulate export in the surface ocean. Global Biogeochem. Cycles 12:297–310 [Google Scholar]
- 52. Millard ES, Halfon E, Minns CK, Charlton CC. 1993. Effect of primary productivity and vertical mixing on PCB dynamics in planktonic model ecosystems. Environ. Toxicol. Chem. 12:931–946 [Google Scholar]
- 53. Witt G. 2002. Occurrence and transport of polycyclic aromatic hydrocarbons in the water bodies of the Baltic Sea. Mar. Chem. 79:49–66 [Google Scholar]
- 54. Passow U, Ziervogel K, Asper V, Diercks A. 2012. Marine snow formation in the aftermath of the Deepwater Horizon oil spill. Environ. Res. Lett. 7:035301. doi:10.1088/1748-9326/7/3/035301 [Google Scholar]
- 55. Gunnison D, Alexander M. 1975. Basis for the resistance of several algae to microbial decomposition. Appl. Microbiol. 29:729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gol'man LP, Mikhaseva MF, Reznikov VM. 1973. Infrared spectra of lignin preparations of pteridophytes and seaweeds. Dokl. Akad. Nauk SSSR 17:1031–1033 [Google Scholar]
- 57. Pastuska G. 1961. Die Kieselgelschicht-Chromatographie von Phenolen und Phenolcarbonsäuren. Fresenius J. Anal. Chem. 179:355–358 [Google Scholar]
- 58. Zelibor JL, Romankiw L, Hatcher PG, Colwell RR. 1988. Comparative analysis of the chemical composition of mixed and pure cultures of green algae and their decomposed residues by 13C nuclear magnetic resonance spectroscopy. Appl. Environ. Microbiol. 54:1051–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Repeta DJ, Hartman NT, John S, Jones AD, Goericke R. 2004. Structure elucidation and characterization of polychlorinated biphenyl carboxylic acids as major constituents of chromophoric dissolved organic matter in seawater. Environ. Sci. Technol. 38:5373–5378 [DOI] [PubMed] [Google Scholar]
- 60. Borde X, Guieysse B, Delgado O, Muñoz R, Hatti-Kaul R, Nugier-Chauvin C, Patin H, Mattiasson B. 2003. Synergistic relationships in algal-bacterial microcosms for the treatment of aromatic pollutants. Bioresour. Technol. 86:293–300 [DOI] [PubMed] [Google Scholar]
- 61. Muñoz R, Guieysse B, Mattiasson B. 2003. Phenanthrene biodegradation by an algal-bacterial consortium in two-phase partitioning bioreactors. Appl. Microbiol. Biotechnol. 61:261–267 [DOI] [PubMed] [Google Scholar]
- 62. Safanova ET, Dmitrieva IA, Kvitko KV. 1999. The interaction of algae with alcanotrophic bacteria in black oil decomposition. Resour. Conserv. Recycling 27:193–201 [Google Scholar]
- 63. Warshawsky D, LaDow K, Schneider J. 2007. Enhanced degradation of benzo[a]pyrene by Mycobacterium sp. in conjunction with green alga. Chemosphere 69:500–506 [DOI] [PubMed] [Google Scholar]