Abstract
The gene encoding β-mannanase (EC 3.2.1.78) BaMan26A from the bacterium Bifidobacterium adolescentis (living in the human gut) was cloned and the gene product characterized. The enzyme was found to be modular and to contain a putative signal peptide. It possesses a catalytic module of the glycoside hydrolase family 26, a predicted immunoglobulin-like module, and two putative carbohydrate-binding modules (CBMs) of family 23. The enzyme is likely cell attached either by the sortase mechanism (LPXTG motif) or via a C-terminal transmembrane helix. The gene was expressed in Escherichia coli without the native signal peptide or the cell anchor. Two variants were made: one containing all four modules, designated BaMan26A-101K, and one truncated before the CBMs, designated BaMan26A-53K. BaMan26A-101K, which contains the CBMs, showed an affinity to carob galactomannan having a dissociation constant of 0.34 μM (8.8 mg/liter), whereas BaMan26A-53K did not bind, showing that at least one of the putative CBMs of family 23 is mannan binding. For BaMan26A-53K, kcat was determined to be 444 s−1 and Km 21.3 g/liter using carob galactomannan as the substrate at the optimal pH of 5.3. Both of the enzyme variants hydrolyzed konjac glucomannan, as well as carob and guar gum galactomannans to a mixture of oligosaccharides. The dominant product from ivory nut mannan was found to be mannotriose. Mannobiose and mannotetraose were produced to a lesser extent, as shown by high-performance anion-exchange chromatography. Mannobiose was not hydrolyzed, and mannotriose was hydrolyzed at a significantly lower rate than the longer oligosaccharides.
INTRODUCTION
Plant β-mannans are abundant cell wall polysaccharides having a β-1,4-linked mannan backbone, in some cases interrupted by glucose units. Backbone mannose can also have appended galactose side groups linked by α-1,6-bonds, such as in galactomannans and in galactoglucomannan, the major softwood hemicellulose (1, 2). Konjac glucomannan and carob and guar gum galactomannans are storage polysaccharides, often used in foods because of their gelling properties (3, 4). β-Mannans can be hydrolyzed by β-mannanases (endo-β-1,4-mannanases; EC 3.2.1.78) which hydrolyze the polymer in a random fashion (5). According to the classification based on sequence similarity (6), known β-mannanases are classified into the glycoside hydrolase (GH) families GH5, GH26, and GH113. These families belong to clan GH-A, in which the enzymes share the (β/α)8-barrel fold, as displayed in the Carbohydrate-Active enZYmes database (www.cazy.org) (7).
As with many polysaccharidases, β-mannanases can be modular, carrying along with catalytic modules such additional modules as carbohydrate-binding modules (CBMs), classified into what to date are 64 families (www.cazy.org) and having the general function of promoting association of the enzyme to the substrate (8). Several β-mannanases produced by terrestrial bacteria and fungi found in environments containing decaying plant material have been characterized in molecular detail (5). For example, the mode of action and the crystal structure have been determined for the GH5 β-mannanases of the bacterium Thermomonospora fusca (9) and the fungus Trichoderma reesei (10–12), the GH26 β-mannanase CfMan26A from the bacterium Cellulomonas fimi (13–16), and the GH5 and GH26 β-mannanases of the bacterium Cellvibrio japonicus (17, 18). Also, the crystal structures of GH26 β-mannanases from Bacillus subtilis strains and from the thermophilic bacterium Alicyclobacillus acidocaldarius (GH113) have been described previously (19–21). The GH5 β-mannanases of the tomato plant and of the blue mussel have also been characterized in detail, and their three-dimensional (3D) structures have been determined (22, 23).
Not only can β-mannans be metabolized in terrestrial and marine environments, but also guar gum galactomannan can be fermented in the human digestive tract (24), and mixed cultures from human feces are able to hydrolyze guar gum galactomannan (25) and glucomannan oligosaccharides (26, 27). The processing and utilization of nondigestible diet polysaccharides by the human gut bacteria are highly important for the health of the host (28, 29). Bifidobacterium is one of the most abundant genera of the gut flora, harboring many species generally regarded as being health beneficial (30). In general, bifidobacteria have the capacity to metabolize various complex carbohydrates (28). At least some of the strains, including Bifidobacterium adolescentis, can grow on glucomannan and glucomannan oligosaccharides (31). However, the putative enzyme(s) responsible for β-mannan depolymerization in the human gut is largely unknown, although Bacteroides ovatus produces β-mannanase activity when grown on galactomannan (32, 33). The lack of sequence information regarding the B. ovatus enzyme(s), however, restricts any bioinformatic analysis or classification as well as any clear identification. An exo-acting β-mannosidase of GH2 from Bacteroides thetaiotaomicron has been well characterized both functionally and structurally (34).
The genome sequencing of several bifidobacteria has made it possible to predict the presence of putative carbohydrate-active enzymes, the majority of which are predicted proteins yet to be characterized functionally (35–37). The aim of the present study was to express and characterize a putative β-mannanase from one of the Bifidobacterium species, to increase the knowledge of the β-mannan catabolism of human gut bacteria. Accordingly, to possibly find a homolog to known mannanases, the sequence of CfMan26A from C. fimi (16) was used as a query in a BLAST search, one that resulted in a hit in the in silico-translated genome of B. adolescentis. The corresponding gene was expressed, and the gene product was identified as a β-mannanase (BaMan26A). The enzyme was characterized and was found to be modular and likely to be cell attached.
MATERIALS AND METHODS
Bacterial strains, media, and vectors.
The Zero Blunt TOPO PCR cloning kit and the electrocompetent Escherichia coli TOP10 strain employed were purchased from Invitrogen, Carlsbad, CA. E. coli BL21(DE3) was purchased from Stratagene, La Jolla, CA. The cells were grown in liquid Luria broth (LB) or on LB agar (15 g/liter) plates at 37°C. When the cells contained the pCR-Blunt II-TOPO vector or the pET28b+ vector (Novagen, Darmstadt, Germany), the medium was supplemented with 30 μg/ml kanamycin.
Cloning.
All of the cloning reagents were from Fermentas (St. Leon-Rot, Germany) unless stated otherwise. The manufacturer's recommendations and protocols were followed. When necessary, a DNA cleanup was performed using the Jetquick DNA cleanup kit (Genomed, Löhne, Germany). The genomic DNA from B. adolescentis ATCC 15703 was purchased from LGC Promochem AB, Teddington, United Kingdom. The B. adolescentis gene encoding the β-mannanase BaMan26A was amplified by PCR using 0.04 μg genomic DNA, Pfu DNA polymerase, and the primers 1 (forward) (5′-ATACCATGGCAAAAACCACAGTCACCAAACTG) and 2 (reverse) (5′-AATCTCGAGTTCGATATCGCCTCCCTTCC). Each of the primers used in this study introduces the restriction sites for NcoI (underlined) for forward primers and XhoI (underlined) for reverse primers. Primers 1 and 2 amplify a fragment encoding the full-length β-mannanase BaMan26A, including the signal peptide. Primer 2 deletes the native stop codon and introduces the Leu and Glu that precede the C-terminal 6×His tag (used for purification by means of affinity chromatography) encoded by the pET28b+ expression vector.
Following PCR, the generated fragment was agarose gel purified and subcloned into a pCR-Blunt II-TOPO vector. Then the target fragment, digested by NcoI/XhoI, was subcloned into pET28b+ to construct the expression plasmid pET28b+BaMan26A. After sequencing (at Eurofins, Ebersberg, Germany), it was shown that the coding sequence was identical to the one deposited in the GenBank database (GeneID 4557264), except that the native stop codon was deleted and was replaced by a His tag-coding sequence in the region corresponding to the C terminus, as expected. pET28b+BaMan26A was transformed into the E. coli BL21(DE3) expression strain to create a strain expressing the full-length BaMan26A with a 6×His tag (the enzyme variant being designated BaMan26A-107K).
The plasmid pET28b+BaMan26A was used as a template to generate a plasmid (pET28b+BaMan26A-NoS) encoding a BaMan26A variant without the predicted signal peptide. Primers 2 and 3 (forward) (5′-ATACCATGGCAGAAGGAAAATCGGCATCC) were used for the PCR from which the product was subcloned into pET28b+ as described above.
pET28b+BaMan26A-NoS was used as a template, together with primers 3 and 4 (reverse) (5′-ATCTCGAGTGAACCGGTGCGGGACAG), so as to generate pET28b+BaMan26A-aa29-971, encoding the β-mannanase without both the N-terminal signal peptide and the putative transmembrane helix (designated BaMan26A-101K). This variant starts with Met, followed by Ala29, and ends with Ser971 (numbered from the native start codon), followed by Leu and Glu from the restriction site and also the 6×His tag. The PCR product was subcloned into pET28b+ as described above.
pET28b+BaMan26A-CatIg, coding for the catalytic and immunoglobulin-like (Ig-like) modules of the β-mannanase (BaMan26A-53K), was generated with the primers 3 and 5 (reverse) (5′-AATCTCGAGTGGTTTGGCACCCAGTTTTAC), using the β-mannanase gene without signal peptide in the pCR-Blunt II-TOPO vector as a template in the PCR. The PCR product was subcloned into pET28b+ as described above. pET28b+BaMan26A-CatIg codes for BaMan26A-53K. Since this variant lacks the predicted N-terminal signal peptide, it starts with Met, followed by Ala29. The C terminus ends with Pro540 (numbering from the native start codon), followed by Leu and Glu, encoded by the restriction site, and the 6×His tag.
Activity assay.
The β-mannanase activity assay was performed with use of dinitrosalicylic acid (DNS), as described by Stålbrand et al. (38). Either 5 g/liter carob galactomannan (locust bean gum from Ceratonia siliqua seeds; Sigma-Aldrich, St. Louis, MO) or 5 g/liter guar gum (Sigma-Aldrich) dissolved in 50 mM Na citrate buffer (pH 5.3) was used as the substrate. The incubations were performed in duplicate at 37°C for 15 min.
Protein expression and purification.
Overnight cultures of E. coli BL21(DE3) containing the appropriate construct of the β-mannanase gene were inoculated in LB medium, and when an optical density at 600 nm (OD600) of 1.0 to 1.2 was reached, expression was induced by use of 0.4 mM isopropyl β-d-1-thiogalactopyranoside (IPTG). The cells were then grown for 1 h and were harvested by centrifugation. The pellet was suspended in binding buffer for affinity chromatography (50 mM Na phosphate, 300 mM NaCl, 10 mM imidazole [pH 8.0]), and a 1 mM concentration of the protease inhibitor phenylmethylsulfonyl fluoride (PMSF) was added. The cells were lysed by use of a French pressure cell (SLM-Aminco, Rochester, NY), and the lysate was centrifuged and filtered through a 0.2-μm filter before being applied to a nickel-nitrilotriacetic acid (Ni-NTA) cartridge or an Ni-NTA column from Qiagen (Hilden, Germany). The column was washed with washing buffer (50 mM Na phosphate [pH 8.0], 300 mM NaCl, and 20 to 33 mM imidazole depending on run). The proteins were eluted with 250 mM imidazole, 300 mM NaCl, and 50 mM Na phosphate, pH 8.0. The fractions were assayed for β-mannanase activity and were analyzed by means of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using 12% or 10% polyacrylamide gels. The gels were stained with Coomassie brilliant blue G-250 (39). The protein concentration was measured by use of a NanoDrop ND-1000 spectrophotometer at 280 nm. The molecular weight and the theoretical absorption coefficient as determined by Protparam (40) were used to correlate the absorbance with the protein concentration. The electrophoretically homogeneous fractions that showed the highest specific activity were pooled, and the buffer was exchanged to 50 mM Na citrate buffer, pH 5.3, by using Amicon Ultra 10-kDa-cutoff centrifugal filter devices (Millipore, Tullagreen, Ireland).
Stability and optimal pH.
To determine the optimal pH for β-mannanase activity, the DNS assay was used with carob galactomannan in the pH range of 3 to 8. For the pH range of 3 to 6, 50 mM Na citrate buffer was used, and for the pH range of 6 to 8, 50 mM Na phosphate buffer was used. To test the stability, the enzyme was incubated at 4°C to 50°C in 50 mM Na citrate buffer of pH 5.3 for 24 h, with and without the addition of 0.05 mg/ml bovine serum albumin (BSA; AppliChem, Darmstadt, Germany). Following incubation, the activity assay was performed as described above.
Hydrolysis of poly- and oligosaccharides.
Mannobiose (M2), mannotriose (M3), mannotetraose (M4), mannopentaose (M5), mannohexaose (M6), low-viscosity carob galactomannan, low-viscosity glucomannan from Amorphophallus konjac (konjac glucomannan), and ivory nut mannan were purchased from Megazyme (Bray, Ireland). Hydrolysis mixtures were prepared, each containing 2.5 g/liter polysaccharide (either carob galactomannan, low-viscosity carob galactomannan, or guar gum or konjac glucomannan) or 4 mM oligosaccharide in 50 mM Na citrate buffer at pH 5.3 together with 0.05 mg/ml BSA. The hydrolysis reactions were carried out with 1 μM enzyme added, except for the konjac glucomannan hydrolysis, to which 0.2 μM enzyme was added. The mixtures were incubated at 30°C. Aliquots were taken out after 30 min, 24 h, and 48 h, were boiled for 3 min, and were stored frozen until analyzed. The controls of the substrate and the enzyme were treated in the same way.
The ivory nut mannan was first washed five times in water, the pellets of it being recovered after each step by centrifugation to remove any traces of soluble saccharides. The ivory nut mannan was then dried overnight at 30°C. Following this, 1.25 g/liter ivory nut mannan in 50 mM Na citrate buffer at pH 5.3 containing 0.05 mg/ml BSA was provided with 0.2 μM enzyme, and incubation was performed as described above but with head-over-tail rotation.
Analysis by thin-layer chromatography (TLC) was carried out with use of aluminum sheets covered by silica gel 60 (Merck, Darmstadt, Germany). 1-Buthanol-ethanol-water at a volume ratio of 10:8:7 was used as mobile phase. Staining was performed as described by Bounias (41), with slight modifications. A mixture of 8.2 g/liter N-(1-naphthyl)ethylenediamine dihydrochloride (Sigma), 8.6% sulfuric acid, and ethanol was poured over the TLC sheet. The sheet was then dried and was baked in an oven at 105°C until color development was complete.
The hydrolysis products were quantified by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) (Dionex, Sunnyvale, CA), as described by Hägglund et al. (10). A Carbopac PA-100 column (Dionex) was used for the separation with an isocratic flow of 78 mM NaOH. Low-viscosity carob galactomannan hydrolyzed for 30 min by BaMan26A-101K was analyzed and then incubated overnight together with α-galactosidase from guar gum (Megazyme, Bray, Ireland) and was analyzed by HPAEC thereafter.
Binding studies.
Affinity gel electrophoresis was used to determine the dissociation constant (Kd) for low-viscosity carob galactomannan (15). Native PAGE gels containing 10% acrylamide and 0, 0.005, 0.0125, 0.05, and 0.1 g/liter low-viscosity carob galactomannan were casted. BSA was used as a nonbinding reference protein. A total of 0.7 μg of each protein was loaded, and the gels were run at 100 V for 240 min at 4°C. Three repeats were made. The gels were stained as described above. Binding toward insoluble polysaccharides was performed as described by Hägglund et al. (10). Washed ivory nut mannan and Avicel cellulose (Avicel PH-101; Fluka, Ireland) were suspended to a concentration of 10 g/liter in 100 mM Na phosphate–50 mM Na citrate buffer at pH 7.0, with 0.2 μM enzyme added to the tubes, which were then incubated at 4°C for 1 h with continued stirring. A negative control containing 0.2 μM enzyme but no substrate was treated in the same way. After incubation, the samples were spun down. The residual β-mannanase activity was measured in the supernatant with use of the DNS assay as described above.
Kinetics.
The kinetic properties of BaMan26A-53K were determined on low-viscosity carob galactomannan. Incubation with 3.6 nM enzyme at 37°C was carried out in duplicate for 15 min (for concentrations of 20 g/liter and 30 g/liter) or for 30 min (for concentrations of 5 g/liter and 10 g/liter). Hydrolysis was stopped by adding DNS, and the samples were analyzed as described above. The hydrolysis rate was calculated in terms of micromolar concentration of substrate hydrolyzed per second and was plotted against the substrate concentration in the hydrolysis mixture to obtain a Michaelis-Menten curve. Km and Vmax were determined by nonlinear regression in GraphPad Prism 5 (GraphPad Software, San Diego, CA).
RESULTS
BLAST search, gene cloning, and recombinant expression of BaMan26A.
A BLAST search (blastp) (42) with the peptide sequence of the modular β-mannanase from C. fimi (CfMan26A; UniProt ID Q9XCV5), studied previously by our group and others (13–16), revealed a potential β-mannanase precursor (UniProt ID A1A278) in the in silico-translated genome sequence of B. adolescentis (GenBank accession no. NC_008618.1). The potential B. adolescentis β-mannanase had the locus tag BAD_1030 and was designated BaMan26A. The predicted length of BaMan26A was 1,002 amino acids, including an N-terminal secretion signal peptide, as detected by the SignalP server (43) (Fig. 1). The gene predicted to encode full-length BaMan26A (BaMan26A-107K) was PCR amplified, cloned, and expressed. β-Mannanase activity was detected in the cell lysate. Recombinant expression thus yielded active BaMan26A.
Fig 1.
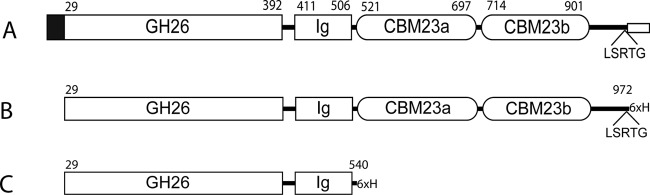
Schematic view of BaMan26A and the recombinant variants characterized in this work. (A) Native BaMan26A (full length); (B) BaMan26A-101K; (C) BaMan26A-53K. The black box represents the signal peptide. GH26 is the catalytic module of glycoside hydrolase family 26. The putative linkers are marked with a black line. The LSRTG motif is indicated. The transmembrane helix sequence followed by 4 charged amino acids is marked with a small box at the C terminus. Numbers above the modules show the predicted starting and ending amino acids of each module (numbered from the native start codon).
Predicted modular organization and cellular location of BaMan26A.
The mature N-terminal part (residues 29 to 392 [Fig. 1]) of BaMan26A showed an identity of 44% with the GH26 catalytic module of CfMan26A (14). A multiple alignment carried out using CLUSTAL W2 (44) with the catalytic modules of CfMan26A and three other GH26 β-mannanases predicted the presence of the two conserved catalytic glutamates in GH26 within the BaMan26A sequence (E205 and E316). The three other β-mannanases were BsMan26A from Bacillus sp. strain JAMB-750 (45) and CjMan26A and CjMan26C from Cellvibrio japonicus (17, 18), with which BaMan26A showed an identity of 35 to 42%. Residues 417 to 517 of BaMan26A also showed 27% identity with the Ig-like module of CfMan26A (14). The Protein Homology/analogY Recognition Engine, version 2.0 (Phyre2) (46), predicted this sequence to be homologous to the C. fimi Ig-like module with 98% confidence. BaMan26A may thus contain an Ig-like module in a manner similar to CfMan26A, although no clear function has been assigned to these modules. Further bioinformatic analysis showed that similar to the case with CfMan26A, the GH26 catalytic and putative Ig-like modules are followed by a predicted CBM of family 23. In contrast to CfMan26A (15, 47), a second putative CBM23 follows the first in the case of BaMan26A. The two putative CBMs of BaMan26A showed 34% and 36% identity with CBM23 of CfMan26A, which has been shown to bind galactomannan but not water-insoluble mannan, xylan, or cellulose (15). An alignment with these three CBMs and the predicted but uncharacterized CBM23 of BsMan26A (45) showed there to be 18 conserved residues (Fig. 2), 5 of them being Trp or Tyr, which are known to often be involved in carbohydrate interactions (8). However, no functions of any of the individual residues in CBM23 have been shown yet.
Fig 2.

Alignment of the two putative repeats of CBM23 from BaMan26A–CBM1 (amino acids 520 to 714) and CBM2 (amino acids 715 to 901) with the CfMan26A CBM23 (15) and putative CBM23 from Bacillus sp. JAMB-750 Man26A (45). Asterisks indicate conserved residues; one or two dots show positions with similarity between groups of amino acids (two dots mark stronger similarity). The alignment was made in CLUSTAL W2.
Toward the C-terminal side of the tandem CBMs of BaMan26A there follow 64 amino acids which lack homology to any known sequence and could comprise a linker (Fig. 1). There follows then a stretch of 22 hydrophobic amino acids predicted to be a transmembrane helix by the TMHMM prediction server (www.cbs.dtu.dk/services/TMHMM-2.0/) (48). This supposed transmembrane helix is followed by four basic residues, which together with the helix could potentially comprise a membrane anchor (49). N terminally from the predicted transmembrane helix there is an LSRTG amino acid sequence which, together with the hydrophobic stretch and the positively charged amino acids, could be a sorting signal. In the sortase mechanism, after the secretion of a protein carrying an LPXTG amino acid sequence, the protein becomes covalently linked to the cell wall by the enzyme sortase (50). Although BaMan26A lacks the exact LPXTG sequence, there are variations of this motif, LSRTG being among them (51). Either way, BaMan26A appears to be cell attached, either through a membrane helix anchor or through attachment to the cell wall mediated by the sortase.
Expression, purification, and basic properties of truncated variants of BaMan26A.
In addition to full-length BaMan26A (BaMan26A-107K), there were also two truncated variants of BaMan26A (both of them lacking the signal peptide) that were likewise cloned and expressed in E. coli: BaMan26A-101K and BaMan26A-53K. The expression of BaMan26A-107K resulted in a considerably (10-fold) lower activity than for BaMan26A-101K and BaMan26-53K. The last two were selected to be purified and to be characterized further.
BaMan26A-101K is truncated right after the LSRTG sequence, to resemble what is predicted to be the mature soluble (neither cell wall nor membrane associated) part of the protein. It contains four modules: catalytic, Ig-like, and the two CBM23 modules, followed by a putative linker consisting of 64 amino acids (Fig. 1B). The other variant to be characterized was BaMan26A-53K, containing only the catalytic and Ig-like modules (Fig. 1C), equivalent to a 50-kDa fragment of CfMan26A (CfMan26A-50K), for which the crystal structure has been determined (14). For CfMan26A, the Ig-like module can be important for the stability of the catalytic module (16). BaMan26A-53K enables the catalytic function of the enzyme to be studied without the influence of CBM23.
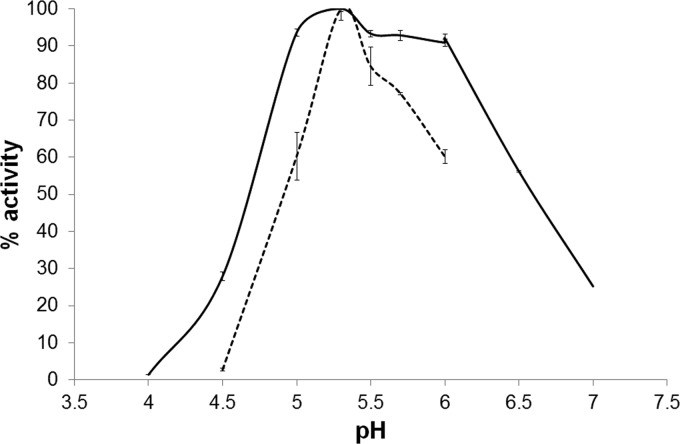
BaMan26A-53K was found to have a narrow pH optimum at pH 5.3 (Fig. 3). BaMan26A-101K also showed a pH optimum at 5.3 but retained over 90% of its activity in the pH range of 5 to 6. After incubation for 24 h in the presence of 0.05 mg/ml BSA at pH 5.3, BaMan26A-53K and BaMan26A-101K retained their activities within the range of 4°C to 37°C. Their activities were lost after incubation for 3 h at 50°C.
Fig 3.
Relative activities of BaMan26A-101K (solid line) and BaMan26A-53K (dashed line) at different pHs. The highest activity obtained in the experiment was set as 100%.
Specific activity and enzyme kinetics.
As determined by the DNS assay, the specific activity of BaMan26A-101K toward carob galactomannan was 340 kat/mol enzyme, whereas that of BaMan26A-53K was 176 kat/mol. On low-viscosity carob galactomannan, the specific activities were 262 kat/mol for BaMan26A-101K and 105 kat/mol for BaMan26A-53K, while on guar gum they were 110 kat/mol and 14 kat/mol, respectively. Using low-viscosity carob galactomannan, the Km for BaMan26A-53K was found to be 21.3 g/liter (standard deviation [SD], 2.9 g/liter), and the kcat was found to be 444 s−1 (SD, 2 s−1).
Products from polysaccharide hydrolysis.
Four different β-1,4-linked mannans were incubated together with BaMan26A-101K and BaMan26A-53K—ivory nut mannan (a water-insoluble homomannan), konjac glucomannan, and carob and guar gum galactomannans—and the hydrolysis products were analyzed. HPAEC chromatograms showed ivory nut mannan to be hydrolyzed to manno-oligosaccharides to a degree of polymerization (DP) of 2 to 4, with the major product being M3 (Table 1). For BaMan26A-101K, the release of M3 was somewhat more prominent than it was for BaMan26A-53K. During the period in which the hydrolysis took place (48 h), BaMan26A-53K also released minor amounts of M5 (data not shown). After 30 min, BaMan26A-53K had converted 7.5% of the mannan, and 15% of it had been converted after 24 h, whereas for BaMan26A-101K, the degrees of conversion were 10% and 32%, respectively.
Table 1.
Hydrolysis products from M3, M5, and ivory nut mannana
| Substrate | Enzyme variant | Time (h) | Concn (mM) of hydrolysis product |
|||
|---|---|---|---|---|---|---|
| M1 | M2 | M3 | M4 | |||
| M3 | BaMan26A-53K | 0.5 | <0.1 | <0.1 | ||
| 24 | 0.93 ± 0.02 | 0.77 ± 0.05 | ||||
| 48 | 1.6 ± 0.09 | 1.1 ± 0.09 | ||||
| BaMan26A-101K | 0.5 | <0.1 | <0.1 | |||
| 24 | 0.65 ± 0.01 | 0.54 ± 0.00 | ||||
| 48 | 1.4 ± 0.01 | 0.94 ± 0.00 | ||||
| M5 | BaMan26A-53K | 0.5 | 0.86 ± 0.12 | 2.3 ± 0.01 | 3.6 ± 0.07 | 0.41 ± 0.11 |
| 48 | 2.4 ± 0.11 | 3.0 ± 0.03 | 1.9 ± 0.02 | |||
| BaMan26A-101K | 0.5 | 2.1 ± 0.01 | 3.3 ± 0.07 | |||
| 48 | 1.6 ± 0.01 | 3.1 ± 0.02 | 2.2 ± 0.02 | |||
| Ivory nut mannan | BaMan26A-53K | 0.5 | 0.038 ± 0.002 | 0.096 ± 0.003 | 0.038 ± 0.001 | |
| 24 | 0.112 ± 0.001 | 0.246 ± 0.001 | 0.020 ± 0.001 | |||
| BaMan26A-101K | 0.5 | 0.030 ± 0.003 | 0.183 ± 0.012 | 0.026 ± 0.002 | ||
| 24 | 0.159 ± 0.008 | 0.608 ± 0.023 | 0.019 ± 0.000 | |||
M3 and M5 were used at 4 mM, and ivory nut mannan was used at 1.25 mg/ml. Where applicable, deviations from the mean of two samples are shown.
Konjac glucomannan was depolymerized to a mixture of glucomanno-oligosaccharides, the dominant product(s) being estimated to represent a DP of 3, as analyzed by use of TLC (Fig. 4A). Interestingly, carob galactomannan was fragmented to smaller products than guar gum galactomannan was (Fig. 4B and C). This may possibly reflect the restrictions brought about by galactose substitutions, which would be more severe in the case of the higher degree of substitution in guar gum galactomannan. The major products obtained from carob galactomannan were estimated to be from a DP of 2 up to a DP of 6 and higher (Fig. 4B). The products of guar gum were estimated to be mainly a DP of 6 and higher (Fig. 4C). HPAEC analysis of the carob galactomannan hydrolyzed showed that in addition to galactomanno-oligosaccharides (as confirmed by α-galactosidase incubation), M3, M2, and M1 were likewise formed, M3 being formed in the highest concentration.
Fig 4.
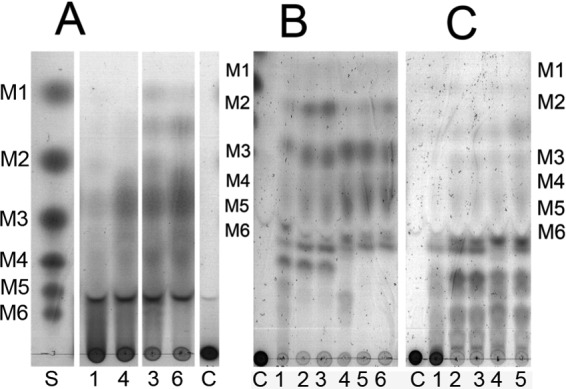
TLC sheets showing hydrolysis products from polysaccharides konjac glucomannan (A), carob galactomannan (B), and guar gum galactomannan (C). Lane S marks the standard of manno-oligosaccharides representing DPs 1 to 6, and lanes C mark control (substrate with no enzyme added). Lanes 1 to 3 contain BaMan26A-53K incubated with substrate for 30 min (lanes 1), 24 h (lanes 2), and 48 h (lanes 3). Lanes 4 to 6 contain BaMan26A-101K incubated with substrate for 30 min (lanes 4), 24 h (lanes 5), and 48 h (lanes 6).
Hydrolysis of manno-oligosaccharides.
Neither of the BaMan26A variants was able to hydrolyze M2, even after 48 h of incubation. The hydrolysis of M3 was considerably slower than that of the higher oligosaccharides (M4, M5, and M6), as judged from the product concentrations after 0.5 h of incubation; see Table 1, in which products of the hydrolysis of M3 and M5 are given. M5 was hydrolyzed mainly to M3 and M2 (Table 1), while M6 was hydrolyzed to M3 as the main product, in addition to M1, M2, M4, and M5. In reference to the results described in this paragraph, no significant difference in the patterns of the hydrolysis products was obvious in comparing BaMan26A-101K with BaMan26A-53K. The results show no clear indication of the occurrence of substantial transglycosylation, in a manner rather similar to that of CfMan26A (14, 52). However, significant transglycosylation has been observed for many other mannanases, such as TrMan5A from T. reesei (11).
Binding to polysaccharides.
BaMan26A-101K showed an affinity for carob galactomannan with a Kd of 8.8 (SD, 0.18) mg/liter or of 0.34 (SD, 0.07) μM, analyzed by affinity gel electrophoresis as done by Stoll et al. (15). In contrast to BaMan26A-101K, BaMan26A-53K and the control protein BSA were not retarded by the presence of carob galactomannan in the gel (data not shown). Both of the BaMan26A variants that were characterized bound to ivory nut mannan. BaMan26A-101K showed 46% residual activity in the supernatant compared with the control, whereas BaMan26A-53K showed 25% residual activity. Neither of the variants bound to Avicel cellulose.
DISCUSSION
Microbial degradation and utilization of carbohydrates in the human gut are of great importance for our health, with such probiotic bacteria as bifidobacteria playing an important role (28, 30). Our study provides new information of relevance to β-mannan conversion by the human gut microbiota. B. adolescentis can grow, for example, on glucose, mannose, and galactose (53) and both hydrolyzed and unhydrolyzed konjac glucomannan (31, 54). Until now, however, the enzymes responsible for mannan saccharification have not been characterized in any detail.
In a manner similar to that of its homolog, the β-mannanase CfMan26A from the soil-living bacterium C. fimi, BaMan26A has a modular organization. Both BaMan26A and CfMan26A have an N-terminal catalytic module of GH family 26, followed by an Ig-like module and CBM23, which in the case of BaMan26A (but not of CfMan26A) is followed by an additional CBM23 in tandem (Fig. 1). Both of these homologs appear to be extracellular and to be cell wall attached, although through different mechanisms. CfMan26A has a putative surface layer homology module for cell attachment (16). For BaMan26A, in contrast, a potential cell wall attachment can occur via either of two mechanisms, one being anchoring to the peptidoglycan of the cell wall by means of a sortase-mediated mechanism, through which the transmembrane helix is cleaved off (50, 55), and the other being through membrane attachment via the C-terminal transmembrane helix. Cell association has been observed for several β-mannanases, for example, the B. ovatus β-mannanase (32), the C. japonicus β-mannanases (17), and the cellulosomal β-mannanases of anaerobic bacteria, such as clostridia. The mechanism involved also differs here. The cellulosomal β-mannanases of cellulolytic anaerobes are generally attached to cell-bound scaffold proteins that harbor several enzymes (56). In contrast, three of the C. japonicus β-mannanases (CjMan26A, CjMan26C, and CjMan5D) appear to be outer-membrane attached (but separate from each other) (17), similar to the situation predicted for B. adolescentis and C. fimi, in the sense of each enzyme not being part of an enzyme complex but, rather, being directly associated with the cell.
BaMan26A-101K had an affinity to carob galactomannan, which was not the case for BaMan26A-53K, which lacks the CBMs. This shows that the predicted CBMs have a combined affinity for the soluble carob galactomannan. At this stage it cannot be determined whether the two CBMs are functional and are similar in affinity or specificity. By comparison, the single CBM23 of CfMan26A has a slightly greater affinity, having a Kd of approximately 0.2 μM (15).
Regarding the catalytic properties of BaMan26A, it is an enzyme that can clearly act in an endo-fashion, releasing oligosaccharides of various lengths from the polymeric substrates (Table 1). The overall pattern of hydrolysis of M4, M5, M6, and ivory nut mannan shows that BaMan26A apparently tends to produce M3 as an initial hydrolysis product, which differs from the profiles of the C. fimi and C. japonicus enzymes mentioned above (14, 17, 18, 57). BaMan26A is considerably slower in hydrolyzing M3 than in hydrolyzing longer oligosaccharides, which is similar to what has been observed for the C. fimi mannanase CfMan26A-50K and the C. japonicus enzyme CjMan26B, for example (14, 57).
The kinetic constants determined by use of carob galactomannan and BaMan26A-53K show a kcat value similar to that of CfMan26A-50K but at the same time an almost 10-fold-higher Km value. An explanation for this may be a greater sensitivity to galactose substitutions for BaMan26A. Restriction by galactose substitutions is also supported by the release of only larger fragments (DP > 6) from the more highly substituted guar gum galactomannan than from carob galactomannan, where fragments with a DP of <6 were more pronounced (Fig. 4B and C). Also, its specific activity toward guar gum galactomannan was found to be 8-fold lower than for carob galactomannan. Many β-mannanases appear to be restricted by galactose substituents (5), including the B. ovatus β-mannanase (32). On the other hand, the presence of backbone glucose in konjac glucomannan possibly does not restrict the activity of BaMan26A, since the products, down to DPs of 2 and 3, are visible in the hydrolysate (Fig. 4A).
BaMan26A is thus far the only identified β-mannanase from the B. adolescentis genome. Compared with the situation in regard to C. japonicus, which has at least 7 β-mannanases with niched functions (17, 18, 57), it appears that B. adolescentis may combine several individual properties in one enzyme. First, BaMan26A appears both to be cell attached (as CjMan26A-C) and to contain CBMs (as CjMan5C). It also shows hydrolytic capacity toward both crystalline ivory nut mannan and soluble mannans, which distinguishes it from CjMan26B, which lacks activity toward ivory nut mannan (57). Furthermore, BaMan26A binds both soluble mannans (via at least one CBM23) and insoluble mannan via as-yet-unidentified motif(s) that is not provided by the CBMs. The ivory nut binding property could possibly be contributed by a secondary sugar binding site (58, 59), present on either the catalytic module or the Ig-like module, that distinguishes BaMan26A from its homolog, CfMan26A from C. fimi, which does not bind ivory nut mannan (15). CjMan26C has a processive mode of action, releasing M2 (17). However, the degree of processivity of BaMan26A, if indeed there is any, remains to be studied. If such is the case, it would appear to not be as stringent as for one of the few other endopolysaccharidases that have been characterized from bifidobacteria, the GH53 endogalactanase from Bifidobacterium longum, which releases galactotriose from galactan (60).
Our study provides new molecular insight into an enzyme potentially responsible for mannan endohydrolysis by a gut bacterium, a property which only rarely has been described for such bacteria. For Bacteroides thetaiotaomicron, an exo-β-mannosidase has been suggested to be involved in mannan degradation (34). The β-mannanase activity of B. ovatus cells, however, appears to degrade galactomannan in a random endo-wise fashion (32), as appears to also be the case for BaMan26A. Assuming that B. adolescentis expresses cell-associated and active BaMan26A, the oligosaccharides would be released close to the cell, potentially beneficial for import and/or for further extracellular or intracellular degradation. In line with this view, additional exo-acting enzymes (β-mannosidases and α-galactosidases) or sugar phosphorylases (35) are likely to be needed to complete the degradation to monomers. An α-galactosidase from B. adolescentis has been expressed recombinantly (61). Its potential activity toward galactosyl substitutions of mannosides has, however, not been reported. Thus, the potential β-mannosidase and α-galactosidase activities of B. adolescentis necessary for monosaccharide release from the BaMan26A products remain to be identified. In addition, there is a possibility that bacterial consortia, rather than individual organisms, contribute to the complete degradation of complex heteromannan substrates in the colon (26).
Genome analyses have thus far indicated only relatively few β-mannanase candidates among the human gut bacteria (35–37). Although it remains to be studied how common these enzymatic activities are among gut bacteria, the B. adolescentis mannanase BaMan26A could potentially be one of the enzymes responsible for gut mannan conversion.
ACKNOWLEDGMENTS
FLÄK (The Swedish Research School of Pharmaceutical Sciences) is thanked for its grant to H.S. Part of this study was supported by grants to H.S. from Formas (The Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning; 213-2011-1620) and Vinnova (The Swedish Agency for Innovation Systems via BIOFORM and the Antidiabetic Food Center). K.K. thanks the Sven and Lily Lawski Foundation for mobility support.
Peter Immerzeel is thanked for his technical help in connection with some of the carbohydrate analyses.
Footnotes
Published ahead of print 12 October 2012
REFERENCES
- 1. Lundqvist J, Teleman A, Junel L, Zacchi G, Dahlman O, Tjerneld F, Stålbrand H. 2002. Isolation and characterization of galactoglucomannan from spruce (Picea abies). Carbohydr. Polym. 48:29–39 [Google Scholar]
- 2. Scheller HV, Ulvskov P. 2010. Hemicelluloses. Annu. Rev. Plant Biol. 61:263–289 [DOI] [PubMed] [Google Scholar]
- 3. Cerqueira MA, Bourbon AI, Pinheiro AC, Martins JT, Souza BWS, Teixeira JA, Vicente AA. 2011. Galactomannans use in the development of edible films/coatings for food applications. Trends Food Sci. Technol. 22:662–671 [Google Scholar]
- 4. Dave V, McCarthy SP. 1997. Review of konjac glucomannan. J. Environ. Polym. Degrad. 5:237–241 [Google Scholar]
- 5. Gilbert HJ, Stålbrand H, Brumer H. 2008. How the walls come crumbling down: recent structural biochemistry of plant polysaccharide degradation. Curr. Opin. Plant Biol. 11:338–348 [DOI] [PubMed] [Google Scholar]
- 6. Henrissat B, Davies G. 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7:637–644 [DOI] [PubMed] [Google Scholar]
- 7. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boraston AB, Bolam DN, Gilbert HJ, Davies GJ. 2004. Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem. J. 382:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hilge M, Gloor SM, Rypniewski W, Sauer O, Heightman TD, Zimmermann W, Winterhalter K, Piontek K. 1998. High-resolution native and complex structures of thermostable beta-mannanase from Thermomonospora fusca—substrate specificity in glycosyl hydrolase family 5. Structure 6:1433–1444 [DOI] [PubMed] [Google Scholar]
- 10. Hägglund P, Eriksson T, Collén A, Nerinckx W, Claeyssens M, Stålbrand H. 2003. A cellulose-binding module of the Trichoderma reesei β-mannanase Man5A increases the mannan-hydrolysis of complex substrates. J. Biotechnol. 101:37–48 [DOI] [PubMed] [Google Scholar]
- 11. Rosengren A, Hägglund P, Anderson L, Pavon-Orozco P, Peterson Wulff R, Nerinckx W, Stålbrand H. 2012. The role of subsite +2 of the Trichoderma reesei β-mannanase TrMan5A in hydrolysis and transglycosylation. Biocatal. Biotransformation 30:338–352 [Google Scholar]
- 12. Sabini E, Schubert H, Murshudov G, Wilson KS, Siika-Aho M, Penttilä M. 2000. The three-dimensional structure of a Trichoderma reesei beta-mannanase from glycoside hydrolase family 5. Acta Crystallogr. D Biol. Crystallogr. 56:3–13 [DOI] [PubMed] [Google Scholar]
- 13. Hekmat O, Lo Leggio L, Rosengren A, Kamarauskaite J, Kolenova K, Stålbrand H. 2010. Rational engineering of mannosyl binding in the distal glycone subsites of Cellulomonas fimi endo-beta-1,4-mannanase: mannosyl binding promoted at subsite-2 and demoted at subsite-3. Biochemistry 49:4884–4896 [DOI] [PubMed] [Google Scholar]
- 14. Le Nours J, Anderson L, Stoll D, Stålbrand H, Lo Leggio L. 2005. The structure and characterization of a modular endo-beta-1,4-mannanase from Cellulomonas fimi. Biochemistry 44:12700–12708 [DOI] [PubMed] [Google Scholar]
- 15. Stoll D, Boraston A, Stålbrand H, McLean BW, Kilburn DG, Warren RAJ. 2000. Mannanase Man26A from Cellulomonas fimi has a mannan-binding module. FEMS Microbiol. Lett. 183:265–269 [DOI] [PubMed] [Google Scholar]
- 16. Stoll D, Stålbrand H, Warren RAJ. 1999. Mannan-degrading enzymes from Cellulomonas fimi. Appl. Environ. Microbiol. 65:2598–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cartmell A, Topakas E, Ducros VMA, Suits MDL, Davies GJ, Gilbert HJ. 2008. The Cellvibrio japonicus mannanase CjMan26C displays a unique exo-mode of action that is conferred by subtle changes to the distal region of the active site. J. Biol. Chem. 283:34403–34413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hogg D, Woo EJ, Bolam DN, McKie VA, Gilbert HJ, Pickersgill RW. 2001. Crystal structure of mannanase 26A from Pseudomonas cellulosa and analysis of residues involved in substrate binding. J. Biol. Chem. 276:31186–31192 [DOI] [PubMed] [Google Scholar]
- 19. Tailford LE, Ducros VMA, Flint JE, Roberts SM, Morland C, Zechel DL, Smith N, Bjornvad ME, Borchert TV, Wilson KS, Davies GJ, Gilbert HJ. 2009. Understanding how diverse beta-mannanases recognize heterogeneous substrates. Biochemistry 48:7009–7018 [DOI] [PubMed] [Google Scholar]
- 20. Yan X-X, An XM, Gui LL, Liang DC. 2008. From structure to function: insights into the catalytic substrate specificity and thermostability displayed by Bacillus subtilis mannanase BCman. J. Mol. Biol. 379:535–544 [DOI] [PubMed] [Google Scholar]
- 21. Zhang YL, Ju JS, Peng H, Gao F, Zhou C, Zeng Y, Xue YF, Li Y, Henrissat B, Gao GF, Ma YH. 2008. Biochemical and structural characterization of the intracellular mannanase AaManA of Alicyclobacillus acidocaldarius reveals a novel glycoside hydrolase family belonging to clan GH-A. J. Biol. Chem. 283:31551–31558 [DOI] [PubMed] [Google Scholar]
- 22. Bourgault R, Oakley AJ, Bewley JD, Wilce MCJ. 2005. Three-dimensional structure of (1,4)-beta-d-mannan mannanohydrolase from tomato fruit. Protein Sci. 14:1233–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Larsson AM, Anderson L, Xu B, Muñoz IG, Uson I, Janson JC, Stålbrand H, Ståhlberg J. 2006. Three-dimensional crystal structure and enzymic characterization of beta-mannanase Man5A from blue mussel Mytilus edulis. J. Mol. Biol. 357:1500–1510 [DOI] [PubMed] [Google Scholar]
- 24. Nyman M, Asp NG, Cummings J, Wiggins H. 1986. Fermentation of dietary fibre in the intestinal tract—comparison between man and rat. Br. J. Nutr. 55:487–496 [DOI] [PubMed] [Google Scholar]
- 25. Tomlin J, Read NW, Edwards CA, Duerden BI. 1986. The degradation of guar gum by a fecal incubation system. Br. J. Nutr. 55:481–486 [DOI] [PubMed] [Google Scholar]
- 26. Albrecht S, van Muiswinkel GC, Xu J, Schols HA, Voragen AG, Gruppen H. 2011. Enzymatic production and characterization of konjac glucomannan oligosaccharides. J. Agric. Food Chem. 59:12658–12666 [DOI] [PubMed] [Google Scholar]
- 27. Asano I, Hamaguchi K, Fujii S, Iino H. 2003. In vitro digestibility and fermentation of mannooligosaccharides from coffee mannan. Food Sci. Technol. Res. 9:62–66 [Google Scholar]
- 28. Gibson GR. 2004. Fibre and effects on probiotics (the prebiotic concept). Clin. Nutr. 1:25–31 [Google Scholar]
- 29. Sears CL. 2005. A dynamic partnership: celebrating our gut flora. Anaerobe 11:247–251 [DOI] [PubMed] [Google Scholar]
- 30. Picard C, Fioramonti J, Francois A, Robinson T, Neant F, Matuchansky C. 2005. Review article: bifidobacteria as probiotic agents—physiological effects and clinical benefits. Aliment. Pharmacol. Ther. 22:495–512 [DOI] [PubMed] [Google Scholar]
- 31. Wang C-H, Lai P, Chen M-E, Chen H-L. 2008. Antioxidative capacity produced by Bifidobacterium- and Lactobacillus acidophilus-mediated fermentations of konjac glucomannan and glucomannan oligosaccharides. J. Sci. Food Agric. 88:1294–1300 [Google Scholar]
- 32. Gherardini FC, Salyers AA. 1987. Purification and characterization of a cell-associated, soluble mannanase from Bacteroides ovatus. J. Bacteriol. 169:2038–2043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Macfarlane GT, Hay S, Macfarlane S, Gibson GR. 1990. Effect of different carbohydrates on growth, polysaccharidase and glycosidase production by Bacteriodes ovatus, in batch and continuous culture. J. Appl. Bacteriol. 68:179–187 [DOI] [PubMed] [Google Scholar]
- 34. Tailford LE, Money VA, Smith NL, Dumon C, Davies GJ, Gilbert HJ. 2007. Mannose foraging by Bacteroides thetaiotaomicron-–structure and specificity of the beta-mannosidase, BtMan2A. J. Biol. Chem. 282:11291–11299 [DOI] [PubMed] [Google Scholar]
- 35. Pokusaeva K, Fitzgerald GF, van Sinderen D. 2011. Carbohydrate metabolism in bifidobacteria. Genes Nutr. 6:285–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. van den Broek LAM, Hinz SWA, Beldman G, Vincken JP, Voragen AGJ. 2008. Bifidobacterium carbohydrases—their role in breakdown and synthesis of (potential) prebiotics. Mol. Nutr. Food Res. 52:146–163 [DOI] [PubMed] [Google Scholar]
- 37. van den Broek LAM, Voragen AGJ. 2008. Bifidobacterium glycoside hydrolases and (potential) prebiotics. Innov. Food Sci. Emerg. Technol. 9:401–407 [Google Scholar]
- 38. Stålbrand H, Siika-aho, Tenkanen MM, Viikari L. 1993. Purification and characterization of 2 beta-mannanases from Trichoderma reesei. J. Biotechnol. 29:229–242 [Google Scholar]
- 39. Lawrence AM, Besir HU. 2009. Staining of proteins in gels with Coomassie G-250 without organic solvent and acetic acid. J. Vis. Exp. doi:10.3791/1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. 2005. Protein identification and analysis tools on the ExPASy server, p 571–607 In Walker JM. (ed), The proteomics protocols handbook. Humana Press, Totowa, NJ [Google Scholar]
- 41. Bounias M. 1980. N-(1-Naphthyl)ethylenediamine dihydrochloride as a new reagent for nanomole quantification of sugars on thin-layer plates by a mathematical calibration process. Anal. Biochem. 106:291–295 [DOI] [PubMed] [Google Scholar]
- 42. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nordahl Petersen T, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8:785–786 [DOI] [PubMed] [Google Scholar]
- 44. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and clustal X version 2.0. Bioinformatics 23:2947–2948 [DOI] [PubMed] [Google Scholar]
- 45. Hatada Y, Takeda N, Hirasawa K, Ohta Y, Usami R, Yoshida Y, Grant WD, Ito S, Horikoshi K. 2005. Sequence of the gene for a high-alkaline mannanase from an alkaliphilic Bacillus sp. strain JAMB-750, its expression in Bacillus subtilis and characterization of the recombinant enzyme. Extremophiles 9:497–500 [DOI] [PubMed] [Google Scholar]
- 46. Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4:363–371 [DOI] [PubMed] [Google Scholar]
- 47. Antov M, Anderson L, Andersson A, Tjerneld F, Stålbrand H. 2006. Affinity partitioning of a Cellulomonas fimi beta-mannanase with a mannan-binding module in galactomannan/starch aqueous two-phase system. J. Chromatogr. A 1123:53–59 [DOI] [PubMed] [Google Scholar]
- 48. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567–580 [DOI] [PubMed] [Google Scholar]
- 49. Cabanes D, Dehoux P, Dussurget O, Frangeul L, Cossart P. 2002. Surface proteins and the pathogenic potential of Listeria monocytogenes. Trends Microbiol. 10:238–245 [DOI] [PubMed] [Google Scholar]
- 50. Ton-That H, Marraffini LA, Schneewind O. 2004. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim. Biophys. Acta 1694:269–278 [DOI] [PubMed] [Google Scholar]
- 51. Kruger RG, Otvos B, Frankel BA, Bentley M, Dostal P, McCafferty DG. 2004. Analysis of the substrate specificity of the Staphylococcus aureus sortase transpeptidase SrtA. Biochemistry 43:1541–1551 [DOI] [PubMed] [Google Scholar]
- 52. Anderson L, Hägglund P, Stoll D, Lo Leggio L, Drakenberg T, Stålbrand H. 2008. Kinetics and stereochemistry of the Cellulomonas fimi beta-mannanase studied using H-1-NMR. Biocatal. Biotransformation 26:86–95 [Google Scholar]
- 53. Degnan BA, Macfarlane GT. 1991. Comparison of carbohydrate substrate preferences in 8 species of bifidobacteria. FEMS Microbiol. Lett. 84:151–156 [DOI] [PubMed] [Google Scholar]
- 54. Al-Ghazzewi FH, Khanna S, Tester RF, Piggott J. 2007. The potential use of hydrolysed konjac glucomannan as a prebiotic. J. Sci. Food Agric. 87:1758–1766 [Google Scholar]
- 55. Schneewind O, Fowler A, Faull KF. 1995. Structure of the cell wall anchor of surface proteins in Staphylococcus aureus. Science 268:103–106 [DOI] [PubMed] [Google Scholar]
- 56. Halstead JR, Vercoe PE, Gilbert HJ, Davidson K, Hazlewood GP. 1999. A family 26 mannanase produced by Clostridium thermocellum as a component of the cellulosome contains a domain which is conserved in mannanases from anaerobic fungi. Microbiology 145:3101–3108 [DOI] [PubMed] [Google Scholar]
- 57. Hogg D, Pell G, Dupree P, Goubet F, Martín-Orúe SM, Armand S, Gilbert HJ. 2003. The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem. J. 371:1027–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cuyvers S, Dornez E, Delcour JA, Courtin CM. 2012. Occurrence and functional significance of secondary carbohydrate binding sites in glycoside hydrolases. Crit. Rev. Biotechnol. 32:93–107 [DOI] [PubMed] [Google Scholar]
- 59. Nielsen MM, Bozonnet S, Seo ES, Mótyán JA, Andersen JM, Dilokpimol A, Abou Hachem M, Gyémánt G, Naested H, Kandra L, Sigurskjold BW, Svensson B. 2009. Two secondary carbohydrate binding sites on the surface of barley alpha-amylase 1 have distinct functions and display synergy in hydrolysis of starch granules. Biochemistry 48:7686–7697 [DOI] [PubMed] [Google Scholar]
- 60. Hinz SW, Pastink MI, van den Broek LA, Vincken JP, Voragen AG. 2005. Bifidobacterium longum endogalactanase liberates galactotriose from type I galactans. Appl. Environ. Microbiol. 71:5501–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hinz SWA, Doeswijk-Voragen CHL, Schipperus R, van den Broek LAM, Vincken JP, Voragen AGJ. 2006. Increasing the transglycosylation activity of alpha-galactosidase from Bifidobacterium adolescentis DSM 20083 by site-directed mutagenesis. Biotechnol. Bioeng. 93:122–131 [DOI] [PubMed] [Google Scholar]