Abstract
The Mediterranean fruit fly (medfly) (Ceratitis capitata) lays eggs in fruits, where larvae subsequently develop, causing large-scale agricultural damage. Within its digestive tract, the fly supports an extended bacterial community that is composed of multiple strains of a variety of enterobacterial species. Most of these bacteria appear to be functionally redundant, with most strains sustaining diazotrophy and/or pectinolysis. At least some of these bacteria were shown to be vertically inherited, but colonization, structural, and metabolic aspects of the community's dynamics have not been investigated. We used fluorescent in situ hybridization, metabolic profiling, plate cultures, and pyrosequencing to show that an initial, egg-borne, diverse community expands throughout the fly's life cycle. While keeping “core” diazotrophic and pectinolytic functions, it also harbors diverse and fluctuating populations that express varied metabolic capabilities. We suggest that the metabolic and compositional plasticity of the fly's microbiota provides potential adaptive advantages to the medfly host and that its acquisition and dynamics are affected by mixed processes that include stochastic effects, host behavior, and molecular barriers.
INTRODUCTION
Insects, probably as all animals, are colonized by microorganisms, of which we still have a very incomplete knowledge (1). These partnerships range from casual associations to complete codependence (2). Obligate bacterial symbionts, or primary symbionts, are mostly endocellular and are often housed in specialized host cells called bacteriocytes. In such close associations, the survivals of the host and primary symbiont are interdependent, and most symbionts are transmitted vertically between generations. In contrast, the location of facultative, or secondary, symbionts within the insect may vary. These facultative bacteria can be beneficial, as they can affect fitness and provide ecological adaptive benefits, but they are not essential for the host's survival (3–6). Secondary symbionts are transmitted vertically or acquired from the environment (6). Some bacterial taxa are found only as primary or secondary symbionts, while others may be one or the other, depending on the insect host (7). Symbionts belonging to the same taxon may therefore act rather differently between hosts or even within one host; as an example, different Wolbachia strains can co-colonize the parasitoid Leptopilina heterotoma in a noncompetitive manner, preferentially locating in different body parts (8). Hence, a true estimate of the diversity of insect symbionts should take into account diversity at the lower taxonomic (strains) or phylogenetic (operational taxonomic units [OTUs] at defined levels of sequence similarity) ranks that reveal underlying levels of interactions.
The Mediterranean fruit fly (medfly), Ceratitis capitata (Diptera: Tephritidae), is a polyphagous insect that seriously damages fruit production worldwide. Females oviposit in unripe fruits, within which the larvae develop, leading to fruit degradation and crop losses. Infestation can lead to total crop failure and to quarantine restrictions (9, 10).
Previous explorations of the medfly's associated bacterial community revealed that the gut microbiota is dominated largely by free-living bacteria of the Enterobacteriaceae, notably by species of Enterobacter, Klebsiella, and Pectobacterium (3, 11–14). These components of the bacterial community remain stable throughout the fly's life cycle and between geographical regions, and some are inherited vertically (3, 12, 14–16). However, significant genotypic variation underlies this apparent taxonomic stability, as a large microdiversity of the 16S rRNA gene marker is observed at the strain level, and alterations in strain distribution are observed during ontogeny and between individuals (3, 14). The very large majority of the symbionts composing this community is able to fix atmospheric nitrogen and to degrade pectin, thereby complementing the fly's nitrogen-deficient diet and possibly providing the necessary energy for nitrogenase activity (17). Nevertheless, alterations in the community structure occur within this functionally homogeneous population during ontogeny (3) and in response to seasonal or geographical changes (12). Despite these observations, albeit detailed, our knowledge of the dynamics of gut colonization during ontogeny is still fragmentary, lacking in both quantitative and qualitative information. Specifically, the colonization dynamics of the insect during development from egg to adult, including changes in microbial abundance, community structure, and community metabolism, have not been investigated.
Based our current knowledge, we hypothesized that the gut is colonized in a successional dynamic process expressed as the expansion of an initial, diverse inoculum, the structure of which is altered during ontogeny; these changes are expressed as shifts in molecular and phenotypic traits that provide metabolic plasticity to the microbiota and potential adaptive advantages to the host. To investigate the metabolic status and the spatial distribution of bacteria in general as well as of specific species within intact medfly host tissues, we implemented fluorescent in situ hybridization (FISH). In addition, to characterize the bacterial community structure during the fly's life cycle, we applied high-throughput sequencing, direct and plate count quantifications, and biochemical tests. We thus describe in detail the spatiotemporal changes in the genetic and phenetic structure of the medfly's gut microbial community and discuss the meaning of species diversity in the microbiota of the insect.
MATERIALS AND METHODS
Bacterial strains.
Enterobacter cloacae DSM 30054T (NCBI accession number AJ251469) and Enterobacter aerogenes DSM 30053T (NCBI accession number FJ971882) were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). Pseudomonas putida KT2400 (NCBI accession number AE015451), Escherichia coli ML35, Enterobacter sp. strain 143 (NCBI accession number AY847158), and Klebsiella oxytoca N8 (NCBI accession number EF645654) were from the laboratory's collection. All strains were maintained on Luria-Bertani (LB) growth medium (Difco). Isolates from larvae, decomposing fruit (rot), and adults were obtained by dilution plating of homogenates onto LB plates incubated at 30°C for 24 to 48 h.
Fly origins and rearing conditions.
Experiments were conducted using wild Ceratitis capitata flies collected from traps containing water and sugar (henceforth referred to as wild flies) or reared from infested fruit. Larvae and pupae were likewise reared from infested fruit. Both flies and fruits were collected at the experimental farm of the Faculty of Agriculture in Rehovot, Israel. Infested fruits were incubated over vermiculite in which the larvae could pupate. Pupae were collected into clear 5-liter cages in a controlled environment (light regimen of 16 h light/8 h dark, temperature of 25°C ± 1.5°C, and relative humidity of 65% ± 10%). Adults were maintained on sugar and solid yeast hydrolysate (sucrose-hydrolysate at a dilution of 3:1; MP Biomedicals, OH) ad libitum until they were used for experimentation. Eggs were obtained by placing surface-sterilized kumquat (Citrus japonica) fruits and female flies into cages, according to methods described previously by Behar et al. (3).
Quantification of total, culturable, and active bacterial populations in the medfly.
Culture-dependent and -independent methods were used to quantify the bacterial populations present at all life stages. The types of samples used (and numbers of samples [n] and individuals per sample [i]) were as follows: eggs (n = 7; i = 5), larvae (n = 6; i = 5), pupae (n = 5; i = 6), 1-day-old adults (n = 6; i = 2), wild adults of an unknown age (n = 10; i = 1), and 30-day-old adult medflies (n = 10; i = 1). The number of individuals per sample was set to provide consistent and replicable results. Adult medflies and larvae were surface sterilized in 70% ethanol for 2 min and then washed in phosphate-buffered saline (PBS). Guts were excised in PBS (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4·7H2O, and 1.8 mM KH2PO4 [pH 7.4]), using sterile forceps under a stereomicroscope (Zeiss, Germany). Pupae were surface sterilized by dipping into 70% ethanol and immediately transferred into sterile distilled water (DW). Excised guts, eggs, and sterilized pupae were transferred into 100 μl PBS and homogenized by using a sterile probe attached to a rotor stator homogenizer (Kimble Chase). The homogenate obtained from each sample was then used to quantify bacterial populations using both dilution plating for culturable bacteria and DAPI (4′,6-diamidino-2-phenylindole) staining for total bacterial cell counts. Serial dilutions were obtained by transferring 50 μl of the homogenized sample into 450 μl sterile DW, mixing the sample with a vortex stirrer, and plating 100 μl of each dilution onto solid LB agar. Direct (total) bacterial cell counts were performed under fluorescence according to a method detailed previously by Ben-Yosef et al. (18). Briefly, Teflon-laminated slides with 6-mm-diameter wells (Electron Microscopy Sciences, Washington, PA) were coated with a 0.1% gelatin solution (Merck) and 0.01% chromium potassium sulfate (Sigma-Aldrich, WI). Four microliters of each homogenized sample was spread within the boundaries of a well. For the quantification of total bacteria, samples were stained directly with 20 μl DAPI (4.6 μg ml−1). After the removal of excess DAPI, samples were mounted with an antifading solution (Citifluor, Canterbury, United Kingdom). One hundred to six hundred bacterial cells were counted in 7 to 50 randomly chosen fields from each slide by using an eyepiece micrometer with a 1-cm diameter and a 100× objective (UPlan Fl 100×/1.30 numerical aperture) mounted onto a BX51 epifluorescence microscope (Olympus, Japan). In addition, the concentrations of total and metabolically active bacterial populations in wild adult midguts (n = 7) were compared, using the same individual flies. Samples were hybridized with 20 μl of the Bacteria-specific probe EUB338-Cy3 (19) prior to counterstaining with DAPI, according to the hybridization method specified below. The average number of bacterial cells per field was used to calculate the total number of bacteria per specimen by using the following formula:
where 28.26 mm2 is the well area, 0.01 mm2 is the micrometer area as seen through the microscope, 100 μl is the volume in which the whole sample was homogenized, and 4 μl is the volume spread onto the slide.
When more than one specimen per sample was used, the above-described result was divided by the number of specimens.
Probes used in this study.
All probes used in this study targeted 16S rRNA. Probe conditions and specifications are detailed in Table S1 in the supplemental material. For probes EUB338 (GCTGCCTCCCGTAGGAGT [E. coli positions 338 to 355] [19]), Ppu646 (CTACCGTACTCTAGCTTG [E. coli positions 646 to 663] [20]), and EnterbactD (TGCTCTCGCGAGGTCGCTTCTCTT [E. coli positions 1251 to 1274] [21]), E. coli ML35, P. putida KT2400, and Enterobacter cloacae DSM 30054T and Enterobacter aerogenes DSM 30053T, respectively, were used as controls for calibrating hybridization conditions, using the conditions detailed in the corresponding reports cited above. Additional probes targeting specific subpopulations present in the medfly were designed.
Oligodeoxynucleotide probe design.
Probes distinguishing between Klebsiella and Enterobacter, two taxa commonly detected in medflies (3, 13, 16), were designed based on 140 partial 16S rRNA gene bacterial sequences, including sequences derived from medfly (GenBank accession numbers AY847157 to AY847186, DQ533879 to DQ533882, DQ533884 to DQ533889, DQ533891 to DQ533895, DQ533897, DQ533898, EF117864, EF117865, EF645648 to EF645662, EU026215, DQ533883, and DQ533896), using the 2009 SILVA bacterial 16S rRNA database (22), which is available online (http://www.arb-silva.de/); the ARB software package (23); and the ARB probe design tool. The probes were optimized to centralize mismatches between the probes and nontarget organisms (24), and accessibility was examined as described previously by Behrens et al. (25). Target specificity was confirmed in silico by using our database as well as the probe match tool in RDP-II (26) (http://rdp.cme.msu.edu/). The result was an “Enterobacter” probe, 6-carboxyfluorescein (FAM6)-Ent615 (AGACTCTAGCCTGCCAGTTT [E. coli positions 615 to 634]), complementary to 90% of the assigned Enterobacter, Citrobacter, and Pantoea sequences from the medfly bacterial database, containing at least one mismatch with sequences assigned as other Enterobacteriaceae species and a “Klebsiella” probe, Cy3-Kox615 (AGACTCCAGCCTGCCAGTTT [E. coli positions 615 to 634]), complementary to 50% of the assigned Klebsiella sequences from the medfly database, containing at least one mismatch to sequences assigned as non-Klebsiella species. In order to prevent probe nontarget hybridization, the FAM6-Ent615 and Cy3-Kox615 fluorescent oligonucleotides were each used together with the nonfluorescent competitor probes cEnt615 and cKox615, respectively (27). Enterobacter sp. 143 and K. oxytoca N8 were used as controls.
FISH.
FISH was used to localize bacteria within the medfly host tissue. Cells were fixed according to methods described previously by Amann et al. (19). Briefly, cells were mixed with cold 4% paraformaldehyde fixative at a ratio of 1:1 (vol/vol) and incubated overnight at 4°C. Cells were pelleted by centrifugation (Hettich Centrifuges) for 10 min at 4°C at 6,000 rpm, and pellets were resuspended in cold PBS. After pelleting again under the same conditions, pellets were resuspended in 50% ethanol in PBS. Samples were stored at −20°C for a maximum of 3 months. Cells were spread onto gelatin-coated slides, as described above.
Fly tissue fixation was carried out according to methods detailed previously by Fukatsu et al. (28) and Gottlieb et al. (29). Eggs, larvae, and wild-type (WT) adults or adults reared from infested fruit were collected into Carnoy's fixative (chloroform, ethanol, and glacial acetic acid at a dilution of 6:3:1 [vol/vol]) 1 day, 1 week, 10 to 15 days, or 30 days following eclosion and incubated at room temperature for 48 h. After fixation, the specimens were decolorized in a 6% hydrogen peroxide solution (30% H2O2 and ethanol at a ratio of 3:50 [vol/vol]) for 48 h. Specimens were then stored in 50% ethanol for up to 2 months. Guts were excised from fixed specimens directly prior to in situ hybridization. Fixed tissue was placed onto a gelatin-coated slide, and specimens were immersed in PBS to prevent them from drying. A hydrophobic barrier pen (Vector Laboratories, CA) was used to contain the specimen, and subsequent solutions were added to the slide.
Optimal FISH conditions were determined with positive- and negative-control strains obtained from the DSMZ or from laboratory stock, as specified above. Probes were also tested on medfly gut homogenates to ensure probe suitability for gut bacteria. Hybridization conditions for probes Kox615 and Ent615 were determined at 46°C with increasing formamide concentrations from 20 to 35% with 5% increments, various hybridization times, and competitor probe/targeted probe ratios of 1:1, 7:3, and 3:2. Probes Cy3-Kox615/cKox615 and FAM6-Ent615/cEnt615 were used at ratios of 1:1 and 2:3, respectively. Finally, probes Cy3-Kox615 and FAM6-Ent615 were used simultaneously at a ratio of 3:7 in hybridizations that lasted 2 h in a hybridization buffer containing 35% formamide.
FISH was performed according to methods described previously by Manz et al. (30): hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.2], 5 mM EDTA, and 0.01% SDS) with the appropriate concentration of deionized formamide (see Table S1 in the supplemental material) and fluorescently labeled oligonucleotide probes at a concentration of 10 pmol/ml were added to the samples at a ratio of 30:1, respectively. Hybridization reactions were performed in a humidity chamber within a hybridization oven (Shel Lab, OR) at 46°C for 2 to 3 h (see Table S1 in the supplemental material), and samples were then washed (102 mM NaCl, 20 mM Tris-HCl [pH 7.2], 10 mM EDTA, 0.01% SDS) for 30 min at 48°C, counterstained with DAPI (1.75 mg · ml−1) or Sybr green (10,000× in dimethyl sulfoxide [DMSO], diluted 1:50 [vol/vol]) (Sigma-Aldrich, WI) for 10 min, mounted in Citifluor (Citifluor, United Kingdom), covered with coverslips, and retained at −20°C until use. No-probe controls were also prepared as a control for sample autofluorescence.
Samples were visualized by using a BX51 epifluorescence microscope with a 100× objective (UPlan Fl 100×/1.30). The fluorochromes Cy5, DAPI, Cy3, and Sybr green/FAM6 were viewed with U-MW1Y2, U-MWU2, U-MNG2, and U-MNB2 mirror units (Olympus, Japan), respectively. Fields were photographed by using a charge-coupled-device (CCD) camera (Digital Sight cooled monochrome; Nikon, Japan) and NIS-elements, edition 3.0, software (Nikon, Japan). Samples were viewed by using a 405-nm diode laser for DAPI, a 488-nm argon ion laser for Sybr green and FAM6, and a 543-nm helium-neon ion laser for Cy3, under a FluoView FV100 (Olympus, United Kingdom) or an IX-81 inverted (Olympus, Japan) confocal microscope, using FV-Viewer or FluoView 500 software, respectively.
Biochemical diversity analyses and amplified rRNA gene restriction analysis.
All biochemical media were acquired from Difco Laboratories (Detroit, MI), with the exception of methyl red (MR)–Voges-Proskauer (MRVP) broth (BBL), which was acquired from Becton Dickinson (Franklin Lakes, NJ). Pure cultures were individually inoculated into the following sterile media: motility indole ornithine (MIO) semisolid agar deeps; Simmons citrate slants; urease broths; carbohydrate broths containing either 10% glucose, lactose, sucrose, or maltose in a phenol red broth base medium; and MRVP broths. Media were incubated at 24°C. Results were recorded after 24 h, with observable turbidity for citrate, urease, and carbohydrate media. MIO tubes were observed for color reactions after 24 h and reincubated for an additional 24 h, a prerequisite for determining the ornithine decarboxylation reaction. Indole production was determined following the addition of Kovac's reagent (Sigma-Aldrich, St. Louis, MO). MRVP media were incubated for 5 days, as recommended by the manufacturer.
Mixed-acid fermentation was determined by adding MR to half of the MRVP culture in separate test tubes. Butylene glycol fermentation (Voges-Proskauer [VP]) was determined by lysing cells in the remaining volume with 40% potassium hydroxide (Sigma-Aldrich) and reacting cytoplasmic contents with alpha naphthol amine (Sigma-Aldrich). All isolates were additionally streaked onto tryptic soy agar plates (Difco) and incubated at 10°C for 24 to 48 h. Diazotrophy (12) and pectinolysis (3)were detected on specific media as described previously by Behar et al.
Amplified rRNA gene restriction analysis was performed as described previously by Ben Ami et al. (13). Briefly, isolates were grouped according to restriction patterns using the enzymes HaeIII (TaKaRa, Japan), RsaI (Fermentas), and EcoRI (TaKaRa, Japan), and a few representatives per group were sequenced for the 16S rRNA gene (Macrogen Inc., Seoul, South Korea). Contigs of the two matching PCR products were assembled by using DNA Baser v2.0.3.24 (Heracle Software, Germany).
Statistical analyses.
The parametric analysis of variance (ANOVA) test, the post hoc Tukey honestly significant difference (HSD) test, and the nonparametric Kruskal-Wallis test were used on bacterial quantification data. The Kruskal-Wallis one-way ANOVA stepwise step-down multiple-comparison test was used to compare bacterial population levels in the different medfly stages. Statistical analysis was carried out by using JMP 7.0 (SAS Institute, NC) or PASW Statistics 18 (SPSS, IL) software.
Pyrosequencing of bacterial 16S rRNA genes.
High-throughput sequencing was performed in order to obtain the species composition of the bacterial community present in larval and adult medflies. Adults were caught on the same apricot tree from which fruits were sampled to obtain the larvae for the experiment, at the experimental farm of the Faculty of Agriculture in Rehovot. The trap was baited with biolure baits, and the flies were sustained on sugar and water until they were collected (less that 24 h). All samples were preserved in 95% ethanol at −20°C until they were processed. The midgut was extracted from five individual adult females, and whole guts from 15 larvae were pooled as three samples. Extracted tissue was subsequently washed in PBS to remove debris and any bacteria not originating in guts. Guts were then homogenized in 100 μl PBS, as described above. Following homogenization, an additional 100 μl of PBS was added to each sample.
Bacterial DNA for PCR was extracted from the samples mentioned above by using the Chemagic DNA Bacteria kit (Chemagen, Germany), according to the manufacturer's instructions. Extracted DNA was rehydrated in 50 μl elution buffer and stored at 4°C for up to 1 month.
A fragment of the 16S rRNA gene was amplified by using primer pair 926F-1392R (E. coli numbering). Pyrosequencing was carried out by the Research and Testing Laboratory (Lubbock, TX), using a Roche 454 FLX genome sequencer system and FLX Titanium reagents (Roche Applied Science, IN) (for additional PCR and sequencing conditions and methods, see reference 31).
Community analysis based on pyrosequencing data.
The 16S rRNA gene reads obtained by pyrosequencing were cleaned by using MOTHUR v1.24 (32). First, FASTA and quality data were extracted from the raw SFF file. Sequences were grouped according to barcode and primer, allowing one mismatch to the barcode and two mismatches to the primer. Denoising was achieved by the AmpliconNoise algorithm (33), removing both 454 sequencing errors and PCR single-base errors. Next, sequences were trimmed to remove barcode and primer sequences, all sequences with homopolymers (i.e., AAAA) longer than 8 bp, and all sequences <200 bp long. All sequences were aligned to the SILVA reference alignment database (32) and filtered so that they all overlapped perfectly (with no overhang or no-database pairs), resulting in 200-bp-long sequences (positions 926 to 1125 [E. coli numbering]). To further reduce sequencing errors, preclustering of the sequences was performed, based on methods described previously by Huse et al. (34). Finally, chimeric reads were removed with the UChime method (35), and all chloroplast, mitochondrion, and “unknown” (i.e., unclassified at the kingdom level) reads were deleted. Pairwise distances were calculated between all DNA reads, which were subsequently clustered into OTUs (operational taxonomic units) at the 0.03 and 0.02 levels, meaning that sequences that displayed >97% and >98% similarities, respectively, with each other were considered the same OTU. Each OTU was phylotyped based on current RDP-II taxonomy (26). The resulting OTUs were arranged into a data matrix where each row was a single sample and each column was a specific OTU; each data point in the matrix represented the abundance of the particular OTU in the particular sample, relativized to the sampling effort (i.e., the number of 454 reads obtained from that sample). Multivariate cluster analysis was performed with PC-ORD v5.32 (MjM Software), with Sorensen distances and flexible beta linkages (β = −0.25).
Phylogenetic analysis.
A detailed phylogenetic analysis of individual genera was performed by using MEGA5 (36), by compiling all the 454 reads from a specific genus or OTU together with all long (>1,200-bp) type sequences of a comparison taxon (i.e., Enterobacteriaceae) available in the RDP-II database. Sequences were aligned by MUSCLE (37), and phylogenetic trees were inferred by the maximum likelihood method based on the Tamura-Nei model (38), such that only the tree with the highest log likelihood is shown. Bootstrap values, i.e., the percentage of trees in which the associated taxa clustered together out of 500 repeats, are shown next to the branches. Trees are drawn to scale, with branch lengths measured as the number of nucleotide substitutions per site.
Nucleotide sequence accession numbers.
Sequences used in the present study are available in the GenBank database under accession numbers DQ533879, DQ533884, DQ53387 to DQ533889, DQ533894 to DQ533897, EF645649, and EF645650. 454 reads are available via the MG-RAST server (http://metagenomics.anl.gov/linkin.cgi?project=2348).
RESULTS
Quantification of metabolically active bacteria.
EUB338 is a Bacteria-specific probe which hybridizes to the 16S rRNA gene. Therefore, signal visibility is dependent on the ribosome number, which in turn depends on metabolic activity (39). To determine the proportion of metabolically active bacteria in the gut, the fraction of total gut bacteria (DAPI stained) containing the appropriate ribosomal content necessary to produce strong signals using Cy3-EUB338 was measured. The total numbers of bacteria (DAPI-stained cells) and of active cells (emitting both a DAPI signal and a EUB338 signal) in the gut were significantly different (F = 11.01; P < 0.016; degrees of freedom [DF] = 1; R2 = 0.98 [by ANOVA]) (see Fig. S1 in the supplemental material). About two-thirds emitted both signals (73%, or 2.86 × 106 out of a total of 3.95 × 106 bacteria · gut−1 [n = 7]), indicating that a large majority of bacterial cells in the gut are engaged in high metabolic activity.
Quantification of bacterial populations during ontogeny.
Both cultivation-dependent and cultivation-independent methods were applied to quantify bacteria present at all stages of the medfly life cycle—eggs, pupae, the larval gut, and the adult midgut—using midguts of both wild adult flies and laboratory-reared flies 1 day or 30 days following eclosion.
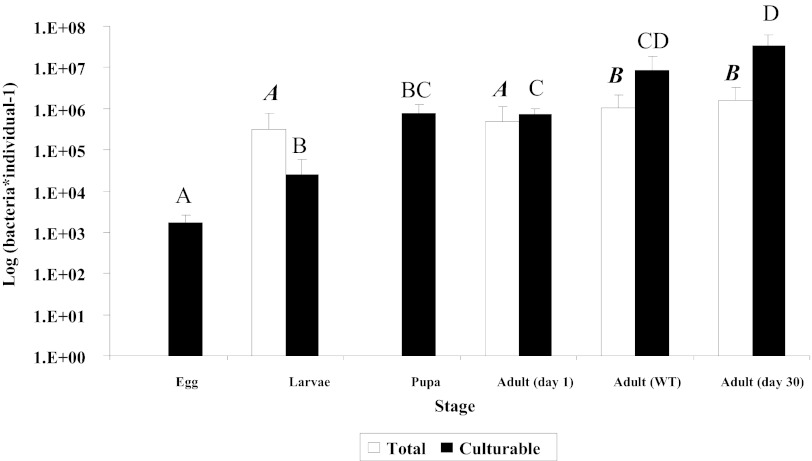
Cultured bacterial population levels changed significantly during ontogeny. Eggs, larvae, adults 1 day following eclosion, and adults 30 days following eclosion belonged to different homogenous subsets (Fig. 1). The pupa subset was similar to the subsets of both larvae and 1-day-old adults. The wild-adult subset was similar to the subsets of both 1-day-old and 30 day-old adults (chi square = 28.33; P < 0.0001; DF = 5; median = 6.6 × 105 cells · midgut−1 [by stepwise Kruskal-Wallis test]). While not as discriminative as culture-based quantification, direct counts yielded similar results (Fig. 1): larvae and 1-day-old adults shared a single subset, which was significantly different from a single subset containing wild adults and 30-day-old laboratory-reared adults (chi square = 9.89; P < 0.02; DF = 3; median = 8.1 × 105 cells · midgut−1 [by stepwise Kruskal-Wallis test]). Low population levels and strong autofluorescence precluded reliable analysis by the direct counting of bacterial cells on the fly eggs and in pupal samples, respectively.
Fig 1.
Culturable and total bacteria per individual medfly during consecutive developmental stages. Black and white columns represent culturable bacteria, as enumerated by dilution plating, and total bacteria, as counted by microscopy, respectively. Midgut bacteria in adults and larvae were counted. Developmental stages not connected by the same letter are significantly different. Boldface italic type refers to white columns, and lightface type refers to black columns. Error bars represent standard deviations.
Spatial and temporal distributions of gut bacteria in eggs, larvae, and adult guts.
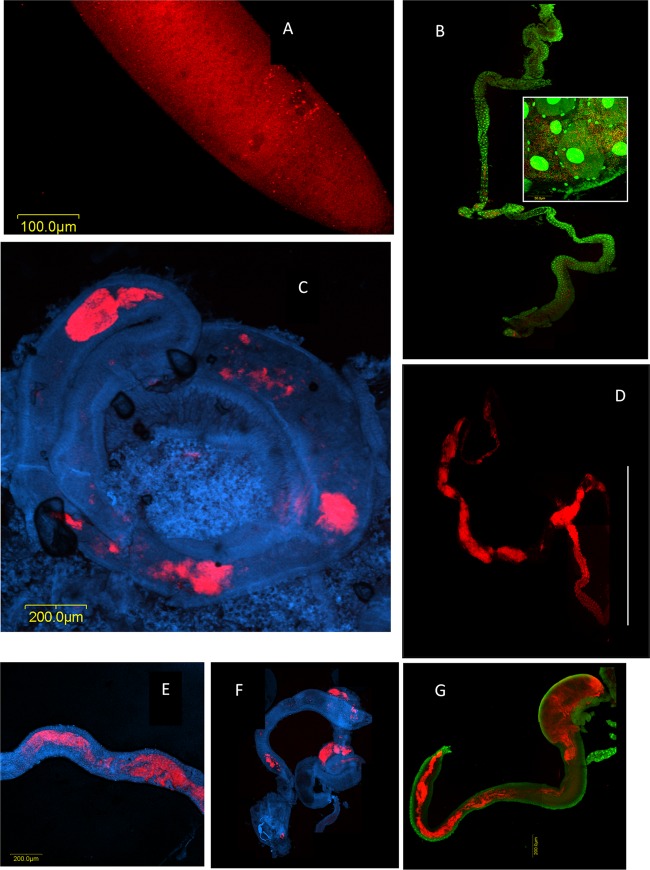
Bacteria were detected only on the surface of eggs that were laid in fruits (Fig. 2A). Signals were not detected from inner egg layers, from eggs that had been laid under sterile condition, or from eggs extracted from the female ovary (by FISH, culture, and 16S rRNA gene amplification) (data not shown). Bacteria-specific signals were detected in midguts of all specimens examined, regardless of age. Signal intensities and distributions, however, varied between individuals of different ages. In larvae, bacterial cells formed dense patches along the gut (Fig. 2B, inset). Midguts of adults on their first day of eclosion were likewise characterized by a patchy distribution (Fig. 2C). In contrast, 30 days following eclosion, intense and contiguous signals were obtained from the midgut, indicating that it was colonized by a dense and confluent mass of bacterial cells (Fig. 2D; see also Fig. S3 in the supplemental material). Accordingly, the extents of colonization of midguts from wild specimens (of unknown ages) varied. In some specimens, colonization was patchy and corresponded to that of 1-day-old adults (Fig. 2F); in others, a more continuous bacterial mass similar to that of 30-day-old adults was observed (Fig. 2G); and in still others, an intermediate extent of colonization was found (Fig. 2E), similar to that observed for 13-day-old flies (see Fig. 4A).
Fig 2.
Bacterial colonization of the medfly during ontogeny, tracked by fluorescent in situ hybridization and confocal microscopy using a Bacteria-directed Cy3-EUB338 oligonucleotide probe applied on laboratory-raised flies and DAPI or Sybr green nucleic acid stain. (A) Egg. Microcolonies on the surface of a medfly egg appear as red dots on the red fluorescent background of the egg chorion. (B) Larval gut. Bacterial growth appears as discontinuous patches along the larval gut. Red, bacterial masses; green, fly gut cell nuclei. (Inset) Within these patches, the bacterial density is high. Red, bacterial cells/microcolonies; green, bacterial cells and fly gut cell nuclei. (C) Adult medfly midgut 1 day after eclosion. Red, bacterial masses. (D) Midgut and hindgut of a laboratory-reared female adult medfly 30 days after eclosion. Continuous bacterial colonization is seen in the midgut as well as in the hindgut. Red, bacterial masses. The line encompasses the hindgut. (E to G) Midguts of field-captured wild specimens showing various extents of gut colonization. (E) Female; (F and G) males. Red, bacterial masses; blue or green, gut cells.
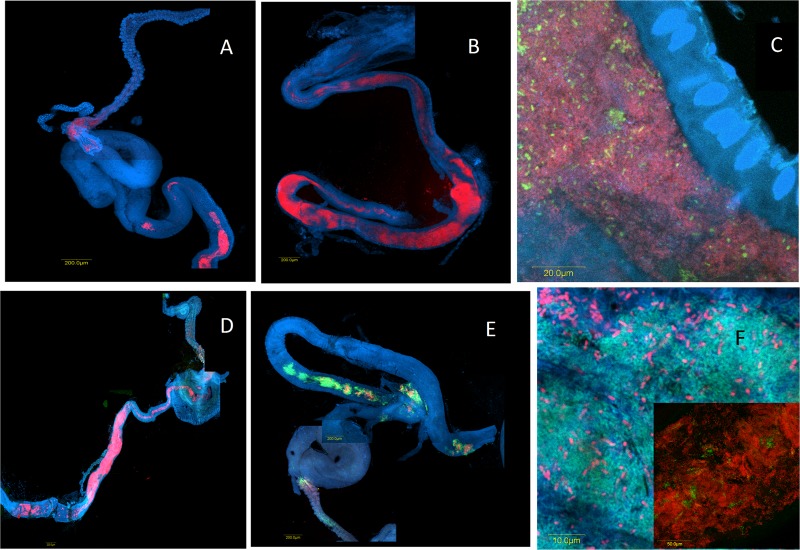
Fig 4.
Colonization of laboratory-reared flies at 13 days (A and C) and 30 days (B and D) following eclosion and of wild-caught flies (E and F) with bacterial populations hybridizing with oligonucleotide probes. (A and B) Klebsiella (Cy3-Kox615) (red); (C) Enterobacter (FAM6-Ent615) (green) and Bacteria (Cy3-EUB3380 (red); (D to F) Enterobacter (FAM6-Ent615) (green) and Klebsiella (Cy3-Kox615) (red). Blue indicates staining by DAPI (nucleic acid stain).
Spatially, most bacteria were located in the central part of the midgut; the density of the bacterial populations gradually decreased toward the foregut or the hindgut. Signals obtained from hindguts were less uniform than those obtained from the midgut and varied between individual flies, as significant bacterial colonization was detected in only several of the tens of specimens examined (Fig. 2D). An examination of the gut wall and midgut lumen revealed clear differences in microbial density. The lumen was densely populated, yet the cell density appeared to be abruptly reduced at the epithelial midgut cell lining (Fig. 3; see also Fig. S3 in the supplemental material), as no bacteria were seen adhering to it. Therefore, the bacterial community appears to be confined to the central portion of the gut by the peritrophic membrane.
Fig 3.
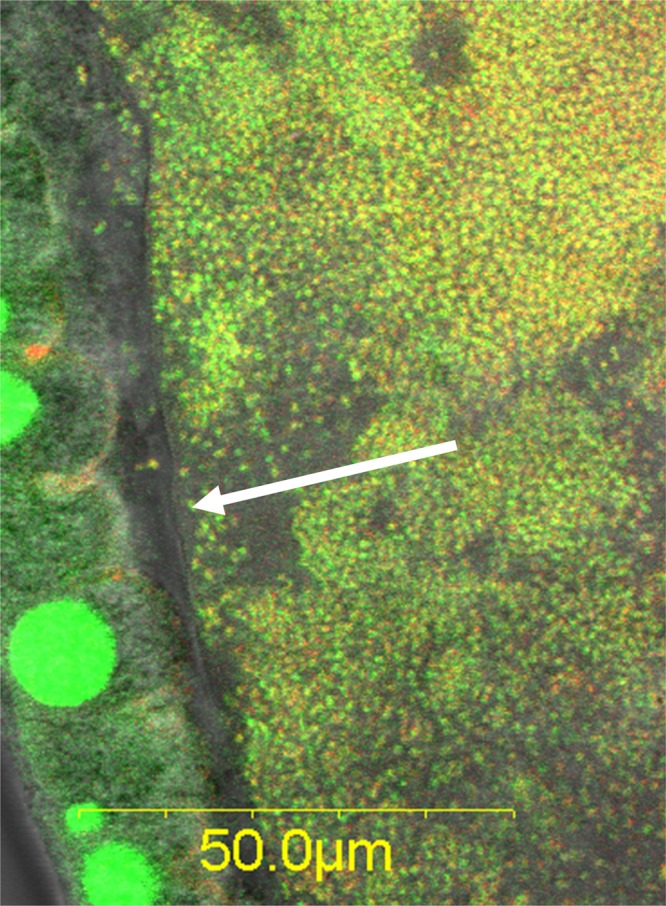
The microbiota is contained in the gut lumen by the peritrophic membrane (arrow), as seen in a wild male. Green, Sybr green nucleic acid stain; red, Cy3-EUB338 Bacteria-specific probe. Bacteria stained with both Sybr green and Cy3 appear yellow. Large green spots are epithelial cell nuclei.
Distribution of specific taxa in larvae and adult guts.
The spatial distribution of various specific bacterial taxa was examined in situ by using specific probes. In the midguts of wild medflies, cells hybridizing with an Enterobacteriaceae-targeted 16S rRNA probe were largely dominant (see Fig. S4 in the supplemental material). When a Klebsiella-specific probe was applied, the dynamics of gut colonization observed for laboratory samples (Fig. 4A and B) were similar to those obtained with a general Bacteria-directed probe (Fig. 2). The Enterobacter-specific FAM6-Ent615 probe, however, yielded only sparse signals, suggesting that these are relatively minor populations (Fig. 4C). This was confirmed when both probes were used together (Fig. 4D; see also Fig. S2 in the supplemental material). However, in wild-caught flies, both groups could form large populations, depending on the individual fly (Fig. 4E and F, inset; see also Fig. S5 in the supplemental material). Microcolonies hybridizing to a pseudomonad-specific probe were observed in one wild-caught specimen (see Fig. S6 in the supplemental material). This is not surprising, as these populations, although persistent in the medfly, exist in very low abundances (40), rendering their in situ detection difficult.
Biochemical diversity analysis.
Phenotypic analysis was performed on 134 isolates from larvae, fruit rot, and 1-day-old adults using 14 biochemical markers, including nitrogen fixation and pectinolysis. This resulted in 19 subgroups differing by at least one parameter (Table 1). Amplified rRNA gene restriction analysis was performed on 67 isolates, revealing 4 different patterns spread over the different subgroups (Table 1). The very large majority of the isolates was pectinolytic (9.7%), diazotrophic (26.9%), or both (56%). The largest phenotypic diversity was observed for the fruit rot samples, which harbored 13 subgroups, followed by 1-day-old adults (6 subgroups) and pupae (4 subgroups). Phenotypic subgroups of the former overlapped subgroups of the two latter age classes, suggesting that strains graduate from one life stage to the next. In each developmental category, one to two subgroups were dominant, representing >45% of the isolates.
Table 1.
Phenotypic groups formed by analyzing 134 isolates originating from fruit rot (53 isolates), pupae (51 isolates), and 1-day-old adults (30 isolates)a
| Detection of marker |
% of isolatesb | Organism(s) determined by amplified rRNA gene restriction analysisc | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N + P | N | P | Ind | Orn | Mot | Glu | Suc | Lac | MR | VP | Urea | Cit | Mal | GLT | ||
| + | − | − | − | + | + | − | + | − | D+ | + | − | + | 39.2 + 1.9 | I, II, III, IV | ||
| + | − | − | − | − | + | + | − | + | − | D+ | + | − | + | 45.1 | ||
| − | + | − | − | − | + | + | − | + | − | D+ | + | − | + | 9.8 | ||
| − | − | − | − | − | − | + | + | − | + | − | D+ | + | − | + | 5.9 + 1.9 | |
| + | + | − | + | + | − | + | + | − | − | − | − | + | 7.5 | II | ||
| + | − | − | + | + | − | − | + | − | WP | + | − | + | 9.4 | II | ||
| + | − | − | + | + | − | − | + | − | WP | − | − | + | 3.8 | II, IV | ||
| + | − | − | + | + | + | − | − | + | − | + | + | + | 3.8 + 3.3 | I | ||
| + | − | − | − | + | + | + | − | − | + | − | + | + | + | 3.8 | ||
| − | − | − | − | − | + | + | + | − | − | + | − | + | + | + | 3.8 | |
| + | + | − | − | + | + | + | − | + | − | + | + | + | 11.5 + 60 | I, II, III, IV | ||
| + | − | + | − | − | + | + | + | − | + | − | + | + | + | 16.6 + 5.7 | ||
| − | + | + | − | − | + | + | + | − | + | − | + | + | + | 10 | ||
| − | − | − | + | − | − | + | + | + | − | + | − | + | + | + | 5.7 | |
| + | − | − | + | + | − | − | + | − | D+ | + | − | + | 28.3 | I, II, III, IV | ||
| + | − | − | − | + | + | − | − | + | − | D+ | + | − | + | 5.7 | ||
| − | + | − | − | + | + | − | − | + | − | D+ | + | − | + | 7.5 | ||
| − | + | − | − | + | + | + | − | − | + | − | + | − | + | 6.6 | ||
| + | + | − | − | + | − | − | + | − | D+ | + | − | + | 3.3 | I | ||
N, atmospheric nitrogen fixation; P, pectinolysis; Ind, indole; Orn, ornithine; Mot, motility; Glu, glucose; Suc, sucrose; Lac, lactose; MR, methyl red; VP, Voges-Proskauer broth; Cit, citrate; Mal, maltose; GLT, growth at low temperature (10°C, recorded on day 5); D+, delayed positive (≥48 h); WP, weakly positive/slight color change at 72 h.
Percentage of isolates from each growth stage (boldface type indicates fruit rot, lightface type indicates pupae, and italic type indicates 1-day-old adults) that match a particular phenotypic profile.
I, Klebsiella oxytoca; II, Pantoea endophytica; III, Pectobacterium cypripedii; IV, Providencia stewartii.
High-throughput analysis of bacterial populations in larval and adult medflies.
16S rRNA gene sequence analysis of DNA extracted from five adult female midguts and from three samples of pooled larva guts was performed by using high-throughput sequencing, providing a detailed landscape of the fly bacterial community. Coverage was complete, standing at values of 0.99 and 1.00 for adult and larval samples, respectively. In adults, 5,000 to 12,000 sequences were obtained per sample, distributed between 5 and 23 OTUs (at 97% similarity). Larval samples yielded 1,700 to 3,200 sequences that included 7 to 13 OTUs. The same single dominant OTU, seemingly related to Kluyvera, prevailed in adults and larvae (samples 1 to 5 and 24 and 25, respectively), composing 81.0 to 99.7% and 99.7% of all sequences, respectively, except for one larval sample, in which a Leuconostoc-related OTU was dominant. A second-most dominant OTU (0.3 to 19% of sequences), found in two adults (samples 3 and 5), was related to the enterobacterium Buttiauxella. Surprisingly, when sequences were clustered into OTUs at a higher (>98%) similarity level, adults and larvae no longer shared the same dominant OTU but were neatly divided between a single “adult” strain (which is dominant in all adults and absent in all larvae) and a single “larval” strain (which is vice versa) (see Fig. S7A and S7B in the supplemental material).
This apparent restricted sequence diversity belied extensive taxonomic diversity: a phylogenetic tree (see Fig. S7C in the supplemental material) built using representative sequences from the dominant Kluyvera-related OTU (defined at a 97% similarity) as well as sequences from defined type strains of enterobacteria included more than 20 enterobacterial genera, most of which appeared in previous analyses performed on isolated strains or on cloned sequences originating from the medfly (3, 11). Thus, due to the taxonomic ambiguity of Enterobacteriaceae and the short length (200 bp) of 454 reads, we cannot determine the enterobacterial genus to which the dominant OTU belongs. Furthermore, when the 200-bp consensus sequence of this OTU was used against the NCBI database to retrieve long (>1,000-bp) 16S rRNA gene sequences at 99% similarity, 370 sequences were obtained. A classification of these sequences based on an alignment of 1,000 bp showed that they formed 207 and 157 OTUs at 99% and 97% similarity levels, respectively; this represents more than a 10-fold increase in OTU diversity, as opposed to the same 370 sequences when cropped to the “original” 200-bp length, confirming that the constrained phylogenetic diversity that we found in the medfly bacterial population was, at least in part, due to the technological bias imposed by 454 read lengths.
DISCUSSION
The transgenerational acquisition of symbionts by insects can occur through a number of mechanisms, such as the direct transfer of intracellular symbionts (e.g., in Buchnera in aphids [e.g., see reference 41]), mother-given “birth presents” of extracellular symbionts (e.g., “Candidatus Ishikawaella” in plataspid stinkbugs [42]), or environmental acquisition from various sources (e.g., Burkholderia in the bean bug [6]). Direct transfer and birth presents are correlated with symbiont-host congruence (43), but symbionts acquired from the environment at each generation do not form coherent groups (44). The Mediterranean fruit fly, Ceratitis capitata, seems to be a “hybrid” case in that at least part of the insect's microbiota is inherited during oviposition as birth presents (3), but as described in this study and in previous studies (see reference 12 and references therein), its community structure is complex and variable. Sources of variation may be internal, originating from different compositions of the birth presents, or external, originating from the acquisition of different environmental bacteria. Bacteria provided in the adult diet easily establish in the fly's gut (13, 16), and specific vertically acquired subpopulations expand during ontogeny (Fig. 2).
High-throughput sequencing of larvae and adult females was performed to examine if the fly's gut microbiota community structure shifts during ontogeny (as previously suggested by Behar et al. [3]). In all samples and at a 97% similarity, one OTU related to the Enterobacteriaceae largely dominated the fly microbiota. However, at a 98% similarity, the larval and adult communities clustered separately (see Fig. S7B in the supplemental material). This means that larvae and adults harbor different members within the Enterobacteriaceae, which, although distinct, are very similar taxonomically. One larval sample was dominated by Leuconostoc, a lactic acid bacterium that can be associated with postharvest fruit decay (45). The origin of this population is not known, but it was postulated previously that medfly larvae can acquire environmental symbionts from rotting fruit in which they develop (3). Changes in bacterial community composition during ontogeny were also apparent from the biochemical analyses, as 19, mostly life-stage-specific, phenotypic subgroups were defined. However, they could not be related to any particular enterobacterial taxon. The polyphyly of the Enterobacteriaceae (46, 47) precludes the taxonomic resolution of either a crude amplified rRNA gene restriction analysis (Table 1), a pyrosequencing approach (see Fig. S7C in the supplemental material), or the design of species-specific probes (see Materials and Methods). This discrepancy may be due to lateral gene transfer, which is extensive in Enterobacteriaceae (48) and enhanced in dense populations. A recent study by Stecher et al. (49) showed that inflamed mammalian guts support higher densities of Enterobacteriaceae than healthy guts; in turn, they engage in more frequent lateral gene transfer. High densities of active enterobacterial populations during all of the fly's life stages, i.e., from rotting fruit to the adult, may thus be conducive to gene transfer and lead to local diversification, providing yet another source of phenotypic diversity. In lygaeid and coreoid stinkbugs as well, a single genotype of environmentally acquired Burkholderia symbionts dominates the insect gut microbiota, yet an underlying phenotypic diversity is also present, as different morphotypes can be cultivated from this seemingly homogenous population (44).
The microbe-encoded genetic diversity of such symbioses is part of the hologenome, the sum of the genetic information of the host and its microbiota; accordingly, it may provide benefits for host adaptation and evolution (50). Recent studies have shown that specific variants of symbionts rapidly spread in insect populations to provide adaptive benefits in response to the environment. For example, insecticide-degrading Burkholderia strains rapidly dominated bean bug microbial communities in fenitrothion-treated soil (6); Rickettsia bellii swept through Southern U.S. whitefly (Bemisia tabaci) populations, increasing host fertility, survival, and the fraction of female progeny (51). Such specific takeovers may be enhanced by host behavior, as in Drosophila melanogaster, where individuals preferentially mated with partners that fed on the same type of food and harbored the same type of gut microbes (52).
The phenotypic plasticity of the medfly's microbiota, as reflected in the variation of almost all phenotypic traits examined, may provide local adaptive advantages in changing environments, such as the extended array of fruits greatly varying in acidity and protein content in which it can feed and develop (53) and the different climates in which it is found (12). Whatever the environment, diazotrophy and pectinolysis may fill “core functions”: they were found in almost all isolates, from all stages (92.6%), a figure nearly identical to that obtained from rotting fruits in a previous study (3), and are rendered essential for the maintenance of the holobiont by the medfly's diet, as they link carbon/energy sources to nitrogen acquisition in an insect that feeds on nitrogen-poor sources (9). Other, as-yet-unknown traits may similarly be vastly conserved across the diverse constituents of the medfly's microbiota.
The female medfly provides larvae with a “survival pack” consisting of diverse pectinolytic and nitrogen-fixing Enterobacteriaceae that are amplified by the host fruit and subsequently maintained throughout the medfly's life (3). At least part of this inoculum was attached to the egg surface, where it formed microcolonies (Fig. 2) (see reference 16), totaling about 1,700 cells per egg. Although ovarian transmission was previously reported (16), we found the eggs to be devoid of bacteria. In the female medfly, the digestive and reproductive systems share an outlet (54), so that eggs are smeared during oviposition. Thus, the bacterial inoculum reaches the fruit through different possible paths: first, ovarian transmission; second, direct fecal contamination of the ovipositor and deposition by mechanical contact; and, third, smearing of the eggs. Subsequently, the microbiotas that develop in the fruit and in the larva provide the inocula that will, in turn, colonize the pupa and then the adult gut. These inocula can be affected by stochastic effects, dispersal limitation (especially for low-abundance populations), historical contingency (the order of arrival), and environmental selection (55). Parameters that could affect transmission and selection include, not exclusively, temperature, humidity, diet, and host-driven (fruit and insect) effects. In the medfly, previous oviposition by a conspecies, as well as fruit size, affects clutch size, i.e., the number of eggs oviposited per site (56, 57). Thus, any of these effects may lead to strikingly different abundances of specific subpopulations at each stage (as observed for adult flies captured at the same location) (Fig. 4).
The reduced size of vertically transmitted symbiont populations creates a bottleneck that can affect population structure and evolution (58) and increase relatedness and intrahost cooperation among symbionts (59). If the size of the population is too small, infection may be lost, while if it is too large, it may hurt the host (60). However, this population is often mixed, as this and other studies (e.g., see reference 61) have shown. Therefore, some mechanisms should exist to overcome homogenization while sieving out unwanted colonizers. Such mechanisms may be at play in the medfly: as stated above, bacteria introduced by feeding established in the fly gut (13, 16) and could be transmitted to the eggs and through two generations (16). These experiments used bacterial strains isolated from fruit flies. In contrast, Escherichia coli, which is seldom detected in medfly samples, did not colonize the medfly digestive tract or any other organ. Even when fed with high-density inocula, it remained confined to the mouth parts of the fly (pseudotrachea of the labelum) at low levels (62). Likewise, some strains of Enterobacter that were detected at all other stages were absent from the eggs (3). This finding suggests a selective role for the host in its microbiota during the various life stages. Host-based selection may be expressed through antimicrobial peptides such as ceratotoxins, which are specifically produced in the accessory glands of mature female medflies and may cover the eggs (63). Even though low concentrations of ceratotoxin killed E. coli and Pseudomonas aeruginosa (a highly pathogenic bacterium in C. capitata [40]), Enterobacter spp. were not affected by the application of >25-times-higher doses (64). Fecal contaminants of eggs obtained in the laboratory were similarly not affected by this peptide (63), suggesting that it may act as a selective chemical barrier to fruit and egg colonization.
The structure and distribution of the microbiota can be regulated by other chemical parameters (e.g., pH and partial oxygen pressure [pO2]) and physical barriers. In termites and polychaete worms, conditions prevailing at particular sites of the digestive system lead to stratified and compartmentalized gut microbiota communities (65–67). In D. melanogaster, the peritrophic membrane was recently shown to protect against intestinal infections by blocking direct contact between intestinal bacteria and the gut epithelium, thus preventing the action of toxins on the latter (68), a situation which may be similar in the medfly (69) (Fig. 3). Host-symbiont relationships in insects are also regulated by molecular interactions that are best understood in D. melanogaster (70). In this fly, gut homeostasis is controlled by cross talk between immune pathways, gut microbiota, and stem cell activity (71). In aged Drosophila flies, the immune system is deficient, and the intestinal epithelium deteriorates. These alterations are associated with excessive intestinal stem cell proliferation, aberrant differentiation, and larger gut populations than in young flies (71), similarly to what we observed for the medfly. It is tempting to suggest that in insects, these processes not only affect abundance but, as in higher animals (72, 73), also lead to changes in the gut community structure.
Supplementary Material
ACKNOWLEDGMENTS
We thank Batia Kaminski for technical help.
This research was partly supported by a Binational Agricultural Research and Development (BARD) grant (3934-06C).
Footnotes
Published ahead of print 26 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02761-12.
REFERENCES
- 1. Weinert LA, Tinsley MC, Temperley M, Jiggins FM. 2007. Are we underestimating the diversity and incidence of insect bacterial symbionts? A case study in ladybird beetles. Biol. Lett. 3:678–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baumann P, Moran NA, Baumann L. 2006. Bacteriocyte-associated endosymbionts of insects, p 403–438 In Dworkin M, Rosenberg E, Schleifer K-H, Stackebrandt E. (ed), The prokaryotes. Springer, New York, NY [Google Scholar]
- 3. Behar A, Jurkevitch E, Yuval B. 2008. Bringing back the fruit into fruit fly-bacteria interactions. Mol. Ecol. 17:1375–1386 [DOI] [PubMed] [Google Scholar]
- 4. Ben Yosef M, Jurkevitch E, Yuval B. 2008. Effect of bacteria on nutritional status and reproductive success of the Mediterranean fruit fly Ceratitis capitata. Physiol. Entomol. 33:145–154 [Google Scholar]
- 5. Kaiwa N, Hosokawa T, Kikuchi Y, Nikoh N, Meng XY, Kimura N, Ito M, Fukatsu T. 2010. Primary gut symbiont and secondary, Sodalis-allied symbiont of the scutellerid stinkbug Cantao ocellatus. Appl. Environ. Microbiol. 76:3486–3494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. 23 April 2012, posting date Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. U. S. A. doi:10.1073/pnas.1200231109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Feldhaar H. 2011. Bacterial symbionts as mediators of ecologically important traits of insect hosts. Ecol. Entomol. 36:533–543 [Google Scholar]
- 8. Mouton L, Henri H, Bouletreau M, Vavre F. 2003. Strain-specific regulation of intracellular Wolbachia density in multiply infected insects. Mol. Ecol. 12:3459–3465 [DOI] [PubMed] [Google Scholar]
- 9. Yuval B, Hendrichs J. 2000. Behavior of flies in the genus Ceratitis (Dacinae: Ceratidini), p 429–457 In Aluja M, Norrbom AL. (ed), Fruit flies Tehpritidae: phylogeny and evolution of behaviour. CRC Press, Boca Raton, FL [Google Scholar]
- 10. Liquido N, Shinoda LA, Cunningham RT. 1991. Host plants of the Mediterranean fruit fly (Diptera: Tephritidae): an annotated world review, p 52 Entomological Society of America, Madison, WI [Google Scholar]
- 11. Behar A, Yuval B, Jurkevitch E. 2005. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol. Ecol. 14:2637–2643 [DOI] [PubMed] [Google Scholar]
- 12. Behar A, Yuval B, Jurkevitch E. 2008. Community structure of the Mediterranean fruit fly microbiota: seasonal and spatial sources of variation. Isr. J. Ecol. Evol. 54:181–191 [Google Scholar]
- 13. Ben Ami E, Yuval B, Jurkevitch E. 2010. Manipulation of the microbiota of mass-reared Mediterranean fruit flies Ceratitis capitata (Diptera: Tephritidae) improves sterile male sexual performance. ISME J. 4:28–37 [DOI] [PubMed] [Google Scholar]
- 14. Marchini D, Dallai RMR, Marri L. 2002. Bacteria associated with the oesophageal bulb of the medfly Ceratitis capitata (Diptera: Tephritidae). Curr. Microbiol. 44:120–124 [DOI] [PubMed] [Google Scholar]
- 15. Lauzon C. 2003. Symbiotic relationships of tephritids, p 115–129 In Bourtzis K, Miller TA. (ed), Insect symbiosis. CRC Press, Boca Raton, FL [Google Scholar]
- 16. Lauzon CR, McCombs SD, Potter SE, Peabody NC. 2009. Establishment and vertical passage of Enterobacter (Pantoea) agglomerans and Klebsiella pneumoniae through all life stages of the Mediterranean fruit fly (Diptera: Tephritidae). Ann. Entomol. Soc. Am. 102:85–95 [Google Scholar]
- 17. Behar A, Ben-Yosef M, Lauzon CR, Yuval B, Jurkevitch E. 2008. Structure and function of the bacterial community associated with the Mediterranean fruit fly, p 251–271 In Bourtzis K, Miller T. (ed), Insect symbiosis. CRC Press, Boca Raton, FL [Google Scholar]
- 18. Ben-Yosef M, Aharon Y, Jurkevitch E, Yuval B. 2010. Give us the tools and we will do the job: symbiotic bacteria affect olive fly fitness in a diet-dependent fashion. Proc. Biol. Sci. 277:1545–1552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kloep F, Manz W, Röske I. 2006. Multivariate analysis of microbial communities in the River Elbe (Germany) on different phylogenetic and spatial levels of resolution. FEMS Microbiol. Ecol. 56:79–94 [DOI] [PubMed] [Google Scholar]
- 21. Ootsubo M, Shimizu T, Tanaka R, Sawabe T, Tajima K, Yoshimizu M, Ezura Y, Ezaki T, Oyaizu H. 2002. Oligonucleotide probe for detecting Enterobacteriaceae by in situ hybridization. J. Appl. Microbiol. 93:60–68 [DOI] [PubMed] [Google Scholar]
- 22. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ludwig W, Klenk H-P. 2005. Overview: a phylogenetic backbone and taxonomic framework for procaryotic systematics, p 49–65 In Brenner DJ, Krieg NR, Staley JT, Garrity GM. (ed), Bergey's manual of systematic bacteriology. Springer, New York, NY [Google Scholar]
- 24. Hugenholtz P, Tyson GW, Blackall LL. 2002. Design and evaluation of 16S rRNA-targeted oligonucleotide probes for fluorescence in situ hybridization. Methods Mol. Biol. 179:29–42 [DOI] [PubMed] [Google Scholar]
- 25. Behrens S, Fuchs BM, Mueller F, Amann R. 2003. Is the in situ accessibility of the 16S rRNA of Escherichia coli for Cy3-labeled oligonucleotide probes predicted by a three-dimensional structure model of the 30S ribosomal subunit? Appl. Environ. Microbiol. 69:4935–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. 2009. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 37:D141–D145 doi:10.1093/nar/gkn879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Manz W, Ludwig ARW, Wagner M, Schleiffer KH. 1992. Phylogenetic oligonucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593–600 [Google Scholar]
- 28. Fukatsu T, Koga R, Smith WA, Tanaka K, Nikoh N, Sasaki-Fukatsu K, Yoshizawa K, Dale C, Clayton DH. 2007. Bacterial endosymbiont of the slender pigeonlouse, Columbicola columbae, allied to endosymbionts of grain weevils and tsetse flies. Appl. Environ. Microbiol. 73:6660–6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gottlieb Y, Ghanim M, Chiel E, Gerling D, Portnoy V, Steinberg S, Tzuri G, Horowitz AR, Belausov E, Mozes-Daube N, Kontsedalov S, Gershon M, Gal S, Katzir N, Zchori-Fein E. 2006. Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae). Appl. Environ. Microbiol. 72:3646–3652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097–1106 [DOI] [PubMed] [Google Scholar]
- 31. Dowd SE, Callaway TR, Wolcott RD, Sun Y, McKeehan T, Hagevoort RG, Edrington TS. 2008. Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125 doi:10.1186/1471-2180-8-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing Mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Quince C, Lanzen A, Davenport R, Turnbaugh P. 2011. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12:38 doi:10.1186/1471-2105-12-38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huse SM, Welch DM, Morrison HG, Sogin ML. 2010. Ironing out the wrinkles in the rare biosphere through improved OTU clustering. Environ. Microbiol. 12:1889–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512–526 [DOI] [PubMed] [Google Scholar]
- 39. Ruimy R, Breittmayer V, Boivin V, Christen R. 1994. Assessment of the state of activity of individual bacterial cells by hybridization with a ribosomal RNA targeted fluorescently labelled oligonucleotidic probe. FEMS Microbiol. Ecol. 15:207–213 [Google Scholar]
- 40. Behar A, Yuval B, Jurkevitch E. 2008. Gut bacterial communities in the Mediterranean fruit fly (Ceratitis capitata) and their impact on host longevity. J. Insect Physiol. 54:1377–1383 [DOI] [PubMed] [Google Scholar]
- 41. Douglas AE. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu. Rev. Entomol. 43:17–37 [DOI] [PubMed] [Google Scholar]
- 42. Hosokawa T, Kikuchi Y, Nikoh N, Shimada M, Fukatsu T. 2006. Strict host-symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS Biol. 4:e337 doi:10.1371/journal.pbio.0040337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moran NA, McCutcheon JP, Nakabachi A. 2008. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 42:165–190 [DOI] [PubMed] [Google Scholar]
- 44. Kikuchi Y, Hosokawa T, Fukatsu T. 2011. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 5:446–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Conn KE, Ogawa JM, Manji BT, Adaskaveg JE. 1995. Leuconostoc mesenteroides subsp mesenteroides, the first report of a coccoid bacterium causing a plant disease. Phytopathology 85:593–599 [Google Scholar]
- 46. Naum M, Brown E, Mason-Gamer R. 2008. Is 16S rDNA a reliable phylogenetic marker to characterize relationships below the family level in the Enterobacteriaceae? J. Mol. Evol. 66:630–642 [DOI] [PubMed] [Google Scholar]
- 47. Naum M, Brown EW, Mason-Gamer RJ. 2011. Is a robust phylogeny of the enterobacterial plant pathogens attainable? Cladistics 27:80–93 [DOI] [PubMed] [Google Scholar]
- 48. Treangen TJ, Rocha EPC. 2011. Horizontal transfer, not duplication, drives the expansion of protein families in prokaryotes. PLoS Genet. 7:e1001284 doi:10.1371/journal.pgen.1001284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stecher B, Denzler R, Maier L, Bernet F, Sanders MJ, Pickard DJ, Barthel M, Westendorf AM, Krogfelt KA, Walker AW, Ackermann M, Dobrindt U, Thomson NR, Hardt W-D. 2012. Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. U. S. A. 109:1269–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol. Rev. 32:723–735 [DOI] [PubMed] [Google Scholar]
- 51. Himler AG, Adachi-Hagimori T, Bergen JE, Kozuch A, Kelly SE, Tabashnik BE, Chiel E, Duckworth VE, Dennehy TJ, Zchori-Fein E, Hunter MS. 2011. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science 332:254–256 [DOI] [PubMed] [Google Scholar]
- 52. Sharon G, Segal D, Ringo JM, Hefetz A, Zilber-Rosenberg I, Rosenberg E. 2010. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 107:20051–20056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Christenson LD, Foote RH. 1960. Biology of fruit flies. Annu. Rev. Entomol. 5:171–192 [Google Scholar]
- 54. Munguira ML, Muniz SFM. 1983. Estudio morfológico del aparato reproductor femenino de Ceratitis capitata Wied (Dipt: Trypetidae). Bol. Serv. Plagas 9:31–43 [Google Scholar]
- 55. Costello EK, Stagaman K, Dethlefsen L, Bohannan BJM, Relman DA. 2012. The application of ecological theory toward an understanding of the human microbiome. Science 336:1255–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. McDonald PT, McInnis DO. 1985. Ceratitis capitata: effect of host fruit size on the number of eggs per clutch. Entomol. Exp. Appl. 37:207–211 [Google Scholar]
- 57. Papaj DR, Roitberg BD, Opp SB, Aluja M, Prokopy RJ, Wong TTY. 1990. Effect of marking pheromone on clutch size in the Mediterranean fruit fly. Physiol. Entomol. 15:463–468 [Google Scholar]
- 58. Mira A, Moran NA. 2002. Estimating population size and transmission bottlenecks in maternally transmitted endosymbiotic bacteria. Microb. Ecol. 44:137–143 [DOI] [PubMed] [Google Scholar]
- 59. Vautrin E, Vavre F. 2009. Interactions between vertically transmitted symbionts: cooperation or conflict? Trends Microbiol. 17:95–99 [DOI] [PubMed] [Google Scholar]
- 60. Hosokawa T, Kikuchi Y, Fukatsu T. 2007. How many symbionts are provided by mothers, acquired by offspring, and needed for successful vertical transmission in an obligate insect-bacterium mutualism? Mol. Ecol. 16:5316–5325 [DOI] [PubMed] [Google Scholar]
- 61. Ferrari J, Vavre F. 2011. Bacterial symbionts in insects or the story of communities affecting communities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 366:1389–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Sela S, Nestel D, Pinto R, Nemny-Lavy E, Bar-Joseph M. 2005. Mediterranean fruit fly as a potential vector of bacterial pathogens. Appl. Environ. Microbiol. 71:4052–4056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Marchini D, Manetti AGO, Rosetto M, Bernini LF, Telford JL, Baldari CT, Dallai R. 1995. cDNA sequence and expression of the ceratotoxin gene encoding an antibacterial sex-specific peptide from the medfly Ceratitis capitata (Diptera). J. Biol. Chem. 270:6199–6204 [DOI] [PubMed] [Google Scholar]
- 64. Marri L, Dallai R, Marchini D. 1996. The novel antibacterial peptide ceratotoxin A alters permeability of the inner and outer membrane of Escherichia coli K-12. Curr. Microbiol. 33:40–43 [DOI] [PubMed] [Google Scholar]
- 65. Friedrich MW, Schmitt-Wagner D, Lueders T, Brune A. 2001. Axial differences in community structure of Crenarchaeota and Euryarchaeota in the highly compartmentalized gut of the soil-feeding termite Cubitermes orthognathus. Appl. Environ. Microbiol. 67:4880–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li M, Yang H, Gu J-D. 2009. Phylogenetic diversity and axial distribution of microbes in the intestinal tract of the polychaete Neanthes glandici. Microb. Ecol. 58:892–902 [DOI] [PubMed] [Google Scholar]
- 67. Nakajima H, Hongoh Y, Usami R, Kudo T, Ohkuma M. 2005. Spatial distribution of bacterial phylotypes in the gut of the termite Reticulitermes speratus and the bacterial community colonizing the gut epithelium. FEMS Microbiol. Ecol. 54:247–255 [DOI] [PubMed] [Google Scholar]
- 68. Kuraishi T, Binggeli O, Opota O, Buchon N, Lemaitre B. 2011. Genetic evidence for a protective role of the peritrophic matrix against intestinal bacterial infection in Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 108:15966–15971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lauzon CR, Potter SE. 2012. Description of the irradiated and nonirradiated midgut of Ceratitis capitata Wiedemann (Diptera: Tephritidae) used for sterile insect technique. J. Pest Sci. 85(2):217–226 [Google Scholar]
- 70. Lemaitre B, Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25:697–743 [DOI] [PubMed] [Google Scholar]
- 71. Buchon N, Broderick NA, Chakrabarti S, Lemaitre B. 2009. Invasive and indigenous microbiota impact intestinal stem cell activity through multiple pathways in Drosophila. Genes Dev. 23:2333–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hongoh Y, Ekpornprasit L, Inoue T, Moriya S, Trakulnaleamsai S, Ohkuma M, Noparatnaraporn N, Kudo T. 2006. Intracolony variation of bacterial gut microbiota among castes and ages in the fungus-growing termite Macrotermes gilvus. Mol. Ecol. 15:505–516 [DOI] [PubMed] [Google Scholar]
- 73. Claesson MJ, Cusack S, O'Sullivan O, Greene-Diniz R, de Weerd H, Flannery E, Marchesi JR, Falush D, Dinan T, Fitzgerald G, Stanton C, van Sinderen D, O'Connor M, Harnedy N, O'Connor K, Henry C, O'Mahony D, Fitzgerald AP, Shanahan F, Twomey C, Hill C, Ross RP, O'Toole PW. 2011. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl 1):4586–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.