Abstract
Polyphosphate (polyP) is a ubiquitous biochemical with many cellular functions and comprises an important environmental phosphorus pool. However, methodological challenges have hampered routine quantification of polyP in environmental samples. We tested 15 protocols to extract inorganic polyphosphate from natural marine samples and cultured cyanobacteria for fluorometric quantification with 4′,6-diamidino-2-phenylindole (DAPI) without prior purification. A combination of brief boiling and digestion with proteinase K was superior to all other protocols, including other enzymatic digestions and neutral or alkaline leaches. However, three successive extractions were required to extract all polyP. Standard addition revealed matrix effects that differed between sample types, causing polyP to be over- or underestimated by up to 50% in the samples tested here. Although previous studies judged that the presence of DNA would not complicate fluorometric quantification of polyP with DAPI, we show that RNA can cause significant interference at the wavelengths used to measure polyP. Importantly, treating samples with DNase and RNase before proteinase K digestion reduced fluorescence by up to 57%. We measured particulate polyP along a North Pacific coastal-to-open ocean transect and show that particulate polyP concentrations increased toward the open ocean. While our final method is optimized for marine particulate matter, different environmental sample types may need to be assessed for matrix effects, extraction efficiency, and nucleic acid interference.
INTRODUCTION
Inorganic polyphosphate (polyP) is a linear polymer of three to several hundred phosphate residues that appears to be found in cells of all organisms (1). It is required for many cellular functions, including bacterial virulence (2–4), biofilm formation (4, 5), quorum sensing (4), competitive fitness in the environment (6), microbial stress responses (3, 7, 8), and survival during stationary phase (7, 9–11). PolyP can also act as a phosphorus (P) store in microbes. PolyP breakdown allows cells to survive during P limitation (12, 13), although some microbes transiently accumulate polyP upon nitrogen and P stress (8, 14, 15). Massive accumulation of polyP by certain bacteria sequesters dissolved P during biological wastewater treatment (16–18).
While polyP is studied in a range of microbiological and environmental fields, the lack of easy quantitative methods has hampered progress. For example, polyP has very rarely been measured in the oceans despite increasing interest in the physiological responses of marine microbes to the ultralow P concentrations frequently found at sea and their biogeochemical implications (14, 19–25). PolyP has been found at nanomolar concentrations, comprising around 10% of total particulate P (26–28). PolyP also plays a role in redox cycling and geological sequestration of P (26, 27) and potentially in phytoplankton iron storage (29).
Common analytical approaches have significant drawbacks. Phosphorus-31 nuclear magnetic resonance spectroscopy requires large sample sizes and may be insensitive to polyP tightly associated with metal cations in solid granules. Moreover, alkaline extractions may incompletely extract and partly hydrolyze polyP (30, 31). Assays based on enzymes of polyP metabolism (either exopolyphosphatase followed by orthophosphate [PO43−] determination [32, 33] or polyP kinase followed by ATP measurement [34, 35]) entail purifying polyP, but purification procedures suffer from low yields (36, 37) and there is no commercial source for the necessary enzymes.
Recently, a new method of quantifying polyP without prior purification, which is termed the in vivo method and is based on the fluorescence of 4′,6-diamidino-2-phenylindole (DAPI), was reported (36, 38, 39). DAPI is a common nucleic acid stain, but its fluorescence spectrum changes when bound to polyP (38). This method has been used to quantify polyP in cell cultures (36, 38), activated sludge samples (36), and marine plankton and sediments (27, 39).
We revisited the in vivo method to quantify polyP in marine plankton samples but found that neither the alkaline extraction method described for freeze-dried plankton and sediment samples (39) nor the freeze-thaw extraction described for fresh bacterial samples (36) gave satisfactory results with our fresh marine plankton samples. We show here that a different extraction procedure is required for fresh samples, that matrix effects can complicate polyP quantification, and that the presence of nucleic acids in natural samples can lead to large overestimates of polyP. We then use our adapted method to gain insights into the importance of polyP in the marine environment.
MATERIALS AND METHODS
Definitions.
We define particulate polyP as polyP in particulate matter, i.e., primarily inside living and dead microbial cells. Extraction is the liberation of polyP from within particles such that it is dissolved in the sample buffer. Purification, in contrast, is the postextraction separation of polyP from all other chemical constituents of the solution matrix. In vivo refers to quantifying polyP after extraction but without purification. While it is possible that some of the extracted polyP might still be present in suspended granules, we believe that our extraction protocol most likely dissolves these (see below). We express all quantities of polyP as nmol of the constituent phosphate residues (thus, 1 nmol polyP yields 1 nmol PO43− upon complete hydrolysis).
Reagents and standard.
PolyP with a chain length of 45 ± 5 residues was from Sigma-Aldrich (S4379). Aliquots were weighed out, dissolved in 20 mM Tris buffer (pH 7.0), diluted as needed, and stored at −20°C in between uses. All other reagents were of analytical or enzyme grade and from Sigma-Aldrich or Fisher Scientific. Proteinase K, lysozyme, and ADP were from Fisher (BP1700, BP535, and A2754, respectively), DNase and RNase cocktails were from Ambion (AM2235 or AM2238 and AM2286, respectively), and DNA and RNA were from Sigma-Aldrich (D1626 and R6750, respectively). All solutions and buffers were prepared with ultrapure (18.2 MΩ cm−1) water. Tris buffer was always 20 mM (pH 7.0) unless otherwise stated. DAPI was stored at −20°C as a 1 mM stock solution and diluted to 100 μM prior to staining, of which 60 μl was added to 500 μl of sample or standard solutions to stain polyP.
Cyanobacterial and marine particle samples.
We used three sample types to test the method. Cyanobacteria (Synechococcus sp. strain WH8102) were grown at 23°C and 30 μmol photons m−2 s−1 in SN medium based on 0.2 μm filtered Sargasso seawater with PO43− added to 45 μM. Cells were harvested from 1- to 1.5-ml aliquots of culture medium in log or late stationary phase by centrifugation (either 4,000 × g for 3 min or 16,000 × g for 5 min, depending on the batch), supernatants were removed, and the pellets were frozen on ice and stored at −20°C. These cultures were later discovered to be nonaxenic, owing to prior bacterial contamination of the stock culture; this has little bearing on the results presented here because our goal with the cultures was simply to obtain aliquots of microbial biomass that we could use in the development of our method. Natural marine particles were collected from 5-m depth in coastal waters of the Vineyard Sound, Cape Cod, via a water intake line on the Woods Hole Oceanographic Institution campus in March 2012. Four liters of water was preconcentrated to ∼350 ml by filtration through a 0.22-μm cellulose acetate bottle top filter (Corning), and 2-ml aliquots were pelleted by centrifugation at 16,100 × g for 10 min. Supernatants were discarded, and pellets were frozen on ice and stored at −20°C. Vineyard Sound water was sampled again in September 2012: replicate 50-ml aliquots were filtered onto 25-mm-diameter Whatman GF/F filters, the back of each filter was blotted dry on a laboratory wipe, and filters were wrapped in aluminum foil, flash-frozen in liquid N2, and stored at −20°C. These filtered samples were used only to assess our measurement precision. Further particle samples were collected from 5-m depth in the North Pacific on a transect between 48.58°N, 125.50°W, and 48.82°N, 128.67°W, in May 2012. Nine hundred milliliters of seawater was filtered onto precombusted (450°C, 7 h) 25-mm GF/F filters. Filters were blotted dry, wrapped in aluminum foil, flash-frozen in liquid N2, and stored at −20°C back on land.
PolyP measurements.
All measurements were made on a Horiba Scientific FluoroMax-4 spectrofluorometer at an excitation wavelength of 415 nm and emission wavelength of 550 nm, with an integration time of 0.5 s. Fluorescence emission spectra were acquired at 415-nm excitation and emission from 430 to 650 nm in 1-nm increments and integrated for 0.1 s. All bandwidths were 5 nm. The fluorescence signal is reported as thousands of counts per second (kcps). Samples and standards were made up to 500 μl in microcentrifuge tubes with Tris buffer, stained with 60 μl 100 μM DAPI, vortexed, incubated for at least 2 min (standards) or 7 min (samples), vortexed again, and measured in a final volume of 3 ml Tris buffer in a quartz fluorescence cuvette (Fisher Scientific; 14-385-918A). Standards to construct a calibration curve (0.2 to 7.0 nmol polyP) and stained Tris buffer blanks were measured with each set of samples; no problems were ever noted with analytical precision or instrument drift. We found that if samples were vortexed twice during incubation with DAPI and mixed well with a pipette in the cuvette prior to measurement, continuous stirring inside the fluorometer (38, 39) was unnecessary.
Purification of polyP.
We tested an adaptation of previously described purification protocols using silica membrane spin columns (33, 34, 36). Owing to disappointing results with commercially available columns, and inspired by prior work (40), we made our own by packing four 7-mm-diameter punches of Sterlitech 0.3-μm-pore-size GF-75 glass fiber filters into spin columns with a 0.2-μm polyvinylidene difluoride (PVDF) membrane filter (Analytical Sales & Services, NJ; product 8506-00). We quantified the purification yield by mixing various amounts of polyP standard in 5 μl Tris with 200 μl 5.5 M guanidinium isothiocyanate (GITC) and 400 μl 100% ethanol after optimizing the GITC/ethanol ratio in preliminary trials. The mixture was vortexed, loaded onto a column, centrifuged (10,000 × g, 1 min), washed twice with a wash buffer (5 mM Tris [pH 7.4], 100 mM NaCl, 5 mM EDTA, 50% [vol/vol] ethanol), and centrifuged twice to completely dry the column (10,000 × g, 1 min), and polyP was eluted five times with 90 μl Tris buffer. Previous experiments had shown that five elutions were necessary and sufficient to recover all polyP. All elutions from each column were combined and made up to 500 μl with Tris buffer.
Extracting polyP from samples.
We tested 15 different protocols for dissolving polyP from samples (Table 1). All procedures were carried out in microcentrifuge tubes (1.5 or 2.0 ml). Boiling was performed by immersing the tubes in a floating tube rack in a beaker of boiling water; 37°C enzymatic digestions were conducted in an Eppendorf Thermomixer with gentle mixing. Some protocols were directly taken from, or inspired by, previous reports, including leaching with 0.25 M NaOH (39), simple freeze-thawing (36), boiling in Tris buffer (41), and use of EDTA (42). We first tested our methods with the Synechococcus samples and then tested those that showed most promise with Vineyard Sound samples. For the Synechococcus samples, boiling and proteinase K treatments were performed for 5 and 10 min, respectively, but for Vineyard Sound samples, these were extended to 10 and 60 min, respectively. Based on the results, we adopted brief boiling and subsequent proteinase K treatment as our “core” extraction protocol and optimized the treatment times to 5 min boiling and 30 min proteinase K in further experiments. Using both Vineyard Sound and Pacific samples, we then tested how many extraction rounds of our core protocol were required to extract all polyP. For the Pacific samples, we took one 7-mm-diameter punch from a GF/F filter for each measurement. All proteinase K treatments were with 10 μl of 20 mg ml−1 proteinase K.
Table 1.
Summary of extraction protocolsa
| Treatment name in Fig. 3 | Summary of treatmentb |
|---|---|
| NaOH leach | 2 20-min leaches with 160 μl 0.25 M NaOH, neutralization with HCl; centrifugation, removal of the supernatant, dilution with 500 μl Tris |
| Lyso/n.ases | 190 μl lysis buffer, sonication, lysozyme, DNase and RNase; centrifugation, removal of the supernatant, addition of 300 μl Tris, centrifugation, removal of the supernatant |
| Lysozyme | 190 μl lysis buffer, sonication, lysozyme; centrifugation, removal of the supernatant, addition of 300 μl Tris, centrifugation, removal of the supernatant |
| Freeze-thaw | Frozen pellets thawed at room temp, addition of 200 μl Tris, vortexing, centrifugation, removal of the supernatant; addition of 300 μl Tris, centrifugation, removal of the supernatant |
| Dry boiled | Sonication of the cell pellet, boiling, sonication; addition of 200 μl Tris, centrifugation, removal of the supernatant; addition of 300 μl Tris, centrifugation, removal of the supernatant |
| Tris boiled | As for “dry boiled,” but addition of the 200 μl Tris before sonication and boiling |
| Dry boil/lyso | Sonication of the pellet, boiling, sonication, addition of 190 μl lysis buffer and lysozyme; centrifugation, removal of the supernatant; addition of 300 μl Tris, centrifugation, removal of the supernatant |
| NaOH boiled | 160 μl 0.25 M NaOH, sonication, boiling, sonication; centrifugation, removal of the supernatant, neutralization with HCl; addition of 250 μl Tris, centrifugation, removal of the supernatant |
| Short Tris boil | As for “Tris boiled,” but only 2 min boiling |
| Long Tris boil | As for “Tris boiled,” but 20 min boiling |
| Nucleases after boiling | 200 μl lysis buffer, sonication, boiling, sonication, DNase and RNase; centrifugation, removal of the supernatant; addition of 300 μl Tris, centrifugation, removal of the supernatant |
| Lysozyme & nucleases after boiling | 200 μl lysis buffer, sonication, boiling, sonication, lysozyme, DNase and RNase; centrifugation, removal of the supernatant; addition of 300 μl Tris, centrifugation, removal of the supernatant |
| Proteinase K after boiling | 200 μl Tris, sonication, boiling, sonication, proteinase K treatment for 10 min at 37°C; centrifugation, removal of the supernatant; addition of 300 μl Tris, centrifugation, removal of the supernatant |
| All enzymes after boiling | 200 μl lysis buffer, sonication, boiling, sonication, lysozyme, DNase and RNase; proteinase K treatment for 10 min at 37°C; centrifugation, removal of the supernatant; addition of 300 μl Tris, centrifugation, removal of the supernatant |
| EDTA | As for “Tris boiled,” but 10 or 50 mM EDTA was added to buffer before boiling for 10 min |
| Proteinase K for Vineyard | As for “Proteinase K after boiling,” but boiled for 10 min and treated with proteinase K for 1 h |
| EDTA proteinase K | As for “Proteinase K for Vineyard,” but 10 or 50 mM EDTA was added to buffer before boiling |
Centrifugation was always for 1 min at 16,100 × g, and sonication was always for 15 s. Boiling was for 5 min for Synechococcus (Fig. 3a and b) and 10 min for Vineyard Sound samples (Fig. 3c).
Lysis buffer, 10 mM Tris (pH 7.0), 100 mM NaCl, 2 mM MgCl2, and 2 mM EDTA; lysozyme, 10 μl of 25 mg ml−1 lysozyme, incubation for 10 min at 37°C; DNase and RNase, 10 units DNase, 100 units RNase T1, and 2.5 units RNase A, incubation for 10 min at 37°C; proteinase K, 10 μl of 20 mg ml−1 proteinase K.
Standard addition experiments.
For the Vineyard Sound and Pacific samples, 10 replicate samples were extracted using the core extraction protocol, each replicate was split exactly in half after vortexing, and one split of each replicate was amended with 0.2 to 5 nmol polyP. For Synechococcus, we found that the late-stationary-phase cultures we harvested contained very large amounts of polyP and thus needed to be diluted very heavily. Therefore, instead of taking 10 replicate samples, we took one single sample, added 330 μl Tris buffer, homogenized it by vortexing vigorously, and dispensed 18 15-μl aliquots into a final volume of 300 μl Tris buffer. After treating these aliquots with our core extraction protocol, nine aliquots were amended with 0.4 to 5 nmol polyP. All solutions were made up to 500 μl with Tris buffer, stained with DAPI, and measured. For this experiment, we performed only a single extraction step for all three sample types.
ADP and nucleic acid interference.
Although interference from nucleic acids and nucleoside triphosphates has been considered negligible (38), we measured a series of fluorescence emission spectra of either Tris blanks, Vineyard Sound samples, or North Pacific samples, with and without added DNA, RNA, or ADP solutions. To test whether treatment with nucleases can mitigate interference from nucleic acids, we measured the fluorescence emission spectra of solutions of 3 μg DNA or RNA (∼9 nmol P) in 500 μl Tris buffer after treating these solutions either with our core polyP extraction protocol or with a 10-min treatment with DNase or RNase cocktail at 37°C after boiling and before proteinase K treatment (20 units DNase or 200 units RNase T1 plus 5 units RNase A). To test whether nucleic acids were problematic in our samples, we first measured fluorescence emission spectra of replicate Vineyard Sound and Pacific samples treated either with our core extraction protocol or with the modified version of the protocol that included a 10-min treatment at 37°C with DNase and RNase cocktail. We then took six replicate samples each of Vineyard Sound, Pacific, and Synechococcus, treating three replicates of each sample type with the core protocol and three with the modified version that included nuclease treatment. Nuclease treatment was optimized to consist of 10 units DNase, 100 units RNase T1, and 2.5 units RNase A.
Effect of enzymes on fluorescence of standard.
We tested the effect of proteinase K and DNase and RNase cocktail additions on the fluorescence signal of a polyP standard solution. Triplicate solutions of 2 nmol polyP in 500 μl Tris were incubated either without enzymes, with proteinase K only, with DNase and RNase cocktail only, or with DNase and RNase cocktail and proteinase K. Incubations were for 40 min at 37°C, but for the treatment with both enzymes, we followed our core extraction protocol of a 10-min incubation with nucleases alone and a further 30-min incubation upon adding proteinase K. Blank solutions without the polyP standard were prepared in triplicate for each treatment and incubated alongside the samples. All solutions were stained with DAPI, and fluorescence was measured.
Quantification of polyP in the North Pacific.
We quantified polyP along the transect in the North Pacific (which crossed a transition zone between two different ecological regimes [43]), using three to five 4-mm punches from each filter. PolyP was extracted using the amended extraction protocol that included nuclease treatment (outlined in Fig. S1 in the supplemental material). Since we discovered that three successive extractions were necessary to extract all polyP from marine particle samples and that there was both a background level of fluorescence (which we ascribe to DAPI interacting with constituents other than polyP in the extract) and detectable fluorescence in extracts even without added DAPI, each sample was extracted four times with 300 μl Tris. Extracts 1 to 3 were combined into one sample to measure polyP, which we call the analytical extract. The fourth extract was used to measure the stained background signal (background extract). The analytical extract was then split into three equal subsamples: one to measure the fluorescence without DAPI added, one to measure the DAPI-stained fluorescence, and the third to measure stained fluorescence with addition of 2 nmol polyP standard to account for matrix effects. The background extract was split into two equal subsamples, of which one was stained with DAPI and the other was left unstained. The signal due to polyP was calculated by correcting the fluorescence in the DAPI-stained split of the analytical extract for unstained background fluorescence, the stained fluorescence in the background extract, and any matrix effect as determined by the standard addition. A detailed description of the calculations is given in the supplemental material, and the method is summarized in a flowchart (see Fig. S1 in the supplemental material).
Chlorophyll a measurement.
Chlorophyll a (Chl a), a proxy for phytoplankton biomass, was quantified along the North Pacific transect fluorometrically after filtering seawater samples onto GF/F filters and extracting them in 90% acetone for 24 h at 4°C in the dark.
Measurement precision.
We determined the analytical precision of the fluorescence measurement on eight separate occasions using 3 to 4 replicate solutions of polyP standard. We then estimated the overall precision in quantifying polyP in field samples with the filtered Vineyard Sound samples, using the method described above for the North Pacific samples. Four replicate sets of two punches (6-mm diameter) were subsampled from each of three GF/F filters, yielding 12 replicates in total. We report precision as the mean and standard deviation (SD) of the replicates.
RESULTS
Measurement of standards.
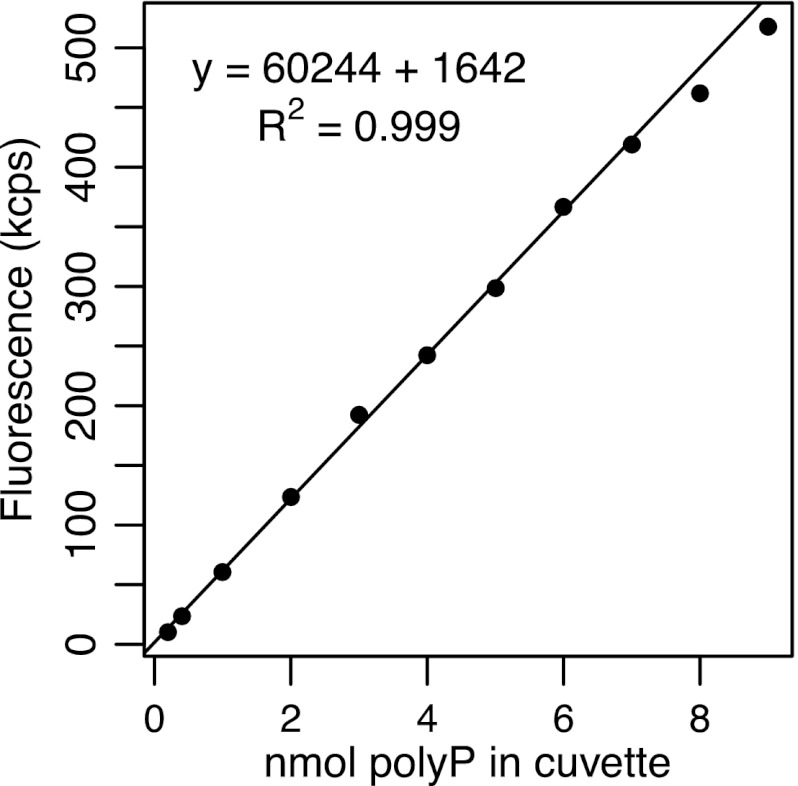
Calibration curves of blank-corrected signal versus amount of polyP were always linear between 0.2 and 7.0 nmol polyP, with an R2 value of ≥0.998 (Fig. 1). Given a final volume of 3 ml, this corresponds to a concentration of 0.067 to 2.3 μM in the cuvette. The fluorescence signal of DAPI-stained Tris buffer blanks typically ranged from 5 to 7 kcps, while 0.2 nmol polyP (0.067 μM) already yielded around 15 kcps.
Fig 1.
Typical calibration curve of the fluorescence signal versus the quantity of polyP in the spectrofluorometer cuvette, in nmol orthophosphate equivalents (1 nmol polyP = 1 nmol PO43− upon complete hydrolysis). Since each measurement was made in 3 ml of solution, the calibration curve spans a concentration range of 0.067 to 2.3 μM polyP.
Purification of polyP.
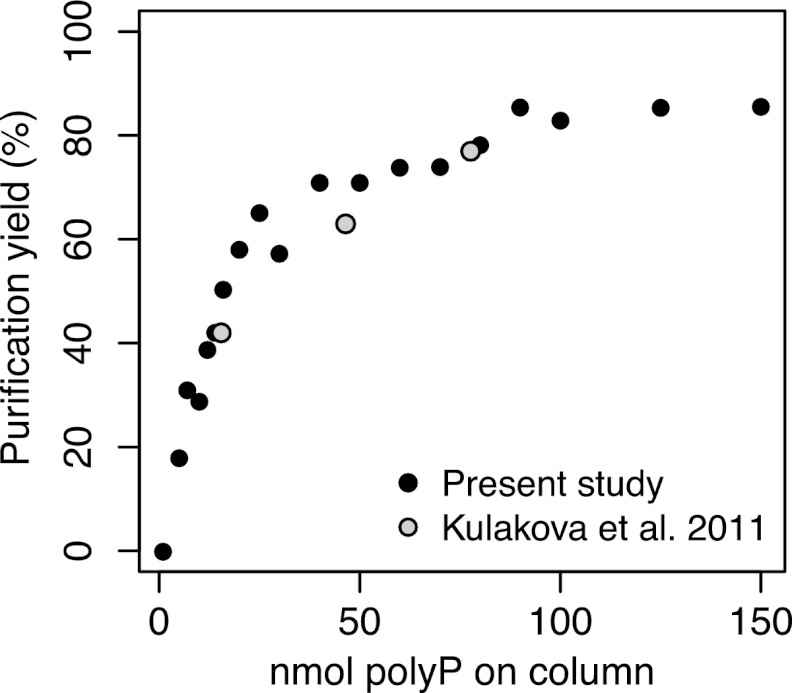
The percentage yield of standard polyP solutions (i.e., the quantity recovered by purification relative to the quantity initially added) upon purification with custom-made silica membrane spin columns increased sharply with the quantity of polyP initially added (Fig. 2). Above 50 nmol polyP, yields were relatively stable, gradually leveling off at 85%. However, the very large quantity of polyP required to achieve high and stable yields makes this an unattractive method for routine quantification. Moreover, if columns without 0.2-μm PVDF filters were used, procedural blanks were often high and variable (≥20 kcps, even with commercial columns such as Qiagen's QiaQuick columns or Epoch Life Science's EconoSpin columns), apparently due to glass particles in the eluent.
Fig 2.
Percentage yield of polyP upon purification on custom-made silica spin columns versus the amount of polyP loaded on the column in 200 μl GITC plus 400 μl ethanol. Results of Kulakova et al. (36) were estimated from the concentrations they reported, assuming a volume of 800 μl for their samples.
Extracting polyP from samples.
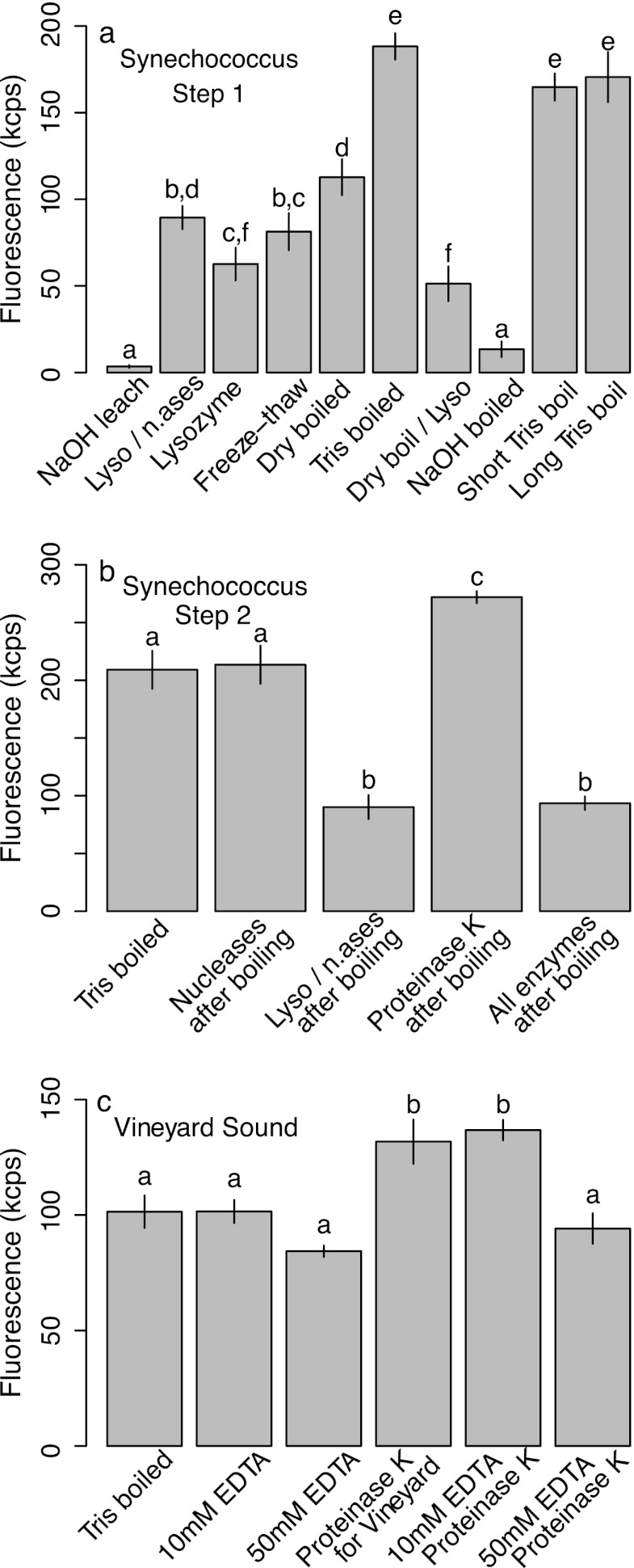
Different polyP extraction protocols (Table 1) yielded significantly different fluorescence signals with replicate samples of the Synechococcus culture (Fig. 3a) (one-way analysis of variance [ANOVA] with post hoc Bonferroni-corrected pairwise t tests; F = 180; degrees of freedom [df] = 10, 33; P < 0.001). Simply boiling the sample for 5 min in Tris buffer gave the best results; leaching or boiling in 0.25 M NaOH gave the lowest results of all treatments. Enzymatic treatment with lysozyme in fact reduced the signal, although this was partially mitigated by DNase and RNase treatment. Both shorter (2-min) and longer (20-min) boiling times resulted in slightly lower signals relative to the 5-min treatment, although not significantly so.
Fig 3.
Fluorescence signals in DAPI-stained sample extracts obtained with a range of extraction protocols in a Synechococcus culture (a and b) and in natural marine particle samples from Vineyard Sound (c). Data in panel a identified boiling as an ideal first extraction step, data in panel b showed that subsequent proteinase K digestion was required, and data in panel c showed that this protocol was suitable for natural environmental samples, too. Data are means and SDs of 4 (a and b) or 3 (c) replicates. Letters indicate significant differences determined after an ANOVA using pairwise t tests with a Bonferroni correction. Treatments are detailed in Table 1. n.ases, nucleases.
We then evaluated a range of enzymatic treatments after an initial 5-min boiling step (Fig. 3b) and confirmed that lysozyme treatment reduced the signal while a 30-min treatment with proteinase K after boiling yielded a higher signal (one-way ANOVA with post hoc Bonferroni-corrected pairwise t tests; F = 167; df = 4, 17; P < 0.001).
Finally, we tested this approach with a natural marine particle sample from Vineyard Sound (Fig. 3c) and found that proteinase K treatment after boiling in Tris buffer resulted in a significantly elevated signal (one-way ANOVA with post hoc Bonferroni-corrected pairwise t tests; F = 34; df = 5, 12; P < 0.001). Addition of 10 mM EDTA to the samples prior to boiling made no significant difference, but addition of 50 mM EDTA led to a significant reduction in signal.
We therefore adopted the following as our “core” protocol: add 500 μl Tris buffer to the sample in a microcentrifuge tube, vortex thoroughly to resuspend, sonicate for 15 s, immerse in boiling water for 5 min, sonicate for 15 s, add 10 μl 20 mg ml−1 proteinase K, and incubate at 37°C for 30 min with mixing at 300 rpm; vortex 2 or 3 times during enzyme treatment then centrifuge for 1 min at 16,100 × g, and remove supernatant to measure dissolved polyP after addition of DAPI.
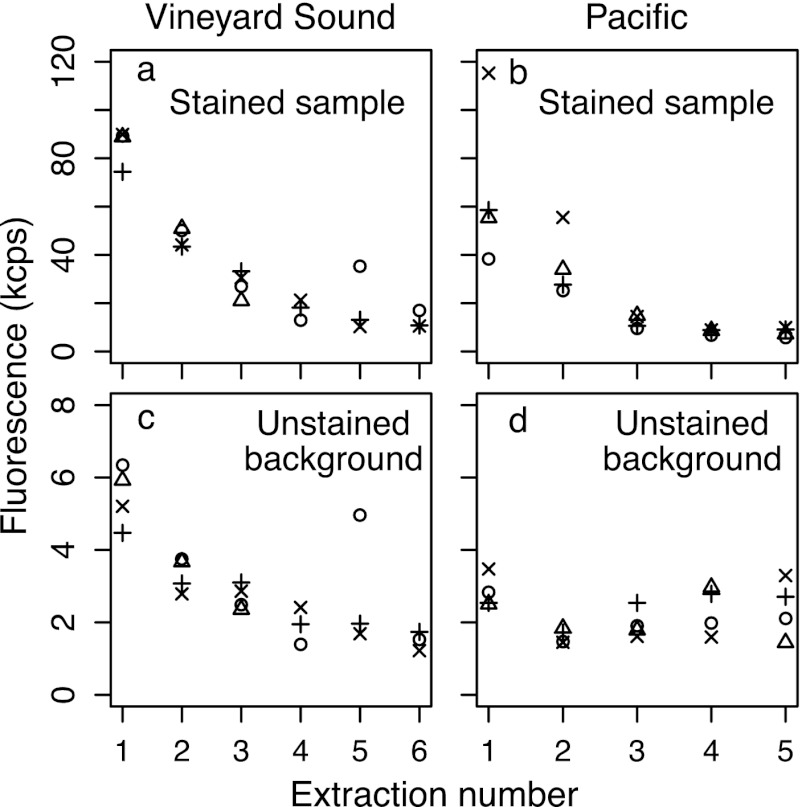
We then tested how many treatment rounds with the core protocol were required to extract all polyP, using Vineyard Sound and Pacific samples. We took four replicate samples from each location and conducted five (Pacific) or six (Vineyard Sound) sequential extractions on each, keeping all extractions separate. After three extractions, a low but detectable level of background fluorescence was reached and remained steady around 10 to 20 kcps, i.e., higher than the 5 to 7 kcps in a stained Tris blank (Fig. 4a and b). Moreover, the extracts had significant background fluorescence even in the absence of DAPI, in this case, 2 to 6 kcps (Fig. 4c and d). Strangely, the variability between replicate Pacific samples in this experiment was very high (Fig. 4b): in other experiments with the Pacific samples, fluorescence in stained extract 1 was always around 100 kcps with low variability (Fig. 5, 6, and 7). While we do not know why the results in Fig. 4b were so atypically variable, they nevertheless show that three extractions are sufficient; a lack of samples prevented us from repeating this experiment.
Fig 4.
Fluorescence in successive extracts of four replicate Vineyard Sound (a and c) and North Pacific (b and d) particle samples. Different plotting symbols distinguish each of the replicates. Extracts were split into equal halves, one of which was stained with DAPI (a and b) and one which was used to measure the unstained background fluorescence (c and d). Note the different y axis scales.
Fig 5.
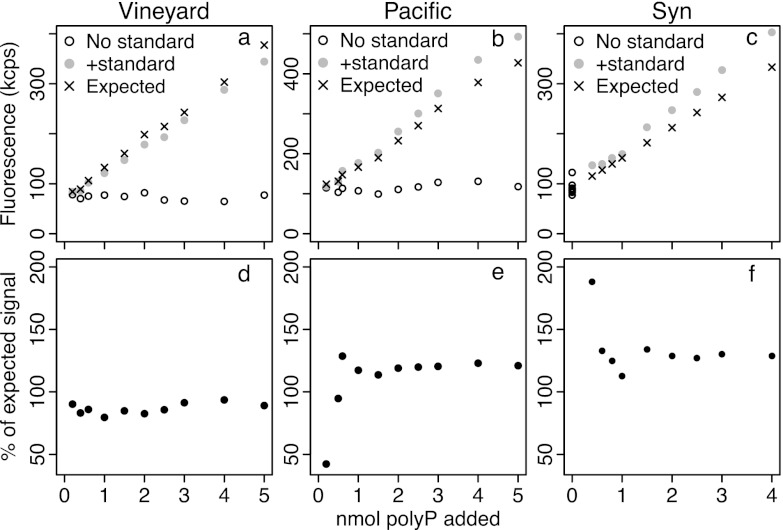
Results of standard addition experiments to test for matrix effects in Vineyard Sound samples (a and d), North Pacific samples (b and e), and a Synechococcus culture (c and f). Top panels show fluorescence measured in DAPI-stained extracts with and without added polyP standard. Each Vineyard Sound and North Pacific sample was extracted once, and the extract was split into two equal aliquots, one of which was left unamended and one which was amended with various amounts of polyP standard. The expected values were calculated based on the fluorescence in the unamended split and the fluorescence expected from each quantity of polyP, as calculated using an external calibration. For the Synechococcus culture, one sample was homogenized and divided into 18 equal aliquots and polyP was extracted from each. Nine were amended with polyP standard, and nine were left unamended; expected values were calculated from the mean fluorescence of the unamended aliquots and the fluorescence expected from the amount of standard added, based on an external calibration curve.
Fig 6.
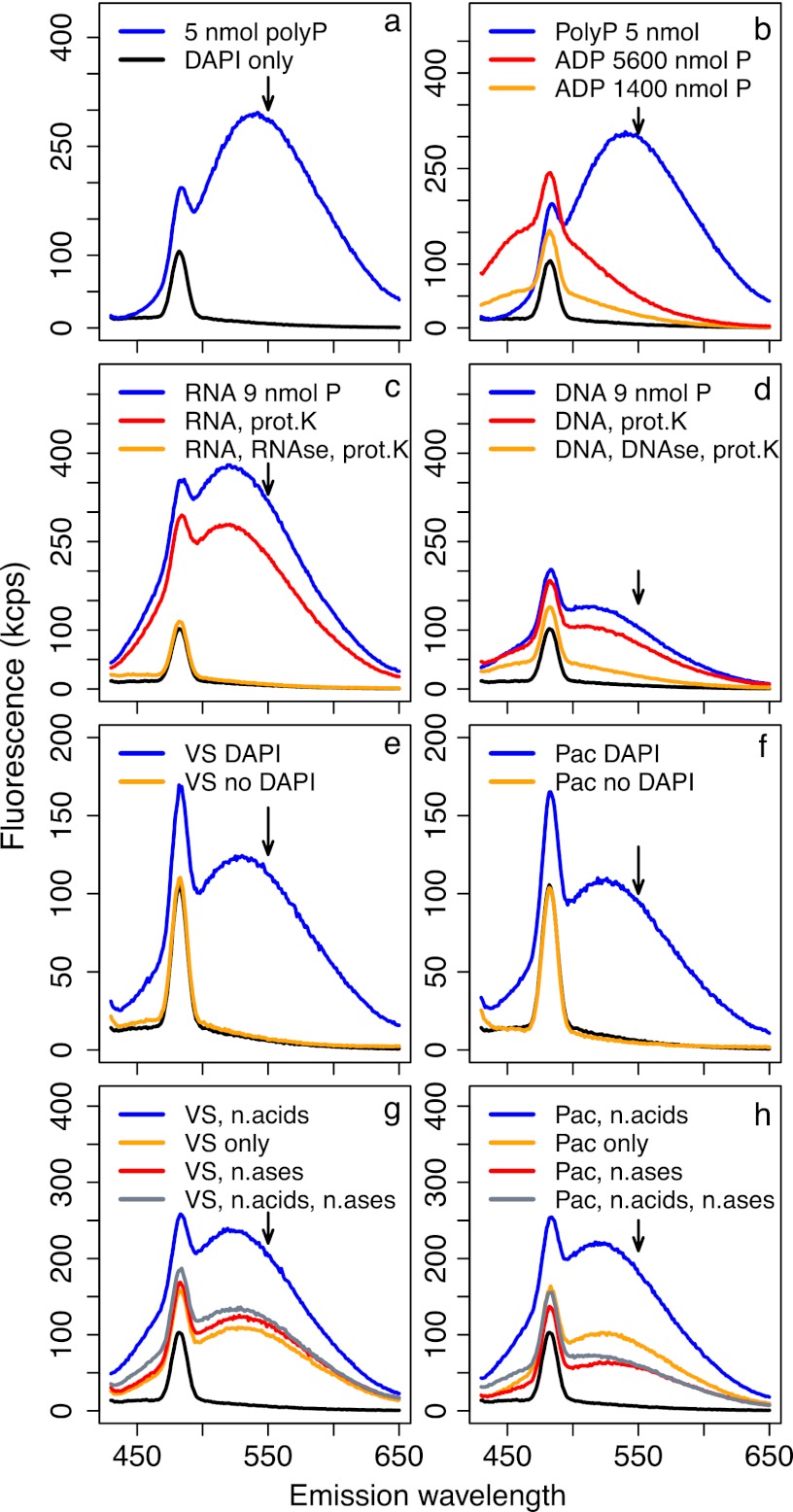
Fluorescence emission spectra under 415-nm excitation; each spectrum corresponds to a single sample. All solutions were stained with DAPI unless otherwise indicated. Arrows indicate the wavelength at which emission is measured for polyP quantification (550 nm), and black lines in all panels show spectra of blank Tris buffer stained with DAPI. Fluorescence of 5 nmol polyP solutions greatly exceeded that of even very concentrated ADP solutions (a and b). However, 3 μg of RNA and DNA (∼9 nmol P) yielded high fluorescence when stained with DAPI (c and d), which was only partly reduced during our core polyP extraction protocol of boiling and proteinase K (prot.K) treatment (red lines) but almost completely reduced when nuclease treatments were incorporated into the extraction protocol (orange lines). Natural particle samples from the Vineyard Sound (e) and North Pacific (f) showed very clean fluorescence spectra with and without DAPI; stained spectra were consistent with polyP-DAPI fluorescence. When Vineyard Sound (g) and North Pacific (h) samples were amended with 1.5 μg each of DNA and RNA (∼4.5 nmol P), fluorescence was strongly increased relative to that of unamended samples (blue versus orange lines, respectively). This increase was completely removed by treatment with DNase and RNase (gray lines). Treatment with DNase and RNase of samples not amended with nucleic acids (n,acids) did not change fluorescence in Vineyard Sound samples (g, red line) but caused a strong decrease in the North Pacific sample (h, red line).
Fig 7.
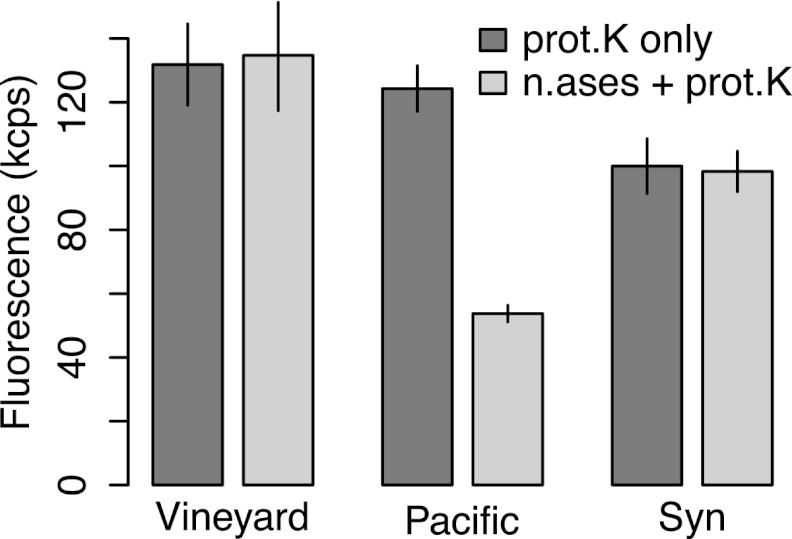
Six replicate samples from the Vineyard Sound, the North Pacific, and the Synechococcus culture were divided into two treatments of three replicates each and extracted either with just boiling and proteinase K treatment or with boiling, DNase and RNase treatment, and proteinase K treatment. Data are means and SDs.
Standard addition experiments.
We used standard addition to test whether matrix effects compromised polyP quantification (see the supplemental material for details). The additional fluorescence signal due to the added standard in our samples was consistently either lower or greater than the signal that the corresponding amount of standard would have in pure solution (Fig. 5). All results obtained with additions of >1 nmol polyP standard were very consistent for each sample type; with additions of <1 nmol, there was more variability, since the increase in fluorescence was small relative to the fluorescence of the unamended samples (Fig. 5d, e, and f). Considering only additions of >1 nmol polyP standard, we determined that matrix effects reduced the signal by 6 to 17% in Vineyard Sound samples and increased the signal by 14 to 21% in Pacific samples and by 27 to 34% in Synechococcus samples. The consistency of our results justifies using a single addition of 2 nmol polyP for routine sample quantification. Overall, not accounting for this matrix effect may cause errors of 10 to 30% in these sample types. However, matrix effects of 41 to 50% were found later with the filtered Vineyard Sound samples (see below).
Fluorescence emission spectra and ADP and nucleic acid interference.
The presence of polyP yielded a single, large, broad peak centered around 540 nm that was not seen in the spectrum of DAPI-stained Tris blanks (Fig. 6a). The presence of 700 to 2,800 nmol ADP (1,400 to 5,600 nmol P) raised the signal at 550 nm but only by a small amount compared to the signal of 5 nmol polyP (Fig. 6b). In contrast, 3 μg RNA (∼9 nmol P) caused a very large peak around 530 nm and a fluorescence signal at 550 nm that was comparable to that of 5 nmol polyP (Fig. 6c). This peak was only slightly reduced when the RNA solution was treated with our core protocol of 5 min boiling and 30 min proteinase K treatment but disappeared completely when the solution was treated for 10 min with an RNase cocktail at 37°C (Fig. 6c). Three micrograms DNA (∼9 nmol P) yielded a peak about half as large as that of RNA, which was largely unaffected by the core extraction protocol but was almost absent after incubation for 10 min with DNase at 37°C (Fig. 6d). The relative brightness of fluorescence at 550 nm with DAPI thus follows the following order: polyP > RNA > DNA ≫ ADP. Although DAPI fluoresces more brightly at shorter wavelengths when bound to DNA than to RNA, it is known that the fluorescence peak of DAPI bound to RNA is shifted to longer wavelengths than when DAPI is bound to DNA (44, 45).
The Vineyard Sound and Pacific samples had very clean fluorescence spectra with and without DAPI (Fig. 6e and f). Addition of 1.5 μg DNA and 1.5 μg RNA to Vineyard Sound (Fig. 6g) and to Pacific (Fig. 6h) samples increased the fluorescence signal at 550 nm strongly relative to that of the simple DAPI-stained extracts (only the 1st extraction was used). This increase completely disappeared if samples amended with DNA and RNA were also treated for 10 min with DNase and RNase at 37°C. While treatment of the pure sample extract with DNase and RNase (i.e., without addition of nucleic acids) did not affect the fluorescence signal at 550 nm for Vineyard Sound or Synechococcus samples (Fig. 6g and 7), the fluorescence measured in Pacific samples was strongly decreased after nuclease treatment (Fig. 6h and 7).
Effect of enzymes on fluorescence of the standard.
Adding proteinase K decreased the fluorescence signal by 15% relative to that of the pure standard solution, adding nucleases increased the signal by 31%, and adding both enzymes increased the signal by 24% (see Fig. S3 in the supplemental material). All blanks, with and without enzymes, ranged only from 5.4 to 6.4 kcps, while treatment means ranged from 80 to 160 kcps. All of these effects are fully accounted for by the standard polyP addition.
Measurement precision.
The analytical precision as determined by repeated analyses of 3 to 4 replicate polyP standard solutions was always in the range of 1 to 3% relative standard deviation (RSD) (see Fig. S3 in the supplemental material). To test the overall precision in quantifying polyP in field samples, we used the replicate filtered Vineyard Sound samples and found that the mean (± SD) concentration of particulate polyP was 0.70 ± 0.19 μM, i.e., an RSD of 27%. Most of the variability was introduced by subtracting the unusually high fluorescence of the DAPI-stained background extracts (the fourth extracts). These samples also had greater matrix effects than all other samples, with fluorescence of added standard polyP reduced by 41 to 50% relative to pure standard solutions. Furthermore, natural heterogeneity in polyP abundance also contributed to the observed imprecision.
Quantification of polyP in the Pacific Ocean.
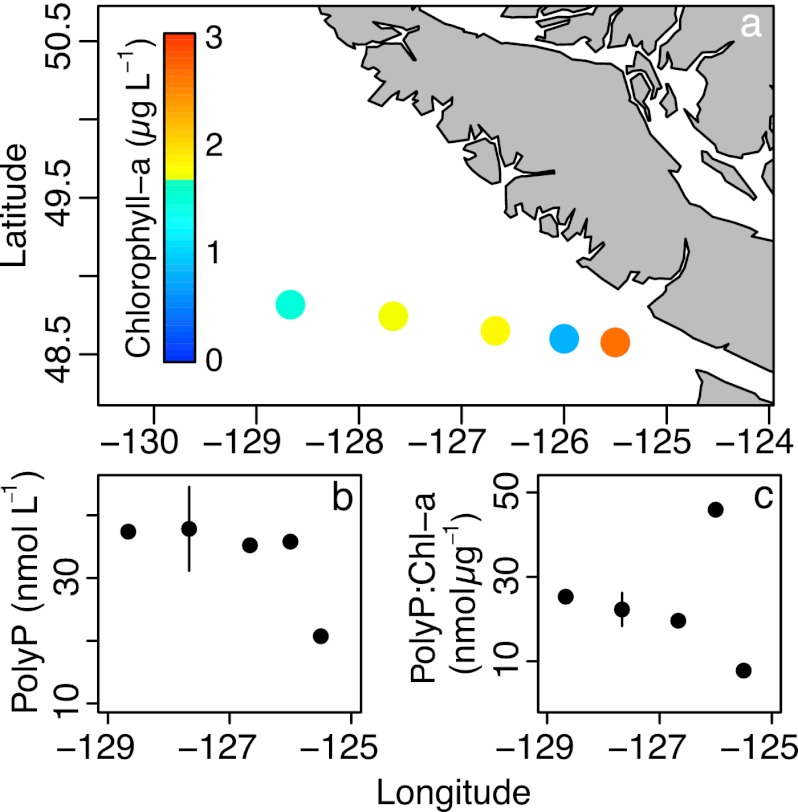
We quantified particulate polyP at five stations along a North Pacific near-shore to offshore transect (Fig. 8a) using our core extraction protocol including a 10-min nuclease treatment. The particulate polyP concentration increased from 21 to about 38 nmol liter−1 (Fig. 8b), while the Chl a concentration decreased with distance from the shore (2.6 to 1.5 μg liter−1). The sample at 48.74°N, 127.67°W (second-most oceanic station), was analyzed twice, and the average and range of the two values are shown. The ratio of polyP to Chl a increased steadily from 7.9 nmol μg−1 at the near-shore station to 26 nmol μg−1 at the offshore station, though a value of 46 was found close to shore at the station with the lowest Chl a (Fig. 8c).
Fig 8.
Particulate polyP was quantified on a transect in the North Pacific after filtering 900 ml of seawater collected at 5-m depth onto GF/F filters. Sampling locations and chlorophyll a concentrations at 5 m are shown in panel a. The concentration of polyP increased from coast to open ocean (b), as did the polyP/chlorophyll a ratio (c). Given the low sample size of this opportunistically sampled data set, we eschewed statistical analysis. The error bars in panels b and c indicate the range of a duplicate polyP measurement of one sample; the data point is the mean of the two measurements.
DISCUSSION
Purification versus in vivo quantification of polyP.
As in previous studies (36, 37), we found that purification of polyP with silica matrices gave unsatisfactory yields. While the relationship between percentage yield and amount of polyP was relatively tight, more than 50 nmol polyP was needed to give high and stable yields, a concentration which might require impractical sample sizes. The one study that examined the chain length dependency of purification yield reported no difference in yield upwards of 45 residues (37). Moreover, our results are remarkably consistent with those of three previous yield measurements (36) despite using a different protocol. While purification appears unsatisfactory for quantifying polyP, it may still be useful in P radiotracer experiments to quantify the amount of label incorporated into polyP: a large quantity of unlabeled carrier polyP can be added immediately prior to purification to ensure a high yield.
Improvements to the in vivo measurement method.
We have tested a broader range of extraction protocols than previous studies; in particular, we are unaware of prior attempts to use enzymatic digestions to extract polyP. We found that 5 min boiling in Tris buffer followed by a 30-min proteinase K treatment was superior to all other methods we tested, both with a Synechococcus culture and with natural marine particles collected from the Vineyard Sound. We posit that this reflects more-efficient dissolution of polyP from granules. Hot water extraction has been used previously to liberate polyP granules (41) and has been described as a good method of obtaining relatively unaltered polyP (1). PolyP can associate with proteins in solution (1, 46, 47), which might prevent binding of DAPI. Moreover, polyP granules contain protein (42, 48, 49), which has been hypothesized to stabilize the granules (42). While the proteases trypsin and chymotrypsin were reported not to dissolve granules, sodium dodecyl sulfate, a nonspecific protein denaturant, did dissolve them (42). Proteinase K has much broader specificity than trypsin and chymotrypsin, and thus, we believe that the primary effect of proteinase K treatment is probably to dissolve granules.
Neither an alkaline leach nor simple freeze-thawing gave very satisfactory results in our experiments, although these have been used by other workers (36, 39). This must reflect inherent differences in sample types, with freeze-dried, ground samples (39) perhaps being more vulnerable to NaOH than our fresh samples. Similarly, the bacterial cultures and sludge samples analyzed by Kulakova et al. (36) were maybe more easily lysed by freeze-thawing than our samples.
Previous work on developing the in vivo polyP quantification method (36, 38) did not discuss the extraction efficiency of extraction protocols, although Diaz and Ingall (39) used two successive alkaline leaches. While our combined boiling, nucleases, and proteinase K treatment was by far the best method of all we tested, our samples still needed three successive extractions until a stable background was reached.
Interference by nucleic acids and nucleoside polyphosphates was judged to be negligible based on experiments with DNA and ATP (38, 39). We ran similar experiments with ADP, DNA, and RNA. While our results support this conclusion for nucleosides and suggest that DNA is probably no major problem, we found that fluorescence of the RNA-DAPI complex is a potentially serious interference. Moreover, nuclease treatment strongly reduced the fluorescence signal in our North Pacific samples but had no effect in the other samples, demonstrating that nucleic acids cannot always be ignored when measuring polyP.
Finally, standard addition revealed a matrix effect in all sample types, which caused the DAPI-polyP signal to be either over- or underestimated, depending on sample type. This result was definitely not caused by a discrepancy between the nominal and actual amounts of polyP added (e.g., due to pipetting errors), since we used the exact same polyP standard solutions for the additions and the standard curve. Furthermore, all solutions and standard additions were made at exactly the same time with the same set of pipettes. Hence, any slight discrepancy between nominal and actual amounts of polyP added to the samples would have been present in the calibration curve, too, and have no net effect on the final result.
We also found that addition of proteinase K and nucleases to pure solutions of polyP standard can decrease or increase the fluorescence signal by up to 30%. Since the enzyme-amended blanks showed no such differences relative to enzyme-free blanks, we ascribe this matrix effect to an interaction between the enzymes and the polyP-DAPI complex. The increase in fluorescence upon addition of nucleases validates our observations with natural particle samples that the matrix can enhance the fluorescence signal. The use of enzymes does not, however, complicate the quantification of polyP, since our method accounts for any matrix effects by standard addition. Moreover, it is striking that while nuclease addition clearly increases fluorescence in a pure standard solution, the signal was not affected in Vineyard Sound or Synechococcus samples (Fig. 7). This might simply indicate that the signal increase due to nucleases was matched by a reduction in fluorescence from nucleic acid digestion. Alternatively, it may be that the sample matrix was already so complex that the presence of nucleases had no further effect, in which case, the matrix effect from the added enzymes would be greatest in pure standard solutions.
In our samples, the matrix effect changed the fluorescence signal by up to 50%, so we recommend standard addition for routine quantification. However, no matrix effect was reported for a marine bacterial culture (39), but to our knowledge, other sample types (e.g., sludge or sediments) have not yet been examined for matrix effects.
Measurement precision.
The analytical precision of our fluorescence measurements was very good, though quantification of replicate field samples had a higher RSD of ±27%. Part of the variability doubtless stems from heterogeneity introduced by subsampling the filters, a known problem in chemical oceanography (50). Furthermore, the DAPI-stained background fluorescence and matrix effects were unusually high in these samples, and extraction and quantification of polyP in such near-shore samples rich in organic and inorganic particles may be inherently less precise than extraction and quantification in open ocean samples.
PolyP in the marine environment.
Our new method has allowed us to assemble a unique data set on total particulate polyP concentrations in a transect across an oceanographic transition zone. Since we used GF/F filters, which have a nominal pore size of 0.7 μm, we expect that the majority of the polyP we measured was derived from phytoplankton, not heterotrophic bacteria. Interestingly, we found that open ocean waters generally contained more particulate polyP by volume and as a ratio to Chl a than near-shore waters and that the PolyP-Chl a ratio spiked at a station within the ecological transition zone (43). A recent metagenomic study concluded that polyP metabolism is more important where external soluble reactive P (SRP) is <0.2 μmol liter−1 (51). SRP was most probably above 0.2 μmol liter−1 throughout our transect, increasing toward the high-nutrient/low-chlorophyll (HNLC) waters of the open subarctic (43).
Thus, the observed increase of polyP with SRP might reflect luxury P storage under abundant supply. Yet it is also possible that higher polyP levels further offshore fulfill important physiological functions—especially since it is not obvious how cells would benefit from a luxury polyP store in a region that has perennially high SRP. In the Sargasso Sea, Trichodesmium colonies showed a clear increase in polyP content with distance from the shore without a concomitant gradient in SRP (28). This was hence hypothesized to reflect a putative increase in stress factors, possibly temperature stress, with distance from the shore (28).
Our transect crossed a region previously described as a transition zone from nutrient-limited near-shore waters that are iron replete to iron-limited HNLC offshore waters that are P replete (43). Mixing of these waters was reported to yield blooms characteristic of an iron-fertilized offshore community, implying that there might be a gradient in nutrient (iron) stress across our transect (43). PolyP might be involved in the luxury storage of iron (29), and it is conceivable that cells further offshore might use polyP to build up an iron store to outlast periods of iron depletion later in the season.
Being an ecological transition zone, the phytoplankton community composition also changed along the transect, with Synechococcus increasing and diatoms decreasing with distance from the shore (43), a trend which might be partly responsible for our observations. However, we suspect that physiological plasticity in cellular polyP levels in response to environmental factors may outweigh community composition effects. Variation in cellular polyP content in the oceans might therefore point to variation in the severity of stress factors. Our data thus underscore the need for more measurements of this important compound in the ocean.
Future application of the method.
Our method is optimized for marine particulate samples and phytoplankton cultures and is feasible for the routine analysis of large sample sets from oceanographic survey programs. We recommend that future applications of the in vivo fluorescence method for quantifying polyP in other types of samples should ensure that the extraction protocol is optimized, with boiling and enzymatic protein digestion holding promise. We suggest that samples be treated with nucleases routinely and that the presence or lack of matrix effects should be verified using standard addition. Moreover, we recommend that successive extractions are made to ensure that all polyP is extracted from the sample.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dan Repeta for granting us usage of his spectrofluorometer, Sara Bender and Ginger Armbrust for sharing chlorophyll data from the North Pacific GEOMICS cruise, Julia Diaz for helpful discussion of polyP extraction protocols, and Dawn Moran, Sheean Haley, Louie Wurch, and Sonya Dyhrman for assistance with culturing Synechococcus. We are grateful to three anonymous reviewers for constructive criticism that improved this paper.
Footnotes
Published ahead of print 26 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02592-12.
REFERENCES
- 1. Kulaev IS, Vagabov VM, Kulakovskaya TV. 2004. The biochemistry of inorganic polyphosphates, 2nd ed Wiley, Chichester, West Sussex, England [Google Scholar]
- 2. Candon HL, Allan BJ, Fraley CD, Gaynor EC. 2007. Polyphosphate kinase 1 is a pathogenesis determinant in Campylobacter jejuni. J. Bacteriol. 189:8099–8108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kim KS, Rao NN, Fraley CD, Kornberg A. 2002. Inorganic polyphosphate is essential for long-term survival and virulence factors in Shigella and Salmonella spp. Proc. Natl. Acad. Sci. U. S. A. 99:7675–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rashid MH, Rumbaugh K, Passador L, Davies DG, Hamood AN, Iglewski BH, Kornberg A. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 97:9636–9641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen W, Palmer RJ, Kuramitsu HK. 2002. Role of polyphosphate kinase in biofilm formation by Porphyromonas gingivalis. Infect. Immun. 70:4708–4715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Silby MW, Nicoll JS, Levy SB. 2009. Requirement of polyphosphate by Pseudomonas fluorescens Pf0-1 for competitive fitness and heat tolerance in laboratory media and sterile soil. Appl. Environ. Microbiol. 75:3872–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kornberg A, Rao NN, Ault-Riché D. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89–125 [DOI] [PubMed] [Google Scholar]
- 8. Rao NN, Liu SJ, Kornberg A. 1998. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J. Bacteriol. 180:2186–2193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brown MRW, Kornberg A. 2008. The long and short of it—polyphosphate, PPK and bacterial survival. Trends Biochem. Sci. 33:284–290 [DOI] [PubMed] [Google Scholar]
- 10. Rao NN, Kornberg A. 1996. Polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178:1394–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shiba T, Tsutsumi K, Yano H, Ihara Y, Kameda A, Tanaka K, Takahashi H, Munekata M, Rao NN, Kornberg A. 1997. Inorganic polyphosphate and the induction of rpoS expression. Proc. Natl. Acad. Sci. U. S. A. 94:11210–11215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jahid IK, Silva AJ, Benitez JA. 2006. Polyphosphate stores enhance the ability of Vibrio cholerae to overcome environmental stresses in a low-phosphate environment. Appl. Environ. Microbiol. 72:7043–7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shirahama K, Yazaki Y, Sakano K, Wada Y, Ohsumi Y. 1996. Vacuolar function in the phosphate homeostasis of the yeast Saccharomyces cerevisiae. Plant Cell Physiol. 37:1090–1093 [DOI] [PubMed] [Google Scholar]
- 14. Dyhrman ST, Jenkins BD, Rynearson TA, Saito MA, Mercier ML, Alexander H, Whitney LP, Drzewianowski A, Bulygin VV, Bertrand EM, Wu ZJ, Benitez-Nelson C, Heithoff A. 2012. The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS One 7:e33768 doi:10.1371/journal.pone.0033768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Perry MJ. 1976. Phosphate utilization by an oceanic diatom in phosphorus-limited chemostat culture and in the oligotrophic waters of the central North-Pacific. Limnol. Oceanogr. 21:88–107 [Google Scholar]
- 16. Günther S, Trutnau M, Kleinsteuber S, Hause G, Bley T, Röske I, Harms H, Müller S. 2009. Dynamics of polyphosphate-accumulating bacteria in wastewater treatment plant microbial communities detected via DAPI (4′,6-diamidino-2-phenylindole) and tetracycline labeling. Appl. Environ. Microbiol. 75:2111–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kulakovskaya TV, Vagabov VM, Kulaev IS. 2012. Inorganic polyphosphate in industry, agriculture and medicine: modern state and outlook. Process Biochem. 47:1–10 [Google Scholar]
- 18. Majed N, Chernenko T, Diem M, Gu AZ. 2012. Identification of functionally relevant populations in enhanced biological phosphorus removal processes based on intracellular polymers profiles and insights into the metabolic diversity and heterogeneity. Environ. Sci. Technol. 46:5010–5017 [DOI] [PubMed] [Google Scholar]
- 19. Martin P, Van Mooy BAS, Heithoff A, Dyhrman ST. 2011. Phosphorus supply drives rapid turnover of membrane phospholipids in the diatom Thalassiosira pseudonana. ISME J. 5:1057–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pujo-Pay M, Conan P, Oriol L, Cornet-Barthaux V, Falco C, Ghiglione JF, Goyet C, Moutin T, Prieur L. 2011. Integrated survey of elemental stoichiometry (C, N, P) from the western to eastern Mediterranean Sea. Biogeosciences 8:883–899 [Google Scholar]
- 21. Thingstad TF, Krom MD, Mantoura RFC, Flaten GAF, Groom S, Herut B, Kress N, Law CS, Pasternak A, Pitta P, Psarra S, Rassoulzadegan F, Tanaka T, Tselepides A, Wassmann P, Woodward EMS, Riser CW, Zodiatis G, Zohary T. 2005. Nature of phosphorus limitation in the ultraoligotrophic eastern Mediterranean. Science 309:1068–1071 [DOI] [PubMed] [Google Scholar]
- 22. Twining BS, Nunez-Milland D, Vogt S, Johnson RS, Sedwick PN. 2010. Variations in Synechococcus cell quotas of phosphorus, sulfur, manganese, iron, nickel, and zinc within mesoscale eddies in the Sargasso Sea. Limnol. Oceanogr. 55:492–506 [Google Scholar]
- 23. Van Mooy BAS, Fredricks HF, Pedler BE, Dyhrman ST, Karl DM, Koblizek M, Lomas MW, Mincer TJ, Moore LR, Moutin T, Rappe MS, Webb EA. 2009. Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458:69–72 [DOI] [PubMed] [Google Scholar]
- 24. Wu JF, Sunda W, Boyle EA, Karl DM. 2000. Phosphate depletion in the western North Atlantic Ocean. Science 289:759–762 [DOI] [PubMed] [Google Scholar]
- 25. Wurch LL, Bertrand EM, Saito MA, Van Mooy BAS, Dyhrman ST. 2011. Proteome changes driven by phosphorus deficiency and recovery in the brown tide-forming alga Aureococcus anophagefferens. PLoS One 6:e28949 doi:10.1371/journal.pone.0028949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diaz J, Ingall E, Benitez-Nelson C, Paterson D, de Jonge MD, McNulty I, Brandes JA. 2008. Marine polyphosphate: a key player in geologic phosphorus sequestration. Science 320:652–655 [DOI] [PubMed] [Google Scholar]
- 27. Diaz JM, Ingall ED, Snow SD, Benitez-Nelson CR, Taillefert M, Brandes JA. 2012. Potential role of inorganic polyphosphate in the cycling of phosphorus within the hypoxic water column of Effingham Inlet, British Columbia. Global Biogeochem. Cycles 26:GB2040 doi:10.1029/2011GB004226 [Google Scholar]
- 28. Orchard ED, Benitez-Nelson CR, Pellechia PJ, Lomas MW, Dyhrman ST. 2010. Polyphosphate in Trichodesmium from the low-phosphorus Sargasso Sea. Limnol. Oceanogr. 55:2161–2169 [Google Scholar]
- 29. Nuester J, Vogt S, Twining BS. 2012. Localization of iron within centric diatoms of the genus Thalassiosira. J. Phycol. 48:626–634 [DOI] [PubMed] [Google Scholar]
- 30. Ahlgren J, De Brabandere H, Reitzel K, Rydin E, Gogoll A, Waldebäck M. 2007. Sediment phosphorus extractants for phosphorus-31 nuclear magnetic resonance analysis: a quantitative evaluation. J. Environ. Qual. 36:892–898 [DOI] [PubMed] [Google Scholar]
- 31. Hupfer M, Glöss S, Schmieder P, Grossart HP. 2008. Methods for detection and quantification of polyphosphate and polyphosphate accumulating microorganisms in aquatic sediments. Int. Rev. Hydrobiol. 93:1–30 [Google Scholar]
- 32. Ruiz FA, Rodrigues CO, Docampo R. 2001. Rapid changes in polyphosphate content within acidocalcisomes in response to cell growth, differentiation, and environmental stress in Trypanosoma cruzi. J. Biol. Chem. 276:26114–26121 [DOI] [PubMed] [Google Scholar]
- 33. Werner TP, Amrhein N, Freimoser FM. 2005. Novel method for the quantification of inorganic polyphosphate (iPoP) in Saccharomyces cerevisiae shows dependence of iPoP content on the growth phase. Arch. Microbiol. 184:129–136 [DOI] [PubMed] [Google Scholar]
- 34. Ault-Riché D, Fraley CD, Tzeng CM, Kornberg A. 1998. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180:1841–1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Motomura K, Hirota R, Ohnaka N, Okada M, Ikeda T, Morohoshi T, Ohtake H, Kuroda A. 2011. Overproduction of YjbB reduces the level of polyphosphate in Escherichia coli: a hypothetical role of YjbB in phosphate export and polyphosphate accumulation. FEMS Microbiol. Lett. 320:25–32 [DOI] [PubMed] [Google Scholar]
- 36. Kulakova AN, Hobbs D, Smithen M, Pavlov E, Gilbert JA, Quinn JP, McGrath JW. 2011. Direct quantification of inorganic polyphosphate in microbial cells using 4′-6-diamidino-2-phenylindole (DAPI). Environ. Sci. Technol. 45:7799–7803 [DOI] [PubMed] [Google Scholar]
- 37. Mullan A, Quinn JP, McGrath JW. 2002. A nonradioactive method for the assay of polyphosphate kinase activity and its application in the study of polyphosphate metabolism in Burkholderia cepacia. Anal. Biochem. 308:294–299 [DOI] [PubMed] [Google Scholar]
- 38. Aschar-Sobbi R, Abramov AY, Diao C, Kargacin ME, Kargacin GJ, French RJ, Pavlov E. 2008. High sensitivity, quantitative measurements of polyphosphate using a new DAPI-Based approach. J. Fluoresc. 18:859–866 [DOI] [PubMed] [Google Scholar]
- 39. Diaz JM, Ingall ED. 2010. Fluorometric quantification of natural inorganic polyphosphate. Environ. Sci. Technol. 44:4665–4671 [DOI] [PubMed] [Google Scholar]
- 40. Borodina TA, Lehrach H, Soldatov AV. 2003. DNA purification on homemade silica spin-columns. Anal. Biochem. 321:135–137 [DOI] [PubMed] [Google Scholar]
- 41. Eixler S, Selig U, Karsten U. 2005. Extraction and detection methods for polyphosphate storage in autotrophic planktonic organisms. Hydrobiologia 533:135–143 [Google Scholar]
- 42. Friedberg I, Avigad G. 1968. Structures containing polyphosphate in Micrococcus lysodeikticus. J. Bacteriol. 96:544–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ribalet F, Marchetti A, Hubbard KA, Brown K, Durkin CA, Morales R, Robert M, Swalwell JE, Tortell PD, Armbrust EV. 2010. Unveiling a phytoplankton hotspot at a narrow boundary between coastal and offshore waters. Proc. Natl. Acad. Sci. U. S. A. 107:16571–16576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kapuscinski J. 1990. Interactions of nucleic acids with fluorescent dyes: spectral properties of condensed complexes. J. Histochem. Cytochem. 38:1323–1329 [DOI] [PubMed] [Google Scholar]
- 45. Tanious FA, Veal JM, Buczak H, Ratmeyer LS, Wilson WD. 1992. DAPI (4′,6-diamidino-2-phenylindole) binds differently to DNA and RNA: minor-groove binding at AT sites and intercalation at AU sites. Biochemistry 31:3103–3112 [DOI] [PubMed] [Google Scholar]
- 46. Yang ZX, Zhou YN, Yang Y, Jin DJ. 2010. Polyphosphate binds to the principal sigma factor of RNA polymerase during starvation response in Helicobacter pylori. Mol. Microbiol. 77:618–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Young N, Bullock S, Orlovich DA, Ashford AE. 1993. Association of polyphosphate with protein in freeze-substituted sclerotia of Sclerotinia minor. Protoplasma 174:134–141 [Google Scholar]
- 48. Jacobson L, Halmann M, Yariv J. 1982. The molecular composition of the volutin granule of yeast. Biochem. J. 201:473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pallerla SR, Knebel S, Polen T, Klauth P, Hollender J, Wendisch VF, Schoberth SM. 2005. Formation of volutin granules in Corynebacterium glutamicum. FEMS Microbiol. Lett. 243:133–140 [DOI] [PubMed] [Google Scholar]
- 50. Peterson ML, Wakeham SG, Lee C, Askea MA, Miquel JC. 2005. Novel techniques for collection of sinking particles in the ocean and determining their settling rates. Limnol. Oceanogr. Methods 3:520–532 [Google Scholar]
- 51. Temperton B, Gilbert JA, Quinn JP, McGrath JW. 2011. Novel analysis of oceanic surface water metagenomes suggests importance of polyphosphate metabolism in oligotrophic environments. PLoS One 6:e16499 doi:10.1371/journal.pone.0016499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.