Abstract
Stable isotope probing of magnetic-bead-captured rRNA (Mag-SIP) indicated clear differences in in situ organic substrate utilization by major microbial groups between the more oxidized (0 to 2 cm) and sulfate-reducing (2 to 5 cm) horizons of marine intertidal sediment. We also showed that cyanobacteria and diatoms may survive by glucose utilization under dark anoxic conditions.
TEXT
The microbial community in marine sediments is highly diverse and consists mainly of microorganisms related only distantly to described isolates, whose functions are therefore difficult to predict (1–3). A recent study indicated a higher functional redundancy in the oxidized top layer of marine sediments, where higher disturbance rates and higher availability of substrates may lead to the formation of a community of fast-growing generalists (4). In contrast, a consortium of microbes consisting of specialized fermenting and sulfate-reducing bacteria is thought to be involved in organic matter degradation under sulfate-reducing conditions (5). However, intermediate metabolites are generally at low concentrations due to their high turnover rates, making in situ identification of the microbial groups utilizing them difficult. We utilized a recently developed stable isotope-probing method based on magnetic bead capturing of specific 16S rRNA and subsequent sensitive 13C analysis of the captured material (Mag-SIP) (6, 7) to show major differences in substrate utilization by predominant microbial groups between the oxidized top layer and sulfate-reducing deeper layer of an intertidal marine sediment.
In this study, sediment cores (internal diameter, 5.2 cm) were collected at an intertidal flat in the Rattekaai area of the Oosterschelde Bay (The Netherlands) in May 2008 and injected with d-[13C]glucose, sodium [13C]propionate, sodium [13C]acetate, or a 13C-labeled alga-derived amino acid mixture (Cambridge Isotope Laboratories, Andover, MA) (98% to 99% 13C). Final substrate concentrations were 0.2 μmol 13C cm−3 for amino acids and 0.9 μmol 13C cm−3 for the other three substrates. These substrates represent both major constituents of the organic matter pool (carbohydrates and amino acids) and the main fermentation products (acetate and propionate) in marine sediments. Cores were incubated (24 h, 14°C) and sectioned in surface layers (0 to 2 cm) and deeper layers (2 to 5 cm), which corresponded to a clear color change of the sediment from brown-yellow to dark gray. A nested set of probes targeting approximately 80% of the rRNA sequences recovered from the two sediment layers was used with the Mag-SIP protocol (7). The testing of the probes for total bacterial rRNA (EUB338), Deltaproteobacteria rRNA (DELTA495a), and Desulfobacteraceae rRNA (Dbact653) was previously described by Miyatake et al. (7). Probe CYA361 was used to target cyanobacterium and chloroplast rRNA (20% formamide [8]). We designed a new specific probe and matching helper probes that target the 16S rRNA of most Beta- and Gammaproteobacteria (specific probe BG553 sequence, CGC CCA GTA ATT CCG ATT [60% formamide]; helper probe BG553_up_help sequence, AAC CGC CTR CGN RCG CTT TA; helper probe BG553_down_help sequence, AAC GCT YGC ACC CTM CTG ATT). In this study, the BG553 probe is basically Gammaproteobacteria specific, as we did not detect Betaproteobacteria-related sequences in any of the clone libraries (see Fig. S1 and S2 in the supplemental material). Optimal formamide concentrations in terms of capture efficiency versus specificity were determined as previously described (7) and resulted in specificity of more than 90% (see Fig. S1 in the supplemental material). Clone libraries of reverse-transcribed 16S rRNA from both the total RNA extracts and from the captured 16S rRNA fractions were constructed as described before (7). In addition, clone libraries of the 16S rRNA gene were made from DNA extracted at pH 7.0 using the same phenol-chloroform protocol as was used for RNA. Nucleotide sequences have been deposited in the GenBank/DDBJ/EMBL database (see below).
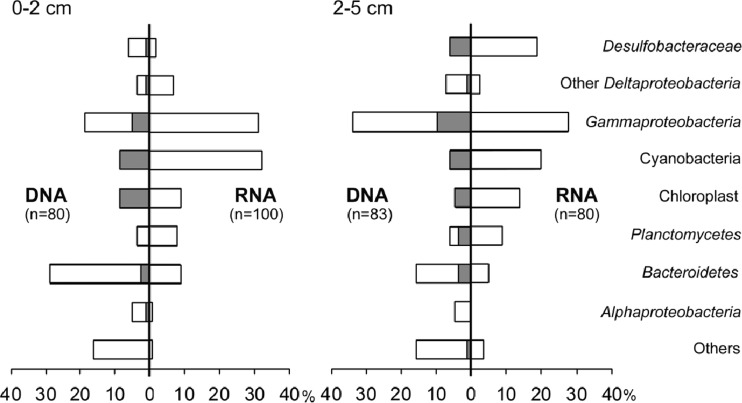
We compared 16S rRNA- and rRNA gene-derived clone libraries, which are considered to represent metabolically active populations and numerically abundant populations, respectively (Fig. 1; see also Fig. S1 in the supplemental material) (9–11). Desulfobacteraceae clones were found in similar (6%) proportions in the rRNA gene libraries corresponding to both depths but were much more abundant in the rRNA library of the deeper layer (19%) than of the surface layer (2%), suggesting that that this group was mainly active in the deeper layer. Gammaproteobacteria clones were found in almost the same proportions in rRNA gene and rRNA libraries from the two layers. Interestingly, cyanobacterial and diatom chloroplast sequences were much more abundant in the rRNA libraries than in the rRNA gene libraries, and nearly all cyanobacterium/chloroplast rRNA gene phylotypes were found in the rRNA libraries (Fig. 1), suggesting that they were viable and actively growing even in dark anoxic sediments. Moreover, in the Mag-SIP incubations, cyanobacteria and diatoms incorporated all the tested 13C substrates in the surface layer and glucose and propionate in the deeper layer (Fig. 2), which clearly shows that they were metabolically active in both layers. Active heterotrophic growth of cyanobacteria and diatoms on glucose under oxic conditions in the dark is well known (12, 13). Under anoxic conditions, many cyanobacteria are able to gain energy from fermentation of storage carbohydrates accumulated during photoautotrophic growth, but fermentation of external organic substrates in free-living cyanobacteria is poorly documented (14). Viability and growth of cyanobacteria under dark anoxic conditions have also been observed in Baltic Sea sediment, based on 16S rRNA libraries and incorporation of bromodeoxyuridine into DNA (15). Recently, Kamp et al. (16) reported that diatoms are able to perform dissimilatory nitrate reduction of intracellular stored nitrate under anoxic dark conditions, which may aid in their survival in anoxic sediments. Many of the cyanobacteria and diatoms in marine sediments may therefore be mixotrophs, suggesting that the functional distinction between phototrophic primary producers and heterotrophic bacteria is blurred, as has also been found for oceanic waters (17).
Fig 1.
Proportion of clones affiliated with major phylogenetic groups among members of each library derived from 16S rRNA gene or reverse-transcribed 16S rRNA in either the surface layer (0 to 2 cm) or the deeper layer (2 to 5 cm). Total numbers of clones sequenced are indicated (n). The shaded parts of the bars in rRNA gene libraries indicate the proportions of clones which were also found in the rRNA library (at the level of ≥97% sequence similarity) in each depth layer.
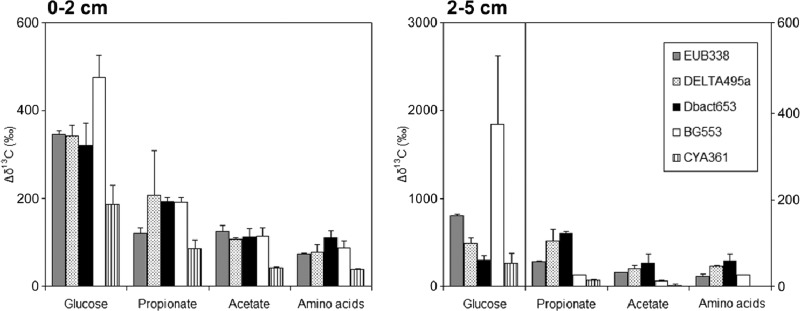
Fig 2.
The increase in δ13C ratios between labeled sediments and unlabeled control sediment (Δδ13C) for the different captured-16S rRNA fractions. Notice that the glucose data for the layer at 2 to 5 cm are plotted on a different axis (left-hand axis) than the data for the other substrates (right-hand axis), as indicated by the vertical line. The probes used in this study targeted bacteria (EUB338), Deltaproteobacteria (DELTA495a), Desulfobacteraceae (Dbact653), Gammaproteobacteria (BG553), and cyanobacteria-diatoms (CYA361). Data for 13C-amino acid-labeled sediment were normalized to the amount of 13C added with the other substrates. Part of the results determined for [13C]glucose, [13C]propionate, and [13C]acetate in the deeper layer are derived from Miyatake et al. (7); all results from the surface layer, amino acid labeling in both layers, and new target organisms (Gammaproteobacteria, cyanobacteria, and diatoms) in both layers are new data. Averages and standard deviations of the results determined for duplicate sediment incubations are presented. Captured rRNA from unlabeled controls had δ13C values between −15‰ and −20‰, within the typical range for marine phytobenthos and bacteria (23), and the δ13C values determined in duplicate analyses of unlabeled controls were within 2‰.
There were strong differences in label incorporation between substrates and microbial groups in the deeper layer (Fig. 2) representing the sulfate-reducing zone of the sediment (18). Gammaproteobacteria clearly showed much higher glucose incorporation than other groups, but they were relatively less important for the other substrates. Desulfobacteraceae, which primarily belonged to the Desulfosarcina-Desulfococcus group (see Fig. S2 in the supplemental material), were main consumers of propionate, acetate, and amino acids (Fig. 2). The Desulfosarcina-Desulfococcus group is ubiquitous and sometimes predominates in microbial communities in anoxic coastal sediments (1, 2, 19), and isolates are complete oxidizing members of the sulfate-reducing bacteria that are able to use a wide range of substrates but typically do not utilize carbohydrates (5, 20). The relatively minor labeling of Desulfobacteraceae with glucose in the deeper layer may suggest some direct incorporation but could also be explained by the use of labeled fermentation products produced by Gammaproteobacteria, which were the dominant glucose consumers (Fig. 2). Webster et al. (21) applied DNA-SIP to study the use of [13C]acetate in anoxic intertidal sediment, and their results partially agree with those of our study, as Desulfobacteraceae were indicated as major consumers of acetate. Epsilonproteobacteria, which were not detected in any of the clone libraries in our study (see Fig. S1 and S2 in the supplemental material), were the dominant consumers of [13C]acetate in another study by the same group on anoxic sediments of the Severn estuary (22). This difference in active community structure may be due to the frequent mixing of the intertidal sediments in the macrotidal Severn estuary or to the low sulfate concentrations detected in the sediment porewater. Our results are in agreement with the model that a consortium of specialized bacteria are involved in anaerobic organic matter degradation and suggest that the major active phylogenetic groups are also the main functional groups in sulfate-reducing marine sediments.
Differences in labeling between groups and substrates were much smaller in the surface layer, indicating limited substrate specialization (Fig. 2). The surface layer also contained the top of the anoxic sediment, and it may well be that the small differences in labeling detected were actually due to incorporation by bacteria in this anoxic part of the surface layer. Based on community dynamics in relation to sediment biogeochemistry and in agreement with our observations, Böer et al. (4) also suggested a higher functional redundancy in the top layer, which was attributed to high disturbance rates and high availability of substrates leading to a community of fast-growing generalists.
In summary, clear shifts between the two layers in the relationship between active phylotypes and substrate incorporation were observed by using Mag-SIP. At the surface, all substrates were evenly utilized by all major groups, indicating limited specialization at the phylogenetic level in this study. In contrast, the major phylogenetic groups were also the main functional groups in the deeper layer, with Gammaproteobacteria dominating glucose utilization and Desulfobacteraceae, specifically members of the Desulfosarcina-Desulfococcus group, important in the utilization of fermentation products. We also showed that the Mag-SIP protocol is sensitive enough to target groups accounting for only 1% to 2% of the total 16S rRNA clones, which means that it can be used to study in situ substrate utilization by dominant environmental clades. Additionally, Mag-SIP labeling results indicate that cyanobacteria and diatoms may survive by glucose utilization under dark anoxic conditions.
Nucleotide sequence accession numbers.
Nucleotide sequences have been deposited in the GenBank/DDBJ/EMBL database under accession numbers GQ449821 to GQ450274.
Supplementary Material
ACKNOWLEDGMENTS
We thank Peter van Breugel, Marco Houtekamer, and Veronique Confurius for assistance with the 13C stable isotope and molecular analysis.
This work was supported by an NWO-VIDI grant to H.T.S.B.
Footnotes
Published ahead of print 19 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02027-12.
REFERENCES
- 1. Bowman JP, McCuaig RD. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69:2463–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gobet A, Boër SI, Huse SM, van Beusekom JEE, Quince C, Sogin ML, Boetius A, Ramette A. 2012. Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J. 6:542–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kemp PF, Aller JY. 2004. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 47:161–177 [DOI] [PubMed] [Google Scholar]
- 4. Böer SI, Hedtkamp SIC, van Beusekom JEE, Fuhrman JA, Boetius A, Ramette A. 2009. Time- and sediment depth-related variations in bacterial diversity and community structure in subtidal sands. ISME J. 3:780–791 [DOI] [PubMed] [Google Scholar]
- 5. Canfield DE, Kristensen E, Thamdrup B. 2005. Aquatic geomicrobiology. Advances in marine biology, vol 48 Elsevier, Amsterdam, The Netherlands: [DOI] [PubMed] [Google Scholar]
- 6. MacGregor BJ, Bruchert V, Fleischer S, Amann R. 2002. Isolation of small-subunit rRNA for stable isotopic characterization. Environ. Microbiol. 4:451–464 [DOI] [PubMed] [Google Scholar]
- 7. Miyatake T, MacGregor BJ, Boschker HTS. 2009. Linking microbial community function to phylogeny of sulfate-reducing Deltaproteobacteria in marine sediments by combining stable isotope probing with magnetic-bead capture hybridization of 16S rRNA. Appl. Environ. Microbiol. 75:4927–4935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schönhuber W, Zarda B, Eix S, Rippka R, Herdman M, Ludwig W, Amann R. 1999. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 65:1259–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Duineveld BM, Kowalchuk GA, Keijzer A, van Elsas JD, van Veen JA. 2001. Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16S rRNA as well as DNA fragments coding for 16S rRNA. Appl. Environ. Microbiol. 67:172–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gaidos E, Rusch A, Ilardo M. 2011. Ribosomal tag pyrosequencing of DNA and RNA from benthic coral reef microbiota: community spatial structure, rare members and nitrogen-cycling guilds. Environ. Microbiol. 13:1138–1152 [DOI] [PubMed] [Google Scholar]
- 11. Mills HJ, Martinez RJ, Story S, Sobecky PA. 2005. Characterization of microbial community structure in Gulf of Mexico gas hydrates: comparative analysis of DNA- and RNA-derived clone libraries. Appl. Environ. Microbiol. 71:3235–3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lewin J, Hellebust JA. 1976. Heterotrophic nutrition of the marine pennate diatom Nitzschia angularis var. affinis. Mar. Biol. 36:313–320 [Google Scholar]
- 13. Smith DJ, Underwood GJC. 2000. The production of extracellular carbohydrates by estuarine benthic diatoms: the effects of growth phase and light and dark treatment. J. Phycol. 36:321–333 [Google Scholar]
- 14. Stal LJ, Moezelaar R. 1997. Fermentation in cyanobacteria. FEMS Microbiol. Rev. 21:179–211 [Google Scholar]
- 15. Edlund A, Hårdeman F, Jansson JK, Sjöling S. 2008. Active bacterial community structure along vertical redox gradients in Baltic Sea sediment. Environ. Microbiol. 10:2051–2063 [DOI] [PubMed] [Google Scholar]
- 16. Kamp A, de Beer D, Nitsch JL, Lavik G, Stief P. 2011. Diatoms respire nitrate to survive dark and anoxic conditions. Proc. Natl. Acad. Sci. U. S. A. 108:5649–5654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eiler A. 2006. Evidence for the ubiquity of mixotrophic bacteria in the upper ocean: implications and consequences. Appl. Environ. Microbiol. 72:7431–7437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Oenema O. 1990. Sulfate reduction in fine-grained sediments in the Eastern Scheldt, Southwest Netherlands. Biogeochemistry 9:53–74 [Google Scholar]
- 19. Ravenschlag K, Sahm K, Knoblauch C, Jørgensen BB, Amann R. 2000. Community structure, cellular rRNA content, and activity of sulfate-reducing bacteria in marine arctic sediments. Appl. Environ. Microbiol. 66:3592–3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Widdel F, Hansen TA. 1992. The dissimilatory sulfate- and sulfur-reducing bacteria, p 583–624 In Balows A, Trüper HG, Dworkin W, Harder W, Schleifer KH. (ed), The prokaryotes, 2nd ed Springer, Berlin, Germany [Google Scholar]
- 21. Webster G, Watt LC, Rinna J, Fry JC, Evershed RP, Parkes RJ, Weightman AJ. 2006. A comparison of stable-isotope probing of DNA and phospholipid fatty acids to study prokaryotic functional diversity in sulfate-reducing marine sediment enrichment slurries. Environ. Microbiol. 8:1575–1589 [DOI] [PubMed] [Google Scholar]
- 22. Webster G, Rinna J, Roussel EG, Fry JC, Weightman AJ, Parkes RJ. 2010. Prokaryotic functional diversity in different biogeochemical depth zones in tidal sediments of the Severn Estuary, UK, revealed by stable-isotope probing. FEMS Microbiol. Ecol. 72:179–197 [DOI] [PubMed] [Google Scholar]
- 23. Boschker HTS, Middelburg JJ. 2002. Stable isotopes and biomarkers in microbial ecology. FEMS Microbiol. Ecol. 40:85–95 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.