Abstract
Background
The p53 protein is activated by genotoxic stress, oncogene expression and during senescence, p53 transcriptionally activates genes involved in growth arrest and apoptosis. p53 activation is regulated by post-translational modification, including phosphorylation of the N-terminal transactivation domain. Here, we have examined how Glycogen Synthase Kinase (GSK3), a protein kinase involved in tumorigenesis, differentiation and apoptosis, phosphorylates and regulates p53.
Results
The 2 isoforms of GSK3, GSK3α and GSK3β, phosphorylate the sequence Ser-X-X-X-Ser(P) when the C-terminal serine residue is already phosphorylated. Several p53 kinases were examined for their ability to create GSK3 phosphorylation sites on the p53 protein. Our results demonstrate that phosphorylation of serine 37 of p53 by DNA-PK creates a site for GSK3β phosphorylation at serine 33 in vitro. GSK3α did not phosphorylate p53 under any condition. GSK3β increased the transcriptional activity of the p53 protein in vivo. Mutation of either serine 33 or serine 37 of p53 to alanine blocked the ability of GSK3β to regulate p53 transcriptional activity. GSK3β is therefore able to regulate p53 function in vivo. p53's transcriptional activity is commonly increased by DNA damage. However, GSK3β kinase activity was inhibited in response to DNA damage, suggesting that GSK3β regulation of p53 is not involved in the p53-DNA damage response.
Conclusions
GSK3β can regulate p53's transcriptional activity by phosphorylating serine 33. However, GSK3β does not appear to be part of the p53-DNA damage response pathway. Instead, GSK3β may provide the link between p53 and non-DNA damage mechanisms for p53 activation.
Background
The p53 tumor suppressor gene is activated during several cellular processes. These include DNA damage caused by Ionizing Radiation and genotoxic agents [1], by expression of activated oncogenes such as ras or myc [2], or during progression of primary cells to senescence [3]. The activation of p53 by these diverse stimuli can initiate either growth arrest or apoptosis depending on the cellular context [1,2,3]. p53 posses sequence-specific DNA binding activity and functions in the cell as a transcriptional regulator. Many p53 regulated genes have been identified [3,4,5], and the majority of the cellular effects of p53 activation can be attributed to the activation of these p53 target genes.
The mechanism of p53 activation in response to either DNA damage or oncogene expression occurs through stabilization of the p53 protein. In unstimulated cells, the mdm2 protein binds to the N-terminal transactivation domain of p53 and targets it for ubiquitin-dependent degradation [6,7]. Activation of p53 requires disruption of the mdm2-p53 interaction to allow p53 accumulation in the cell. 2 distinct mechanisms for p53 activation have so far been elucidated. The expression of oncogenes such as ras in untransformed cells stimulates transcription of the p14Arf gene [2]. p14Arf binds to and sequesters mdm2, allowing free p53 protein to accumulate in the cells [8]. Activation of p53 by DNA damage is also brought about by inhibition of p53-mdm2 interaction. The product of the Ataxia Telangiectasia gene, the ATM protein kinase [9], directly phosphorylates serine 15 of the p53 protein in response to Ionizing Radiation [10,11]. In addition, ATM phosphorylates and activates chk2 kinase [12]. Activated chk2 can then directly phosphorylate serine 20 of p53 [13,14]. ATM therefore controls the phosphorylation of serines 15 and 20 of p53. In addition, DNA damage increases the phosphorylation of serines 33 and 37 of p53 through an ATM-independent mechanism [15,16,17,18]. These DNA damage-induced phosphorylations of p53 block the binding of mdm2 to the N-terminal of the p53 protein [18]. Thus phosphorylation of the p53 protein in response to DNA damage or expression of p14Arf prevents mdm2 binding and p53 protein then accumulates in the cell.
Although stabilization of the p53 protein is the initial step in p53 activation, subsequent steps, including activation of p53's DNA binding activity and changes in p53's transcriptional activity, are also involved. For example, p53's DNA binding activity is increased by the DNA damage-induced acetylation of the C-terminal of p53 [16,19], and this acetylation requires the prior phosphorylation of the N-terminal of p53 [16]. In addition, phosphorylation of the N-terminal transactivation domain of p53 may be required to stimulate transcriptional activation of p53 target genes. Multiple phosphorylation sites have been detected in the N-terminal of p53, including serines 6, 9, 15, 20, 33, 37 and 46 [10,11,12,13,14, 20,21,22]. While phosphorylation of serines 15 and 20 of p53 are clearly dependent on the ATM and chk2 protein kinases [10,11,12,13,14], the kinases responsible for phosphorylation of the remaining serine residues in vivo is not clear.
The activation of p53 by DNA damage or oncogenes such as ras results in either growth arrest or apoptosis of the affected cell. In this study, we have examined how Glycogen Synthase Kinase 3β (GSK3β), a protein kinase involved in tumorigenesis, differentiation and apoptosis, regulates the function of p53 [23,24]. GSK3β phosphorylates several transcription factors, including NFATc and HSF1 [24,25,26,27]. GSK3β is constitutively active in resting cells but is inhibited when cells are exposed to growth factors [24,28].
GSK3 inhibition occurs when the p110-PI 3-kinase/Protein Kinase B (PKB) pathway is activated by growth factors [23,24,25,29]. Activated PKB then phosphorylates GSK3β, inhibiting GSK3 kinase activity [29]. This activation of the p110-PI 3-kinase/PKB pathway, and inhibition of GSK3, delivers a strong anti-apoptotic signal to the cell [23,24,25]. Given the well characterized role of p53 in apoptosis [3,4,5], we examined if GSK3p participates in the regulation of the p53 protein.
GSK3 phosphorylates the consensus sequence Ser-X-X-X-Ser(P), where the C-terminal serine residue is already phosphorylated [24,28]. Thus GSK3 only phosphorylates target proteins which have already been phosphorylated by a separate, priming kinase. p53 contains 5 potential GSK3 phosphorylation sites, 3 in the N-terminal transactivation domain and 2 in the C-terminal regulatory domain. Here, we show that GSK3β, but not GSK3α, can phosphorylate serine 33 of p53 in vitro when serine 37 is already phosphorylated. Further, GSK3β can increase p53's transcriptional activity in vivo, and this activation is lost when serine 33 is mutated to alanine.
Results
The p53 protein contains several serine residues which are located within potential GSK3 phosphorylation sites. Protein kinases which can phosphorylate p53 within these predicted GSK3 sites include MAP kinase, Protein Kinase A, Protein Kinase C, Casein Kinase II, Jun kinase (JNK) and DNA-dependent Protein Kinase (DNA-PK) [30,31,32,33,34,35]. These kinases were examined to determine if they can act as the priming kinase for either of the 2 isoforms of GSK3, GSK3α and GSK3β. The general protocol was to incubate purified priming kinases with p53-GST fusion protein and ATP for 5 h, then heat inactivate the priming kinase. Preliminary experiments indicated that each of the tested kinases was able to phosphorylate p53-GST under the experimental conditions (data not shown). Aliquots of the prephosphorylated p53-GST were then incubated with or without recombinant GSK3α or GSK3β and 32P-ATP to measure p53 phosphorylation.
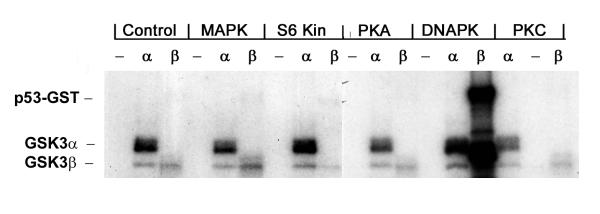
p53-GST preincubated in buffer alone, and then exposed to the heat inactivation protocol was not phosphorylated by either GSK3α or GSK3β (fig 1, Control). Autophosphorylation of GSK3α and GSK3β can be seen (figure 1). Unphosphorylated p53-GST is therefore not a substrate for GSK3α or GSK3β in vitro. p53-GST was then prephosphorylated with MAP kinase, S6 kinase, Protein Kinase A, DNA-PK and Protein Kinase C (fig 1) or Casein Kinase II and JNK1/JNK2 (data not shown). Following incubation in each of the primary kinases, the prephosphorylated p53-GST was then incubated with 32P-ATP and either GSK3α or GSK3β. Prephosphorylation of p53-GST by MAP kinase, S6 kinase, Protein Kinase A or Protein Kinase C (fig 1) or Casein Kinase II or JNK1 or JNK 2 (data not shown) failed to create a phosphorylation site for either GSK3α or GSK3β. However, prephosphorylation of p53-GST by DNA-PK resulted in strong phosphorylation of p53 by GSK3β (fig 1) but not GSK3α. GST protein alone incubated with DNA-PK was not phosphorylated by GSK3β (data not shown). Since there was no significant phosphorylation of p53-GST by GSK3α (fig 1), this indicates that phosphorylation of p53 by DNA-PK, in vitro, creates a site for GSK3β to phosphorylate p53.
Figure 1.
Phosphorylation of p53 by GSK3β. p53-GST (2 μg) was incubated in the absence (Control) or presence of the indicated protein kinase as described in methods. Primary kinases were then heat inactivated (65°C/15 min). Aliquots of the phosphorylated p53-GST were then incubated in the absence (-), or presence of recombinant GSK3α (α) or GSK3β (β) and 10 μCi 32P-ATP. Phosphorylated p53-GST (0.25 μg total p53-GST was detected by SDS-PAGE followed by auto-radiography. MAPK, MAP kinase; S6 Kin, S6 Kinase; PKA, Protein Kinase A; PKC, Protein Kinase C.
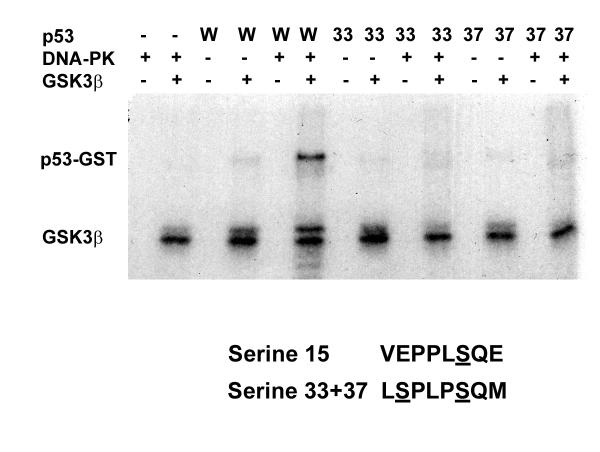
Next, we set out to identify the exact amino-acid(s) within the p53 protein which were phosphorylated by GSK3β. p53 is phosphorylated by DNA-PK at serines 15 and 37 [32]. Of these 2 sites, the sequence around serine 37 contains a predicted GSK3 phosphorylation site at serine 33 (figure 2, underlined). Serine 33 and 37 of p53 were individually mutated to alanine, and p53-GST fusion proteins containing these mutations prepared. The ability of DNA-PK to create a GSK3β phosphorylation site on these p53 proteins was then examined.
Figure 2.
GSK3β phosphorvlates serine 33 of p53. wtp53-GST (W), p53-GSTS33A (33) or p53-GSTS37A (37) were preincubated in buffer (-) or DNA-PK (+) for 5 h in the presence of excess ATP. Following heat inactivation of the DNA-PK, aliquots of the phosphorylated p53-GST fusion proteins were incubated for 30 min without (-) or with (+) GSK-3β. Position of p53-GST and GSK3β is indicated. The sequence of p53 around serines 15 and 37 is shown below the figure.
In the absence of p53, no phosphorylation by GSK3β was detected (fig 2). Unphosphorylated p53-GST was not a substrate for GSK3β, whereas p53-GST prephosphorylated by DNA-PK was (fig 2). Mutation of serine 33 of p53 to alanine (S33A) blocked the ability of GSK3β to phosphorylate p53, indicating that serine 33 is the likely target for GSK3β (fig 2, 33). Similarly, mutation of serine 37, which abolishes the DNA-PK phosphorylation site, blocks phosphorylation of p53 by GSK3β. This indicates that p53 must be phosphorylated on serine 37 by DNA-PK before it can be phosphorylated at serine 33 by GSK3β.
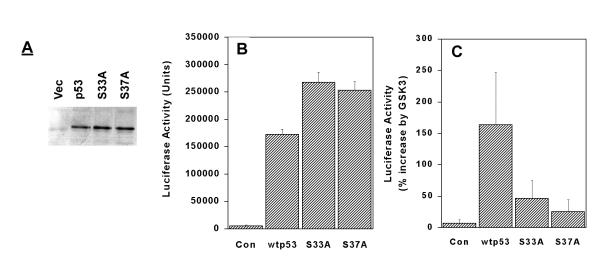
To examine the in vivo function of these in vitro phosphorylations, wtp53 and p53 with the S33A and S37A mutations were sub-cloned into the expression vector pcDNA3.1. These were then expressed in the human osteosarcoma cell line SAOS-2, which does not express endogenous p53 protein [33]. First, we analyzed the level of expression of each of the p53 proteins following transient expression in SAOS-2 cells. In fig 3A, approx equal amounts of wtp53, p53S33A and p53S37A were detected by western blot, indicating that they were expressed at similar levels. In fig 3B, we examined the transcriptional activity of these p53 constructs. A p53-reporter construct, p50-2, which specifically responds to wtp53 by increasing transcription of the luciferase gene, was used [described in 33]. In fig 3B, SAOS-2 cells transiently expressing vector (Con), showed minimal activation of the p53-reporter construct. Cells expressing wtp53 showed significant activation, as did cells expressing both the S33A and S37A mutations. Both the S33A and S37A mutations displayed slightly higher basal levels of transcriptional activity than the wtp53 protein.
Figure 3.
GSK3β regulates p53 transcriptional activity.(A) SAOS-2 cells were transiently transfected with vector (pcDNA3.1), or expression vectors for wtp53, wtp53S33A (S33A) or wtp53S37A (S37A). p53 expression was detected by western blotting. (B+C). SAOS-2 cells were transiently transfected with p50-2, a p53-responsive luciferase reporter construct, and 50 ng of vector (Con), wtp53, wtp53S33A (S33A), or wtp53S37A (S37A) as indicated, β-galactosidase activity from pCMV-Gal was used to adjust for transfection efficiency. In (B), the actual transcriptional activity of each p53 construct is shown. In (C), cells were cotransfected with an expression vector for GSK3β (pcDGSKSβ ; 800 ng). The transcriptional activity was calculated by dividing p53 activity in the presence of GSK3b by p53 activity in the absence of GSK3b and expressing the answer as a percentage. 0% = no increase
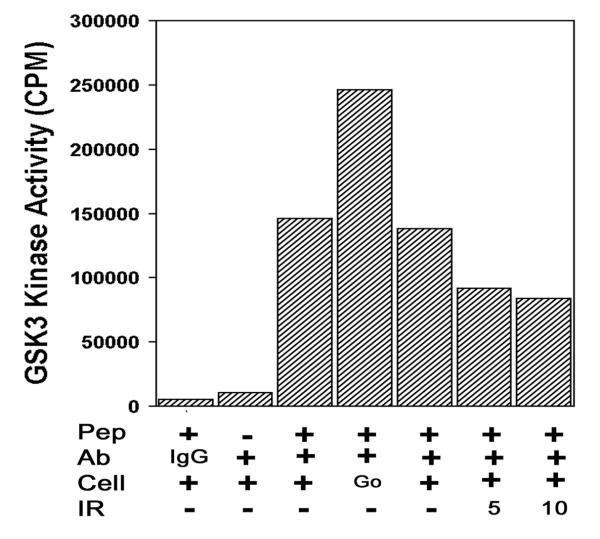
Serines 33 and 37 are located within the N-terminal transcriptional activation domain of the p53 protein. To determine if GSK3β regulates p53's transcriptional activity in vivo, a wild type GSK3β expression vector was cotransfected with either wtp53 or p53 with mutations in serines 33 or 37. The ability of GSK3β to activate each construct was calculated by expressing the p53 transcriptional activity in the presence of GSK3β as a percentage of that observed in the absence of GSK3β. On this scale, no activation by GSK3β yields a zero percent increase in p53 transcriptional relative to p53 alone. GSK3β alone (fig 3C, Con) did not significantly increase the activity of the luciferase reporter construct. When GSK3β was cotransfected with wtp53, p53-dependent activity from the luciferase reporter construct was increased by 160% compared to wtp53 alone. To determine if this activation of p53 transcriptional activity required the GSK3β phosphorylation site at serine 33, wtp53 with serine to alanine mutations at either positions 33 or 37 were cotransfected with GSK3β. Mutation of either serine 33 or 37 significantly reduced the ability of GSK3β to upregulate p53 transcriptional activity. This is consistent with the phosphorylation data in fig 1 and 2, which indicated that serine 37 phosphorylation is required for the subsequent phosphorylation of serine 33 by GSK3β. These results suggests that GSK3β may be a physiological regulator of the p53 protein. GSK3β is constitutively active in resting cells, and is inactivated by PKB through the growth factor dependent p110-PI 3-kinase pathway [25,28,29]. Thus the activity of GSK3β in growing cells is less than that in cells arrested in Go. p53 can be activated by several distinct pathways, including DNA damage. If GSK3β is involved in the activation of p53 by DNA damage, then GSK3β is predicted to be upregulated in response to DNA damage. In fig 4, GSK3β kinase activity was monitored by immunoprecipitating GSK3β and then measuring the ability of the immunoprecipitated protein to phosphorylate a specific GSK3β peptide substrate. In fig 4, the omission of either the peptide or the GSK3β antibody from the assay resulted in minimal phosphorylation of the substrate peptide (fig 4). When both GSK3β antibody and substrate peptide were employed, high levels of GSK3β-dependent kinase activity were detected. When cells were incubated for 24 h in low serum (0.5%) to induce growth arrest in Go, the levels of GSK3β activity were increased compared to asynchronously growing cells (fig 4, Go). When asynchronously growing cells were exposed to 5Gy or 10Gy of Ionizing Radiation to cause DNA damage, the levels of GSK3β activity were reduced. Similar results were seen using p53-GST as the substrate for GSK3β phosphorylation in the kinase assay (data not shown). This implies that the phosphorylation of serine 33 of p53 by GSK3β would be decreased in cells exposed to DNA damage, but elevated in cells growth arrested in Go.
Figure 4.
Ionizing Radiation inhibits GSK3β kinase activity. SAOS-2 cells were immunoprecipitated with anti-GSK3 antibody and incubated with CREB phosphopeptide substrate. Total CPM incorporated into the substrate peptide are shown. Pep: Assays carried out with or without peptide. Ab: Immunoprecipitation carried out with either IgG or anti-GSK3 antibody (+). Cell: Cells were either growing asynchronously (+) or preincubated for 24 h in 0.5% Serum to induce quiescence (Go). IR: Cells were exposed to 5 or 10Gy of Ionizing Radiation and allowed to recover for 60 min.
Discussion
Several p53 kinases were examined for their ability to create in vitro GSK3 phosphorylation sites on p53. Phosphorylation of p53 by DNA-PK created a phosphorylation site for GSK3β, but not for GSK3α. GSK3α and GSK3β have 98% homology within the kinase domain, although regions N- and C-terminal to this are less well conserved [36]. GSK3α and GSK3β have similar substrate specificity in vivo, and are regulated in parallel in response to growth factors [24,27,28,29]. However, disruption of GSK3β in mice results in embryonic lethality and impaired NFκ B function [37], indicating that GSK3α cannot substitute for GSK3β in this model system. GSK3α and GSK3β therefore have overlapping cellular functions, but each isoform also regulates distinct signaling pathways. Our results clearly show that p53 phsophorylation is specific for GSK3β.
The GSK3β phosphorylation site was identified by mutagenesis as serine 33 of p53, and we were able to show that this was dependent on prior phosphorylation of serine 37 by DNA-PK. We also examined if GSK3β regulates in vivo p53 function through a mechanism involving serines 33 and 37 of p53. Previous studies have shown that phosphorylation of serines 15, 20, 33 and 37 of p53 block the interaction of p53 with mdm2, leading to stabilization and accumulation of p53 protein in the cell [10,11,13,14,15,18]. mdm2 binding is dependent on the phosphorylation status of serine 20 of p53 [15], although phosphorylation of serines 15, 33 and 37 also play a role. We did not detect any significant difference in the level of expression of p53 with single mutations in either serines 33 or 37 when compared to wtp53, indicating that single point mutations in serines 33 or 37 do not greatly alter p53 stability.
A key function of p53 is the transcriptional activation of genes which regulate growth arrest and apoptosis [1,2,3,4]. Individual mutation of either serine 33 or 37 slightly increased the basal transcriptional activity of the p53 protein. This is in keeping with observations made by other groups, who demonstrated that single or multiple mutations in p53 phosphorylation sites has minimal effect on the basal transcriptional activity of the p53 protein [38,39,40]. We also determined if GSK3β could regulate p53 transcriptional activity in vivo. GSK3β is constitutively active in resting cells, but exhibits lower activity in asynchronously growing cells [24]. To increase the activity of GSK3β, we co-transfected GSK3β with either wtp53 or p53 bearing serine to alanine mutations at positions 33 or 37. GSK3β increased the transcriptional activity of wtp53, but not of p53 with mutations in either serine 33 or 37. Therefore both serine 33 and serine 37 are required for GSK3β to activate p53 transcriptional activity in vivo. GSK3β regulates many stress activated transcription factors. For example, GSK3β is required for activation of NFκ B [37], but inhibits activation of Heat Shock Factor-1 [27]. Our results indicate that GSK3β may also be involved in the activation of the p53 protein as well.
A key question is whether DNA-PK or some other kinase phosphorylates serine 37 of p53 in vivo. DNA-PK is 460 kd DNA-activated protein kinase which participates in the cellular response to DNA damage [41]. DNA-PK is involved in DNA strand-break repair, and can phosphorylate serines 15 and serine 37 of p53 in vitro [32]. Some reports indicate that DNA-PK is required for the activation of p53 [42], but recent genetic studies have shown clearly that DNA-PK is not required for p53 activation by Ionizing Radiation [43]. Whether DNA-PK is required for p53 activation in response to other stimuli is not known. A more likely candidate is the Atr protein kinase, a kinase related to DNA-PK [44], which can regulate the phosphorylation of serine 37 of p53 in response to DNA damage. The present data indicates that transcriptional activation of p53 by GSK3β requires both serines 33 and 37, but it does not allow us to determine if phosphorylation of serine 33 is dependent on phosphorylation of serine 37 in vivo. It is possible that, in vivo, GSK3β directly phosphorylates serine 33 independently of serine 37, but that both residues must be phosphorylated for transcriptional activation to occur in vivo. Future studies will address this issue.
p53 is activated by multiple pathways, including DNA damage and oncogene activation [1,2,3]. If GSK3β is required for the activation of p53 by DNA damage, GSK3β activity should be regulated by DNA damage. However, when cells were exposed to Ionizing Radiation, GSK3β kinase activity was inhibited rather than enhanced, implying decreased GSK3β-dependent phosphorylation of p53 after DNA damage. Serine 33 of p53 is also phosphorylated by other kinases, including the CDK7-cyclin H complex [45] and p38 MAPK [46]. p38 MAPK is involved in p53 activation by genotoxic stress [46], and may be responsible for p53 phosphorylation at serine 33 in response to DNA damage. GSK3β-dependent phosphorylation of p53 at serine 33 may be part of other p53-regulatory pathways, such as oncogene activation or apoptosis, which are not directly activated by DNA-damage. For example, activation of the p110-PI 3-kinase/PKB pathway delivers an anti-apoptotic signal to the cell [23,25,47]. Further, activation of the p110-PI 3-kinase/PKB is associated with inhibition of both p53 dependent apoptosis [48] and p53 transcriptional activity [49,50]. Although PKB has many down stream targets which may regulate these effects [47] a key target of PKB is GSK3β [29]. Phosphorylation of GSK3β by PKB inhibits GSK3β kinase activity [29]. This would be predicted to block phosphorylation of serine 33 of p53 by GSK3β, decreasing p53 transcriptional activity and therefore reducing the transcription of p53-regulated growth and pro-apoptotic proteins. However, clarification of the potential role of GSK3β in regulating p53 activation by non-DNA damage pathways will require additional study.
Conclusions
This study demonstrates that GSK3β, but not GSK3α, can directly phosphorylate serine 33 of p53 when serine 37 of p53 is already phosphorylated. GSK3β can increase p53's transcriptional activity in vivo, and this activation requires serines 33 and 37 of the p53 protein. Thus GSK3β may phosphorylate and activate p53 in vivo. However, GSK3β is not part of the p53-DNA damage response pathway. Instead, GSK3β may provide the link between p53 and non-DNA damage mechanisms for p53 activation, such as oncogene activation.
Materials and methods
Phosphorylation Reactions
Wild type p53 or p53 bearing mutations in serines 33 or 37 were subcloned into pGEX-2T GST fusion vector (Pharmacia, NJ) and p53-GST purified as previously described [33]. p53-GST protein (2 μg; measured using Bio-Rad Protein assay Kit, Biorad, CA) was incubated with MAP kinase (Erk2, 10 Units, New England Biolabs, MA), Protein Kinase A (Catalytic sub-unit, 5 Units, Calbiochem, CA), S6 kinase (0.2 Units, Upstate Biotechnology, NY), Protein Kinase C (O.1 mUnits, Roche Molecular Biochemicals, IN) or DNA-PK (20 Units, Promega Corp, Wl) for 5 h at 30°C in the following buffers (40 μl final volume). MAP kinase and Protein Kinase A: 20 mM Hepes pH 7.2/10 mM Na-Acetate/30 mM MgCl2/0.2 mM EDTA/1 mM EDTA/I0 μM ATP. S6 Kinase: 20 mM MOPS pH 7.2/30 mM MgCl2/5 mM EGTA/1 mM DTT/I0 μM ATP. DNA-PK:25 mM Hepes pH7.5/150 mM KCI/10 mM MgCl2/20% Glycerol/0.1% NP40/20 μM ZnCl2/1 mM DTT/250 ng DNA/4.2 mM spermidine/ 10 μM ATP. PKC: 20 mM Hepes pH 7.4/ 20 mM MgCl2/ 0.2 mM EGTA/ 1 mM CaCl2/1.5 μg phosphatidylserine. Samples were then incubated at 65°C for 20 min to inactivate kinases, and 25% of the reaction incubated for 30 min with GSK3α or GSK3β in GSK3 kinase buffer (KGB Buffer: 8 mM MOPS, pH7.2/ 10 mM MgCl2/ 0.2 mM EDTA/ 5 μM ATP/10 μCi 32P-ATP) in a final volume of 40 μl. Reactions were terminated by the addition of SDS sample buffer and p53-GST phosphorylation detected by SDS-PAGE and auotradiography. Equal amounts of p53-GST (0.25 μg) were analyzed by SDS-PAGE and equal loading confirmed by coomassie blue staining of the SDS-PAG.
GSK3 immunokinase assay
SAOS-2 cells (5 × 106) were lyzed in 0.5 ml of GLB buffer (50 mM Tris pH7.5/1 mM EDTA/1 mM EGTA/0.5% NP40/0.5 M NaCI/1 mM DTT/1 mM PMSF/leupeptin/aprotinin/600 μM Na3VO4/50 mM NaF). Extracts were cleared by centrifugation and incubated with GSK3 antibody (1 μg; Upstate Biotech, NY) prebound to Sepharose A/G agarose beads for 2 h. The beads were washed in 4 × 1 ml of GLB buffer and then in 2 × 1 ml of KGB buffer. Kinase reactions contained 5 μM ATP/15 μCi 32P-ATP/0.5 μg PhosphoCREB peptide in 30 μl of KGB. After incubate for 10 min at room temperature, the beads were collected by centrifugation and 20 μl of the reaction mix spotted onto circles of P81 paper (Whatman, USA). The P81 paper was washed in 4 × 10 ml changes of 100 mM phosphoric acid, dried and counted. PhosphoCREB peptide (sequence KRREILSRRPS(P)YR) was obtained from New England Biolabs, MA.
Mutagenesis was carried out using the Altered Sites Mutagenesis System (Promega, Wl) as previously described by us [33]. Human wild type p53 or p53 bearing mutations in serines 33 or 37 were inserted into the BamH1 site of the pcDNA3.1 expression vector (Invitrogen, CA).
Luciferase reporter assays were carried out in the p53 null cell line SAOS-2 using the p53 specific luciferase reporter construct p50-Luc, and pCMV-β-galatosidase to control for transfection efficiency. Cells were transfected using Lipofectin (Gibco-BRL) containing p50-Luc (1 μg), pCMV-Gal (1 μg) and pcDNAp53 (50 ng) in a final volume of 400 μl as described in the manufacturers protocol.
Acknowledgments
Acknowledgements
We thank J. Woodgett for GSK3β cDNA. This work was supported by grant number CA64585 to BDP and funds from the AJCRT foundation.
Contributor Information
Gaetan A Turenne, Email: ATurenne@kendallstrategies.com.
Brendan D Price, Email: brendan_price@dfci.harvard.edu.
References
- Canman CE, Lim D-S. The role of ATM in DNA damage responses to cancer. Oncogene. 1998;17:3301–3308. doi: 10.1038/sj.onc.1202577. [DOI] [PubMed] [Google Scholar]
- Lowe SW. Activation of p53 by Oncogenes. Endocr Relat. 1999;6:45–48. doi: 10.1677/erc.0.0060045. [DOI] [PubMed] [Google Scholar]
- Lundberg AS, Hahn WC, Gupta P, Weinberg RA. Genes involved in senescence and immortalization. Curr Opin Cell Biol. 2000;12:705–709. doi: 10.1016/S0955-0674(00)00155-1. [DOI] [PubMed] [Google Scholar]
- Vousden KH. p53:death star. Cell. 2000;103:691–694. doi: 10.1016/S0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2001;108:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Kubbutat MCG, Jones SN, Vousden KH. Regulation of p53 stability by mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeosis NJ, Silverman A, AIIand L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho RA. The lnk4a tumor suppressor gene product p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell. 1998;92:713–723. doi: 10.1016/S0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- Meyn MS. Ataxia Telangiectasia, cancer and the pathobiology of the ATM gene. Clin Genet. 1999;55:289–304. doi: 10.1034/j.1399-0004.1999.550501.x. [DOI] [PubMed] [Google Scholar]
- Banin S, Moyal L, Shieh S-Y, Taya Y, Anderson CW, Chessa L, Smorodinsky Nl, Prives C, Reiss Y, Shiloh Y, Ziv Y. Enhanced phosphorylation of p53 byATM in response to DNA damage. Science. 1998;281:1674–1678. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- Canman CE, Lim D-S, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1682. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- Shieh S-Y, Ahn J, Tamai K, Taya Y, Prives C. The human homologs of checkpoint kinases Chk1 and Cds1 (chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 2000;14:289–300. [PMC free article] [PubMed] [Google Scholar]
- Chebab NH, Malikzay A, Appel M, Halozonetis TD. Chk2/hCds1 functions as a DNA damage checkpoint in G1 by stabilizing p53. Genes Dev. 2000;14:278–288. [PMC free article] [PubMed] [Google Scholar]
- Shieh S-Y, Taya Y, Prives C. DNA damage-inducible phosphorylation of p53 at N-terminal sites including a novel site, Ser20, requires tetramerization. EMBO J. 1999;18:1815–1823. doi: 10.1093/emboj/18.7.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi K, Herrera JE, Saito S, Miki T, Bustin M, Vassilev A, Anderson CW, Appella E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino-terminus of p53. Genes Dev. 1997;11:3471–3481. doi: 10.1101/gad.11.24.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh S-Y, Ikeda M, Taya Y, Prives C. DNA-damage induced phosphorylation of p53 alleviates inhibition by mdm2. Cell. 1997;91:325–334. doi: 10.1016/S0092-8674(00)80416-X. [DOI] [PubMed] [Google Scholar]
- Gu W, Roeder RG. Activation of p53-sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/S0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- Steegenga WT, van der Eb AJ, Jochemsen AG. How phosphorylation regulates the activity of p53. JMol Biol. 1996;263:103–113. doi: 10.1006/jmbi.1996.0560. [DOI] [PubMed] [Google Scholar]
- Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102:849–862. doi: 10.1016/S0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- Higashimto H, Saito S, Tong XH, Hong A, Sakaguchi K, Appella E, Anderson CW. Human p53 is phosphorylated on serines 6 and 9 in response to DNA damage-inducing agents. J Biol Chem. 2000;275:23199–23203. doi: 10.1074/jbc.M002674200. [DOI] [PubMed] [Google Scholar]
- Franke TF, Kaplan DR, Cantley LC. PI3K: Downstream Aktion blocks apoptosis. Cell. 1997;88:435–437. doi: 10.1016/S0092-8674(00)81883-8. [DOI] [PubMed] [Google Scholar]
- Kirn L, Kimmel AR. GSK3, a master switch regulating cell-fate specification and tumorigenesis. Curr Opin Gen Dev. 2000;10:508–514. doi: 10.1016/S0959-437X(00)00120-9. [DOI] [PubMed] [Google Scholar]
- Kennedy SG, Wagner AJ, Conzen SD, Jordan J, Bellacosa A, Tsichlis PN, Hay N. The PI 3-kinase/Akt signaling pathway delivers an anti-apoptotic signal. Genes Dev. 1997;11:701–713. doi: 10.1101/gad.11.6.701. [DOI] [PubMed] [Google Scholar]
- Neal JW, Clipstone NA. Glycogen Synthase kinase-3 inhibits the DNA binding activity of NFATc. J Biol Chem. 2000;276:3666–3673. doi: 10.1074/jbc.M004888200. [DOI] [PubMed] [Google Scholar]
- Xavier LJ, Mercier PA, McLoughlin CM, AIi A, Woodgett JR, Ovsenek N. Glycogen Synthase Kinase 3β negatively regulates both DNA-Binding and transcriptional activities of Heat Shock factor-1. J Biol Chem. 2000;275:29147–29152. doi: 10.1074/jbc.M002169200. [DOI] [PubMed] [Google Scholar]
- Eldar-Finkelman H, Argast GM, Foord B, EH Fischer, EG Krebbs. Expression and characterization of glycogen synthase kinase-3 mutants and their effect on glycogen synthase activity in intact cells. Proc NatI Acad. 1996;93:1028–10233. doi: 10.1073/pnas.93.19.10228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DAE, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- Meek DW, Simon S, Kikkawa U, W Eckhart. The p53 tumor suppresser protein is phosphorylated at serine 389 by casein kinase II. EMBO J. 1994;9:3253–3260. doi: 10.1002/j.1460-2075.1990.tb07524.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Campbell DG, Caudwell FB, Meek DW. Phosphorylation of the tumor suppressor p53 by mitogen activated kinases. J Biol Chem. 1994;269:9253–9260. [PubMed] [Google Scholar]
- Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, Anderson CW. Human DNA activated protein kinase phosphorylates serines 15 and 37 in the amino terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youmell M, Park SJ, Basu S, Price BD. Regulation of the p53 protein by protein kinase Cα and protein kinase Cε. Biochem Biophys Res Comm. 1998;245:514–518. doi: 10.1006/bbrc.1998.8471. [DOI] [PubMed] [Google Scholar]
- Adler V, Pincus MR, Minamoto T, Fuchs SY, Bluth MJ, Brandt-Rauf T, Friedman FK, Robison RC, Chen JM, Wang XW, Harris CC, Ronai Z. Conformation-dependent phosphorylation of p53. Proc NatI Acad Sci. 1997;94:1686–1691. doi: 10.1073/pnas.94.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SY, Adler V, Buschman T, Yin Z, Wu X, Jones SN, Ronai Z. JNK targets p53 ubiquitination and degradation in nonstressed cells. Genes Dev. 1998;12:2658–2663. doi: 10.1101/gad.12.17.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodgett JR. Molecular cloning and expression of glycogen synthase kinase-3/Factor A. EMBO J. 1990;9:2431–2438. doi: 10.1002/j.1460-2075.1990.tb07419.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeflich KP, Luo J, Ruble EA, Tsao M-S, Jin U, JR Woodgett. Requirement for glycogen synthase kinase-3β in cell survival and NF-κ B activation. Nature. 2000;406:86–90. doi: 10.1038/35017574. [DOI] [PubMed] [Google Scholar]
- Unger T, Sionov RV, Moallen E, Yee CL, Howley PM, Oren M, Haupt Y. Mutations in serines 15 and 20 of human p53 impair its apoptotic activity. Oncogene. 1999;18:3205–3212. doi: 10.1038/sj.onc.1202656. [DOI] [PubMed] [Google Scholar]
- Ashcroft M, Kubbutata HG, Vousden KH. Regulation of p53 function and stability by phosphorylation. Mol Cell Biol. 1999;19:1751–1758. doi: 10.1128/mcb.19.3.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blattner C, Tobiasch E, Litfen M, Rahmsdorf HJ, Herrlich P. DNA damage induced p53 stabilization: no indication for an involvement of p53 phosphorylation. Oncogene. 1999;18:1723–1732. doi: 10.1038/sj.onc.1202480. [DOI] [PubMed] [Google Scholar]
- Featherstone C, Jackson SP. DNA-dependent protein kinase gets a break: its role in repairing DNA and maintaining genomic integrity. Br J Cancer. 1999;80 Suppl(1):14–19. [PubMed] [Google Scholar]
- Woo RA, McLure KG, Lees-Miller SP, Rancourt DE, Lee PW. DNA-dependent protein kinase acts upstream of p53 in response to DNA damage. Nature. 1998;394:700–704. doi: 10.1038/29343. [DOI] [PubMed] [Google Scholar]
- Jimenez GS, Bryntesson F, Torres-Arzayus Ml, Priestly A, Beche M, Sakaguchi K, Appella E, Jeggo PA, Taccioli GE, Wahl GM, Hubank F. DNA-dependent protein kinase is not required for the p53-dependent response to DNA damage. Nature. 1999;400:81–83. doi: 10.1038/21913. [DOI] [PubMed] [Google Scholar]
- Tibbetts RS, Brumbaugh KM, Williams JM, Sarkaria JN, Cliby WA, Shieh S-Y, Taya Y, Prives C, Abraham RT. A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev. 1999;13:152–157. doi: 10.1101/gad.13.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayaraman L, Tamai K, Taya Y, Prives C, ZQ Pan. p53 is phosporylated by CDK7-cyclin H in a p36mat1 dependent manner. Mol Cell Biol. 1997;17:7220–7229. doi: 10.1128/mcb.17.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Prieto R, Rojas JM, Taya Y, Gutkind JS. A role for p38 mitogen-activated protein kinase pathway in the transcriptional activation of p53 on genotoxic stress by chemotherapeutic agents. Cancer Research. 2000;60:2464–2472. [PubMed] [Google Scholar]
- Datta SR, Brunet A, Greenberg ME. Cellular survival: a play in three akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- Mazzoni IE, Said FA, Aloyz R, Miller FD, Kaplan D. Ras regulates sympathetic neuron survival by suppressing the p53-mediated cell death pathway. J Neuro. 1999;19:9716–9727. doi: 10.1523/JNEUROSCI.19-22-09716.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbatini P, McCormick F. Phosphoinositide 3-OH kinase (PI3K) and PKB/Akt delay the onset of p53-mediated, transcriptionally dependent apoptosis. J Biol Chem. 1999;274:24263–24269. doi: 10.1074/jbc.274.34.24263. [DOI] [PubMed] [Google Scholar]
- Yamaguchi A, Tamatani M, Matsuzuki H, Namikawa K, Kiyama H, Vitek MP, Mitsuda N, Tohyama M. Akt activation protects hippocampal neurons from apoptosis by inhibiting transcriptional activity of p53. J Biol Chem. 2001;276:5256–5264. doi: 10.1074/jbc.M008552200. [DOI] [PubMed] [Google Scholar]