Abstract
Marine microbial communities are complex and dynamic, and their ecology impacts biogeochemical cycles in pelagic ecosystems. Yet, little is known about the relative activities of different microbial populations within genetically diverse communities. We used rRNA as a proxy for activity to quantify the relative specific activities (rRNA/ribosomal DNA [rDNA or rRNA genes]) of the eubacterial populations and to identify locations or clades for which there are uncouplings between specific activity and abundance. After analyzing 1.6 million sequences from 16S rDNA and rRNA (cDNA) libraries from two euphotic depths from a representative site in the Pacific Ocean, we show that although there is an overall positive relationship between the abundances (rDNAs) and activities (rRNAs) among populations of the bacterial community, for some populations these measures are uncoupled. Different ecological strategies are exemplified by the two numerically dominant clades at this site: the cyanobacterium Prochlorococcus is abundant but disproportionately more active, while the heterotrophic SAR11 is abundant but less active. Other rare populations, such as Alteromonas, have high specific activities in spite of their low abundances, suggesting intense population regulation. More detailed analyses using a complementary quantitative PCR (qPCR)-based approach of measuring relative specific activity for Prochlorococcus populations in the Pacific and Atlantic Oceans also show that specific activity, but not abundance, reflects the key drivers of light and nutrients in this system; our results also suggest substantial top-down regulation (e.g., grazing, viruses, or organismal interactions) or transport (e.g., mixing, immigration, or emigration) of these populations. Thus, we show here that abundance and specific activity can be uncoupled in open ocean systems and that describing both is critical to characterizing microbial communities and predicting marine ecosystem functioning and responses to change.
INTRODUCTION
The application of massively parallel DNA sequencing to marine microbial ecology has increasingly led to identification of the breadth of bacterioplankton phylogenetic and genomic diversity (1–3). In general, the most abundant environmental sequence types have been assumed to be the most active, both on a per-cell basis and as members of the community, and thus they are assumed to drive nutrient and energy cycling, although there are some important exceptions (e.g., N2 fixers). Robust tools that can determine the activities of specific phylotypes in the environment have not been widely applied, even though the ability to measure in situ activity will address key questions in microbial ecology and biogeochemistry, including the environmental variables that favor specific taxa (4), the role of predation in regulating bacterioplankton abundances (5), and the relative importance of specific taxa in biogeochemical cycling (6), among others.
In culture- and genome-based studies of a few model organisms, marine microbes are often partitioned into oligotrophs (e.g., Prochlorococcus and “Candidatus Pelagibacter”), which grow slowly on low levels of nutrients, and copiotrophs (e.g., Roseobacter, Alteromonas), which require high concentrations of nutrients and may display boom-or-bust population dynamics (7). Although clearly delineated oligotrophs and copiotrophs are useful as a binary classification, reality may reflect a gradient in responsiveness to environmental conditions (8). Some copiotrophs may cycle between rare and abundant states, while others remain rare in spite of high apparent growth rates due to predation pressure (9), and oligotrophs may be more dynamic than previously thought (7). Thus, combining measures of specific activity and abundance can provide a more complete assessment of the bacterioplankton community structure and the impacts of its members on biogeochemical cycling and microbial ecology.
Under steady-state growth and in the absence of selective predation by viruses and grazers, the most active bacteria would be the most abundant, with higher growth rates leading to more biomass (10). Based on these assumptions, investigators have often assumed that rare bacteria are composed of slow-growing or dormant bacteria that act as a “seed bank,” becoming abundant when positively selected for by the environment (11). Yet, there is increasing evidence that rare bacteria may be disproportionately active relative to their abundances (9, 12). Observations of uncoupled specific activity and abundance in a coastal ocean time series and in lakes have suggested that apparent equilibrium in these environments masks a dynamic system and that some low-abundance taxa may contribute disproportionately to ecological and biogeochemical processes relative to their abundances (9, 12).
Historically, the activities of marine microbes have been assessed at the community level by measuring the incorporation of labeled precursors (13, 14). However, this technique cannot determine the activities of specific populations, which may differ from those of the total community. Other phylum-specific measurements often require incubations that may distort microbial populations due to “bottle effects,” incorporation of substrates, or development of specific probes or primers. To address this gap, there is a growing trend of obtaining phylotype-associated measurements of specific activity by quantifying rRNA and ribosomal DNA (rDNA or rRNA genes) that reflect the activities and abundances, respectively, of specific ribotypes (9, 12). Cellular ribosome abundances have been shown to correlate well with growth rates of marine microbes in culture (15, 16), with some noted exceptions (17, 18), and without the incubation or substrate incorporation requirement of many other approaches. However, there are limitations to using this technique to infer an in situ growth rate, as the rRNA content per cell is dependent on the cell size, the growth rate, a taxon-specific relationship with growth, and likely other clade-specific and environmental variables (17). While the abundance of rDNA copies per cell (i.e., rDNA operons per cell) can vary by roughly 1 order of magnitude between taxa (19), the copies of rRNA per cell have an even wider range (from 0 in a dead cell to >10,000 copies in live cells [20]). Further, in rRNA and rDNA library-based approaches, the number of reads for each taxon observed is relative to reads for other organisms in that library rather than an absolute measure of abundance. Many comparisons of rRNA and rDNA have been based on relatively small libraries that are potentially biased in their interpretation because of insufficient sampling depth. These considerations complicate direct comparisons between rDNA and rRNA in library sequencing (21).
Taking advantage of the strengths of this approach while acknowledging that this technique, like others, has limitations, we have applied the ratio of rRNA to rDNA for a given ribotype as a proxy for bacterium-specific activity, which likely reflects the rates of biomass- and non-biomass-producing cellular processes. The strength of measuring rRNA/rDNA ratios comes not from a single time point but in comparing measurements across time and space to identify the drivers of activity and abundance for these organisms at representative locations in the ocean. In contrast to a previous study, in which specific activity was examined using a coastal time series (9), we examined open ocean stations and variability in specific activities with depth. We focused on determining specific activity in the cyanobacterium Prochlorococcus, which is important as a model organism and as the dominant clade in the tropical and subtropical open ocean environments. As Prochlorococcus is an organism with well-known nutrient requirements and light-driven physiology, measuring Prochlorococcus specific activity allowed us to identify potential uncoupling of abundance and specific activity in the environment. Overall, this study allowed us to examine the specific activities of different populations of bacteria across their broad genetic diversity to better understand their respective roles in biologically driven ocean processes.
MATERIALS AND METHODS
Sample collection.
Seawater for 16S rRNA and rDNA library construction was collected at two depths (25 and 100 m) during Hawaii Ocean Time-Series (HOT) cruise 215 at Station ALOHA (22°45′N, 158°W) on 25 September 2009, 2215 coordinated universal time (UTC). Bacterioplankton samples for DNA and RNA extraction were collected as described previously (3, 22), with slight modifications. Briefly, seawater was prefiltered through a 125-mm Whatman glass fiber (GF)/A filter to remove large eukaryotes. For DNA extraction, each sample was collected onto a 0.22-μm Sterivex filter cartridge (Millipore), covered with 2 ml of preservation buffer (50 mM Tris [pH 8.3], 40 mM EDTA, and 0.75 M sucrose) and stored at −80°C. In total, 11 and 19 liters of seawater were filtered for DNA samples from the 25-m and 100-m depths, respectively. The material for RNA extraction was collected by duplicate filtration of 1 liter of seawater from the same sample used for DNA collection on 0.22-μm Durapore filters (Millipore). After filtration, each filter was immediately immersed in 1 ml of RNAlater (Ambion) and frozen at −80°C until extraction. Environmental variables were obtained from the HOT website (http://hahana.soest.hawaii.edu/hot/hot-dogs).
Additional depth profiles for Prochlorococcus quantitative PCR (qPCR) and quantitative reverse transcription-PCR (qRT-PCR) were taken at a station in the equatorial Pacific (2°S, 155°W) on 2 September 2006, 0207 UTC, and a station in the Sargasso Sea (30°43′N, 72°41′W) on 24 May 2010, 1832 UTC. At each station, discrete water samples at ∼10 depths in the upper ∼200 m were taken using a conductivity-temperature-depth (CTD) rosette system. At the equatorial Pacific site, macronutrient samples were collected in bottles, frozen, and then analyzed on shore using an Astoria AutoAnalyzer as described previously (23). The detection limit for nitrate is 0.05 μmol liter−1. At the Sargasso Sea site, nitrate was measured using an in situ UV spectrophotometer (ISUS) nitrate sensor (Satlantic) mounted on the CTD rosette (24, 25). The ISUS nitrate detection limit is 0.5 μmol liter−1, and values below the detection limit were reported as zero. To obtain the material for qPCR, 100 ml of seawater was filtered through a 0.22-μm polycarbonate filter under low vacuum conditions (more than −3 × 104 Pa), washed with 3 ml of preservation solution (10 mM Tris, 100 mM EDTA, 0.5 M NaCl [pH 8.0]), and frozen at −80°C until extraction. In obtaining samples for Prochlorococcus qRT-PCR, 100-ml samples were taken in duplicate, and each filter was preserved in 600 μl of RLT solution (Qiagen) with 1% β-mercaptoethanol and frozen at −80°C until extraction.
Nucleic acid extraction for 16S rRNA and rDNA libraries.
The preservation solution in the Sterivex cartridge was collected and concentrated using an Amicon Ultra 10-kDa filter unit (Millipore) and combined with the filter. DNA extraction was performed according to the manufacturer's instructions (DNeasy tissue kit; Qiagen) with the addition of bead beating (0.2 g, 0.1-mm Zr beads for 3 min at 4,800 rpm). Total RNA was extracted as described previously (22), with some modifications. Briefly, the RNAlater solution was concentrated to less than 100 μl using an Amicon Ultra 10-kDa filter unit and then returned to the vial with a filter. Then, 0.2 g of 0.1-mm Zr beads was added to the vial, and the sample was lysed by bead beating at 4,800 rpm for 2 min at 4°C. Total RNA was isolated from the lysate using an RNeasy MinElute column (Qiagen) following the manufacturer's instructions and treated with DNase using a Turbo DNA-free kit (Ambion). cDNA was synthesized using a SuperScript II reverse transcriptase system (Invitrogen) with random hexamer primers.
Amplicon library preparation.
16S gene fragments were amplified by PCR from environmental DNA and cDNA templates using universal primers 926F and 1392R plus adaptors for 454 pyrosequencing. Triplicate PCRs (20 μl) for each library included 0.25 U JumpStart Taq (Sigma), 1× JumpStart PCR buffer, 0.05 μM each primer, 200 μM each deoxynucleoside triphosphate (dNTP), and 300 μg/ml bovine serum albumin (BSA). The reaction mixture was thermocycled at 95°C for 3 min, 25 cycles at 95°C for 30 s, 55°C for 45 s, and 72°C for 90 s, followed by a final extension of 10 min at 72°C. PCR products were separated by electrophoresis on Tris-acetate-EDTA (TAE) agarose gels and purified using a MinElute gel extraction kit (Qiagen). Finally, 454 library preparation and sequencing were conducted at the Joint Genome Institute (JGI) in Walnut Creek, CA.
Analysis of 16S rRNA and rDNA tag libraries.
A total of 1.6 million raw sequences were obtained by sequencing PCR amplicons for the four libraries described above. Sequences were processed using QIIME 1.2.1 (SVN1920) (26). Briefly, the sequences were filtered for length (300 ≥ length ≤ 540 bp), quality score (mean, >30), and number of homopolymer runs (<8). Sequencing errors were minimized by using Denoiser 0.91 for titanium reads, by chimera removal using ChimeraSlayer, and by defining operational taxonomic units (OTUs) at 97% identity using the UCLUST algorithm. OTUs were removed with only one occurrence in all four libraries, with a eukaryotic or archaeal best BLAST hit (refseq_genomic) or an E value of >10−30 against the SILVA (v106) database (27). This analysis yielded a total of 861,659 sequences after preprocessing; to control for variation in the number of reads per library, the libraries were each subsampled 75,000 times (21). For the 50 most abundant OTUs, a representative sequence was hand aligned using ARB (28) and a maximum-likelihood tree constructed using PhyML (default parameters) with 100 bootstrap replicates (29) and a lane mask.
Prochlorococcus ecotype qPCR and qRT-PCR DNA extraction.
Filters for qPCR were shaken without beads in a bead beater (BioSpec) for 2 min at 4,800 rpm with 650 μl of 10 mM Tris (pH 8.0) buffer, followed by incubation at 95°C for 15 min. The resulting lysate was used as the template for qPCRs in parallel with serially diluted standards of known cell concentrations. For qRT-PCR, individual samples were lysed with 0.2 g of zirconium beads in the bead beater at 4,800 rpm, alternating bead beating (30 s) and cooling on ice, for a total of 2 min of bead beating. Total RNA was extracted from the sample using an RNeasy minicolumn (Qiagen) with DNase digestion according to the manufacturer's protocol. cDNA was synthesized using random hexamer primers from the extracted RNA using an iScript cDNA synthesis kit (Bio-Rad) and used as the template in qPCRs following the same protocol as that for the genomic DNA qPCR.
Prochlorococcus ecotype-specific qPCR and qRT-PCR.
For environmental DNA samples, qPCR was performed using primers targeting the 23S rRNA designed for specific ecotypes. Ecotype (e)MIT9312 (eHL-II) was amplified as described previously (23, 30), and eMIT9313 (eLL-IV) was amplified using the modified primers 23S-678F (5′-CGAGTCTGAATAGGGCGATC-3′) and 23S-1210R (5′-CTCCCCTACCATTTAACAAG-3′) and thermocycled at 95°C for 15 min, 45 cycles at 95°C for 15 s, 62°C for 30 s, and 72°C for 30 s, followed by a final extension of 5 min at 72°C. The specificities of the eMIT9313 primers were verified using lab isolates of Prochlorococcus and Synechococcus as well as environmental clone libraries. For synthesized cDNA, qPCR was performed using 23S rRNA gene primers for eMIT9312 (eHL-II) (30) and eMIT9313 (eLL-IV) (23S-678F and 23S-1210R). The relative abundances and 23S rRNA gene expression levels of Prochlorococcus ecotypes from environmental DNA and cDNA pools, respectively, were determined by comparing the threshold cycle (CT) value with serially diluted culture standards. Because the template preparation process for the standards (30) differs from that for the environmental DNA and cDNA samples, the qPCR results are presented as unitless relative values.
Nucleotide sequence accession number.
The sequences reported here have been deposited in the GenBank short-read archive under accession number SRA058462. Oceanographic data are available at http://bcodmo.org.
RESULTS AND DISCUSSION
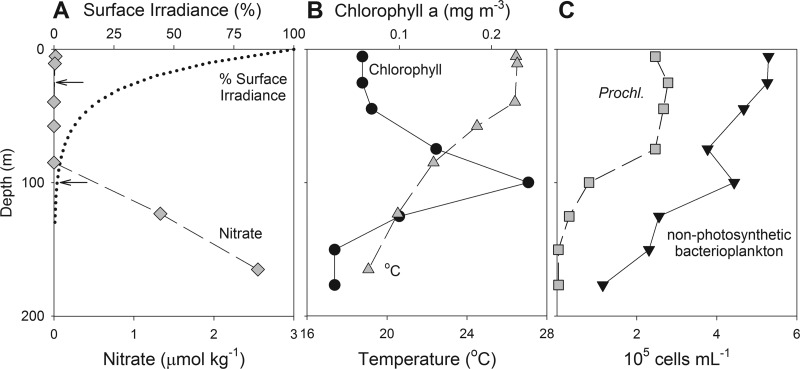
To quantify the abundances and specific activities of diverse bacterioplankton in the open ocean, we sampled DNA and RNA from a well-characterized oceanographic station in the North Pacific Ocean (ALOHA) in conjunction with the Hawaii Ocean Time-Series in September 2009. At this location, the alphaproteobacterium SAR11 and the cyanobacterium Prochlorococcus are generally the dominant clades throughout the year (31, 32). However, the surface waters (0 to 200 m) of station ALOHA are highly stratified in key environmental variables such as temperature, light, and nitrate, resulting in vertical gradients in biological properties such as chlorophyll concentration and Prochlorococcus and nonphotosynthetic bacterioplankton abundances (Fig. 1), among others. To examine the bacterioplankton community in detail, we used 16S rDNA libraries to quantify the diversity and relative abundances of specific ribotypes coupled with 16S rRNA libraries to link their phylogenetic identities to environmental activities. Large-scale 16S ribosomal DNA and RNA tag libraries (mean fragment length, 390 bp) covering the variable regions V6 to V8 were constructed from samples collected at two depths (25 m and 100 m), which correspond to the upper photic zone and the deep chlorophyll maximum, respectively (Fig. 1, arrows).
Fig 1.
Vertical variability of key physical, chemical, and biological variables at station ALOHA. (A) Irradiance (percentage of surface) and nitrate (μmol kg−1). Arrows indicate the depths of samples chosen for the bacterioplankton diversity analysis. (B) Chlorophyll a (mg m−3) and temperature (°C). (C) Cellular abundance was measured by flow cytometry for Prochlorococcus and nonphotosynthetic bacterioplankton (cells ml−1).
A total of ∼1.6 million reads (630 Mbp of sequence) were obtained among the four libraries and carefully edited for quality (see Materials and Methods), netting 861,659 sequences for subsequent analyses. In contrast with the high diversity and extreme “rare biosphere” observed by others (33), our rarefaction collector curves of each library (see Fig. S1 in the supplemental material) plateau to a maximal value (Good's coverage, ≥99%), indicating that the community's diversity is well constrained. Additionally, the number of OTUs (operational taxonomic units, here defined at 97% identity) observed in each library (219 to 575 OTUs) (see Table S1) is similar to values predicted for coastal seawater in a well-curated 16S rDNA clone library (1). Nevertheless, a substantial fraction of the OTUs from each library (16 to 38%) are unique to that library, suggesting that further sampling may yield additional OTUs (see Fig. S2 in the supplemental material), even though most of the library-specific OTUs are in low abundances (<1% total). The majority of OTUs are unique to each depth (53% at 25 m, 66% at 100 m), indicating substantial microbial biogeography even in the upper 100 m of the open ocean (see Fig. S2). These distinct communities form within the water column even though similar ecological and biogeochemical processes are thought to occur at both depths, albeit at much different rates.
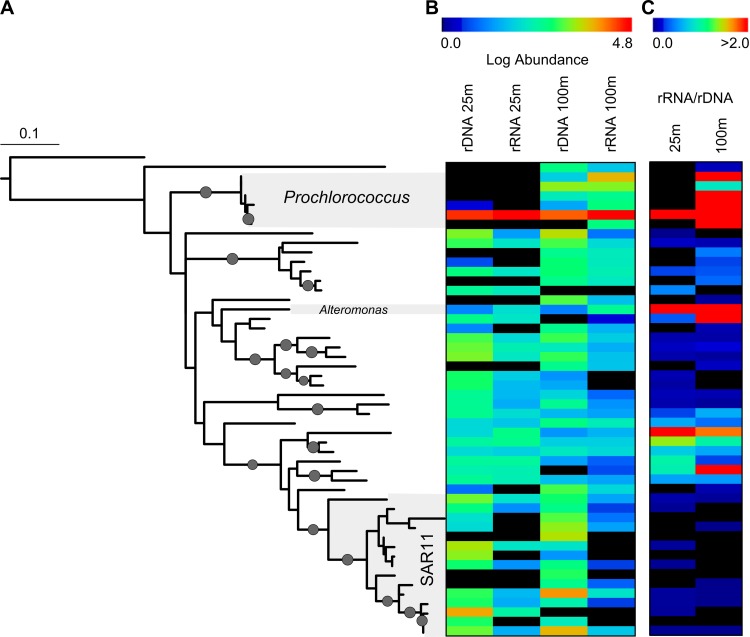
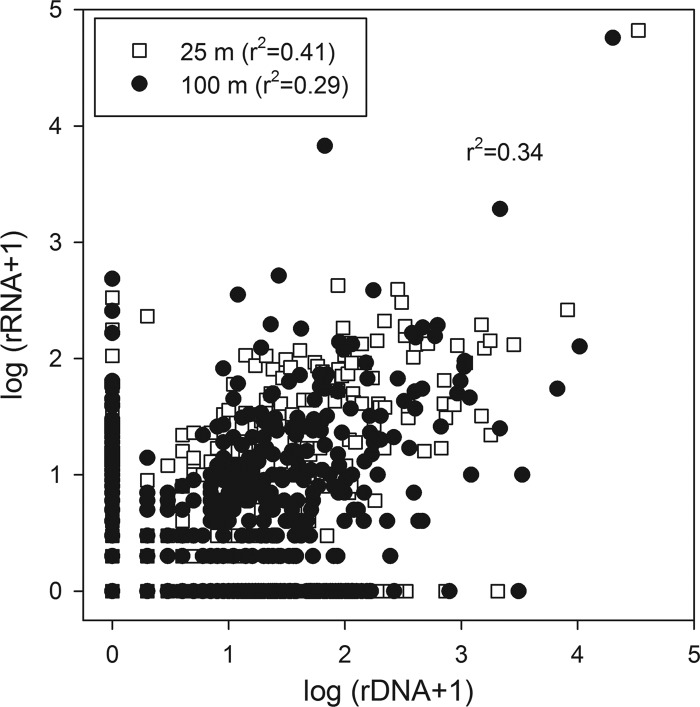
In addition to differences in the taxonomic composition between depths, the relative abundances of OTUs change substantially both between depths and between DNA (abundance) and RNA (activity) libraries at the same depth. For example, among the 50 most abundant OTUs for the combined data set (representing 90% of the total sequences), most OTUs are present in all four libraries (Fig. 2), and they represent the broad phylogenetic groups common to many open ocean environments, including Prochlorococcus, SAR11, and Roseobacter (see Table S2 in the supplemental material). However, the relative abundances of these OTUs can vary substantially between depths, such as for the most abundant Alphaproteobacteria rDNA clade (OTU 1506), which is substantially more abundant in the rDNA library at 25 m than at 100 m (Fig. 2). OTUs can also vary in their relative rank abundances in rDNA and rRNA libraries, leading to dramatic differences in the rRNA/rDNA ratios among OTUs both within a given depth and between depths (Fig. 2C). Differences in the rRNA/rDNA ratios can be further skewed by dissimilarities in the evenness of diversity among the libraries; rDNA libraries have significantly (P = 0.013) higher Shannon indices (3.13 ± 0.41) than those of the rRNA libraries (1.12 ± 0.35), demonstrating a disproportionate effect of a small number of OTUs on relative activity. Due to differing numbers of copies per cell of rRNA and rDNA and the dependence of quantification on the abundance of other OTUs in the library, 1:1 ratios in rRNA and rDNA libraries would not be expected even when activity was highly coupled with abundance for a specific OTU. However, if abundance and activity are related, we would expect the relative rank of an OTU to be the same within a given depth, and in fact at both depths the rRNA and rDNA rank abundances were positively related (Fig. 3) (Kendall's and Spearman's P < 0.005, 25-m Kendall's tau = 0.1646, n = 470; 100-m tau = 0.0834, n = 642). In spite of an overall relationship between ranks in the rDNA and rRNA libraries, deviations in correspondences of relative ranks between rDNA and rRNA libraries can be used to identify OTUs for which there are uncouplings of abundance and activity. Also, while ∼50% of the bacterioplankton assemblage is shared between depths, the abundances (rDNA), activities (rRNA), and specific activities (rRNA/rDNA) of ribotypes vary dramatically.
Fig 2.
Phylogenetic relationship of bacterial diversity with activity and abundance measurements for 16S rRNA and rDNA libraries from station ALOHA at 25 m and 100 m. (A) Maximum-likelihood phylogenetic tree of the partial 16S rRNA sequences for the 50 most abundant OTUs (97% identity) in equally subsampled 16S rRNA and rDNA libraries (75,000 members). Bootstrap percentages of >80 for a given branch are indicated by a small circle. (B) Heat map showing the log abundance (log [observations + 1]) for each OTU from the tree in panel A for RNA and DNA libraries. (C) Ratio of RNA to DNA (rRNA/[rDNA + 1]) showing the specific activity of each OTU for libraries from 25 and 100 m.
Fig 3.
Relationship between activity [log (rRNA + 1)] and abundance [log (rDNA + 1)] at 25 m (open squares) and 100 m (closed circles) of each OTU. To eliminate bias, correlations are for log rRNA and log rDNA and therefore limited to OTUs where both rRNA and rDNA were present.
The numerically dominant OTU across all libraries was most similar to Prochlorococcus, which is consistent with previous molecular diversity studies and flow cytometry counts (Fig. 2) (3). Despite the low maximal growth rate (<1 day−1) and small cell size (34) that typically correspond to a low ribosome requirement for cellular maintenance (20), Prochlorococcus dominates both abundance and activity libraries, suggesting an important role in the ecology and biogeochemistry of the bacterioplankton community, as observed previously (3, 35). The second-most-abundant OTU across libraries was most similar to “Candidatus Pelagibacter,” which is consistent with the SAR11 clade being one of the most abundant clades in the surface ocean (31, 36). However, this OTU, as well as other closely related Alphaproteobacteria, was strikingly underrepresented in the rRNA libraries (Fig. 2), and in rank order abundance it was 11 positions lower in rank in the rRNA than in the rDNA libraries at both depths. These results suggest that SAR11-like clades display less activity per cell than other bacterioplankton populations, with potentially broader implications for their ecological and biogeochemical importance in the open ocean. This underrepresentation in libraries of activity (i.e., rRNA) may be due to a low growth rate and the extremely small cytosol volume of cells in the SAR11 clade, which is roughly 1 order of magnitude less than that of Prochlorococcus cells (37). Alternatively, these results may reflect a low point of activity in a temporally or spatially dynamic population (9).
Similar to studies in other aquatic environments (9, 12), we observed that OTUs often dismissed as “weeds”—low-abundance species that are easily cultured and thought to be unimportant in natural environments—are disproportionately abundant in rRNA libraries. The resulting high rRNA/rDNA ratio for such weeds suggests that although these OTUs are not numerically abundant among the marine microbial milieu, they are highly active, and their populations may be regulated by tightly coupled grazing or viral pressure (38, 39) or, alternatively, that their prevalence cycles between abundance and rarity. Interestingly, one low-abundance OTU with high specific activity (rRNA/rDNA > 7) that ranked at least 18 positions higher in the rRNA libraries than libraries at both depths is related most closely to Alteromonas, a genus shown to enhance Prochlorococcus growth under conditions of oxidative stress (40, 41). Thus, this clade's ecological niche and success, despite low abundance, could be due to a mutualistic relationship with Prochlorococcus. These results highlight that specific activity in the marine environment may be independent from abundance and that interactions between taxa in complex ocean environments may guide productivity over simple bottom-up (resource-limited) or top-down (predation/viral lysis) pressures.
Uncoupling of specific activity and abundance in ecotypes of Prochlorococcus.
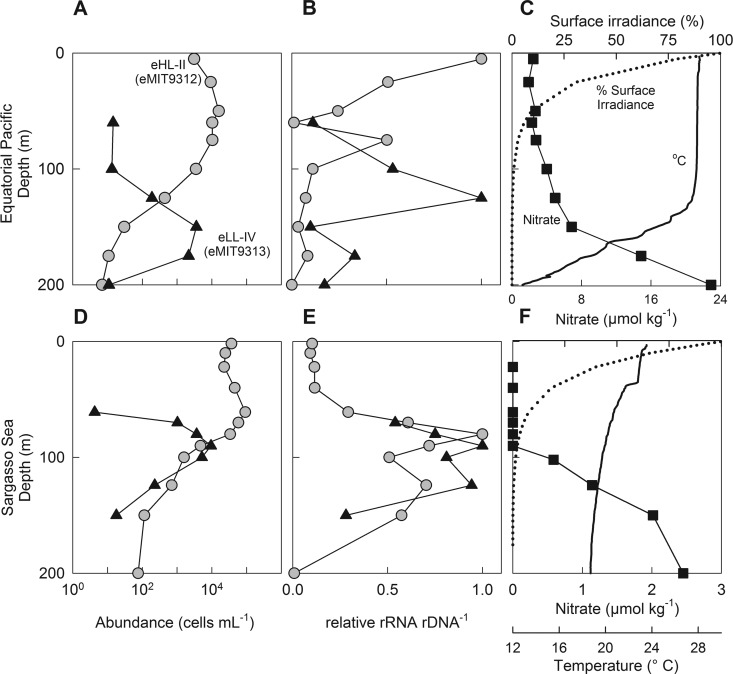
Due to the abundance and biogeochemical importance of Prochlorococcus in the open ocean and the difficulty in resolving ecologically distinct Prochlorococcus clades using partial 16S rDNA sequences (e.g., Fig. 2), we applied an alternative approach to examine this group in greater detail. Here, we used 23S rRNA-targeted qPCR primers to quantitatively measure the abundances (rDNA) and activities (rRNA) of two dominant ecotypes of Prochlorococcus (high-light eMIT9312/eHL-II and low-light eMIT9313/eLL-IV) for two open ocean depth profiles in relation to environmental variables. In both the Sargasso Sea and the equatorial Pacific stations, the two ecotypes exhibited maxima in cellular abundance with depth that are consistent with proposed physiological and genetic characteristics: eHL-II is adapted to high-light and low-nutrient regimes, whereas eLL-IV is adapted to low-light and higher-nutrient regimes (Fig. 4A and D) (42, 43). The abundances obtained with this technique are comparable to those from qPCR studies of ecotype abundances using well-established qPCR primers (23, 32, 44), and the most prevalent clades were captured in this analysis (Fig. 1 and 4). While the abundance data (rDNA) aligned with the expected light-driven physiology of the clades, we saw distinct results in the specific activities that differed between locations (Fig. 4). In both ocean depth profiles, the maximal specific activity largely coincided with abundance for eLL-IV, suggesting a relatively strong coupling of population abundance and specific activity. However, the abundances and specific activity of eHL-II were dramatically uncoupled with depth in the Sargasso Sea (but not the equatorial Pacific) (Fig. 4B and E), with a maximal relative activity at ∼75 m but a fairly constant cell abundance throughout the upper mixed layer (Fig. 4D and E). The differences in eHL-II specific activities among the locations could be explained by environmental nutrient distributions: the station in the equatorial Pacific has nutrients at the surface, where eHL-II can fully take advantage of its adaptation to high light, whereas at the station in the Sargasso Sea, nutrients are below detection near the surface, and eHL-II specific activity is maximal close to the nutricline, even though light energy is much less favorable at that depth. Yet, the abundance of eHL-II remains high near the surface in the Sargasso Sea, a finding that that may be attributable to mixing from a deeper source population, an uncoupling of growth and grazing, or other processes resulting in a non-steady-state condition.
Fig 4.
Prochlorococcus abundance and activity with associated physicochemical variability versus depth for two representative locations in the equatorial Pacific (A, B, and C) and the Sargasso Sea (D, E, and F). Abundance profiles (A and D) of the two dominant ecotypes, high-light eMIT9312/eHL-II and low-light eMIT9313/eLL-IV, measured using qPCR of 23S rDNA (cells ml−1). Normalized specific activity (B and E) of eHL-II and eLL-IV ecotypes measured using qPCR of 23S rRNA and rDNA. Profiles of physicochemical variability (C and F), including irradiance (percentage of surface), temperature (°C), and nitrate (μmol kg−1).
As was observed in the Sargasso Sea, all of the distinguishable subpopulations of Prochlorococcus in the North Pacific (station ALOHA) had maximal specific activities at the 100-m depth (Fig. 2), even though the total Prochlorococcus abundance was highest near the surface (Fig. 1). Thus, the observations at station ALOHA were also consistent with a deep nutricline limiting Prochlorococcus productivity but not cell numbers in the surface ocean (Fig. 2). Regardless of the precise mechanism, measurements of specific activity (and not abundance) were most consistent with the observed nutrient and light distributions. Thus, specific activity measurements identify the locations or time points when a resource-limitation-only model (bottom-up control) for Prochlorococcus populations (and likely other bacterioplankton) does not apply and provide a potential methodology for resolving unexplained biogeography or other ecological patterns in genetically distinct groups of bacterioplankton (45).
Implications of uncoupled specific activity and abundance.
Marine microbiology has progressed dramatically from assessing the abundance and diversity of the bacterioplankton (16S rDNA surveys) to making functional assignments using genomics and metagenomics. There are now methods that can relate the presence of an organism or gene to its expression or activity in that environment (rRNA qRT-PCR, metatranscriptomics) (46). Our results suggest that low-abundance microbes may be disproportionately active in certain environments (Alteromonas) and that some of the most abundant microbes (e.g., SAR11) may have low metabolic activity. Comparisons of rRNA and rDNA levels in situ have the potential to estimate environmental growth rates and activity without the biases associated with incubations or culturing. Although absolute quantification of a specific target is not possible in gene libraries, qRT-PCR of rRNA (cDNA) and qPCR of rDNA provide estimates of specific activities for clades of organisms and are complementary to library-based approaches that allow intercomparison among many groups. We observed uncoupling of abundance and specific activity of Prochlorococcus in the Sargasso Sea depth profile, which highlights deficiencies in our understanding of marine microbial ecology and population structure. If the specific activity is higher for eHL-II deeper in the water column, was this population in the process of becoming more abundant, or do other factors such as predation limit its abundance at depth? Techniques that allow us to investigate the activities and abundances of specific ecotypes in situ will also allow us to examine the environmental factors that structure bacterioplankton populations and link the relative abundances and specific activities of marine bacterioplankton populations. Future studies should include a broad suite of methodologies that assess different metrics of microbial activity (e.g., ATP, carbon uptake, growth rate) to examine how rRNA-based activity is related to these other measurements.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded in part by funds from the U.S. National Science Foundation (number OCE10-31064 to Z.I.J.) and the Gordon and Betty Moore Foundation (to D.M.K.). Sequencing was conducted in part by the U.S. Department of Energy Joint Genome Institute and was supported by the Office of Science of the U.S. Department of Energy under contract no. DE-AC02-05CH11231.
The funders had no role in study design, data collection and analysis, the decision to publish, or preparation of the manuscript.
We thank Brett Updyke for assistance with sampling and Brian Binder for valuable discussion. We also acknowledge helpful comments from the anonymous reviewers.
Footnotes
Published ahead of print 19 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02155-12.
REFERENCES
- 1. Acinas SG, Klepac-Ceraj V, Hunt DE, Pharino C, Ceraj I, Distel DL, Polz MF. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551–554 [DOI] [PubMed] [Google Scholar]
- 2. Coleman ML, Chisholm SW. 2010. Ecosystem-specific selection pressures revealed through comparative population genomics. Proc. Natl. Acad. Sci. U. S. A. 107:18634–18639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard N-U, Martinez A, Sullivan MB, Edwards R, Brito BR, Chisholm SW, Karl DM. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311:496–503 [DOI] [PubMed] [Google Scholar]
- 4. Johnson ZI, Zinser ER, Coe A, McNulty NP, Woodward EMS, Chisholm SW. 2006. Niche partitioning among Prochlorococcus ecotypes along ocean-scale environmental gradients. Science 311:1737–1740 [DOI] [PubMed] [Google Scholar]
- 5. Apple JK, Strom SL, Palenik B, Brahamsha B. 2011. Variability in protist grazing and growth on different marine Synechococcus isolates. Appl. Environ. Microbiol. 77:3074–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Strom SL. 2008. Microbial ecology of ocean biogeochemistry: a community perspective. Science 320:1043–1045 [DOI] [PubMed] [Google Scholar]
- 7. Polz MF, Hunt DE, Preheim SP, Weinreich DM. 2006. Patterns and mechanisms of genetic and phenotypic differentiation in marine microbes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 361:2009–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lauro FM, McDougald D, Thomas T, Williams TJ, Egan S, Rice S, DeMaere MZ, Ting L, Ertan H, Johnson J, Ferriera S, Lapidus A, Anderson I, Kyrpides N, Munk AC, Detter C, Han CS, Brown MV, Robb FT, Kjelleberg S, Cavicchioli R. 2009. The genomic basis of trophic strategy in marine bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:15527–15533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Campbell BJ, Yu L, Heidelberg JF, Kirchman DL. 2011. Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl. Acad. Sci. U. S. A. 108:12776–12781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pedrós-Alió C. 2012. The rare bacterial biosphere. Ann. Rev. Mar. Sci. 4:449–466 [DOI] [PubMed] [Google Scholar]
- 11. Lennon JT, Jones SE. 2011. Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 9:119–130 [DOI] [PubMed] [Google Scholar]
- 12. Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. U. S. A. 107:5881–5886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fuhrman J, Azam F. 1982. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar. Biol. 66:109–120 [Google Scholar]
- 14. Kirchman D, K'Nees E, Hodson R. 1985. Leucine incorporation and its potential as a measure of protein synthesis by bacteria in natural aquatic systems. Appl. Environ. Microbiol. 49:599–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kemp PF, Lee S, Laroche J. 1993. Estimating the growth rate of slowly growing marine bacteria from RNA content. Appl. Environ. Microbiol. 59:2594–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Worden AZ, Binder BJ. 2003. Growth regulation of rRNA content in Prochlorococcus and Synechococcus (marine cyanobacteria) measured by whole-cell hybridization of rRNA-targeted peptide nucleic acids. J. Phycol. 39:527–534 [Google Scholar]
- 17. Kerkhof L, Kemp P. 1999. Small ribosomal RNA content in marine Proteobacteria during non-steady-state growth. FEMS Microbiol. Ecol. 30:253–260 [DOI] [PubMed] [Google Scholar]
- 18. Morgenroth E, Obermayer A, Arnold E, Brühl A, Wagner M, Wilderer P. 2000. Effect of long-term idle periods on the performance of sequencing batch reactors. Water Sci. Technol. 41:105–113 [Google Scholar]
- 19. Acinas SG, Marcelino LA, Klepac-Ceraj V, Polz MF. 2004. Divergence and redundancy of 16S rRNA sequences in genomes with multiple rRNA operons. J. Bacteriol. 186:2629–2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fegatella F, Lim J, Kjelleberg S, Cavicchioli R. 1998. Implications of rRNA operon copy number and ribosome content in the marine oligotrophic ultramicrobacterium Sphingomonas sp. strain RB2256. Appl. Environ. Microbiol. 64:4433–4438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gihring TM, Green SJ, Schadt CW. 2012. Massively parallel rRNA gene sequencing exacerbates the potential for biased community diversity comparisons due to variable library sizes. Environ. Microbiol. 14:285–290 [DOI] [PubMed] [Google Scholar]
- 22. Frias-Lopez J, Shi Y, Tyson GW, Coleman ML, Schuster SC, Chisholm SW, DeLong EF. 2008. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. U. S. A. 105:3805–3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Johnson ZI, Shyam R, Ritchie AE, Lin Y, Mioni C, Lance VP, Murray JW, Zinser ER. 2010. The effects of iron- and light-limitation on phytoplankton communities of deep chlorophyll maxima of the Western Pacific Ocean. J. Mar. Res. 68:1–26 [Google Scholar]
- 24. Johnson KS. 2010. Simultaneous measurements of nitrate, oxygen, and carbon dioxide on oceanographic moorings: observing the Redfield ratio in real time. Limnol. Oceanogr. 55:615–627 [Google Scholar]
- 25. Johnson KS, Coletti LJ. 2002. In situ ultraviolet spectrophotometry for high resolution and long-term monitoring of nitrate, bromide and bisulfide in the ocean. Deep Sea Res. Part 1 Oceanogr. Res. Pap. 49:1291–1305 [Google Scholar]
- 26. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35:7188–7196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guindon S, Gascuel O. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696–704 [DOI] [PubMed] [Google Scholar]
- 30. Ahlgren NA, Rocap G, Chisholm SW. 2006. Measurement of Prochlorococcus ecotypes using real-time PCR reveals different abundances of genotypes with similar light physiologies. Environ. Microbiol. 8:441–454 [DOI] [PubMed] [Google Scholar]
- 31. Eiler A, Hayakawa DH, Church MJ, Karl DM, Rappé MS. 2009. Dynamics of the SAR11 bacterioplankton lineage in relation to environmental conditions in the oligotrophic North Pacific subtropical gyre. Environ. Microbiol. 11:2291–2300 [DOI] [PubMed] [Google Scholar]
- 32. Malmstrom RR, Coe A, Kettler GC, Martiny AC, Frias-Lopez J, Zinser ER, Chisholm SW. 2010. Temporal dynamics of Prochlorococcus ecotypes in the Atlantic and Pacific oceans. ISME J. 4:1252–1264 [DOI] [PubMed] [Google Scholar]
- 33. Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U. S. A. 103:12115–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johnson ZI, Lin Y. 2009. Prochlorococcus: approved for export. Proc. Natl. Acad. Sci. U. S. A. 106:10400–10401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu DY, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, LI K, Kravitz S, Heidelberg JF, Utterback T, Rogers YH, Falcon LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. 2007. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 5:398–431 doi:10.1371/journal.pbio.0050077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morris RM, Rappe MS, Connon SA, Vergin KL, Siebold WA, Carlson CA, Giovannoni SJ. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806–810 [DOI] [PubMed] [Google Scholar]
- 37. Sowell SM, Wilhelm LJ, Norbeck AD, Lipton MS, Nicora CD, Barofsky DF, Carlson CA, Smith RD, Giovanonni SJ. 2009. Transport functions dominate the SAR11 metaproteome at low-nutrient extremes in the Sargasso Sea. ISME J. 3:93–105 [DOI] [PubMed] [Google Scholar]
- 38. Landry MR, Kirchman DL. 2002. Microbial community structure and variability in the tropical Pacific. Deep Sea Res. Part 2 Top. Stud. Oceanogr. 49:2669–2693 [Google Scholar]
- 39. Suttle CA. 2007. Marine viruses—major players in the global ecosystem. Nat. Rev. Microbiol. 5:801–812 [DOI] [PubMed] [Google Scholar]
- 40. Morris JJ, Johnson ZI, Szul MJ, Keller M, Zinser ER. 2011. Dependence of the cyanobacterium Prochlorococcus on hydrogen peroxide scavenging microbes for growth at the ocean's surface. PLoS One 6:e16805 doi:10.1371/journal.pone.0016805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Morris JJ, Kirkegaard R, Szul MJ, Johnson ZI, Zinser ER. 2008. Facilitation of robust growth of Prochlorococcus colonies and dilute liquid cultures by “helper” heterotrophic bacteria. Appl. Environ. Microbiol. 74:4530–4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moore LR, Rocap G, Chisholm SW. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464–467 [DOI] [PubMed] [Google Scholar]
- 43. Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, Post AF, Hagemann M, Paulsen I, Partensky F. 2009. Ecological genomics of marine picocyanobacteria. Microbiol. Mol. Biol. Rev. 73:249–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zinser ER, Coe A, Johnson ZI, Martiny A, Fuller NJ, Scanlan DJ, Chisholm SW. 2006. Prochlorococcus ecotype abundances in the North Atlantic Ocean revealed by an improved quantitative PCR method. Appl. Environ. Microbiol. 72:723–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fuhrman JA. 2009. Microbial community structure and its functional implications. Nature 459:193–199 [DOI] [PubMed] [Google Scholar]
- 46. Gifford SM, Sharma S, Rinta-Kanto JM, Moran MA. 2011. Quantitative analysis of a deeply sequenced marine microbial metatranscriptome. ISME J. 5:461–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.