Abstract
The gray mold fungus Botrytis cinerea is a major threat to fruit and vegetable production. Strawberry fields usually receive several fungicide treatments against Botrytis per season. Gray mold isolates from several German strawberry-growing regions were analyzed to determine their sensitivity against botryticides. Fungicide resistance was commonly observed, with many isolates possessing resistance to multiple (up to six) fungicides. A stronger variant of the previously described multidrug resistance (MDR) phenotype MDR1, called MDR1h, was found to be widely distributed, conferring increased partial resistance to two important botryticides, cyprodinil and fludioxonil. A 3-bp deletion mutation in a transcription factor-encoding gene, mrr1, was found to be correlated with MDR1h. All MDR1h isolates and the majority of isolates with resistance to multiple fungicides were found to be genetically distinct. Multiple-gene sequencing confirmed that they belong to a novel clade, called Botrytis group S, which is closely related to B. cinerea and the host-specific species B. fabae. Isolates of Botrytis group S genotypes were found to be widespread in all German strawberry-growing regions but almost absent from vineyards. Our data indicate a clear subdivision of gray mold populations, which are differentially distributed according to their host preference and adaptation to chemical treatments.
INTRODUCTION
The necrotrophic ascomycete Botrytis cinerea Pers. [teleomorph Botryotinia fuckeliana (de Bary) Whetzel] causes gray mold disease on more than 200 mostly dicotyledonous plant species (1). In temperate climates under humid conditions, the gray mold fungus leads to serious losses in yield and quality on numerous crops, particularly fruits, vegetables, and ornamental flowers. B. cinerea is the major representative of the genus Botrytis, which currently comprises more than 20 species. Whereas most Botrytis spp. have a narrow host range and infect monocotyledonous plants, B. cinerea belongs to a distinct phylogenetic clade of species which infect mostly dicotyledonous plants (2).
In Germany, five different classes of fungicides with different modes of action against gray mold in soft-fruit cultures are currently registered, namely, the anilinopyrimidines (cyprodinil, pyrimethanil, and mepanipyrim), the hydroxyanilide fenhexamid, the phenylpyrrole fludioxonil, the carboxamide boscalid, and the quinone outside inhibitors (QoIs) pyraclostrobin, azoxystrobin, and trifloxystrobin. Two benzimidazole-type compounds, benomyl and carbendazim, have been widely used in the past but abandoned for use against gray mold because of widespread resistance (3). The registration of the dicarboximide iprodione, another fungicide against Botrytis, for German soft-fruit production has recently expired. Due to its enormous reproduction rate by conidiospores and its high genetic adaptability, B. cinerea has been classified as a high-risk pathogen regarding fungicide resistance (3, 4). The most commonly observed mechanism of resistance to site-specific fungicides is target site resistance, which is caused by mutations that alter the fungicide target protein (5). For example, mutations in the β-tubulin gene at codon 198 or 200 are well-known to be responsible for high levels of benzimidazole resistance in a variety of fungi (5–7). Field strains with high levels of fenhexamid resistance (called HydR3) have been shown to carry mutations in the erg27 gene, encoding a 3-keto reductase involved in ergosterol biosynthesis (8, 9). Studies with Alternaria alternata and B. cinerea have revealed that boscalid resistance is caused by different amino acid alterations in the succinate dehydrogenase gene (10–12). Resistance to anilinopyrimidines has also been reported in B. cinerea (13), but the molecular target and resistance-related mutations are not yet known. Field resistance to QoIs in B. cinerea and many other plant pathogens is usually caused by a G143A amino acid substitution in the gene encoding cytochrome b (11, 14, 15). Resistance to dicarboximides (e.g., iprodione) has been correlated with mutations in the B. cinerea osmosensing histidine kinase Bos1 (BcBos1) (16, 17). In order to reduce the risk of resistance development in gray mold, it is recommended to avoid the repeated use of the same fungicide but instead to use mixtures of or to alternate between fungicides with different modes of action during one season (4, 18).
Compared to target site modifications, which usually lead to high levels of specific resistance to a single class of fungicides, other mechanisms of fungicide resistance are considered less important in plant-pathogenic fungi (5). In French and German vineyards, B. cinerea populations with multidrug resistance (MDR) phenotypes have been observed at high frequencies (19, 20). MDR is based on overexpression of drug efflux transporters and confers different levels of resistance to diverse fungicides and drugs. MDR causes serious problems in drug therapy against human-pathogenic Candida spp. and bacteria (21, 22), but its role in lowering the efficacy of fungicides against gray mold is not yet clear (19). In B. cinerea, three MDR phenotypes were distinguished; i.e., MDR1 (AniR2) strains show partial resistance to fludioxonil and cyprodinil, MDR2 (AniR2) strains have partial resistance to fenhexamid, cyprodinil, and iprodione, and MDR3 strains combine the fungicide resistance spectra of MDR1 and MDR2 strains (23). The MDR1 phenotype is correlated with gain-of-function point mutations in the zinc cluster transcription factor Mrr1, leading to constitutive overexpression of the atrB-encoded efflux pump (19). The MDR2 phenotype, in contrast, results from a rearrangement in the promoter of the mfsM2 gene, encoding another efflux transporter, caused by insertion of a retroelement-derived sequence. This rearrangement leads to strong constitutive expression of mfsM2 (19, 24).
On cultivated strawberries, gray mold is a major disease worldwide. In contrast to German vineyards, which are usually treated with botryticides no more than twice per season, strawberry fields are sprayed weekly during the flowering period, resulting in multiple treatments. Seasonal spraying programs therefore frequently include repeated usage of the same fungicides or fungicide classes. In the last few years, failures in chemical control of gray mold have been reported by German strawberry growers under conditions of high disease pressure. Our recent work has shown that fungicide resistance frequencies in B. cinerea isolates from northern Germany soft-fruit production can be very high (9, 25, 26). In this study, we have evaluated the prevalence and distribution of fungicide resistance in gray mold populations in different strawberry-growing regions of Germany. High frequencies of fungicide resistance and of a novel, stronger variant of the MDR1 phenotype, called MDR1h, were observed. The majority of strawberry isolates, including all MDR1h isolates, were genetically distinct from vineyard populations of B. cinerea. Phylogenetic analyses indicated that they belong to a previously unknown genotype of the gray mold fungus distinct from but closely related to both B. cinerea and B. fabae.
MATERIALS AND METHODS
Fungicide treatments.
Commercial strawberry fields in different parts of Germany sampled for gray mold isolates had been treated with a range of botryticides, including a product with the active ingredients pyraclostrobin and boscalid (Signum; BASF Crop Protection, Limburgerhof, Germany), fenhexamid (Teldor; Bayer Crop Science, Monheim, Germany), fludioxonil and cyprodinil (Switch; Syngenta Agro GmbH, Maintal, Germany), iprodione (Rovral; BASF), trifloxystrobin (Flint; Bayer), and azoxystrobin (Ortiva; Syngenta).
Botrytis strains used in this work.
Botrytis cinerea strains T4 and B05.10 are laboratory strains with known genome sequences (27). B. calthae strains MUCL1089 (origin, Belgium) and MUCL2830 (Canada) were kindly provided by J. A. L. van Kan (University of Wageningen). B. fabae reference strains CBS109.57 (MUCL395; The Netherlands), MUCL98 (Spain), MUCL7923 (Belgium), and MUCL1104 (Italy) were obtained from BCCM (Brussels, Belgium). B. pseudocinerea strains VD110 and VD256 (France) (28) and B. fabae strain 11002 were kindly provided by A.-S. Walker.
Collection of Botrytis field isolates and fungicide sensitivity assays.
A total of 173 gray mold isolates were sampled from fungicide-treated strawberry fields between 2008 and 2011 in northern (60 isolates), central (38 isolates), and southern (75 isolates) Germany (see Table S1 in the supplemental material). Isolates from northern Germany were from the two major production areas, Stader Geest and Langförden/Vechta, isolates from central Germany were from Koblenz-Ahrweiler, and isolates from southern Germany were from the Breisgau region (see Fig. S1 and Table S1 in the supplemental material). Most isolates were collected from infected fruits between May and July, after the end of fungicide applications, by tipping sterile cotton buds into areas with gray mold. Isolates from Langförden/Vechta were harvested from fruit mummies in October 2011. After transferring the mummies into the laboratory, they were incubated in humid boxes for several days until new sporulation occurred. Spores were transferred onto HA agar plates (10 g liter−1 malt extract, 4 g liter−1 glucose, 4 g liter−1 yeast extract, 15 g liter−1 agar, pH 5.5). After 2 to 5 days of incubation at 20°C under ambient light conditions, agar pieces were cut out from the periphery of the spreading mycelium or conidia were taken from sporulating areas using sterile forceps, and the fungus was transferred to a new agar plate. The isolates were further cultivated for 8 to 10 days until sporulation.
To search for the presence of group S isolates on grapevine berries, 50 gray mold isolates were collected in 2010 from six vineyards in the German Wine Road region (described in references 19 and 20), 62 isolates were collected from a vineyard in Trier in 2011 (Mosel wine-growing region), and 28 isolates were collected from a vineyard in Meckenheim in 2011 (German Wine Road).
To detect fungicide resistance, conidia from sporulating agar plates were washed off with 10 ml sterile water using a glass spatula. The conidia were filtered through glass wool into 10-ml tubes, centrifuged at 3,000 × g for 3 min, resuspended at a defined concentration of conidia, and stored at 4°C for up to 10 days until use. To identify the major specific resistance phenotypes, tests on solid agar medium containing the following discriminatory fungicides at the indicated concentrations were performed (20, 26): carbendazim at 5 mg liter−1, fenhexamid at 3 mg liter−1, iprodione at 7.5 mg liter−1, boscalid at 7.5 mg liter−1, cyprodinil at 8 mg liter−1, and azoxystrobin at 25 mg liter−1 (with 100 mg liter−1 salicyl hydroxamic acid). Isolates with MDR phenotypes (19) were routinely identified by microscopic plate assays. In contrast to non-MDR isolates, MDR1 and MDR1h isolates were able to fully germinate on 0.03 mg liter−1 cyprodinil and on 0.2 mg liter−1 fludioxonil. MDR1h isolates were distinguished from MDR1 isolates by their ability to germinate on 1 mg liter−1 fludioxonil. MDR2 and MDR3 isolates, which could both grow on 0.6 mg liter−1 fenhexamid and 50 mg liter−1 cycloheximide, were not observed in any of the strawberry isolates tested (not shown). All MDR isolates but none of the non-MDR isolates were able to germinate on 5 mg liter−1 tolnaftate (23). For most fungicide tests, HA agar plates were used. For fludioxonil, cyprodinil, and tolnaftate, 0.5% sucrose agar was used (26). For boscalid, plates containing YBA medium were used (19). Up to 20 different strains were inoculated on each fungicide plate, using 5-μl droplets of a suspension containing 2 × 105 spores ml−1. Germination was evaluated microscopically after 16 h of incubation. Mycelial growth was visually recorded after 48 h (72 h for boscalid) of incubation. Quantitative determination of sensitivity values for cyprodinil and fludioxonil was performed by analysis of mycelial growth inhibition in microtiter plates containing 0.1 ml liquid medium, as described in reference 19, except that MICs instead of 50% effective concentrations were determined.
Genetic characterization of Botrytis isolates.
Extraction of genomic DNA was performed as described previously (29) using conidia from sporulating agar plates. To distinguish the isolates genetically, two types of molecular markers were used. For PCR-restriction fragment length polymorphism (RFLP) analysis of the 18S/28S rRNA intergenic spacer (IGS) region, a PCR with primers IGS1a and IGS1b was performed, and the PCR product was digested with HinfI as previously described (30). For detection of B. pseudocinerea (group I) isolates, a 1,171-bp fragment of the B. cinerea homolog of hch was amplified using primers 262 and 520L and digested with HhaI (28, 31). Identification of Botrytis group S isolates was based on the 21-bp insertion in mrr1, using the primer pair Mrr1_spez_F and Mrr1_spez_R. For analysis of the B. cinerea mating types, the MAT1-1-specific primers Mat1F/Mat1R and the MAT1-2-specific primers BcHMG-F/BcHMG-R were used (27). To identify B. cinerea MDR1h isolates carrying a 3-bp deletion in the mrr1 gene, the primer pair Mrr1_spez_F/Mrr1-Pira was used. After performing PCR, followed by digestion with HypCH4V, the PCR product of 460 bp from MDR1h isolates was cut into fragments of 280, 134, 35, and 11 bp, while the 463-bp PCR product from non-MDR1h isolates yielded fragments of 280, 172, and 11 bp. This difference was resolved by electrophoresis in a 2% agarose gel. All primers are listed in Table S4 in the supplemental material.
Gene expression studies.
For analysis of atrB expression, quantitative reverse transcription-PCR (qRT-PCR) was performed as described previously (19), using the primers shown in Table S4 in the supplemental material.
Phylogenetic analyses.
For sequence analysis, genes were amplified from genomic Botrytis DNA with the following primers, shown in Table S4 in the supplemental material. The DNA of mrr1 was amplified in two parts: primers Mrr1_atg and TF1-2_n were used for the first part (fragment size, 1.5 kb), whereas for the second part (fragment size, 1.8 kb), primers TF1-3_n and TF1-4 were used. The nested primers TF1-5 and TF1-2 (part 1) and TF1-3 and TF1-6 (part 2) were used for sequencing. The following primer pairs were used for amplification and sequencing: for fg1020, FG1020-F and FG1020-R (fragment size, 0.8 kb) (32); for hsp60, Hsp60-F and Hsp60-R (fragment size, 1.2 kb) (2); for msf547, MS547-F and MS547-F (fragment size, 0.9 kb) (28); and for nep2, Nep2(−200)for and Nep2(+1147)rev (fragment size, 1.3 kb) (33). For the alignments, the length of the aligned sequences was at least 79% of the total alignment length, except for mrr1 and nep2, where individual sequences covered only 32% (mrr1) and 56% (nep2) of the total length, respectively.
Nucleotide sequences were aligned using the MUSCLE program (34) and manually optimized in the Seaview program (35). Maximum-likelihood analyses were carried out using the RAxML (version 7.3.0) program (36), partitioning the data by implementing a separate GTR (general time reversible) model (37) with gamma correction and a proportion of invariable sites for each codon position. Bootstrap frequencies (38, 39) were calculated with 1,000 replicates, and only branches that received bootstrap frequencies higher than 70% were considered significantly supported.
Nucleotide sequence accession numbers.
The nucleotide sequences generated in this study have been deposited in GenBank and are listed in Table 1.
Table 1.
GenBank accession numbers of sequences determined in this study and used for phylogenetic studies
| Strain | GenBank accession no. for the indicated gene |
||||
|---|---|---|---|---|---|
| mrr1 | ms457 | fg1020 | hsp60 | nep2 | |
| D09_K_4-01 | JX266764 | JX266726 | JX266738 | JX266716 | JX266751 |
| G09_S33 | JX266769 | JX266729 | JX266741 | JX266719 | JX266754 |
| D10_B_F1-06 | JX266766 | JX266727 | JX266739 | JX266717 | JX266752 |
| D10_B_F3-05 | JX266767 | JX266728 | JX266740 | JX266718 | JX266753 |
| D08_H_8-15 | JX266777 | JX266733 | JX266746 | JX266721 | NDa |
| S10_C1 | JX266770 | JX266730 | JX266742 | ND | JX266755 |
| D08_H_8-04 | ND | JX266725 | JX266737 | JX266715 | JX266750 |
| N11_S_E06 | JX266780 | ND | ND | JX266723 | JX266759 |
| N11_S_E08 | JX266772 | ND | ND | JX266720 | JX266757 |
| D06_1-30 | ND | JX266724 | ND | JX266714 | JX266748 |
| MUCL1104 | ND | JX266731 | JX266744 | ND | JX266758 |
| MUCL2830 | JX266781 | JX266734 | JX266747 | ND | ND |
| VD256 | JX266779 | ND | ND | JX266722 | ND |
| D06_5-16 | JX266760 | ND | JX266735 | ND | ND |
| MUCL395 | JX266773 | ND | JX266745 | ND | ND |
| N11_K_W08b | JX266761 | ND | ND | ND | JX266749 |
| N11_K_W11 | JX266771 | ND | ND | ND | JX266756 |
| D04_57 | JX266776 | ND | ND | ND | ND |
| D08_H_8-17 | JX266778 | ND | ND | ND | ND |
| G09_S2 | JX266768 | ND | ND | ND | ND |
| D09_K_2-3 | JX266763 | ND | ND | ND | ND |
| D09_K_A31 | JX266765 | ND | ND | ND | ND |
| N11_S_E15 | JX266762 | ND | ND | ND | ND |
| MUCL98 | JX266774 | ND | ND | ND | ND |
| MUCL7923 | JX266775 | ND | ND | ND | ND |
| 11002 | JX266732 | ND | ND | ND | ND |
| G09_S10 | ND | JX266736 | ND | ND | ND |
| D10_K_S11-02 | ND | ND | JX266743 | ND | ND |
ND, sequence not determined.
RESULTS
Botrytis strawberry isolates show high frequencies of fungicide resistance.
The resistance patterns of 173 Botrytis isolates collected between 2008 and 2011 in strawberry-growing regions in different parts of Germany were analyzed (Fig. 1; see Fig. S1 in the supplemental material). Isolates with no resistance to any of the seven tested fungicides were only rarely found. At all sampling sites, fungicide-resistant Botrytis isolates were retrieved at various but overall high frequencies. Although benzimidazoles have not been regularly used in German strawberry fields since 1976, carbendazim-resistant isolates were observed at frequencies of 43.3%, 44.7%, and 48.0% in northern, central, and southern Germany, respectively (see Table S1 in the supplemental material). Resistance to iprodione, which was commonly used against gray mold in strawberries until 2009, was also found at high frequencies of 40.0% (north), 76.3% (central), and 45.3% (south). Resistance to fenhexamid was found in 45.0%, 78.9%, and 44.0% of the isolates in northern, central, and southern Germany, respectively. For boscalid, which has been registered for use in German strawberry fields since the 2007 season, resistance was observed in isolates obtained in 2010 in northern Germany and 2011 in southern Germany at frequencies of 13.3% and 22.7%, respectively. Strains resistant to the QoI fungicide azoxystrobin were observed at very high frequencies of 83.3% (north), 76.3% (central), and 84.0% (south). Remarkably, all 25 boscalid-resistant isolates were also resistant to azoxystrobin, presumably reflecting the mixture of these two fungicide classes in the only registered boscalid-containing fungicide, Signum. Specific resistance to cyprodinil was observed at frequencies of 25.0%, 68.4%, and 56.0%, respectively. In accordance with previous reports, no highly resistant isolates (growing at 3 mg liter−1 fludioxonil) were retrieved from the fields.
Fig 1.
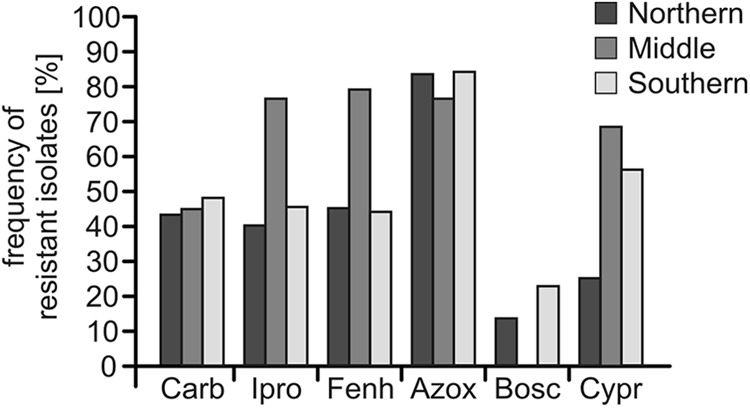
Frequencies of gray mold isolates with specific resistance to six fungicides from different sampling sites in northern (n = 60), central (n = 38), and southern (n = 75) Germany. Frequencies are shown for carbendazim (Carb), iprodione (Ipro), fenhexamid (Fenh), azoxystrobin (Azox), boscalid (Bosc), and cyprodinil (Cypr).
A stronger MDR phenotype is widespread among strawberry gray mold isolates.
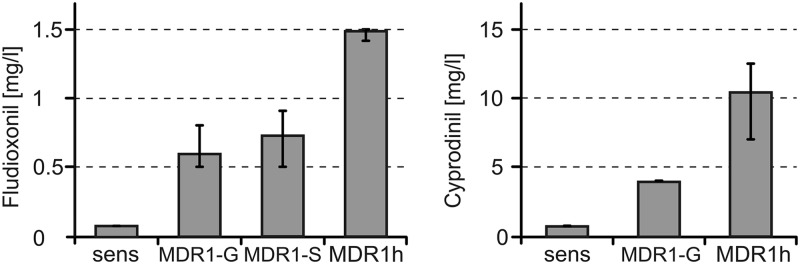
In French and German wine-growing regions, B. cinerea isolates with three distinct MDR phenotypes have been characterized (19, 20). In contrast, none of the isolates described in Table S1 in the supplemental material and none of more than 500 other strawberry isolates tested possessed the MDR2 or MDR3 phenotypes (data not shown). However, a high frequency of strawberry isolates with MDR1-like phenotypes, characterized by partial resistance to fludioxonil and cyprodinil and high resistance to tolnaftate, was observed (19). Many of these isolates appeared to be more tolerant to fludioxonil and cyprodinil than previously characterized MDR1 isolates. This was confirmed by quantitative evaluation of resistance levels for selected isolates (Fig. 2; see Fig. S2 in the supplemental material). One group of isolates had resistance levels similar to those of previously characterized MDR1 isolates of vineyards carrying different MDR1-related mutations in mrr1 (19) and were confirmed to be MDR1. Another group of isolates, however, grew at 2- to 3-fold higher concentrations of either fludioxonil or cyprodinil (in the absence of specific cyprodinil resistance) and were therefore called MDR1h (Fig. 2). For routine distinction between MDR1 and MDR1h isolates, we developed agar plate tests with 0.4 mg liter−1 fludioxonil, which allowed significantly stronger germination and growth of MDR1h than MDR1 isolates (see Fig. S2 in the supplemental material).
Fig 2.
Sensitivity levels of Botrytis cinerea isolates to fludioxonil (left) and cyprodinil (right). MIC values (mg liter−1) are shown. Sensitive (sens) isolates (n = 3), MDR1-G (grape) isolates (n = 13), MDR1-S (strawberry) isolates (n = 13; fludioxonil only), and MDR1h strawberry isolates (n = 8) were tested. Bars indicate the ranges of MIC values obtained.
MDR1h isolates were found to be widely distributed in German strawberry fields, with frequencies being between 31.4% (north) and 60.5% (central) (see Table S1 in the supplemental material). MDR1 phenotypes were also commonly found, with frequencies being between 21.1% (central) and 41.7% (north). While isolates with the MDR1 phenotype have previously been shown to be common in French and German vineyards (19), isolates with the MDR1h phenotype have not been described in vineyards. In order to check for the possible presence of MDR1h phenotypes in vineyards, we reexamined 45 vineyard isolates previously classified as MDR1 in liquid fungicide assays. While some of these isolates had resistance levels that were higher than the average levels for MDR1 isolates, resistance in none of the vineyard isolates reached the levels of resistance in strawberry MDR1h isolates (data not shown). These results suggest that MDR1h phenotypes are restricted to strawberry fields.
MDR1h is correlated with extreme overexpression of a drug efflux transporter gene and with a 3-bp deletion in the transcription factor gene mrr1.
To find a molecular correlation with the MDR1h phenotype, we compared the expression of atrB encoding the ABC efflux transporter in MDR1h, MDR1, and non-MDR isolates. As shown in Fig. 3, atrB transcript levels of MDR1 and MDR1h isolates were strongly increased compared to those of non-MDR strains. Furthermore, atrB mRNA levels in most MDR1h isolates were severalfold higher than those in MDR1 isolates. The increased levels of resistance to fludioxonil and cyprodinil of MDR1h isolates are therefore correlated with increased atrB overexpression (Fig. 2 and 3).
Fig 3.
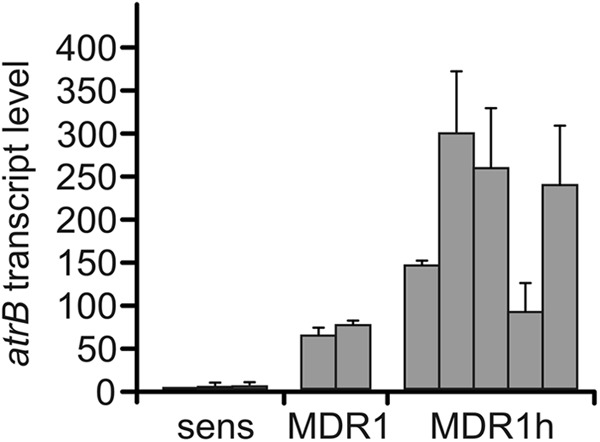
Transcript levels of atrB in sensitive, MDR1, and MDR1h strains, as determined by qRT-PCR analysis. Values shown are relative to the level in sensitive strain B05.10, which was set equal to 1. Sensitive isolates (B05.10, G09_S33, D08_H_8-04), MDR1 isolates (D06_5-16, D06_3-4), and MDR1h isolates (D08_H_8-16, D09_K_4-01, D09_K_1-01, D10_B_S1-24, D10_B_F3-05) were analyzed. Mean values from three experiments and standard deviations are shown.
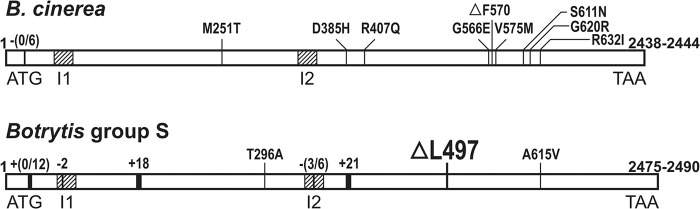
MDR1 strains have been shown to contain different mutations in the mrr1 coding region (19). Surprisingly, the mrr1 sequences of the MDR1h isolates, as well as those of several sensitive strawberry gray mold isolates, were found to be remarkably different from the published mrr1 sequences of B. cinerea. Previously, mrr1 sequences of B. cinerea strains isolated from German and French vineyards have been shown to be highly similar to each other (>99.8% identity), and they differ by only a single nucleotide between the two sequenced B. cinerea strains, T4 and B05.10 (19). In contrast, a comparison of the sequences of strain T4 and several strawberry isolates revealed a nucleotide divergence of 4 to 5%, as well as several small insertions and deletions (see Fig. S3 in the supplemental material). This was the first evidence that these strawberry isolates are genetically distinct from B. cinerea and that they belong to a novel clade, called Botrytis group S, as described below. Comparison of mrr1 sequences of a selected pair of a non-MDR isolate and an MDR1h strawberry isolate revealed only a single polymorphism resulting in an amino acid exchange, namely, a 3-bp deletion in the MDR1h isolate (Fig. 4; see Fig. S4 in the supplemental material). This deletion was found in the mrr1 sequences of eight MDR1h isolates, and in seven of these isolates, it was the only mutation that resulted in an amino acid change compared to the sequence of non-MDR strains. In contrast, the 3-bp deletion was found in none of the 33 tested MDR1 and non-MDR Botrytis group S isolates (data not shown). To corroborate the correlation between the 3-bp deletion and the MDR1h phenotype, a primer pair that allowed detection of the deletion by PCR followed by restriction digestion was designed. Out of 74 MDR1h isolates analyzed, all were found to contain the 3-bp deletion, while none of 33 MDR1 and non-MDR strawberry isolates contained this deletion. These data indicate that the 3-bp deletion is responsible for the high levels of atrB overexpression and the MDR1h phenotype. Sequence analysis of mrr1 from MDR1 group S isolates further revealed two different amino acid exchanges that were correlated with the MDR1 phenotype (Fig. 4; see Table S2 in the supplemental material).
Fig 4.
Comparison of the mrr1 coding region in B. cinerea and Botrytis group S isolates. The boldface numbers indicate the numbers of nucleotides; I1 and I2 indicate introns. Insertions in the Botrytis group S mrr1 sequence relative to the B. cinerea sequence are indicated by numbers with plus signs, and deletions are indicated by numbers with minus signs. MDR1-related amino acid exchanges and one MDR1-related deletion (ΔF570) described previously (19) or identified in this study (cf. Table S2 in the supplemental material) are shown. ΔL497 indicates a deletion found in all MDR1h isolates analyzed. The sequence comparison and numbering of amino acids are based on the B. cinerea strain T4 sequence (see Fig. S2 in the supplemental material).
MDR1h and specific fungicide resistance types are often coupled.
The frequent occurrence of MDR1-type or MDR1h-type resistance corresponded to a high proportion of isolates with specific resistance to multiple fungicides on the basis of target gene mutations. One isolate (D11_H_Bl.119.05) had accumulated MDR1h and specific resistance to six of seven of the botryticides tested (all botryticides tested except fludioxonil). Furthermore, 33 isolates showed simultaneous resistance to five botryticides, as did MDR1h isolates. Twenty-nine of these isolates were sensitive only to boscalid, the most recently introduced fungicide. These data illustrate the high frequencies of multiple-fungicide resistance in strawberry fields. Interestingly, MDR1h isolates were found to contain a significantly higher accumulation of specific resistances than MDR1 and non-MDR isolates (Fig. 5).
Fig 5.
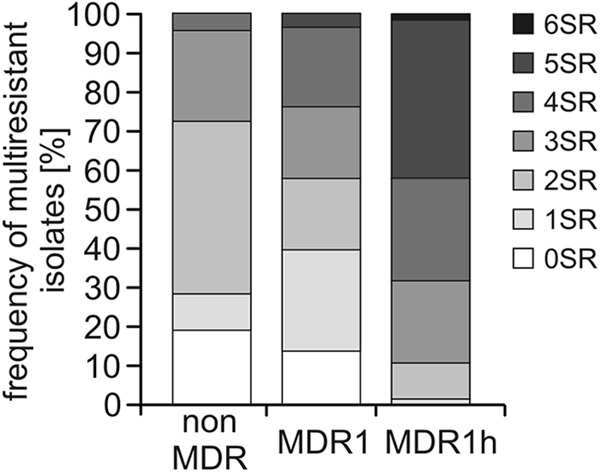
Accumulation of specific resistances (SR; where the numbers indicate the number of fungicides to which the isolates are resistant) in isolates with no MDR (n = 43) or with the MDR1 (n = 54) or MDR1h (n = 76) phenotype from strawberry fields in northern, central, and southern Germany.
Many strawberry gray mold isolates represent a novel clade closely related to but distinct from B. cinerea.
Botrytis pseudocinerea was originally reported to occur in small populations in French vineyards (28, 40). To detect the presence of B. pseudocinerea in strawberry fields, we analyzed the sequences of isolates in the polymorphic region of the hch gene (31). On the basis of this test, three isolates from northern Germany showed the B. pseudocinerea-specific hch PCR-RFLP (HhaI) digestion pattern. In contrast to the majority of strawberry gray mold isolates, all B. pseudocinerea isolates were sensitive to the tested fungicides (see Table S1 in the supplemental material).
PCR-RFLP of the 18S/28S rRNA intergenic spacer region (IGS-RFLP) is a useful tool to estimate the genetic diversity of B. cinerea populations (30, 41). When the IGS-RFLP (HinfI) patterns of strawberry isolates were compared with those of isolates collected from German vineyards, significant differences were found (see Table S3 in the supplemental material). For example, pattern 2, which was found in 49% of vineyard isolates, was never observed in strawberry isolates, whereas pattern 3, which was very rare in vineyard isolates, dominated (58.4%) in strawberry isolates, in particular, among MDR1h strains (88.7%). Within strawberry isolates, the patterns between B. cinerea and Botrytis group S isolates were significantly different from each other. Taken together, these data indicated that the strawberry isolates analyzed in this study were genetically different from vineyard isolates.
As described above, sequencing of mrr1 from several strawberry isolates revealed considerable differences from published B. cinerea mrr1 sequences (Fig. 4; see Fig. S3 in the supplemental material). Because of their apparent genetic separation from common B. cinerea strains and their discovery in strawberry fields, we preliminarily referred to these isolates as Botrytis group S or group S. To determine their phylogenetic relationship to other Botrytis spp. related to B. cinerea, we also sequenced the mrr1 genes of B. fabae, B. pseudocinerea, and B. calthae. Like B. cinerea, these species infect only or mainly dicotyledonous host plants in nature and are phylogenetically separated from the other, predominantly monocot host-specific Botrytis species (2). To take into account possible genetic variation due to different geographic origins, we included in the analysis three isolates from Greece, five isolates from Norway, and one MDR1h isolate from Spain (see Table S2 in the supplemental material). On the basis of the mrr1 sequences, all group S isolates clustered as a distinct clade more closely related to B. fabae than to B. cinerea (Fig. 6; see Fig. S5 in the supplemental material). Because mrr1 belongs to a multigene family of fungal zinc finger transcription factors and might be under positive selection in fungicide-treated strawberry fields, its value for phylogenetic studies is limited. Therefore, we compared the sequences of four other genes previously used for taxonomic studies with Botrytis spp.: fg1020 (40), encoding a putative ubiquitin conjugation factor, and ms547 (40), encoding an RNA helicase-like protein, which also separated the sequences of group S isolates from those of B. cinerea and B. fabae with high bootstrap values. When hsp60 (2), encoding a conserved heat shock protein, was analyzed, B. cinerea and group S isolates had almost identical sequences, while B. fabae sequences were distinct in three nucleotide positions. nep2, encoding a necrosis-inducing protein (33, 42), distinguished B. fabae, B. cinerea, and six out of eight group S isolates, while two group S isolates had nep2 sequences almost identical to the B. cinerea nep2 sequence (see Fig. S5 in the supplemental material). A combined analysis with all five genes supported the phylogenetic separation of group S, B. cinerea, and B. fabae (Fig. 6; see Fig. S6 in the supplemental material).
Fig 6.
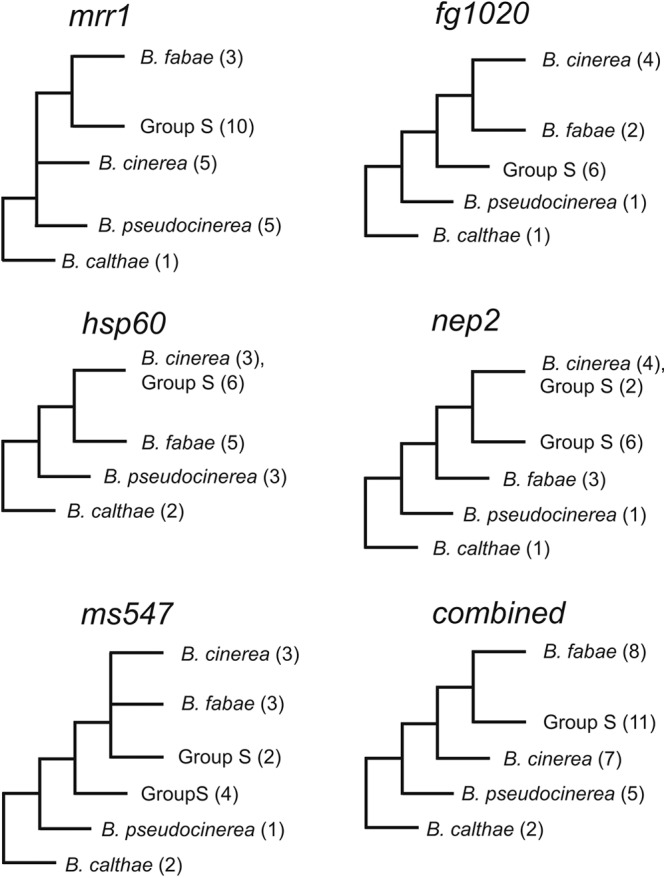
Simplified cladograms based on maximum likelihood analysis of the nucleotide sequence alignments of mrr1, fg1020, hsp60, nep2, and ms547 and on a combined analysis including all five genes from gray mold isolates belonging to different genetic groups and species of Botrytis. Branches receiving bootstrap support lower than 70% were collapsed. Numbers in parentheses indicate the numbers of specimens sequenced within each clade. Full trees for the individual genes are shown in Fig. S4 in the supplemental material, and those for the combined analysis are shown in Fig. S6 in the supplemental material.
Group S gray mold isolates are predominant in German strawberry-growing regions but largely absent from vineyards.
Alignment of mrr1 sequences from 10 group S and 26 B. cinerea isolates revealed the presence of small insertions and deletions (indels) in the coding region and in the two introns that distinguished all group S isolates from all B. cinerea isolates (Fig. 4). The 21-bp indel was used for a PCR-based assay for rapid identification of the strawberry isolates belonging either to group S or to B. cinerea. On the basis of this assay, group S isolates were found to be dominating over B. cinerea isolates in all strawberry-growing regions (74.0%, on average) (Fig. 7). All MDR1h isolates were classified as group S, while MDR1 and sensitive isolates were classified either as group S or as B. cinerea. Even if MDR1h isolates were excluded, group S isolates still represented a major strawberry gray mold population. Nevertheless, the distribution of group S isolates was variable in different fields. For example, two fields in central Germany (M1c and M1d; see Table S1 in the supplemental material) contained only group S isolates (n = 9 each), whereas another field (S1d) contained 35% group S and 65% B. cinerea isolates (n = 17). We also analyzed 78 grapevine berry isolates collected from vineyards in the German Wine Road region in 2010 and 2011 and 62 isolates from a vineyard in the Mosel wine-growing region for the presence of the 21-bp insertion. Except for a single group S isolate, all vineyard isolates were found to be B. cinerea. Taken together, group S isolates are commonly found on strawberries, while they are almost absent on grapevine berries.
Fig 7.
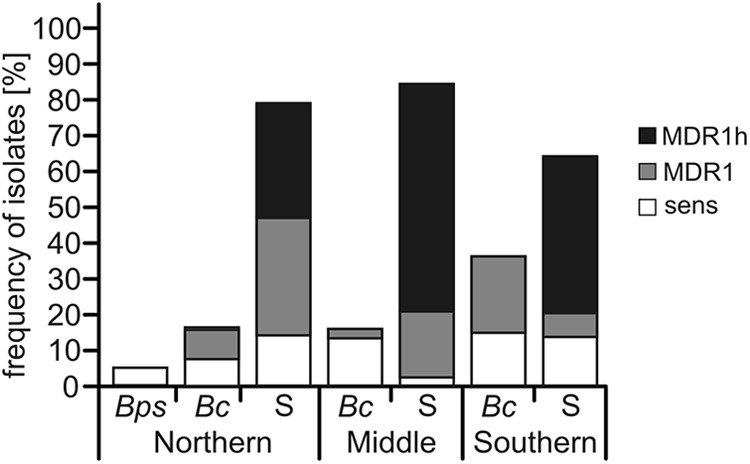
Distribution of different gray mold populations in strawberry fields in northern (n = 60), central (n = 38), and southern Germany (n = 75). Bps, B. pseudocinerea; Bc, B. cinerea; S, Botrytis group S.
We also analyzed the distribution of the two mating types among the strawberry gray mold isolates, using a primer pair specific to B. cinerea, MAT1-1 and MAT1-2 (27). In all growing regions, both mating types were commonly observed in both B. cinerea and group S populations, indicating that sexual reproduction within each of the two groups occurred regularly (see Table S1 in the supplemental material).
DISCUSSION
In view of the high frequency of anti-Botrytis treatments in commercial strawberry fields and the high-risk status of this pathogen, the occurrence of fungicide resistance in these fields is not surprising (25). The strawberry fields from which isolates were recovered had received different spraying programs. Most of the treatments, however, included one application of boscalid and pyraclostrobin (Signum) and fenhexamid (Teldor) and two or three treatments with cyprodinil and fludioxonil (Switch). Accordingly, resistance frequencies were high for most fungicides, with two exceptions (Fig. 1; see Table S1 in the supplemental material). Similar to other reports, we did not recover isolates with high levels of resistance to fludioxonil, which is probably due to the low fitness of such isolates (4, 10). Resistance to boscalid was observed only at low to moderate frequencies in our isolates collected in 2010 and 2011, probably because of the short history of boscalid application in German strawberry fields since 2007. Boscalid resistance can therefore be expected to increase during the next few years in soft-fruit production, as has been observed for gray mold populations from French and German vineyards and in Greek fruit-growing areas during the first few years after the introduction of this fungicide (11, 12, 14, 20).
In addition to specific resistances, a large proportion of the gray mold isolates showed MDR-like phenotypes. MDR1, which has been observed in both vineyards and strawberry fields, provides partial resistance to fludioxonil and cyprodinil and therefore is likely to be selected by the widely used fungicide Switch, which contains cyprodinil and fludioxonil. In the German Wine Road region, MDR1 phenotypes have previously been shown to result from different point mutations in mrr1, resulting in constitutive activation of the Mrr1 transcription factor (19). We did not observe great differences in the levels of sensitivity between MDR1 isolates carrying different mutations, and these isolates behaved similarly to many strawberry isolates classified as MDR1 (Fig. 2) (19). The discovery of the MDR1h phenotype, which has levels of resistance to fludioxonil and cyprodinil more than 2-fold higher than those of MDR1, was therefore surprising. In contrast, the MDR2 and MDR3 phenotypes, which are widely distributed in French and German vineyards, have not been found so far in any of the strawberry isolates. One reason for this might be the unusual promoter rearrangement mutations in mfsM2 that have been shown to be responsible for the MDR2 and MDR3 phenotypes (19). For one of the MDR2-related mutations, evidence was obtained by population genetic studies that it had probably occurred in a single progenitor cell in a French vineyard, and its MDR2 progeny then spread over French and German wine-growing regions (24). The reciprocal absence of MDR1h in vineyards and of MDR2/MDR3 in strawberry fields suggests a restricted distribution of gray mold populations, which might be explained by differential host adaptation and/or reduced genetic exchange (see below).
All MDR1h isolates contained an identical 3-bp deletion in the mrr1 coding region (Fig. 4; see Fig. S4 in the supplemental material). Using a PCR-based assay that allowed specific detection of the deletion, we observed a perfect correlation of the MDR1h phenotype and the presence of the deletion in all 73 MDR1h isolates analyzed. These data suggest a causal relationship between the L497 deletion (ΔL497) in Mrr1 and the MDR1h phenotype. To obtain a final proof, transfer of the mrr1 allele with ΔL497 (mrr1ΔL497) into a sensitive strain should be shown to confer the MDR1h phenotype, similar to the mrr1 allele with the V-to-M change at position 575 (mrr1V575M), which converted a sensitive strain to an MDR1 strain (19).
Isolates with the MDR1h phenotype were found to be widely distributed in German strawberry fields, representing between 31.4% and 60.5% of the population (Fig. 7). Besides providing partial resistance to anilinopyrimidines, a major benefit of the MDR1h phenotype for the fungus is likely to be partial resistance to fludioxonil, which compensates for the inability of Botrytis to develop effective target site resistance mutations against this fungicide. Interestingly, MDR1h isolates showed a significantly higher accumulation of specific fungicide resistances than either MDR1 or non-MDR isolates (Fig. 5). This indicates that MDR1h isolates might represent a distinct subpopulation of gray mold isolates in strawberry fields. We are currently investigating whether there is evidence for a clonal origin of the mrr1(ΔL497) allele, followed by spread over different strawberry-growing regions, similar to the case for the MDR2-related mfsM2 rearrangement mutation described above. The data presented in this work and in reference 25 demonstrate that the gray mold fungus can accumulate multiple resistances, including partial MDR-type resistance, to all classes of fungicides that are currently available against Botrytis in Europe. This could increasingly compromise chemical control of this key pathogen of soft fruits.
Among 173 strawberry isolates tested, we identified 3 B. pseudocinerea isolates in northern Germany. In contrast to most other strawberry gray mold isolates, none of these isolates showed any resistance to the tested fungicides. This observation indicates that, similar to the situation in vineyards (28, 40), B. pseudocinerea does not occur as a major pathogen on strawberries. Early studies suggested that population diversity in B. cinerea was correlated with the presence or absence of two transposable elements named Flipper and Boty (41, 43). Later studies based on microsatellite and IGS-RFLP markers, however, demonstrated that transposons alone were not useful for distinguishing between populations (5, 30, 44). So far, attempts at employing population genetics for the differentiation of subpopulations of B. cinerea according to their host preferences have yielded inconsistent results. Evidence in favor of such a differentiation was reported in some studies (44, 45), while no such support was obtained in others (5, 46, 47). In the present investigation, we have obtained evidence for the existence of a major, previously unknown gray mold subpopulation in German strawberry fields, provisionally called Botrytis group S, which is genetically distinct from B. cinerea sensu stricto. This conclusion is based on several lines of evidence: (i) the IGS-RFLP patterns of the strawberry isolates, in particular, those belonging to Botrytis group S, were clearly different from those of vineyard isolates (see Table S3 in the supplemental material). (ii) Significant differences in the sequences of various genes were detected between several Botrytis group S and B. cinerea isolates, whereas these sequences were relatively homogeneous within each group even between isolates from diverse geographic origins (Fig. 6). (iii) Botrytis group S isolates were common and usually dominant in all German strawberry-growing regions (Fig. 7) but rarely found in vineyards, indicating that their host preference differs from that of B. cinerea. On the basis of sequence comparisons, group S isolates were found to be closely related to B. cinerea isolates pathogenic to diverse hosts and to B. fabae isolates pathogenic only to the Fabaceae (48). The cladograms obtained with five genes differed and showed considerable differences (Fig. 6). The combined tree resembled the mrr1 tree due to the larger size of the sequenced fragments and the higher diversity of mrr1 than the other genes. The exact taxonomic position of Botrytis group S remains to be resolved by more comprehensive sequence comparisons. The similarity of the genetic profiles of group S isolates from widely different geographical origins throughout Europe indicates that they might be sexually isolated from other clades. We are currently investigating the possibility of genetic exchange between B. cinerea and group S isolates by population genetic studies and laboratory crosses.
Taken together, our study has revealed a high frequency of fungicide resistance and the existence of two distinct genotypes in German strawberry fields. Because of the restriction of the MDR1h phenotype to group S isolates and the higher accumulation of fungicide resistance mutations in MDR1h isolates, the overall fungicide resistance patterns were strikingly different between group S and B. cinerea isolates. This indicates that fungicide resistance development in field populations occurs independently in different gray mold genotypes and underlines the practical importance of an understanding of the genetic structure of pathogen field populations. PCR assays for the 21-bp indel and other genetic markers will be useful to rapidly identify the Botrytis populations and their fungicide resistance patterns in the field and help to predict the best choices of fungicides that remain effective for gray mold control.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to G. Karaoglanidis, A.-S. Walker, M. Hellmann, A.-P. Entrop, and U. Dederichs for providing gray mold isolates and to Z. Naoshin, S. Bergstein, and A. Fuchs for help with gray mold sampling, isolation, and analyses.
Footnotes
Published ahead of print 19 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02655-12.
REFERENCES
- 1. Williamson B, Tudzynski B, Tudzynski P, van Kan JAL. 2007. Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8:561–580 [DOI] [PubMed] [Google Scholar]
- 2. Staats M, van Baarlen P, van Kan JAL. 2005. Molecular phylogeny of the plant pathogenic genus Botrytis and the evolution of host specificity. Mol. Biol. Evol. 22:333–346 [DOI] [PubMed] [Google Scholar]
- 3. Leroux P, Fritz R, Debieu D, Albertini C, Lanen C, Bach J, Gredt M, Chapeland F. 2002. Mechanisms of resistance to fungicides in field strains of Botrytis cinerea. Pest Manag. Sci. 58:876–888 [DOI] [PubMed] [Google Scholar]
- 4. Brent KJ, Hollomon DW. 2007. Fungicide resistance: the assessment of risk. Fungicide Resistance Action Committee, Brussels, Belgium: http://www.frac.info/frac/publication/anhang/FRAC_Mono2_2007.pdf [Google Scholar]
- 5. Ma ZH, Michailides TJ. 2005. Advances in understanding molecular mechanisms of fungicide resistance and molecular detection of resistant genotypes in phytopathogenic fungi. Crop Prot. 24:853–863 [Google Scholar]
- 6. Leroux P, Clerjeau M. 1985. Resistance of Botrytis cinerea Pers and Plasmopara viticola (Berk and Curt.) Berl and de Toni to fungicides in French vineyards. Crop Prot. 4:137–160 [Google Scholar]
- 7. Park SY, Jung OJ, Chung YR, Lee CW. 1997. Isolation and characterization of a benomyl-resistant form of beta-tubulin-encoding gene from the phytopathogenic fungus Botryotinia fuckeliana. Mol. Cells 7:104–109 [PubMed] [Google Scholar]
- 8. Fillinger S, Leroux P, Auclair C, Barreau C, Al Hajj C, Debieu D. 2008. Genetic analysis of fenhexamid-resistant field isolates of the phytopathogenic fungus Botrytis cinerea. Antimicrob. Agents Chemother. 52:3933–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weber RWS. 2010. Occurrence of Hyd R3 fenhexamid resistance among Botrytis isolates in northern German soft fruit production. J. Plant Dis. Prot. 117:177–179 [Google Scholar]
- 10. Avenot HF, Sellam A, Karaoglanidis G, Michailides TJ. 2008. Characterization of mutations in the iron-sulphur subunit of succinate dehydrogenase correlating with boscalid resistance in Alternaria alternata from California pistachio. Phytopathology 98:736–742 [DOI] [PubMed] [Google Scholar]
- 11. Leroux P, Gredt M, Leroch M, Walker AS. 2010. Exploring mechanisms of resistance of respiratory inhibitors in field strains of Botrytis cinerea, the causal agent of gray mold. Appl. Environ. Microbiol. 76:6615–6630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Veloukas T, Leroch M, Hahn M, Karaoglanidis GS. 2011. Detection and molecular characterization of boscalid-resistant Botrytis cinerea isolates from strawberry. Plant Dis. 95:1302–1307 [DOI] [PubMed] [Google Scholar]
- 13. Leroux P, Chapeland F, Desbrosses D, Gredt M. 1999. Patterns of cross-resistance to fungicides in Botryotinia fuckeliana (Botrytis cinerea) isolates from French vineyards. Crop Prot. 18:687–697 [Google Scholar]
- 14. Bardas GA, Veloukas T, Koutita O, Karaoglanidis GS. 2010. Multiple resistance of Botrytis cinerea from kiwifruit to SDHIs, QoIs and fungicides of other chemical groups. Pest Manag. Sci. 66:967–973 [DOI] [PubMed] [Google Scholar]
- 15. Samuel S, Papayiannis LC, Leroch M, Veloukas T, Hahn M, Karaoglanidis GS. 2011. Evaluation of the incidence of the G143A mutation and cytb intron presence in the cytochrome bc-1 gene conferring QoI resistance in Botrytis cinerea populations from several hosts. Pest Manag. Sci. 67:1029–1036 [DOI] [PubMed] [Google Scholar]
- 16. Cui W, Beever RE, Parkes SL, Templeton MD. 2004. Evolution of an osmosensing histidine kinase in field strains of Botryotinia fuckeliana (Botrytis cinerea) in response to dicarboximide fungicide usage. J. Phytopathol. 94:1129–1135 [DOI] [PubMed] [Google Scholar]
- 17. Ma ZH, Yan LY, Luo Y, Michailides TJ. 2007. Sequence variation in the two-component histidine kinase gene of Botrytis cinerea associated with resistance to dicarboximide fungicides. Pestic. Biochem. Physiol. 88:300–306 [Google Scholar]
- 18. Staub T. 1991. Fungicide resistance—practical experience with antiresistance strategies and the role of integrated use. Annu. Rev. Phytopathol. 29:421–442 [DOI] [PubMed] [Google Scholar]
- 19. Kretschmer M, Leroch M, Mosbach A, Walker AS, Fillinger S, Mernke D, Schoonbeek HJ, Pradier JM, Leroux P, de Waard MA, Hahn M. 2009. Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. PLoS Pathog. 5:e1000696 doi:10.1371/journal.ppat.1000696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leroch M, Kretschmer M, Hahn M. 2011. Fungicide resistance phenotypes of Botrytis cinerea isolates from commercial vineyards in South West Germany. J. Phytopathol. 159:63–65 [Google Scholar]
- 21. Prasad R, Kapoor K. 2005. Multidrug resistance in yeast Candida. Int. Rev. Cytol. 242:215–248 [DOI] [PubMed] [Google Scholar]
- 22. Rouveix B. 2007. Clinical implications of multiple drug resistance efflux pumps of pathogenic bacteria. J. Antimicrob. Chemother. 59:1208–1209 [DOI] [PubMed] [Google Scholar]
- 23. Chapeland F, Fritz R, Lanen C, Gredt M, Leroux P. 1999. Inheritance and mechanisms of resistance to anilinopyrimidine fungicides in Botrytis cinerea (Botryotinia fuckeliana). Pestic. Biochem. Physiol. 64:85–100 [Google Scholar]
- 24. Mernke D, Dahm S, Walker AS, Lalève A, Fillinger S, Leroch M, Hahn M. 2011. Two promoter rearrangements in a drug efflux transporter gene are responsible for the appearance and spread of multidrug resistance phenotype MDR2 in Botrytis cinerea isolates in French and German vineyards. J. Phytopathol. 101:1176–1183 [DOI] [PubMed] [Google Scholar]
- 25. Weber RWS. 2011. Resistance of Botrytis cinerea to multiple fungicides in northern German small-fruit production. Plant Dis. 95:1263–1269 [DOI] [PubMed] [Google Scholar]
- 26. Weber RWS, Hahn M. 2011. A rapid and simple method for determining fungicide resistance in Botrytis. J. Plant Dis. Prot. 118:17–25 [Google Scholar]
- 27. Amselem J, Cuomo CA, van Kan JAL, Viaud M, Benito EP, Couloux A, Coutinho PM, de Vries RP, Dyer PS, Fillinger S, Fournier E, Gout L, Hahn M, Plummer KM, Pradier JM, Quévillon E, Sharon A, Simon A, ten Have A, Tudzynski B, Tudzynski P, Andrew M, Anthouard V, Beever RE, Beffa R, Benoit I, Bouzid O, Brault B, Chen Z, Choquer M, Collémare J, Cotton P, Danchin EG, Da Silva C, Gautier A, Giraud C, Giraud T, Gonzalez C, Grossetete S, Güldener U, Henrissat B, Howlett BJ, Kodira C, Kretschmer M, Lappartient A, Leroch M, Levis C, Mauceli E, Neuvéglise C, Oeser B, Pearson M, Poulain J, Pousserau N, Quesneville H, Rascle C, Schumacher J, Ségurens B, Sexton A, Silve E, Sirven C, Soanes DM, Talbot NJ, Templeton M, Yandava C, Yarden O, Zeng Q, Rollins JA, Lebrun MH, Dickman M. 2011. Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230 doi:10.1371/journal.pgen.1002230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Walker AS, Gautier AL, Confais J, Martinho D, Viaud M, Le Pêcheur P, Dupont J, Fournier E. 2011. Botrytis pseudocinerea, a new cryptic species causing grey mould in French vineyards in sympatry with Botrytis cinerea. Phytopathology 101:1433–1445 [DOI] [PubMed] [Google Scholar]
- 29. Möller EM, Bahnweg H, Sandermann H, Geiger HH. 1992. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 20:6115–6116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kretschmer M, Hahn M. 2008. Fungicide resistance and genetic diversity of Botrytis cinerea isolates from a vineyard in Germany. J. Plant Dis. Prot. 115:214–219 [Google Scholar]
- 31. Fournier E, Levis C, Fortini D, Leroux P, Giraud T, Brygoo Y. 2003. Characterisation of the Bc-hch, the Botrytis cinerea homolog of the Neurospora crassa het-c vegetative incompatibility locus, and its use as a population marker. Mycologia 95:252–261 [PubMed] [Google Scholar]
- 32. Marthey S, Aguileta G, Rodolphe F, Gendrault A, Giraud T, Fournier E, Lopez-Villavicencio M, Gautier A, Lebrun MH, Chiapello H. 2008. FUNYBASE: a fungal phylogenomic database. BMC Bioinformatics 9:456 doi:10.1186/1471-2105-9-456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Staats M, van Baarlen P, Schouten A, van Kan JAL, Bakker FT. 2007. Positive selection in phytotoxic protein-encoding genes of Botrytis species. Fungal Genet. Biol. 44:52–63 [DOI] [PubMed] [Google Scholar]
- 34. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Galtier N, Gouy M, Gautier C. 1996. SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput. Appl. Biosci. 12:543–548 [DOI] [PubMed] [Google Scholar]
- 36. Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690 [DOI] [PubMed] [Google Scholar]
- 37. Rodríguez F, Oliver JL, Marín A, Medina JR. 1990. The general stochastic model of nucleotide substitution. J. Theor. Biol. 142:485–501 [DOI] [PubMed] [Google Scholar]
- 38. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Mol. Evol. 39:783–791 [DOI] [PubMed] [Google Scholar]
- 39. Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57:758–771 [DOI] [PubMed] [Google Scholar]
- 40. Fournier E, Giraud T, Albertini C, Brygoo Y. 2005. Partition of the Botrytis cinerea complex in France using multiple gene genealogies. Mycologia 97:1251–1267 [DOI] [PubMed] [Google Scholar]
- 41. Giraud T, Fortini D, Levis C, Leroux P, Brygoo Y. 1997. RFLP markers show genetic recombination in Botryotinia fuckeliana (Botrytis cinerea) and transposable elements reveal two sympatric species. Mol. Biol. Evol. 14:1177–1185 [DOI] [PubMed] [Google Scholar]
- 42. Schouten A, van Baarlen P, van Kan JAL. 2008. Phytotoxic Nep1-like proteins from the necrotrophic fungus Botrytis cinerea associated with membranes and the nucleus of plant cells. New Phytol. 177:493–505 [DOI] [PubMed] [Google Scholar]
- 43. Giraud T, Fortini D, Levis C, Lamarque C, Leroux P, Lobuglio Y, Brygoo Y. 1999. Two sibling species of the Botrytis cinerea complex, transposa and vacuma, are found in sympatry on numerous host plants. J. Phytopathol. 89:967–973 [DOI] [PubMed] [Google Scholar]
- 44. Fournier E, Giraud T. 2008. Sympatric genetic differentiation of a generalist pathogenic fungus Botrytis cinerea, on two different host plants, grapevine and bramble. J. Evol. Biol. 21:122–132 [DOI] [PubMed] [Google Scholar]
- 45. Muñoz G, Hinrichsen P, Brygoo Y, Giraud T. 2002. Genetic characterisation of Botrytis cinerea populations in Chile. Mycol. Res. 106:594–601 [Google Scholar]
- 46. Cettul E, Rekab D, Locci R, Firrao G. 2008. Evolutionary analysis of endopolygalacturonase-encoding genes of Botrytis cinerea. Mol. Plant Pathol. 9:675–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moyano C, Alfonso C, Gallego J, Raposo R, Melgarejo P. 2003. Comparison of RAPD and AFLP marker analysis as a means to study the genetic structure of Botrytis cinerea populations. Eur. J. Plant Pathol. 109:515–522 [Google Scholar]
- 48. Jarvis WR. 1977. Botryotinia and Botrytis species; taxonomy, physiology and pathogenicity. Monograph no. 15 Canadian Department of Agriculture, Ottawa, ON, Canada [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.