Abstract
Listeria monocytogenes, the causative agent of human listeriosis, is known for its ability to withstand severe environmental stresses. The glutamate decarboxylase (GAD) system is one of the principal systems utilized by the bacterium to cope with acid stress, a reaction that produces γ-aminobutyrate (GABA) from glutamate. Recently, we have shown that GABA can accumulate intracellularly under acidic conditions, even under conditions where no extracellular glutamate-GABA exchange is detectable. The GABA shunt, a pathway that metabolizes GABA to succinate, has been described for several other bacterial genera, and the present study sought to determine whether L. monocytogenes has this metabolic capacity, which, if present, could provide a possible route for succinate biosynthesis in L. monocytogenes. Using crude protein extracts from L. monocytogenes EGD-e, we show that this strain exhibits activity for the two main enzyme reactions in the GABA shunt, GABA aminotransferase (GABA-AT) and succinic semialdehyde dehydrogenase (SSDH). Two genes were identified as candidates for encoding these enzyme activities, argD (GABA-AT) and lmo0913 (SSDH). Crude protein extracts prepared from a mutant lacking a functional argD gene significantly reduced GABA-AT activity, while an lmo0913 mutant lost all detectable SSDH activity. The deletion of lmo0913 increased the acid tolerance of EGD-e and showed an increased accumulation of intracellular GABA, suggesting that this pathway plays a significant role in the survival of this pathogen under acidic conditions. This is the first report of such a pathway in the genus Listeria, which highlights an important link between metabolism and acid tolerance and also presents a possible compensatory pathway to partially overcome the incomplete tricarboxylic acid cycle of Listeria.
INTRODUCTION
Listeria monocytogenes is a food-borne pathogen that causes listeriosis, a disease with a mortality rate of up to 30% (1), which has recently shown an increase in the number of reported cases across Europe (2). Almost 99% of cases are associated with a contaminated food source (3), and as such, L. monocytogenes is of major concern to the food industry. This bacterium is quite resilient to environmental stresses, including an ability to grow under acidic conditions with pH values as low as 4.5 and to survive at pH values as low as 2.0 for extended periods (4–6). This characteristic is important for its pathogenicity because it allows the bacterium to survive the acidic environment in various foods and in the stomach of the potential host.
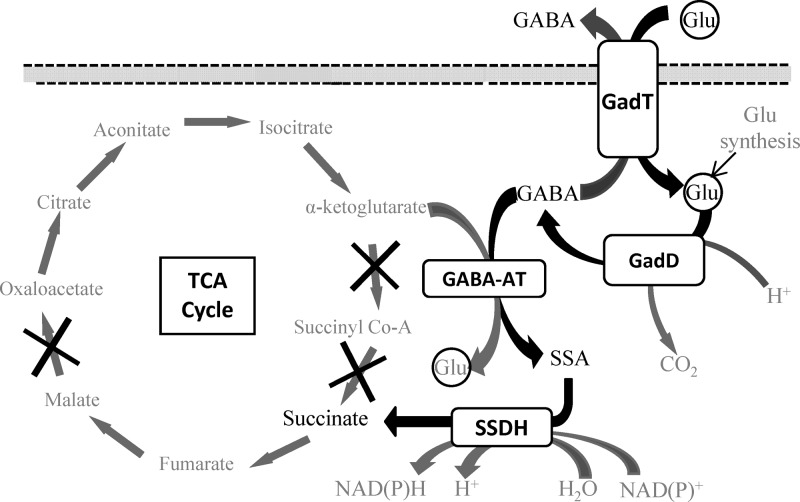
To counteract this threat, strains of Listeria can employ a variety of acid resistance mechanisms, including the arginine deiminase system (7), the adaptive acid tolerance response (ATR) (8), and the glutamate decarboxylase (GAD) system (9). It is clear, however, that there is a degree of strain-to-strain variation in terms of the ability to cope with an acid challenge (9–11). The GAD system operates to maintain the internal pH homeostasis of the bacteria. Upon acid exposure, an extracellular molecule of glutamate (Glu) is taken up by an antiporter (GadT) and decarboxylated intracellularly by a Glu decarboxylase enzyme (GadD) to form γ-aminobutyric acid (GABA). This reaction consumes a proton, thereby increasing the intracellular pH. GABA is then exported via GadT in exchange for a further molecule of Glu (Fig. 1). L. monocytogenes can possess up to three genes encoding Glu decarboxylases (gadD1, gadD2, and gadD3) and two genes encoding antiporters (gadT1 and gadT2), arranged into three operons (gadD1T1, gadT2D2, and gadD3) (12); however, strains of serotype 4 lack gadD1T1. Recent work from our laboratory has demonstrated the existence of the intracellular GAD system (GADi), which converts intracellular Glu to GABA and contributes significantly to acid resistance (11, 13). The activity of this system results in the accumulation of high levels of GABAi under acidic conditions even in the absence of GABA export.
Fig 1.
Glutamate decarboxylase system and GABA shunt pathway of L. monocytogenes. Shown is the proposed model for the metabolism of GABAi in L. monocytogenes. Glui is decarboxylated to GABAi by GadD. The GABA can either be exported by GadT in exchange for another Glu or enter the GABA shunt pathway. Here GABAi donates its amino group to α-ketoglutarate via a transaminase enzyme (GABA-AT), resulting in the formation of SSA and Glui. The SSA is then oxidized to succinate by a dehydrogenase (SSDH). The incomplete TCA cycle of L. monocytogenes is shown, with the missing steps marked with an “X.” The GABA shunt pathway can provide an alternative source of succinate for the bacteria.
In Escherichia coli, GABA is used as both carbon and nitrogen sources and is metabolized via the GABA shunt pathway (Fig. 1) (14, 15). This pathway incorporates two further enzymes downstream from Glu decarboxylase. A GABA/α-ketoglutarate aminotransferase (GABA-AT) (GabT) removes the amino group from GABA to form succinic semialdehyde (SSA) and Glu. The SSA is then oxidized through the activity of succinic semialdehyde dehydrogenase (SSDH) (GabD) to form succinate (16). While the GABA shunt pathway has not been extensively studied in bacteria, it is thought to play a role in Glu metabolism, anaplerosis, and antioxidant defense (17, 18). The use of arginine, ornithine, and agmatine as nitrogen sources by E. coli relies on the GABA shunt pathway, as these are converted first to putrescine and subsequently to GABA (19). In plants and mammals, the pathway has been described as an alternative route to produce succinate, bypassing two enzymes of the tricarboxylic acid (TCA) cycle, namely, α-ketoglutarate dehydrogenase and succinyl coenzyme A (CoA) synthetase (Fig. 1) (16, 20). For L. monocytogenes, the GABA shunt may represent a potential route to compensate for the incomplete TCA cycle identified through sequencing and biochemical analysis (21, 22). Genome sequencing has shown an absence of genes encoding α-ketoglutarate dehydrogenase, succinyl-CoA synthetase, and malate dehydrogenase (Fig. 1). Work reported previously by Eisenreich et al. (23) supported this with biochemical data showing that oxaloacetate is not derived from α-ketoglutarate by the TCA cycle. Therefore, the importance of this pathway cannot be overlooked when trying to establish a complete metabolic profile of this bacterium.
The aim of this study was to determine if L. monocytogenes possesses the two metabolic steps comprising the GABA shunt pathway. Second, strains with disruptions or deletions in genes predicted to be involved in this pathway (argD and lmo0913) were analyzed for effects on the respective activities in order to confirm their roles. Finally, due to the fact that the GAD system and the GABA shunt are linked, the main metabolites of the pathway were quantified in the cell in response to acid treatment.
MATERIALS AND METHODS
Bacterial strains and growth.
L. monocytogenes wild-type strain EGD-e and its isogenic lmo0913 (in-frame deletion) (Δlmo0913) and argD (insertional disruption with pLSV101) (argD::pLSV101) mutants (24) were used in this study. L. monocytogenes wild-type strain 10403S along with its isogenic 10403S Δlmrg_02013 mutant were also used. Strains were grown on brain heart infusion (BHI) agar plates for 24 to 48 h at 37°C. Cultures grown overnight were set up by inoculating a single colony into 25 ml of BHI broth in 250-ml conical flasks with continuous shaking at 37°C. E. coli strains BW25113 and JW2636 were used as reference strains for the GABA shunt. These strains were grown in Luria-Bertani (LB) medium for 18 h at 37°C when needed and stored on LB agar plates. All strains and relevant properties are listed in Table 1.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant property(ies) | Source or reference |
|---|---|---|
| Strains | ||
| L. monocytogenes | ||
| EGD-e | Serovar 1/2a, wild type | S. Foster |
| EGD-e argD::pLSV101 mutant | pLSV101 plasmid insertion into the argD gene | K. Schauer |
| EGD-e Δlmo0913 mutant | EGD-e with an in-frame 1,407-bp deletion of the lmo0913 gene | This study |
| EGD-e Δlmo0913 pKAK0913 mutant | Deletion of lmo0913 complemented with a cloned copy of the full lmrg_02013 gene | This study |
| 10403S | Serovar 1/2a, wild type | K. Boor |
| 10403S Δlmrg_02013 mutant | 10403S with an in-frame 1,407-bp deletion of the lmrg_02013 gene | 26 |
| 10403S Δlmrg_02013 pKAK0913 mutant | Deletion of lmrg_02013 complemented with a cloned copy of the full lmrg_02013 gene | 26 |
| E. coli | ||
| K-12 BW25113 | Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) lambda− rph-1 Δ(rhaD-rhaB)568 hsdR514 | NBRP (Japan), E. coli |
| JW2636 | BW25113 ΔgabD | NBRP (Japan), E. coli |
| JW2636::pKAK0913 | JW2636 possessing a full copy of lmrg_02013 from 10403S cloned into pKSV7 | This study |
| Plasmid | ||
| pKAK0913 | pKSV7 shuttle vector carrying a full copy of lmrg_02013 used to complement deletion of lmo0913 or gabD | 26 |
Generation of mutant and recombinant strains.
Mutants were generated by the splicing by overlap extension (SOEing) PCR method and allelic replacement, as previously described (25). The deletion of lmrg_02013 in 10403S was reported previously (26); however, the deletion of lmo0913 in EGD-e was newly generated by using the primers listed in Table 2. Plasmid pKAK0913, as described previously by Abram et al. (26), was used to complement the deletions of lmo0913 in EGD-e, lmrg_02013 in 10403S, and gabD in JW2636. This plasmid carries a full copy of lmrg_02013 from 10403S. Positive transformants were selected through growth on BHI agar supplemented with chloramphenicol (10 μg ml−1) or LB agar supplemented with ampicillin (100 μg ml−1) and confirmed by PCR. PCR amplification of DNA for use in cloning and downstream work was carried out by using the high-fidelity Velocity DNA polymerase (Bioline), while screening was carried out by using Biotaq DNA polymerase (Bioline).
Table 2.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| lmo0913 A | CGGAATTCGGAGCAGTTTTTGTTAGCC |
| lmo0913 B | TGCTGTTTCTTTAATACTCAAAAATACCACTCC |
| lmo0913 C | AGTATTAAAGAAACAGCAATCCAAGTGAAATTC |
| lmo0913 D | CGGAATTCAAGTGTGCCATAAGTTGC |
| lmo0913 For | GGTTACTATTCATGGGCAG |
| lmo0913 Rev | CATCCTGTTATCCCTCC |
| lmo0913 Comp-F | AGGGATCCGGAGCAGTTTTTGTTAGCC |
| lmo0913 Comp-R | CGGGATCCAAGTGTGCCATAAGTTGC |
GABA-AT and SSDH assays.
Crude protein assays to detect the presence of GABA-AT and SSDH activities were carried out based on an assay described previously by Bartsch et al. (27) but modified in order to run it in a 96-well-plate format.
Protein extraction.
To prepare crude protein extracts, a culture of bacteria was grown overnight in 200 ml of BHI (L. monocytogenes) or LB (E. coli) medium at 37°C with continuous shaking. The pellet was retained after centrifugation at 12,000 × g for 10 min, followed by two wash steps in wash buffer (10 mM NaCl, 10 mM Na3O4P [pH 7.0]). Pellets were then resuspended in 4 ml of a sonication buffer optimized for their downstream application. For GABA-AT, the sonication buffer comprised 20 mM Na3O4P, 0.01 mM pyridoxal phosphate, 5 mM mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1 mM dithiothreitol (DTT) (pH 7.0). For SSDH, it comprised 100 mM sodium phosphate, 1 mM DTT, 1 mM PMSF, and 9% glycerol (pH 9.5). Samples were then sonicated on ice by six 30-s pulses at a 16-μm amplitude, allowing 30 s of rest between each pulse to lyse the cells, followed by centrifugation at 6,000 × g for 10 min and retention of the supernatant. The supernatant was then centrifuged a final time at 15,000 × g for 15 min to remove any remaining cell debris. Protein concentrations for both assays were normalized to 7 mg ml−1 after determining each sample concentration by using the Bio-Rad DC protein assay (Bio-Rad).
GABA-AT assay.
One hundred microliters of protein was added to 1,000 μl reaction mix (85 mM Tris base, 15 mM Tris-HCl, 0 to 100 mM GABA, 5 mM α-ketoglutarate, 0.02% bovine serum albumin [BSA]) and incubated at 37°C for 1 h. In a 96-well plate, the SSA produced by the reaction was measured by the use of a GABase assay described previously by O'Byrne et al. (28), using 80 mM 2-aminoethyl hydrogen sulfate for the detection of SSA. Plates were incubated at 37°C in a Sunrise absorbance reader (Tecan, Salzburg, Austria), with optical density at 340 nm (OD340) readings being recorded every 60 s by Magellan software (Tecan, Salzburg, Austria). Any background signal detected with 0 mM GABA was subtracted to highlight the increase in the level of SSA production.
SSDH assay.
NADP to a final concentration of 1 mM was added to 2 ml of a normalized protein preparation (7 mg ml−1), and 150 μl of this mix was added to 100 μl of SSA (0 to 1.0 mM) in a 96-well plate. Plates were incubated at 37°C in a Tecan Sunrise absorbance reader, with OD340 readings being recorded every 60 s by Magellan software.
GABase assays.
GABAi, extracellular GABA (GABAe), as well as intracellular SSA concentrations were measured as previously described (13, 28). Cultures were grown overnight at 37°C with aeration in BHI medium. Prior to the GABA measurements, the pH of the culture was lowered to 4.0 with HCl. Extractions were made after 1 h of pH treatment. Non-HCl-treated cultures were used as negative controls.
Acid survival assays.
Cultures of bacteria were grown overnight at 37°C in BHI medium. The pH of these cultures was lowered to pH 2.5 by using 2.8 M HCl. Samples were taken every 20 min for 1 h and serially diluted in phosphate-buffered saline (PBS). Dilutions were plated in triplicate onto BHI agar and incubated overnight at 37°C. Colonies were counted to determine the number of surviving cells.
Measurement of Glu and succinate concentrations.
Glu and succinate concentrations were measured in the same samples tested for GABA and SSA. Both methods were performed by the use of enzymatic kits supplied by Roche Bio-Pharm. The methods were adapted to fit a 96-well-plate format. All reagents were scaled down from use in a 3-ml (final volume) reaction mixture to use in a 300-μl (final volume) reaction mixture. Standards of either monosodium glutamate (Sigma) or sodium succinate (BDH) were used to establish a standard curve for the enzymatic reaction. One hundred microliters of sample or standards (0 to 1.0 mM) was added to 200 μl of kit reagents and incubated at 25°C (Glu) or 37°C (succinate) in a 96-well plate in a Tecan Sunrise absorbance reader, with the OD500 (Glu) or OD340 (succinate) being recorded every 60 s by Magellan software. Concentrations of the metabolites were determined based on the standard curve generated from known sample concentrations.
Statistical analysis of results.
Experiments were carried out with three biological replicates and at least two technical replicates for each sample. Significant differences between samples tested were determined by using a paired Student t test. Results were considered significant when they possessed a P value of <0.05. Error bars indicating standard deviations from the means are displayed on graphs.
RESULTS
L. monocytogenes possesses activity for both enzymes of the GABA shunt.
The sequences corresponding to GabT and GabD from E. coli were used to search the L. monocytogenes genome for homologues that might encode GABA aminotransferase (GABA-AT) and succinic semialdehyde dehydrogenase (SSDH) activities, respectively. BLAST analysis showed that ArgD from L. monocytogenes EGD-e shared 52% similarity and 34% identity to GabT. In L. monocytogenes EGD-e, the predicted SSDH is Lmo0913, with 67% similarity and 49% identity to GabD. The corresponding gene product in L. monocytogenes 10403S is LMRG_02013, with 68% similarity and 49% identity. These two proteins, ArgD and Lmo0913/LMRG_02013, appeared frequently throughout the published sequences of L. monocytogenes. Twenty-four out of 25 strains searched possessed a homologue of Lmo0913/LMRG_02013 with >98% identity, while 14 out of 25 possessed an ArgD homologue with >93% identity. Most notable is LO28, which does not appear to have similar proteins. Evidence suggested that lmo0913 produces a monocistronic transcript (29) with a termination site predicted immediately downstream. The transcription of lmo0913 appears to be σB dependent (26).
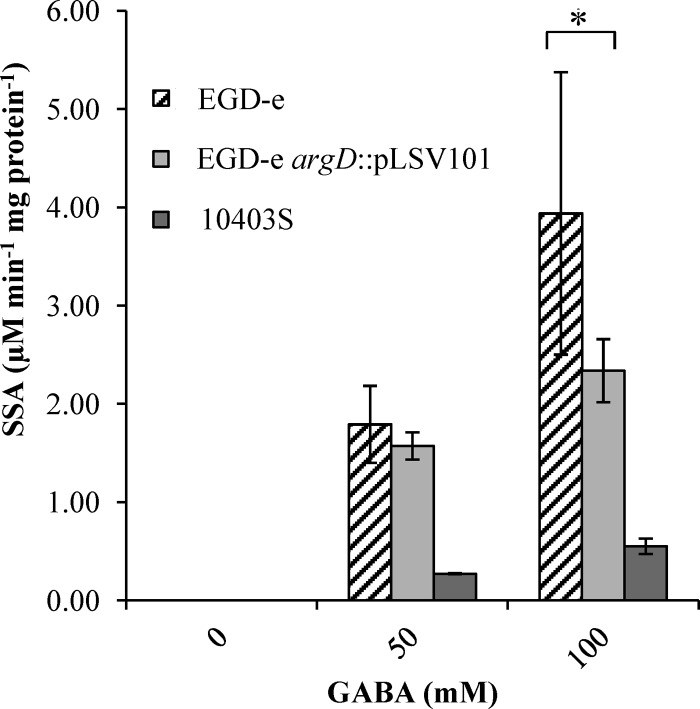
The first step in the GABA shunt is the conversion of GABA to SSA. Crude protein extracts were tested for their abilities to produce SSA from GABA, as described above. The coincubation of these protein preparations (7 mg ml−1) for 1 h with GABA (50 and 100 mM) resulted in the production of SSA (1.79 and 3.94 μM SSA min−1 mg protein−1, respectively) (Fig. 2). In wild-type strain EGD-e, the concentration of SSA produced increased in proportion to the concentration of GABA added. When a mutant with a disruption in a putative GABA-AT gene (argD) was used, the level of production of SSA was reduced (1.57 and 2.34 μM SSA min−1 mg protein−1, respectively; P value of <0.05), indicating an involvement of this gene in the metabolic step. The effect of the disruption of argD was more apparent at a higher concentration of 100 mM GABA, whereas at 50 mM GABA, the difference did not appear to be significant. The reduced SSA levels in the absence of argD suggest that an alternative GABA-AT activity is also present in the cell extracts. GABA-AT activity was also observed for the 10403S strain but at much lower levels than for EGD-e (0.55 μM SSA min−1 mg protein−1 with 100 mM GABA).
Fig 2.
GABA-AT activity of L. monocytogenes. Values for GABA-AT activities of crude cell extracts harvested after growth overnight (16 h) and incubated with either 0, 50, or 100 mM GABA are shown. Values are means of data from three individual culture replicates. Errors bars shown display the deviations of values seen between each replicate sample. The asterisk indicates a statistically significant difference (P < 0.05), as determined by a Student t test.
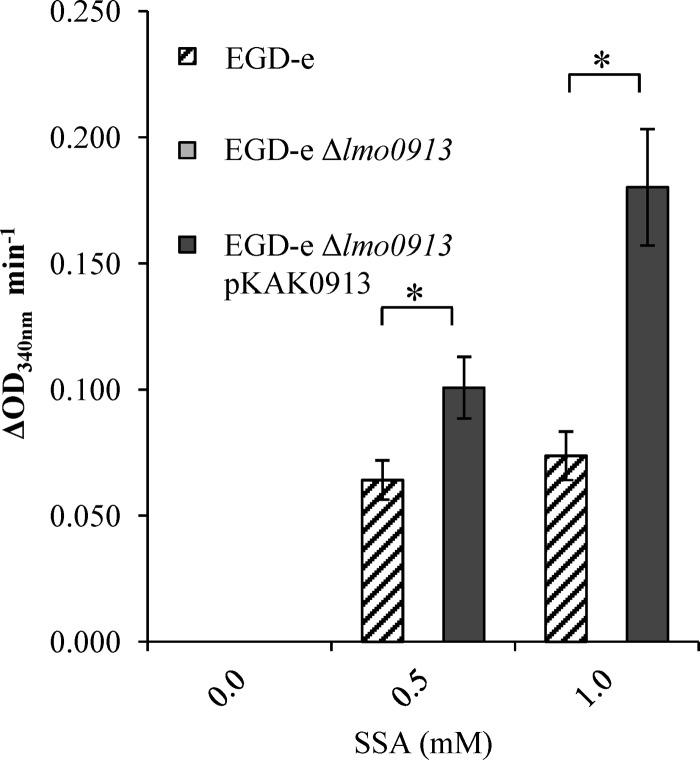
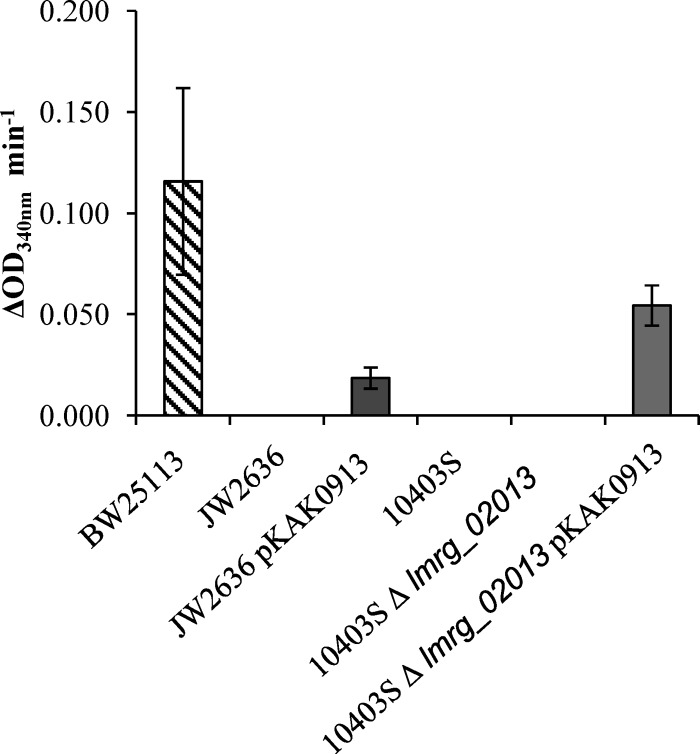
The oxidation of SSA to succinate is the second step in the GABA shunt. Crude protein extracts were tested for SSDH activity, the enzyme that carries out this step. The production of NADPH (the by-product of the SSDH reaction) was seen in the EGD-e protein extract that was incubated with SSA (Fig. 3). The production of NADPH did not occur in the absence of the substrate, and its level of production increased in proportion to the concentration of the substrate (SSA). The deletion of the putative SSDH-encoding gene, lmo0913, resulted in a complete loss of activity for this reaction. To determine if Lmo0913 acts directly as an SSDH, the corresponding gene from 10403S was cloned into pKSV7, generating plasmid pKAK0913, and this plasmid was used to transform two strains of L. monocytogenes carrying a deletion of lmo0913 or lmrg_02013: the EGD-e Δlmo0913 and 10403S Δlmrg_02013 mutants. An E. coli strain lacking a functional SSDH (ΔgabD) was also transformed with this construct. Crude protein extracts of these strains were analyzed for SSDH activity. E. coli BW25113 extracts were found to have SSDH activity when incubated with SSA (Fig. 4). The disruption of the SSDH of E. coli, gabD, resulted in a complete loss of this activity. The complementation of this mutant with lmrg_02013 from 10403S using pKAK0913 partially restored the SSDH activity.
Fig 3.
SSDH activity of L. monocytogenes. Crude cell extracts of EGD-e and the EGD-e Δlmo0913 and EGD-e Δlmo0913 pKAK0913 mutants were incubated with either 0, 0.5, or 1.0 mM SSA. NADPH production was measured as the OD340. Values are the means of data from three individual culture replicates. Errors bars shown display the deviations of values seen between each replicate sample. Asterisks indicate a statistically significant difference (P < 0.05), as determined by a Student t test.
Fig 4.
SSDH activity in E. coli and L. monocytogenes 10403S. Crude cell extracts of either E. coli K-12, JD23574, JD23574::pKAK0913, 10403S, or the 10403S Δlmrg_02013 or 10403S Δlmrg_02013 pKAK0913 mutant were harvested after 16 to 18 h of growth and incubated with 1.0 mM SSA. NADPH production was measured as the OD340. Values are the means of data from two individual culture replicates, each measured twice. Errors bars shown display the deviations of values seen between each replicate sample.
No SSDH activity was detected in extracts from either strain 10403S or the Δlmrg_02013 derivative of this strain; however, the complementation of the knockout mutant with lmrg_02013 resulted in the production of NADPH. As expected, the complementation of the Δlmo0913 mutation in EGD-e restored the SSDH activity in this strain. It is interesting to note that LMRG_02013 from 10403S has six amino acid differences from Lmo0913 in EGD-e (deletions of the V2, F3, and L4 amino acids, as well as the E251/248D, G312/309A, and D352/349E substitutions). These data showed that Lmo0913/LMRG_02013 is responsible for the SSDH activity in L. monocytogenes and further suggest that the baseline SSDH activity varies between strains (Fig. 3 and 4).
Acid exposure affects intracellular concentrations of GABA shunt intermediates.
The levels of Glu did not appear to significantly change in response to acid in both EGD-e and pLSV101::argD cells (Table 3); however, the deletion of lmo0913 resulted in a reduction in the levels of Glu from 0.53 mM to 0.45 mM (P value of 0.03). As expected, GABAi concentrations were affected in EGD-e in response to acid. Both EGD-e and the pLSV101::argD strain showed 2.6- and 7.5-fold increases in GABAi levels, reaching concentrations of 1.1 and 0.6 mM, respectively. However, a dramatic 9.2-fold increase in the GABAi concentration was observed for the Δlmo0913 mutant, reaching a concentration of ∼3.5 mM. This, along with a significant decrease in the Glui concentration, suggests a highly active GADi system in this strain. The increase seen in the SSA concentration before and after acid treatment for EGD-e was not significant, and the increase in the concentration seen for the argD::pLSV101 strain was close to the detectable range of the assay, but the increase for the Δlmo0913 mutant was higher, as would be expected for a gene involved in the breakdown of SSA. The succinate concentration increased to a small extent for both EGD-e and the argD::pLSV101 strain after acid treatment; however, the increase for the Δlmo0913 mutant was not significant (P value of 0.165).
Table 3.
Intracellular concentrations of intracellular metabolites in stationary-phase cells
| Strain | Mean concn, mM (SD) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Glutamate |
GABA |
SSA |
Succinate |
|||||
| pH 7.0 | pH 4.0 | pH 7.0 | pH 4.0 | pH 7.0 | pH 4.0 | pH 7.0 | pH 4.0 | |
| EGD-e | 0.55 (±0.06) | 0.53 (±0.05) | 0.43 (±0.05) | 1.14 (±0.68)a | 0.41 (±0.41) | 0.52 (±0.52) | 0.24 (±0.05) | 0.44 (±0.07)a |
| argD::pLSV101 | 0.59 (±0.02) | 0.48 (±0.06) | 0.51 (±0.02) | 0.60 (±0.01)a,b | 0.24 (±0.02) | 0.34 (±0.02)a | 0.17 (±0.01) | 0.34 (±0.04)a |
| Δlmo0913 | 0.53 (±0.01) | 0.45 (±0.02)a,b | 0.35 (±0.06) | 3.24 (±1.83)a,b | 0.53 (±0.53) | 0.61 (±0.61)a | 0.24 (±0.05) | 0.54 (±0.24) |
Statistically significant difference between acid-treated and non-acid-treated cells.
Statistically significant difference between wild-type and mutant strains under the conditions tested. A difference was identified to be statistically significant by using a paired Student t test (P < 0.05).
Lmo0913 affects acid resistance of L. monocytogenes EGD-e.
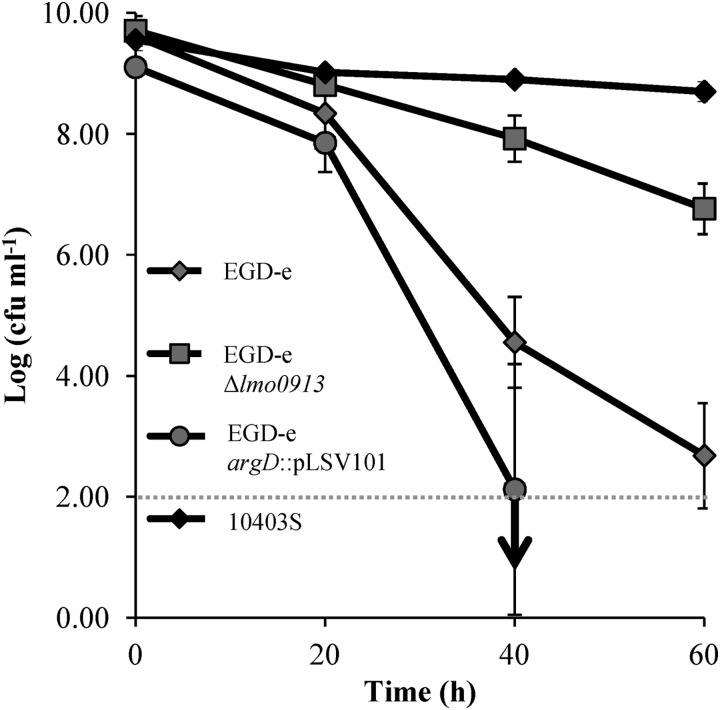
EGD-e showed a reduction in cell numbers by ∼8 log cycles, and the argD mutant strain demonstrated a similar reduction (Fig. 5). However, the Δlmo0913 mutant showed a dramatic increase (∼5 log cycles) in acid resistance compared to the acid resistance of the wild type. The increase in the acid resistance of the Δlmo0913 mutant reached levels just short of those achieved by 10403S, a highly-acid-resistant strain that utilizes both the GADi and GADe systems with great efficiency (13). Thus, a strong correlation existed between GABAi accumulation and survival at a low pH. Attempts to complement this resistant phenotype failed, however, as the transformed strain (EGD-e Δlmo0913 pKSV7::lmo0913) did not grow as well as the other strains and thus gave an incomparable acid resistance phenotype (data not shown). This growth defect was not caused by the presence of the lmo0913 gene on the plasmid, since strains containing pKSV7 also grew more slowly than the wild type.
Fig 5.
Acid survival of L. monocytogenes EGD-e and GABA putative shunt system mutants. Stationary-phase EGD-e and GABA shunt mutants were challenged at pH 2.5. Cell counts were taken every 20 min. Values are the means of data from three individual cultures, with the cell counts for each culture being the means of counts from three platings. The detection limit of the assay is highlighted by the dotted line, while the downward-pointing arrow indicates that subsequent counts dropped below this level.
DISCUSSION
We have previously shown that L. monocytogenes possesses a GADi system that leads to GABAi accumulation under acidic conditions (11). Here we report that this GABA can be metabolized by ArgD and Lmo0913 via the GABA shunt pathway. As the GABA shunt is coupled to the GAD system, which is the principal mechanism of acid tolerance in L. monocytogenes, we investigated the acid tolerance phenotypes of both the argD::pLSV101 and Δlmo0913 strains. Interestingly, these mutants yielded quite contrasting acid tolerance phenotypes. While the disruption of argD slightly hindered the cells' ability to cope with acid treatment, the deletion of lmo0913 increased survival by over 5 log cycles (Fig. 5). This significant increase highlights a role for the GABA shunt in acid tolerance. The acid-resistant phenotype of the Δlmo0913 mutant was accompanied by 9.2-fold and 1.1-fold increases in GABAi and SSA concentrations, respectively (Table 3). The increases in the concentrations of these metabolites are likely to be caused by a diminished capacity for GABA metabolism in this mutant (Fig. 3). Interestingly, EGD-e does not utilize the GADe system but only the GADi system. Recently, we demonstrated that GADi contributes to acid resistance independently of the antiport carried out by GADe (11). The fact that the deletion of the lmo0913 gene is able to convert EGD-e from one of the weakest strains in terms of acid resistance to a moderate one (2 logs lower than that of 10403S, which is the most acid-resistant strain that we have identified, and 5 logs higher than that of EGD-e) (Fig. 5) through an increase in the GABAi concentration underpins the importance of this system in acid resistance.
If the disruption of the GABA shunt by the deletion of lmo0913 is responsible for the GABAi buildup and, subsequently, acid resistance, a similar result might be expected for the argD mutant. This, however, was not the case, suggesting that in this mutant, the transamination activity observed is probably carried out by an enzyme that compensates for the loss of argD. GsaB has 46% similarity and 28% identity to GabT, a GABA-AT (30). It was shown previously that for several bacteria, multiple copies of GABA shunt genes exist (31, 32), and this may well be the case for L. monocytogenes. BLAST searches for a secondary SSDH also revealed a likely candidate in Lmo0383, which has 54% similarity and 33% identity to the corresponding SSDH (GabD) in E. coli. Further work to characterize the roles of both gsaB and lmo0383 are under way to test their roles in the GABA shunt pathway.
When the SSDH activities of EGD-e and 10403S were compared, it was interesting that no SSDH activity was detectable under the conditions used for 10403S. The difference between the two strains may suggest differing degrees of importance of the GABA shunt between strains (although both belong to lineage II and share the same serotype, serotype 1/2a). Strain-to-strain variation is also exemplified by the fact that previously, a deletion of lmrg_02013 in 10403S did not confer acid resistance but instead resulted in a mild sensitivity to acid (26). As mentioned above, the main difference between the two strains is that EGD-e utilizes only GADi, and therefore, unlike 10403S, which utilizes both GADi and GADe, it must rely solely on the GABA shunt to metabolize the GABA generated during acid stress (11). Thus, it appears that the elevated levels of SSDH activity in EGD-e might be related to the larger GABAi pools that exist in this strain during acid stress.
The existence of the GABA shunt pathway has significant implications for our understanding of the overall metabolism of L. monocytogenes. The genome sequence of L. monocytogenes lacks a complete set of enzymes for the classical TCA cycle model (21), and this is consistent with biochemical measurements (22). Two of the enzymes missing from L. monocytogenes are α-ketoglutarate dehydrogenase and succinyl-CoA synthetase, which are responsible for the conversion of α-ketoglutarate to succinate (Fig. 1). The GABA shunt pathway appears to serve different species of bacteria in a range of ways, from sporulation (33) to nitrogen metabolism (30). For L. monocytogenes, it is possible that the pathway can in part substitute for the lack of a functional TCA cycle. It is important to note, however, that the GABA shunt does not provide a means of completing the TCA cycle, since malate dehydrogenase, required to convert malate to oxaloacetate, is also absent from L. monocytogenes (21, 23). According to data reported previously by Trivett and Meyer (34), L. monocytogenes lacks a functional succinate dehydrogenase, required to convert succinate to fumarate. This presents yet another disruption in the classical TCA cycle and would indicate a dead end for the succinate produced by the GABA shunt pathway. However, their experiments did show activity for fumarate reductase. Evidence was presented previously which showed that fumarate reductase can compensate for a loss of succinate dehydrogenase (35, 36). This could putatively convert succinate to fumarate. From here, fumarate can enter the arginine biosynthetic pathway and thus provides a means of recycling the carbon for further biosynthesis. It is also important to note that while the pathway is a means of biosynthesizing succinate, the activity of the SSDH enzyme may be important for generating NAD(P)H that could be coupled to electron transport, as is the case for plants (37). This provides a means of energy production for the cell.
In summary, we have shown that L. monocytogenes EGD-e does have the capacity to metabolize GABAi via the steps of the GABA shunt. As we have shown SSDH activity for both Lmo0913 and LMRG_02013, we propose that these proteins be renamed GabD. The accumulation of GABAi appears to be important for the survival of the bacteria under conditions of acid stress. As seen for the lmo0913 mutant strain, a high-level accumulation of GABAi leads to an increase in the survival rate at a low pH. The regulation of the metabolism of GABA may therefore be an important aspect of this survival response. A greater understanding of this pathway may in the long term give new insights into the mechanisms of acid tolerance in this pathogen which can be used to devise improved food safety regimes. While additional studies are required to elucidate both secondary enzymes in the pathway and the regulatory mechanisms involved, we have shown that the GABA shunt pathway is functional in L. monocytogenes, providing this pathogen with a means to synthesize the TCA intermediate succinate.
ACKNOWLEDGMENTS
We are grateful to colleagues in the Bacterial Stress Response Group and in Microbiology at NUI Galway for helpful discussions and for critical comments on the manuscript. We thank K. Schauer for providing the argD::pLSV101 insertion knockout and the NBRP (Japan) for the E. coli strains used in this study.
The research was supported by a Science Foundation Ireland SIRG award to K. A. G. Karatzas (grant number 09/SIRG/B1570).
Footnotes
Published ahead of print 12 October 2012
REFERENCES
- 1. Farber JM, Peterkin PI. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Allerberger F, Wagner M. 2010. Listeriosis: a resurgent foodborne infection. Clin. Microbiol. Infect. 16:16–23 [DOI] [PubMed] [Google Scholar]
- 3. Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cole MB, Jones MV, Holyoak C. 1990. The effect of pH, salt concentration and temperature on the survival and growth of Listeria monocytogenes. J. Appl. Microbiol. 69:63–72 [DOI] [PubMed] [Google Scholar]
- 5. McClure PJ, Roberts TA, Oguru PO. 1989. Comparison of the effects of sodium chloride, pH and temperature on the growth of Listeria monocytogenes on gradient plates and in liquid medium. Lett. Appl. Microbiol. 9:95–99 [Google Scholar]
- 6. Young KM, Foegeding PM. 1993. Acetic, lactic and citric acids and pH inhibition of Listeria monocytogenes Scott A and the effect on intracellular pH. J. Appl. Bacteriol. 74:515–520 [PubMed] [Google Scholar]
- 7. Ryan S, Begley M, Gahan CGM, Hill C. 2009. Molecular characterization of the arginine deiminase system in Listeria monocytogenes: regulation and role in acid tolerance. Environ. Microbiol. 11:432–445 [DOI] [PubMed] [Google Scholar]
- 8. Davis MJ, Coote PJ, O'Byrne CP. 1996. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology 142:2975–2982 [DOI] [PubMed] [Google Scholar]
- 9. Cotter PD, Gahan CG, Hill C. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465–475 [DOI] [PubMed] [Google Scholar]
- 10. Adrião A, Vieira M, Fernandes I, Barbosa M, Sol M, Tenreiro RP, Chambel L, Barata B, Zilhao I, Shama G, Perni S, Jordan SJ, Andrew PW, Faleiro ML. 2008. Marked intra-strain variation in response of Listeria monocytogenes dairy isolates to acid or salt stress and the effect of acid or salt adaptation on adherence to abiotic surfaces. Int. J. Food Microbiol. 123:142–150 [DOI] [PubMed] [Google Scholar]
- 11. Karatzas K-AG, Suur L, O'Byrne CP. 2012. Characterization of the intracellular glutamate decarboxylase system: analysis of its function, transcription, and role in the acid resistance of various strains of Listeria monocytogenes. Appl. Environ. Microbiol. 78:3571–3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cotter PD, Ryan S, Gahan CGM, Hill C. 2005. Presence of GadD1 glutamate decarboxylase in selected Listeria monocytogenes strains is associated with an ability to grow at low pH. Appl. Environ. Microbiol. 71:2832–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Karatzas K-AG, Brennan O, Heavin S, Morrissey J, O'Byrne CP. 2010. Intracellular accumulation of high levels of γ-aminobutyrate by Listeria monocytogenes 10403S in response to low pH: uncoupling of γ-aminobutyrate synthesis from efflux in a chemically defined medium. Appl. Environ. Microbiol. 76:3529–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dover S, Halpern YS. 1972. Control of the pathway of γ-aminobutyrate breakdown in Escherichia coli K-12. J. Bacteriol. 110:165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dover S, Halpern YS. 1972. Utilization of γ-aminobutyric acid as the sole carbon and nitrogen source by Escherichia coli K-12 mutants. J. Bacteriol. 109:835–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fait A, Fromm H, Walter D, Galili G, Fernie AR. 2008. Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci. 13:14–19 [DOI] [PubMed] [Google Scholar]
- 17. de Carvalho LPS, Ling Y, Shen C, Warren JD, Rhee KY. 2011. On the chemical mechanism of succinic semialdehyde dehydrogenase (GabD1) from Mycobacterium tuberculosis. Arch. Biochem. Biophys. 509:90–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feehily C, Karatzas KAG. 27 September 2012. Role of glutamate metabolism in bacterial responses towards acid and other stresses. J. Appl. Microbiol. doi:10.1111/j.1365-2672.2012.05434.x [DOI] [PubMed] [Google Scholar]
- 19. Schneider BL, Ruback S, Kiupakis AK, Kasbarian H, Pybus C, Reitzer L. 2002. The Escherichia coli gabDTPC operon: specific γ-aminobutyrate catabolism and nonspecific induction. J. Bacteriol. 184:6976–6986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bouché N, Fromm H. 2004. GABA in plants: just a metabolite? Trends Plant Sci. 9:110–115 [DOI] [PubMed] [Google Scholar]
- 21. Glaser P, Frangeul L, Buchrieser C, Rusniok C, Amend A, Baquero F, Berche P, Bloecker H, Brandt P, Chakraborty T, Charbit A, Chetouani F, Couvé E, de Daruvar A, Dehoux P, Domann E, Domínguez-Bernal G, Duchaud E, Durant L, Dussurget O, Entian KD, Fsihi H, García-del Portillo F, Garrido P, Gautier L, Goebel W, Gómez-López N, Hain T, Hauf J, Jackson D, Jones LM, Kaerst U, Kreft J, Kuhn M, Kunst F, Kurapkat G, Madueno E, Maitournam A, Vicente JM, Ng E, Nedjari H, Nordsiek G, Novella S, de Pablos B, Pérez-Diaz JC, Purcell R, Remmel B, Rose M, Schlueter T, Simoes N, Tierrez A, Vázquez-Boland JA, Voss H, Wehland J, Cossart P. 2001. Comparative genomics of Listeria species. Science 294:849–852 [DOI] [PubMed] [Google Scholar]
- 22. Eisenreich W, Dandekar T, Heesemann J, Goebel W. 2010. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat. Rev. Microbiol. 8:401–412 [DOI] [PubMed] [Google Scholar]
- 23. Eisenreich W, Slaghuis J, Laupitz R, Bussemer J, Stritzker J, Schwarz C, Schwarz R, Dandekar T, Goebel W, Bacher A. 2006. 13C isotopologue perturbation studies of Listeria monocytogenes carbon metabolism and its modulation by the virulence regulator PrfA. Proc. Natl. Acad. Sci. U. S. A. 103:2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Joseph B, Mertins S, Stoll R, Schär J, Umesha KR, Luo Q, Müller-Altrock S, Goebel W. 2008. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J. Bacteriol. 190:5412–5430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horton RM, Ho SN, Pullen JK, Hunt HD, Cai Z, Pease LR. 1993. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Methods Enzymol. 217:270–279 [DOI] [PubMed] [Google Scholar]
- 26. Abram F, Starr E, Karatzas KAG, Matlawska-Wasowska K, Boyd A, Wiedmann M, Boor KJ, Connally D, O'Byrne CP. 2008. Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl. Environ. Microbiol. 74:6848–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bartsch K, von Johnn-Marteville A, Schulz A. 1990. Molecular analysis of two genes of the Escherichia coli gab cluster: nucleotide sequence of the glutamate:succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD). J. Bacteriol. 172:7035–7042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Byrne CP, Feehily C, Ham R, Karatzas KAG. 2011. A modified rapid enzymatic microtiter plate assay for the quantification of intracellular γ-aminobutyric acid and succinate semialdehyde in bacterial cells. J. Microbiol. Methods 84:137–139 [DOI] [PubMed] [Google Scholar]
- 29. Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, Barthelemy M, Vergassola M, Nahori MA, Soubigou G, Régnault B, Coppée JY, Lecuit M, Johansson J, Cossart P. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956 [DOI] [PubMed] [Google Scholar]
- 30. Dover S, Halpern YS. 1974. Genetic analysis of the γ-aminobutyrate utilization pathway in Escherichia coli K-12. J. Bacteriol. 117:494–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Buell CR, Joardar V, Lindeberg M, Selengut J, Paulsen IT, Gwinn ML, Dodson RJ, Deboy RT, Durkin AS, Kolonay JF, Madupu R, Daugherty S, Brinkac L, Beanan MJ, Haft DH, Nelson WC, Davidsen T, Zafar N, Zhou L, Liu J, Yuan Q, Khouri H, Fedorova N, Tran B, Russell D, Berry K, Utterback T, Van Aken SE, Feldblyum TV, D'Ascenzo M, Deng W-L, Ramos AR, Alfano JR, Cartinhour S, Chatterjee AK, Delaney TP, Lazarowitz SG, Martin GB, Schneider DJ, Tang X, Bender CL, White O, Fraser CM, Collmer A. 2003. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc. Natl. Acad. Sci. U. S. A. 100:10181–10186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kurihara S, Kato K, Asada K, Kumagai H, Suzuki H. 2010. A putrescine-inducible pathway comprising PuuE-YneI in which γ-aminobutyrate is degraded into succinate in Escherichia coli K-12. J. Bacteriol. 192:4582–4591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aronson JN, Borris DP, Doerner JF, Akers E. 1975. γ-Aminobutyric acid pathway and modified tricarboxylic acid cycle activity during growth and sporulation of Bacillus thuringiensis. Appl. Microbiol. 30:489–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trivett T, Meyer E. 1971. Citrate cycle and related metabolism of Listeria monocytogenes. J. Bacteriol. 107:770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guest JR. 1981. Partial replacement of succinate dehydrogenase function by phage- and plasmid-specified fumarate reductase in Escherichia coli. J. Gen. Microbiol. 122:171–179 [DOI] [PubMed] [Google Scholar]
- 36. Hirsch CA, Rasminsky M, Davis BD, Lin ECC. 1963. A fumarate reductase in Escherichia coli distinct from succinate dehydrogenase. J. Biol. Chem. 238:3770–3774 [PubMed] [Google Scholar]
- 37. Busch K, Piehler J, Fromm H. 2000. Plant succinic semialdehyde dehydrogenase: dissection of nucleotide binding by surface plasmon resonance and fluorescence spectroscopy. Biochemistry 39:10110–10117 [DOI] [PubMed] [Google Scholar]