Abstract
The detection of environmental enterococci has been determined primarily by using culture-based techniques that might exclude some enterococcal species as well as those that are nonculturable. To address this, the relative abundances of enterococci were examined by challenging fecal and water samples against a currently available genus-specific assay (Entero1). To determine the diversity of enterococcal species, 16S rRNA gene-based group-specific quantitative PCR (qPCR) assays were developed and evaluated against eight of the most common environmental enterococcal species. Partial 16S rRNA gene sequences of 439 presumptive environmental enterococcal strains were analyzed to study further the diversity of enterococci and to confirm the specificities of group-specific assays. The group-specific qPCR assays showed relatively high amplification rates with targeted species (>98%), although some assays cross-amplified with nontargeted species (1.3 to 6.5%). The results with the group-specific assays also showed that different enterococcal species co-occurred in most fecal samples. The most abundant enterococci in water and fecal samples were Enterococcus faecalis and Enterococcus faecium, although we identified more water isolates as Enterococcus casseliflavus than as any of the other species. The prevalence of the Entero1 marker was in agreement with the combined number of positive signals determined by the group-specific assays in most fecal samples, except in gull feces. On the other hand, the number of group-specific assay signals was lower in all water samples tested, suggesting that other enterococcal species are present in these samples. While the results highlight the value of genus- and group-specific assays for detecting the major enterococcal groups in environmental water samples, additional studies are needed to determine further the diversity, distributions, and relative abundances of all enterococcal species found in water.
INTRODUCTION
For more than a century the microbiological quality of environmental waters has been assessed using fecal indicator bacteria (FIB). While fecal coliforms and Escherichia coli are still widely used in environmental monitoring, enterococci are becoming a frequent target, as they can be used to estimate health risks in both recreational marine waters and bodies of freshwater. The Enterococcus genus includes more than 20 species, many of which are commonly associated with different mammals and birds, while some species have been isolated from nonfecal sources (1). Studies looking at the enterococci diversity in environmental waters have identified most strains as Enterococcus faecalis, Enterococcus faecium, Enterococcus casseliflavus, Enterococcus hirae, Enterococcus durans, and Enterococcus mundtii (2–4). These findings have relied on the isolation of enterococcal strains on selective culturing medium (5), followed by their classification, which may involve biochemical (6, 7) and molecular (8) techniques. Culture-based techniques are also used in regulatory activities to estimate the densities of enterococci in environmental waters. Since none of the enterococcal media available can be used to discriminate between the different species, their densities are recorded as general enterococcus counts. Information on the environmental prevalence of enterococcal species is not only relevant for confirming the presence of fecal enterococci, but it has also been suggested that it can help identify primary fecal pollution sources (9). Different fecal sources can contribute to the pollution of environmental waters, and each of them carries different health risks (10). The general consensus is that human fecal sources are associated with higher risks, particularly due to host-specific pathogens, such as enteric protozoa and viruses. However, nonhuman pollution sources are increasingly receiving attention by those in the health risk community, in light of recent outbreaks in which they are implicated as the most likely sources (11) and due to their relevance in beach closures, where the economic impact can be significant.
A quantitative PCR (qPCR) assay, Entero1, was used recently to estimate the levels of enterococci in recreational waters (12). Originally developed by Ludwig and Schleifer (13), the Entero1 assay targets the 23S rRNA gene. In most bacterial species, rRNA genes are present in multiple copies per genome, and therefore, targeting such genes in environmental samples can improve assay sensitivity due to their lower detection limits. However, less sequencing information is available for the 23S rRNA gene than for the 16S rRNA gene, precluding robust in silico validation. As a result, validation of the Entero1 assay has relied on testing the assay against a relatively small number of environmental strains isolated from a limited number of different geographic locations (12, 14). Moreover, similar to selective enterococcal media, the Entero1 assay cannot be used to determine which of the major enterococcal groups are present in a given sample.
To address some of these issues, we compared the relative occurrences and abundances of environmental and fecal enterococci using the Entero1 assay and several 16S rRNA gene-based group-specific PCR assays, most of which were developed as part of this study. Due to their reported prevalences in the environment, three of the major fecal enterococcal groups (E. faecalis, E. faecium, and E. casseliflavus) were targeted by the group-specific assays. The study was conducted by challenging the assays against fecal samples from diverse hosts and environmental waters with a history of fecal pollution. We also identified 439 strains isolated from surface water samples using 16S rRNA gene sequence analysis.
MATERIALS AND METHODS
Bacterial strains.
The following strains were used as positive and negative controls: E. casseliflavus (ATCC 25788), Enterococcus dispar (ATCC 51266), E. durans (ATCC 19432), E. faecalis (ATCC 19433), E. faecium (ATCC 19434), Enterococcus gallinarum (ATCC 49573), E. hirae (ATCC 8043), Enterococcus pseudoavium (ATCC 49372), Aeromonas eucrenophila (ATCC 23309), Escherichia coli (ATCC 25922), Legionella sainthelensi (ATCC 35248), Proteus vulgaris (ATCC 13315), Salmonella enterica serovar Typhimurium (ATCC 14028), Shigella flexneri (ATCC 29903), Staphylococcus aureus (ATCC 29213), Catellicoccus marimammalium, Citrobacter freundii, E. coli O157:H7, Escherichia hermannii, Klebsiella pneumoniae, and Pseudomonas aeruginosa. Additionally, 439 presumptive Enterococcus sp. strains isolated on Enterococcus indoxyl-β-d-glucoside (mEI) agar (15) were used for evaluating the enterococcal assays. The latter strains were isolated from environmental waters collected from 15 U.S. states (AZ, CO, FL, GA, KS, MD, MN, MT, NJ, NV, NY, OK, WA, WV, and WY). The identities of the environmental enterococcal strains were confirmed using 16S rRNA gene sequencing analysis.
Environmental sample collection and DNA extraction.
The environmental monitoring values of the group-specific assays were tested against water samples (n = 311) collected from different locations in California, North Carolina, and Puerto Rico. The water samples from California and North Carolina were collected from estuarine sites that are primarily impacted by gulls, whereas the water samples from Puerto Rico were collected from sites within the Rio Grande de Arecibo watershed which are presumably impacted by cattle, humans, and wildlife. Additionally, the assays were challenged against fecal samples (n = 497) from 4 domesticated animals (goat, horse, monkey, and pig), 13 wildlife species (chipmunk, coyote, fox, marmot, yellow-bellied marmot, mule, mule deer, rabbit, jackrabbit, raccoon, snowshoe hare, squirrel, and ground squirrel), and 7 avian species (chicken, duck, guinea fowl, gull, pelican, swan, and turkey). The water samples (100 ml each) were collected and filtered onto polycarbonate membranes (0.4-μm pore size, 47-mm diameter) (GE Water and Process Technologies, Trevose, PA). The fecal samples were collected aseptically, transferred to sterile tubes, and transported to the laboratory in ice coolers. The frozen filters and fecal samples were shipped overnight on dry ice to the U.S. Environmental Protection Agency, Cincinnati, OH, and stored at −80°C until further processing. DNA extraction from the filters and fecal samples was performed using the PowerSoil DNA isolation kit (Mo Bio Laboratories, Carlsbad, CA) according to the manufacturer's protocols. Avian fecal samples (i.e., gull and turkey) from France were extracted using the FastDNA spin kit for soil (MP Biomedical, Illkirsh, France) according to the supplier's instructions, except that an additional wash using the salt/ethanol wash solution (SEWS-M) reagent was performed. The DNA concentrations were measured using a NanoDrop ND-1000 UV spectrophotometer (NanoDrop Technologies, Wilmington, DE). The DNA extracts were stored at −20°C until further processing.
Sequencing analyses.
The sequences from 16S rRNA gene PCR products that were generated using universal bacterial primers (8F, 5′-AGAGTTTGATCCTGGCTCAG-3′, and 787R, 5′-CGACTACCAGGGTATCTAAT-3′) were used to determine the identities of the 439 environmental isolates from mEI cultures and reference bacteria. Briefly, PCR assays were performed in 25 μl using the polymerase TaKaRa Ex Taq (TaKaRa Bio, Inc.) in a Tetrad2 thermal cycler (Bio-Rad, Hercules, CA) under the following cycling conditions: an initial denaturation step at 95°C for 5 min and 25 cycles of 1 min at 95°C, 1 min at 56°C, and 1 min at 72°C. The PCR products were sequenced in both directions in the Children's Hospital DNA Core Facility (Cincinnati, OH) using an Applied Biosystems Prism 3730XL DNA analyzer. The raw gene sequences were processed using Sequencher software (Gene Codes, Ann Arbor, MI). For the 16S rRNA gene sequences, homology searches of DNA sequences in the GenBank (nonredundant [NR]) database were undertaken with the National Center for Biotechnology Information (NCBI) BLASTn program (http://www.ncbi.nlm.nih.gov/BLAST/) (16).
Assay development and performance evaluation.
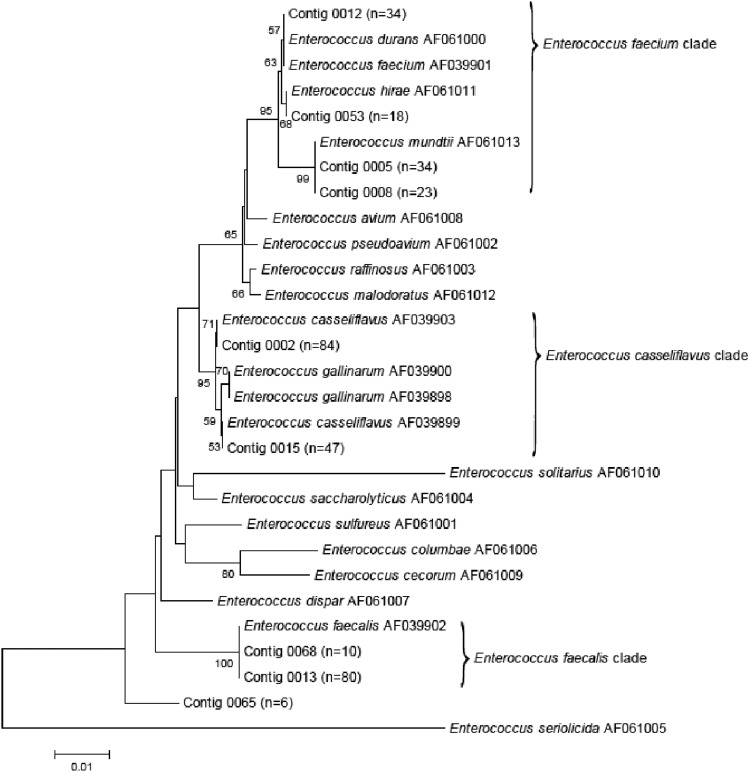
Fifteen different assays were tested in this study: five genus-specific assays, three E. faecalis-specific assays, three E. casseliflavus-specific assays, and four E. faecium-specific assays (Table 1). Eleven assays were tested as conventional PCR assays, and four were tested as qPCR (TaqMan-based) assays (one genus-specific, Entero1, and one for each of the enterococcal groups, Faecalis1, Casseli1, and Faecium1). The Entero1 and Faecalis1 qPCR assays were developed and evaluated in previous studies (12, 17). To develop new enterococcal assays, a phylogenetic tree that included the 16S rRNA gene sequences from reference enterococcal strains (8) and environmental strains was generated using a neighbor-joining algorithm in ARB (18). Unique phylogenetic clades were identified (Fig. 1), and candidate primers were then chosen to target three major environmental clades (E. faecalis, E. faecium, and E. casseliflavus) using the primer design algorithm in ARB (Table 1). Additionally, 16S rRNA gene enterococcal sequences were used to design two group-specific qPCR assays using the Primer Express software (Applied Biosystems, Foster City, CA) (Table 1). The assays were optimized through the use of temperature gradients and were tested for their specificities and sensitivities against the reference bacterial strains and environmental enterococcus isolates described above. The applicabilities of the PCR and qPCR assays in environmental monitoring were also evaluated against the aforementioned set of water and fecal samples.
Table 1.
Summary of oligonucleotide primers and probes for PCR and TaqMan qPCR
| Target organism(s) | Assaya | Primer | P sequence (5′ to 3′) | Ta (°C)b | Size (bp)c | Reference or source |
|---|---|---|---|---|---|---|
| Enterococcus spp. | Entero1 | ECST748F | AGAAATTCCAAACGAACTTG | 60 | 92 | 18 |
| ENC854R | CAGTGCTCTACCTCCATCATT | |||||
| GPL813TQ | 6FAM-TGGTTCTCTCCGAAATAGCTTTAGGGCTA-TAMRA | |||||
| Ent1 | Ent151F | ACACTTGGAAACAGGTGC | 65 | 243 | This study | |
| Ent376R | TCGGTCAGACTTKCGTCC | |||||
| Ent2 | Ent151F | ACACTTGGAAACAGGTGC | 65 | 445 | This study | |
| Ent578R | TTAAGAAACCGCCTGCGC | |||||
| Ent3 | Ent240F | TGCATTAGCTAGTTGGTG | 63 | 356 | This study | |
| Ent578R | TTAAGAAACCGCCTGCGC | |||||
| Ent4 | Ent376F | GGACGMAAGTCTGACCGA | 65 | 220 | This study | |
| Ent578R | TTAAGAAACCGCCTGCGC | |||||
| Enterococcus faecalis | Faecalis1 | FaecalF | CGCTTCTTTCCTCCCGAGT | 60 | 143 | 32 |
| FaecalR | GCCATGCGGCATAAACTG | |||||
| FaecalP | 6FAM-CAATTGGAAA GAGGAGTGGCGGACG-TAMRA | |||||
| Faecalis2 | Ent151F | ACACTTGGAAACAGGTGC | 64 | 318 | This study | |
| Faecal449R | AGTTACTAACGTCCTTGTTC | |||||
| Faecalis3 | Ent240F | TGCATTAGCTAGTTGGTG | 63 | 229 | This study | |
| Faecal449R | AGTTACTAACGTCCTTGTTC | |||||
| Enterococcus casseliflavus | Casseli1 | CasselF | GGAGCTTGCTCCACCGAA | 60 | 132 | This study |
| CasselR | TTTCTTCCATGCGGAAAATAGT | |||||
| CasselP | 6FAM-CGAACGGGTGAGTAACACGTGGGTAA-TAMRA | |||||
| Casseli2 | Cassel190F | GGAAGAAAGTTGAAAGGC | 60 | 204 | This study | |
| Ent376R | TCGGTCAGACTTKCGTCC | |||||
| Casseli3 | Cassel190F | GGAAGAAAGTTGAAAGGC | 60 | 406 | This study | |
| Ent578R | TTAAGAAACCGCCTGCGC | |||||
| Enterococcus faecium | Faecium1 | CiumF | TTCTTTTTCCACCGGAGCTT | 60 | 141 | This study |
| CiumR | AACCATGCGGTTTYGATTG | |||||
| CiumP | 6FAM-AGTAACACGTGGGTAACCTGCCCATCAGA-TAMRA | |||||
| Faecium2 | Cium84F | TGCTCCACCGGAAAAAGA | 63 | 174 | This study | |
| Ent240R | CACCAACTAGCTAATGCA | |||||
| Faecium3 | Cium84F | TGCTCCACCGGAAAAAGA | 64 | 310 | This study | |
| Ent376R | TCGGTCAGACTTKCGTCC | |||||
| Faecium4 | Cium84F | TGCTCCACCGGAAAAAGA | 65 | 512 | This study | |
| Ent578R | TTAAGAAACCGCCTGCGC |
Entero1 targets the 23S rRNA gene, whereas the other assays target the 16S rRNA gene.
Optimum annealing temperatures determined using temperature-gradient PCR.
Approximate product size determined from in silico data.
Fig 1.
Unrooted neighbor-joining tree of 16S rRNA gene sequences obtained from Enterococcus environmental isolates. The number of sequences for each contig is included in parentheses, and the contigs of fewer than 5 sequences are not presented in the phylogenetic tree. The reference bacteria with their GenBank accession numbers and 1,000-replicate bootstrap values are shown in the tree. The bootstrap values reported are the percentages greater than 50%. The scale bar corresponds to 0.01 changes per nucleotide.
For the conventional PCR assays, all water and fecal samples were tested as described previously (19), with the following modifications: 0.5 to 1 ng/μl of DNA extracts was used as a template, and 10-fold dilutions of each DNA extract were used to test for PCR inhibition. The PCR assays were performed in 25 μl using TaKaRa Ex Taq (TaKaRa Bio, Inc.) in a Bio-Rad Tetrad2 Peltier thermal cycler (Bio-Rad, Hercules, CA) under the following cycling conditions: an initial denaturation step at 95°C for 5 min and 25 cycles of 1 min at 95°C, 1 min at optimum annealing temperature (Table 1), and 1 min at 72°C. The PCR products were visualized in 1.5% agarose gels using GelStar nucleic acid gel stain (Lonza, Rockland, ME).
The TaqMan qPCR assays were performed in 25-μl reaction mixtures containing 1× TaqMan universal PCR master mix with AmpErase uracil-N-glycosylase (Applied Biosystems, Foster City, CA), 0.2 μg/μl bovine serum albumin, 0.2 μM (final concentration) of each primer, and a 6-FAM (6-carboxyfluorescein)-labeled hydrolysis probe. The amplification protocol involved an initial incubation step at 50°C for 2 min to activate uracil-N-glycosylase, followed by 10 min of incubation at 95°C to activate AmpliTaq Gold enzyme, and then 40 cycles of 95°C for 15 s and 60°C for 1 min. The qPCR assays were performed using a 7900 HT Fast real-time sequence detector (Applied Biosystems, Foster City, CA). All assays were performed in triplicate in MicroAmp Optical 96-well reaction plates with MicroAmp Optical Caps strips (Applied Biosystems, Foster City, CA). The PCR data were analyzed using ABI's Sequence Detector software (version 2.2.2). Four independent standard curves for each qPCR assay were generated by plotting the threshold cycle (CT) values against the numbers of target copies corresponding to serially diluted plasmid standards purchased from Integrated DNA Technologies (IDT; Coralville, Iowa). The target copy numbers (T) were estimated by the equation T = [D/(PL × 660)] × 6.022 × 1023, where D (g/μl) is plasmid DNA concentration and PL (in base pairs) is plasmid length. Each standard curve was generated from at least five 10-fold plasmid dilutions in triplicate. The percent amplification efficiencies were calculated by the instrument manufacturer's instructions (Applied Biosystems). Two no-template controls per PCR plate were used to check for cross-contamination.
Venn diagrams.
The relationships among the genus- and species-specific qPCR assays against fecal and water samples were determined using Venn diagrams as described previously (20). Briefly, two Venn diagrams were constructed sequentially: the first diagram was used for calculating the prevalences of three species-specific markers, and the second diagram was used to establish the relationship between the genus-specific assay and the three species-specific assays combined.
Nucleotide sequence accession numbers.
The representative sequences were deposited in GenBank under the following accession numbers: JQ804941 to JQ804949.
RESULTS AND DISCUSSION
Rationale for assay development.
Phylogenetic trees that included sequences from reference and environmental enterococcal strains were generated to identify the 16S rRNA gene sequences that could be used to develop multiple enterococcal species-specific assays (Fig. 1). This approach indicated that it was difficult to develop assays that discriminated E. faecium from E. mundtii, E. durans, E. hirae, and E. dispar and E. casseliflavus from E. gallinarum. However, nonribosomal genes can be used to discriminate between different enterococcal species (21, 22). However, only a handful of nonribosomal genes have been used in environmental studies to detect or identify enterococci (14, 23, 24). More importantly, the sequence database for the function-specific genes of environmental enterococci and other phylogenetically related genera is much more limiting than that for the 16S and 23S rRNA genes. Moreover, sequence conservancy in functional genes is considerably lower than that in rRNA genes, which explains why it is difficult to develop genus- and group-specific assays unless comprehensive sequence databases are developed.
Identification of environmental strains.
Based on the 16S rRNA gene sequence analyses of the 439 environmental isolates used in this study, approximately 91% were identified as Enterococcus spp., whereas others were classified as nonenterococci (7%) or unclassified bacteria (2%). These results are in agreement with other studies using mEI agar as the isolation medium for environmental enterococci (15, 25), although Nayak et al. (26) reported relatively lower false-positive rates (1.6%) in subtropical waters. The study by Nayak et al. was based on 61 strains isolated from two lakes on two different dates, which may explain the lower false-positive rate.
Based on the sequence identities of the environmental isolates tested in our study, the most dominant enterococcal species were E. casseliflavus (34%), E. faecalis (25%), and E. mundtii (15%), while E. faecium and E. hirae were identified to a lesser extent (5%) (Table 2). Several enterococcal species have been detected in environmental waters, but their overall prevalences have varied considerably. For example, Mote et al. (25) found that the most dominant enterococcal species were E. faecalis (31%), E. mundtii (31%), and E. casseliflavus (16%), while E. faecium and E. gallinarum were identified less frequently (10% and 4%, respectively). Moore et al. (4) and Grammenou et al. (27) also found different environmental enterococcal species, but E. faecalis and E. faecium were the most dominant species in many water samples. In spite of these differences, these results clearly indicate that multiple enterococcal species can be present in the same water sample. The differences in the occurrences of enterococcal species may be associated with different in situ growth and environmental survival rates (28) and with preferential host distributions of different enterococcal species in different animals. Other studies have suggested that that the environmentally relevant enterococcal species detected in this study are found in a wide variety of hosts (29). Altogether, these data suggest that the identification of enterococcal species might not be an adequate approach to fecal source identification.
Table 2.
Classification of environmental isolates from mEI cultures using 16S rRNA gene sequencing
| Bacteria | No. (%) of isolates | GenBank accession no.a |
|---|---|---|
| Enterococcus casseliflavus | 152 (34) | DQ333294.1 |
| Enterococcus faecalis | 111 (25) | AB534553.1 |
| Enterococcus mundtii | 68 (15) | NR_024906.1 |
| Enterococcus faecium | 23 (5) | EU003447.1 |
| Enterococcus hirae | 20 (5) | Y17302.1 |
| Enterococcus spp. | 21 (5) | NR_036922.1, NR_037082.1, and NR_042054.1 |
| Aerococcus spp. | 24 (5) | HM582941.1 |
| Lactococcus garvieae | 3 (0.7) | AY699289.1 |
| Pediococcus pentosaceus | 4 (0.9) | CP000422.1 |
| Streptococcus gallolyticus subsp. pasteurianus | 4 (0.9) | AB457024.1 |
| Unclassified | 9 (2) | Not available |
| Total | 439 (100) |
All sequences for enterococcus isolates are >99% identical to GenBank reference sequences.
In this study, four nonenterococcal species were identified (i.e., >99% identical to reference sequences) among the environmental isolates, namely, Aerococcus sp., Lactococcus garvieae, Pediococcus pentosaceus, and Streptococcus gallolyticus subsp. pasteurianus. Other studies have reported on the presence of some of these genera in mEI medium. For example, Maraccini et al. (30) showed that most nonenterococcal mEI isolates were identified as Aerococcus viridans (17%), with fewer isolates identified as Streptococcus mutans, S. gallolyticus, Leuconostoc spp., and Pediococcus acidilactici. The samples from the latter study were collected within a 3-day period from one marine site. While it is not known how predominant these nonenterococcal species are during an entire beach season, these data suggest that some nonenterococcal species may be highly abundant in recreational marine waters, potentially resulting in the overestimation of enterococcus densities when using culture-based methods. In another study, Viau and Peccia (31) showed that mEI medium also supported the growth of bacteria from biosolids that were identified as Bacillus spp., Vagococcus spp., and Desemzia incerta. As biosolids and animal fecal waste (i.e., treated manure) are used in agricultural activities, the results from these studies suggest that nonenterococcal species might interfere with the culture-based methods that are used to estimate fecal pollution levels. In our study, the bacterial strains found in the water tested were isolated from waters presumed to be impacted by wastewater treatment plants and, to a lesser extent, by agricultural activities, although wildlife fecal pollution sources cannot be ruled out. Our results suggest further that culture-based methods can support the growth of nonenterococcal species present in freshwater samples and that additional studies are needed to determine better the identities and prevalences of these nontargeted species in fecal and water samples.
Validation of genus-specific enterococcal PCR assays.
The specificities of the Enterococcus genus- and group-specific PCR assays were evaluated against a subset of the enterococcal strains sequenced in this study. This subset (n = 153) included several strains from the most common Enterococcus species identified in this study, nonenterococcal species obtained from culture collections (n = 13), and nonenterococcal strains isolated from this study (n = 4) (Table 3). All of the genus-specific assays successfully amplified the enterococcus-type strains (from the ATCC). Additionally, four of the genus-specific assays generated positive signals with more than 97% of the environmental strains tested in this study and in most cases cross-amplified relatively few non-enterococcal strains (0 to 24%) (Table 3). Two of the assays, Ent2 and Ent3, did not show cross-amplification with nonenterococcal strains and therefore may prove useful as confirmatory tests. However, Ent2 only detected 59% of the enterococcal strains tested, suggesting that it cannot be used as a stand-alone enterococcal assay.
Table 3.
Number (percentage) of positives by the Enterococcus assays against environmental isolates and ATCC strains
| Target organism(s) | Assay | No. (%) of positives |
||||||
|---|---|---|---|---|---|---|---|---|
| Enterococcus casseliflavus (n = 50)a | Enterococcus faecalis (n = 39) | Enterococcus faecium (n = 11) | Enterococcus hirae (n = 5) | Enterococcus mundtii (n = 40) | Other enterococcal species (n = 8)b | Nonenterococcal species (n = 17)c | ||
| Enterococcus spp. | Entero1c | 50 (100) | 39 (100) | 11 (100) | 5 (100) | 40 (100) | 8 (100) | 1 (5.9) |
| Ent1 | 50 (100) | 39 (100) | 11 (100) | 5 (100) | 39 (98) | 7 (88) | 4 (24) | |
| Ent2 | 33 (66) | 19 (49) | 8 (73) | 4 (80) | 18 (45) | 8 (100) | 0 (0) | |
| Ent3 | 50 (100) | 37 (95) | 11 (100) | 4 (80) | 39 (98) | 8 (100) | 0 (0) | |
| Ent4 | 50 (100) | 39 (100) | 11 (100) | 5 (100) | 40 (100) | 8 (100) | 2 (12) | |
| Enterococcus faecalis | Faecalis1d | 5 (10) | 39 (100) | 2 (18) | 0 (0) | 6 (15) | 1 (13) | 0 (0) |
| Faecalis2 | 0 (0) | 16 (41) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Faecalis3 | 0 (0) | 25 (64) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Enterococcus casseliflavus | Casseli1d | 49 (98) | 0 (0) | 1 (9.1) | 1 (20) | 0 (0) | 0 (0) | 0 (0) |
| Casseli2 | 29 (58) | 4 (10) | 1 (9.1) | 0 (0) | 4 (10) | 0 (0) | 0 (0) | |
| Casseli3 | 48 (96) | 0 (0) | 1 (9.1) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Enterococcus faecium | Faecium1d | 2 (4.0) | 0 (0) | 11 (100) | 5 (100) | 40 (100) | 8 (100) | 0 (0) |
| Faecium2 | 27 (54) | 2 (5.1) | 10 (91) | 3 (60) | 35 (88) | 8 (100) | 0 (0) | |
| Faecium3 | 30 (60) | 1 (2.6) | 11 (100) | 5 (100) | 28 (70) | 8 (100) | 0 (0) | |
| Faecium4 | 0 (0) | 0 (0) | 11 (100) | 4 (80) | 19 (48) | 7 (88) | 0 (0) | |
All enterococcal species were identified by NCBI BLAST, with the exception of E. casseliflavus, the sequences of which are nearly identical to those of E. gallinarum.
Sequences of the isolates are affiliated with the E. faecium clade (see Fig. 1).
Seven ATCC strains (Aeromonas eucrenophila, Escherichia coli, Legionella sainthelensi, Proteus vulgaris, Salmonella enterica serovar Typhimurium, Shigella flexneri, and Staphylococcus aureus), six laboratory strains (Catellicoccus marimammalium, Citrobacter freundii, Escherichia coli O157:H7, Escherichia hermannii, Klebsiella pneumoniae, and Pseudomonas aeruginosa), and four environmental strains (Aerococcus species, Lactococcus garvieae, Pediococcus pentosaceus, and Streptococcus pasteurianus).
TaqMan qPCR assays.
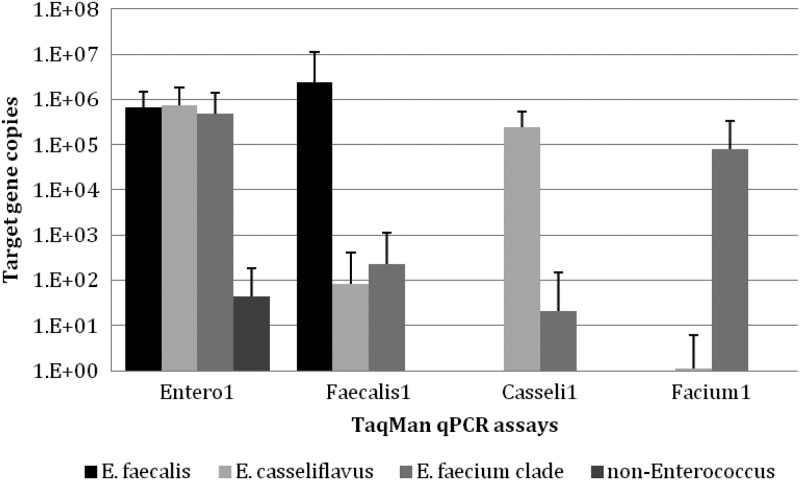
Interestingly, the Entero1 assay showed a low level of cross-amplification with C. marimammalium. Several gull-specific assays target the C. marimammalium 16S rRNA gene. Indeed, the signals with these gull assays have frequently been detected in environmental waters with a history of gull fecal contamination (20, 32). Signals detected against C. marimammalium DNA were approximately 4 orders of magnitude lower than those against the enterococcal species tested (Fig. 2), suggesting that C. marimammalium may not contribute significantly to false-positive signals. Entero1 cross-amplification signals have been observed with other lactobacilli species (33). These results are relevant to environmental monitoring, as the Entero1 assay has been proposed as an alternative method for the rapid detection of Enterococcus spp. in recreational waters (34). As the overestimation of the Entero1 assay due to nontargeted bacteria could result in unnecessary beach closures, additional studies are needed to determine more accurately the levels of false-positive signals in recreational settings. Future studies also need to determine whether these nontargeted populations are present in environmental waters frequently enough to interfere with risk assessment models.
Fig 2.
Mean copy numbers of target markers against environmental isolates of Enterococcus species and non-Enterococcus bacteria. To calculate the mean concentrations, the values below detection limits were treated as 0. The error bars represent 1 SD.
Validation of group-specific enterococcal PCR assays.
Overall, the group-specific assays indicated a relatively high amplification with targeted enterococcal species and low cross-amplification with nonenterococcal species (Table 3). Noteworthy is that the Faecalis2 and Faecalis3 assays showed 100% specificities and amplified the E. faecalis type strain, but they did not amplify some of the E. faecalis environmental strains, suggesting that they may be used in limited cases as group-specific assays. The Faecalis1 TaqMan qPCR assay successfully amplified all of the tested E. faecalis strains (n = 39) and did not cross-react with seven non-E. faecalis ATCC strains, as was observed previously (17). Although the Faecalis1 assay cross-reacted with a low number of non-E. faecalis environmental strains (Table 3), the signal intensities of these nontarget bacteria were more than 4 orders of magnitude lower than those of E. faecalis strains (Fig. 2). Tracking signal intensities will be important to determine the value of these assays in environmental applications. As cross-amplification signals are relatively low for some of these assays, the scenarios showing high levels of environmental signals are likely to be the result of true-positive signals rather than false-positive signals, unless cross-amplification targets are present in high abundance in a given environmental sample. This assumption needs to be tested with these newly developed assays and with most published FIB-targeting assays.
The E. casseliflavus-specific assays showed relatively high specificities (i.e., low cross-amplification rates against nontarget species) compared to the other group-specific assays. The Casseli1 TaqMan qPCR assay successfully amplified 98% (49/50) of E. casseliflavus environmental strains and showed 1.7% (2/120) cross-amplification with nontarget species. Moreover, the Casseli1 and the Casseli3 assays did not cross-amplify with any of the non-Enterococcus bacteria tested in this study. The Casseli2 assay showed the least specificity and sensitivity (i.e., higher cross-amplification rate with nontarget species and lower amplification rate with E. casseliflavus).
Most E. faecium assays primarily amplified E. faecium, E. durans, E. hirae, and E. mundtii strains. This is compatible with the facts that these species formed a cohesive clade and that it is difficult to differentiate these species using 16S rRNA gene sequences (Fig. 1). Specifically, the Faecium1 TaqMan qPCR assay amplified E. faecium, E. hirae, and E. mundtii strains. The Faecium2 and Faecium3 assays cross-reacted with E. casseliflavus, whereas the Faecium4 assay showed the best specificity (i.e., lower cross-amplification rate with E. casseliflavus) (Table 3). Thus, the E. faecium assays developed in this study might be used as a multispecies-specific assay. Future studies should focus on assessing the value of the conventional PCR assays developed in this study for use as qPCR assays.
Detection of enterococci in fecal and environmental water samples.
The Entero1, Faecalis1, Casseli1, and Faecium1 assays were used in more studies based on the overall specificity and sensitivity results and the fact that they can provide quantification data. Specifically, the assays were used to investigate the presence and abundances of enterococci in 497 fecal samples collected from four different geographic locations and from 24 different animals and in 311 environmental water samples collected from California, North Carolina, and Puerto Rico (Table 4). To our knowledge, this represents the largest study in which different enterococcal species have been detected from fecal samples via PCR assays without the need for an enrichment step.
Table 4.
Detection of enterococcal species in different fecal and water samples using TaqMan qPCR assays
| Sample type | Sampling location(s) | No. of samples | No. (%) of positive samples with Entero1 (G) | No. (%) of positive samples with Faecalis1 (A) | No. (%) of positive samples with Casseli1 (B) | No. (%) of positive samples with Faecium1 (C) | Relationship between different assays (% positive)a |
||
|---|---|---|---|---|---|---|---|---|---|
| n(S) | n(G ∩ S) | n(G ∪ S) | |||||||
| Fecal | |||||||||
| Goat | Puerto Rico | 32 | 32 (100) | 18 (56) | 7 (22) | 30 (94) | 32 (100) | 31 (97) | 32 (100) |
| Horse | Puerto Rico | 28 | 28 (100) | 7 (25) | 3 (11) | 22 (79) | 24 (86) | 24 (86) | 28 (100) |
| Monkey | Puerto Rico | 9 | 9 (100) | 7 (78) | 9 (100) | 6 (67) | 9 (100) | 9 (100) | 9 (100) |
| Pig | Puerto Rico | 30 | 30 (100) | 26 (87) | 26 (87) | 29 (97) | 30 (100) | 30 (100) | 30 (100) |
| Wildlifeb | California | 77 | 61 (79) | 13 (17) | 9 (12) | 39 (51) | 49 (64) | 49 (64) | 61 (79) |
| Chicken | Puerto Rico | 35 | 35 (100) | 24 (69) | 19 (54) | 31 (89) | 34 (97) | 34 (97) | 35 (100) |
| Duck | Puerto Rico | 16 | 16 (100) | 9 (56) | 13 (81) | 15 (94) | 16 (100) | 16 (100) | 16 (100) |
| Guinea fowl | Puerto Rico | 11 | 11 (100) | 1 (9.1) | 2 (18) | 6 (55) | 6 (55) | 6 (55) | 11 (100) |
| Gullc | California, Delaware, and France | 220 | 108 (49) | 82 (37) | 10 (4.5) | 33 (15) | 87 (40) | 83 (38) | 112 (51) |
| Pelican | California | 10 | 10 (100) | 10 (100) | 7 (70) | 9 (90) | 10 (100) | 10 (100) | 10 (100) |
| Swan | Puerto Rico | 22 | 22 (100) | 6 (27) | 11 (50) | 18 (82) | 19 (86) | 19 (86) | 22 (100) |
| Turkey | France and Puerto Rico | 7 | 7 (100) | 2 (29) | 6 (86) | 7 (100) | 7 (100) | 7 (100) | 7 (100) |
| Total fecal | 497 | 369 (74) | 205 (41) | 122 (25) | 245 (49) | NAd | NA | NA | |
| Water | |||||||||
| Estuarine water | California | 65 | 55 (85) | 24 (37) | 3 (4.6) | 24 (37) | 31 (48) | 31 (48) | 55 (85) |
| Estuarine water | North Carolina | 109 | 107 (98) | 68 (62) | 9 (8.3) | 34 (31) | 76 (70) | 76 (70) | 107 (98) |
| Surfacee | Puerto Rico | 137 | 69 (50) | 32 (23) | 4 (2.9) | 24 (18) | 37 (27) | 33 (24) | 73 (53) |
| Total water | 311 | 231 (74) | 124 (40) | 16 (5.1) | 82 (26) | NA | NA | NA | |
Results were calculated using a Venn diagram approach, where n(∪) is the total number of samples, n(A) is the number of positive samples with Faecalis1, n(B) is the number of positive samples with Casseli1, n(C) is the number of positive samples with Faecim1, and n(G) is the number of positive samples with Entero1; n(S) = n(A ∪ B ∪ C).
Thirteen different animals: chipmunk, coyote, fox, marmot, yellow-bellied marmot, mule, mule deer, rabbit, jackrabbit, raccoon, snowshoe hare, squirrel, and ground squirrel.
Three species of gull from California (Larus californicus), Delaware (Larus atricilla and Larus smithsonianus), and France (Larus argentatus).
NA, not available.
Water samples were collected from 12 sampling locations in the Arecibo watershed, Puerto Rico, between September 2010 and January 2011 (representing 13 sampling events).
The range of quantification (ROQ) for the Entero1 and Faecalis1 qPCR assays was 101 to 106 DNA copies per reaction. For the Faecium1 and the Casseli1 assays, 10 copies per reaction were below the detection limit of the assay; therefore, the ROQ of these assays was determined to be from 102 to 106 DNA copies. In order to evaluate assay sensitivities, four independent standard curves were used to calculate the percent amplification efficiency average. The Entero1 assay showed the greatest amplification efficiency, followed by the Faecalis1, Faecium1, and Casseli1 assays (averages ± SD, 94.8 ± 0.8, 90.9 ± 1.1, 88.5 ± 2.1, and 85.2 ± 1.3, respectively). All of the no-template controls were negative, indicating the absence of cross-contamination in the qPCR experiments.
Approximately 74%, 41%, 25%, and 49% of the fecal samples were positive for the Entero1, Faecalis1, Casseli1, and Faecium1 markers, respectively (Table 4). However, when excluding the gull samples, the number of positive samples for enterococci increased to 44% for the Casseli1 marker and 74 to 94% for the other markers, clearly suggesting that enterococci are normal inhabitants of most of the hosts tested here. The results of group-specific assays showed that different enterococcal species coinhabit most hosts, although the high prevalences of multiple enterococcal species were evident in some hosts more than others. For example, each of the three group-specific markers was detected in more than 87% of pig feces, while a specific group predominated in gulls, horses, and wildlife. The prevalence of the Entero1 marker [i.e., n(G)] was in agreement with the combined number of positive signals [i.e., n(S) = n(A ∪ B ∪ C)] determined by the three species-specific markers in fecal samples (Table 4). In other words, combining the results from the genus- and group-specific assays [i.e., n(G ∪ S)] did not increase the number of enterococcus-positive samples in most feces types, with the exception of gull fecal samples, in which an increased prevalence was observed [i.e., n(G ∪ S) > n(G) > n(S)]. There are two scenarios that might explain the lower prevalences of the species-specific markers in gull feces. First, it is possible that there are environmental enterococcal species that are detected by the Entero1 assay but not detected with the group-specific assays tested in this study. This suggests that additional group- or species-specific assays are needed to study further the abundances and dynamics of these species in fecal samples and perhaps in environmental waters impacted by gulls. This may be important if these nontargeted enterococcal species are noted to be important in recreational waters. A second scenario relates to the Entero1 assay cross-reacting with some of the indigenous non-Enterococcus bacteria; this may be the case for C. marimammalium, which resides in gull feces and for which signals have been detected in gull-impacted waters. If the latter situation is of any significance, the Entero1 assay may overestimate enterococcal levels but show a positive correlation with the presence of gull feces contamination. Thus, further validation of the specificity of Entero1 against a broad range of non-Enterococcus bacteria is needed; this is particularly the case for members of the Lactobacillales family, as overestimation due to false-positive signals is relevant in scenarios in which molecular assays are used as an alternative to culture-based assays that are used to monitor recreational water quality.
Most water samples tested in this study (74%) contained detectable enterococcal signals. In general terms, among the group-specific assays used, E. faecalis was detected more frequently (40%) in the water samples than E. faecium (26%) and E. casseliflavus (5.1%), regardless of the sample origin. The prevalences of the genus- and group-specific enterococcal assays in estuarine water samples from California and North Carolina were higher than those in tropical surface water samples (Table 4). The estuarine water samples tested in this study have historically been impacted by gulls, while surface waters in Puerto Rico are primarily impacted by wastewater and cattle fecal sources and, to a lesser extent, by domesticated animal sources, such as chickens, pigs, horses, and goats. Interestingly, the Casseli1 marker was seldom detected in gull fecal samples, which could explain the relatively low prevalence of the Casseli1 marker in the temperate water samples. However, the fact that the Casseli1 marker was also seldom detected in tropical waters not impacted by gulls suggests that the low detection rate of E. casseliflavus may not be indicative of low levels of waterfowl in environmental waters. The data suggest that some of the major enterococcal species are cosmopolitan (i.e., present in various hosts); therefore, the use of microbial source-tracking (MST) methods targeting enterococcal species might be difficult to justify in source allocation applications.
Unlike fecal samples, the prevalences of the Entero1 marker [i.e., n(G)] were higher than the combined numbers of positive signals determined by three species-specific markers [i.e., n(S) = n(A ∪ B ∪ C)] in all tested water samples from three different geographical locations (Table 4). Several factors could have contributed to these results. For example, some of the numerically dominant species were not detected with the group-specific assays used in this study. This means that species such as Enterococcus raffinosus, Enterococcus saccharolyticus, Enterococcus avium, E. pseudoavium, and Enterococcus cecorum might be present in some of these samples and contributed significantly to the genus-specific signals. Using Slanetz-Bartley agar, one study showed that a relatively high number of E. raffinosus, E. avium, and E. saccharolyticus strains were isolated from environmental waters (35), while in another study, 25% of the isolates were nontypical enterococcal species and were only classified as Enterococcus sp. (36). Grammenou et al. (27) also isolated E. avium from water samples, but the strains represented approximately 2% of all enterococcus isolates. Altogether, these results suggest that mEI (the medium used in our study) favors the growth of some enterococci, which explains why E. faecalis, E. casseliflavus, and E. faecium are often isolated from mEI plates. On the other hand, Suzuki et al. (37) recently showed that E. faecalis and E. faecium combined did not represent more than 32% of the mEI isolates from five Japanese rivers, potentially implicating the prevalences of other enterococcal species.
An alternative explanation for the differences in prevalence between Entero1 and the collective group-specific markers is that novel enterococci might have also been responsible for a fraction of the signals in the water samples. Indeed, novel enterococcal species have been identified in recent years from water (38–40) and fecal (41, 42) samples. While the relative abundances of novel enterococcal species in water samples are unknown, these results indicate that there is a need for further investigation of enterococcal diversity in both fecal and environmental samples. The results also suggest that some enterococcal species might be more adapted to persist outside of the gut environment than others (2), which might lead to the adaptation of fecal bacteria to secondary habitats (43). The latter scenario has important implications in conventional microbial water quality monitoring and in microbial source-tracking applications using enterococci as targeted populations.
Conclusion.
Overall, the results from this study are in agreement with previously published data demonstrating that animals frequently implicated in the fecal contamination of environmental waters shed different enterococcal species in their feces. This study also suggests that while three major enterococcal groups (i.e., E. faecalis, E. faecium, and E. casseliflavus) tend to dominate in fecally contaminated waters, additional enterococcal species may be present and yet not detected with the currently available genus- and group-specific qPCR assays. Better understandings of the molecular diversity and the occurrence of enterococcal species in fecal samples and environmental waters will be critical in the future evaluation studies of conventional and molecular detection methods used in the application of ambient microbial water quality recommendations. The approach used herein is also suitable when studying the fate and transport of targeted microbial groups in environmental waters and therefore in the improvement of current quantitative microbial risk assessment models. Future studies are needed to determine whether enterococcal species (group)-specific assays correlate better with risks than genus-specific assays and can then be of value in public health and environmental monitoring studies.
ACKNOWLEDGMENTS
We thank Jill Hoelle and Laura Boczek for providing the bacterial strains we used as reference strains and Brandon Iker for technical assistance. We also thank Dana Kolpin and Ed Furlong (U.S. Geological Survey) for access to the water samples we used for the isolation of some of the environmental enterococcal strains.
The U.S. Environmental Protection Agency, through its Office of Research and Development, funded and managed or partially funded and collaborated in the research described herein. This work has been subjected to the agency's administrative review and has been approved for external publication. Any opinions expressed in this paper are those of the authors and do not necessarily reflect the views of the agency; therefore, no official endorsement should be inferred. Any mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print 19 October 2012
REFERENCES
- 1. Muller T, Ulrich A, Ott EM, Muller M. 2001. Identification of plant-associated enterococci. J. Appl. Microbiol. 91:268–278 [DOI] [PubMed] [Google Scholar]
- 2. Badgley BD, Nayak BS, Harwood VJ. 2010. The importance of sediment and submerged aquatic vegetation as potential habitats for persistent strains of enterococci in a subtropical watershed. Water Res. 44:5857–5866 [DOI] [PubMed] [Google Scholar]
- 3. Ferguson DM, Moore DF, Getrich MA, Zhowandai MH. 2005. Enumeration and speciation of enterococci found in marine and intertidal sediments and coastal water in southern California. J. Appl. Microbiol. 99:598–608 [DOI] [PubMed] [Google Scholar]
- 4. Moore DF, Guzman JA, McGee C. 2008. Species distribution and antimicrobial resistance of enterococci isolated from surface and ocean water. J. Appl. Microbiol. 105:1017–1025 [DOI] [PubMed] [Google Scholar]
- 5. American Public Health Association 2005. Standard methods for the examination of water and wastewater, 21st ed American Public Health Association, Washington, DC [Google Scholar]
- 6. Facklam R, Elliott JA. 1995. Identification, classification, and clinical relevance of catalase-negative, Gram-positive cocci, excluding the streptococci and enterococci. Clin. Microbiol. Rev. 8:479–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Manero A, Vilanova X, Cerda-Cuellar M, Blanch AR. 2002. Characterization of sewage waters by biochemical fingerprinting of Enterococci. Water Res. 36:2831–2835 [DOI] [PubMed] [Google Scholar]
- 8. Patel R, Piper KE, Rouse MS, Steckelberg JM, Uhl JR, Kohner P, Hopkins MK, Cockerill FR, Kline BC. 1998. Determination of 16S rRNA sequences of enterococci and application to species identification of nonmotile Enterococcus gallinarum isolates. J. Clin. Microbiol. 36:3399–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wheeler AL, Hartel PG, Godfrey DG, Hill JL, Segars WI. 2002. Potential of Enterococcus faecalis as a human fecal indicator for microbial source tracking. J. Environ. Qual. 31:1286–1293 [DOI] [PubMed] [Google Scholar]
- 10. Schoen ME, Ashbolt NJ. 2010. Assessing pathogen risk to swimmers at non-sewage impacted recreational beaches. Environ. Sci. Technol. 44:2286–2291 [DOI] [PubMed] [Google Scholar]
- 11. Gardner TJ, Fitzgerald C, Xavier C, Klein R, Pruckler J, Stroika S, McLaughlin JB. 2011. Outbreak of campylobacteriosis associated with consumption of raw peas. Clin. Infect. Dis. 53:26–32 [DOI] [PubMed] [Google Scholar]
- 12. Haugland RA, Siefring SC, Wymer LJ, Brenner KP, Dufour AP. 2005. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559–568 [DOI] [PubMed] [Google Scholar]
- 13. Ludwig W, Schleifer KH. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556–562 [DOI] [PubMed] [Google Scholar]
- 14. Maheux AF, Bissonnette L, Boissinot M, Bernier JL, Huppe V, Berube E, Boudreau DK, Picard FJ, Huletsky A, Bergeron MG. 2011. Method for rapid and sensitive detection of Enterococcus sp. and Enterococcus faecalis/faecium cells in potable water samples. Water Res. 45:2342–2354 [DOI] [PubMed] [Google Scholar]
- 15. Messer JW, Dufour AP. 1998. A rapid, specific membrane filtration procedure for enumeration of enterococci in recreational water. Appl. Environ. Microbiol. 64:678–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Santo Domingo JW, Siefring SC, Haugland RA. 2003. Real-time PCR method to detect Enterococcus faecalis in water. Biotechnol. Lett. 25:261–265 [DOI] [PubMed] [Google Scholar]
- 18. Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Forster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, Konig A, Liss T, Lussmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ryu H, Tran H, Ware M, Iker B, Griffin S, Edge T, Newmann N, Villegas EN, Santo Domingo JW. 2011. Application of leftover sample material from waterborne protozoa monitoring for the molecular detection of Bacteroidales and fecal source tracking markers. J. Microbiol. Methods 86:337–343 [DOI] [PubMed] [Google Scholar]
- 20. Ryu H, Griffith J, Hill S, Edge TA, Toledo-Hernandez C, Gonzalez-Nieves J, Santo Domingo JW. 2012. Comparison of gull feces-specific assays targeting the 16S rRNA genes of Catellicoccus marimammalium and Streptococcus spp. Appl. Environ. Microbiol. 78:1909–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jackson CR, Fedorka-Cray PJ, Barrett JB. 2004. Use of a genus- and species-specific multiplex PCR for identification of enterococci. J. Clin. Microbiol. 42:3558–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vermette CJ, Russell AH, Desai AR, Hill JE. 2010. Resolution of phenotypically distinct strains of Enterococcus spp. in a complex microbial community using cpn60 universal target sequencing. Microb. Ecol. 59:14–24 [DOI] [PubMed] [Google Scholar]
- 23. Ahmed W, Sidhu JP, Toze S. 2012. Speciation and frequency of virulence genes of Enterococcus spp. isolated from rainwater tank samples in Southeast Queensland, Australia. Environ. Sci. Technol. 46:6843–6850 [DOI] [PubMed] [Google Scholar]
- 24. Scott TM, Jenkins TM, Lukasik J, Rose JB. 2005. Potential use of a host associated molecular marker in Enterococcus faecium as an index of human fecal pollution. Environ. Sci. Technol. 39:283–287 [PubMed] [Google Scholar]
- 25. Mote BL, Turner JW, Lipp EK. 2012. Persistence and growth of the fecal indicator bacteria enterococci in detritus and natural estuarine plankton communities. Appl. Environ. Microbiol. 78:2569–2577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nayak BS, Badgley B, Harwood VJ. 2011. Comparison of genotypic and phylogenetic relationships of environmental Enterococcus isolates by BOX-PCR typing and 16S rRNA gene sequencing. Appl. Environ. Microbiol. 77:5050–5055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Grammenou P, Spiliopoulou I, Sazakli E, Papapetropoulou M. 2006. PFGE analysis of enterococci isolates from recreational and drinking water in Greece. J. Water Health 4:263–269 [PubMed] [Google Scholar]
- 28. Lleo MM, Bonato B, Tafi MC, Signoretto C, Boaretti M, Canepari P. 2001. Resuscitation rate in different enterococcal species in the viable but non-culturable state. J. Appl. Microbiol. 91:1095–1102 [DOI] [PubMed] [Google Scholar]
- 29. Layton BA, Walters SP, Lam LH, Boehm AB. 2010. Enterococcus species distribution among human and animal hosts using multiplex PCR. J. Appl. Microbiol. 109:539–547 [DOI] [PubMed] [Google Scholar]
- 30. Maraccini PA, Ferguson DM, Boehm AB. 2012. Diurnal variation in Enterococcus species composition in polluted ocean water and a potential role for the enterococcal carotenoid in protection against photoinactivation. Appl. Environ. Microbiol. 78:305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Viau E, Peccia J. 2009. Evaluation of the enterococci indicator in biosolids using culture-based and quantitative PCR assays. Water Res. 43:4878–4887 [DOI] [PubMed] [Google Scholar]
- 32. Lu J, Santo Domingo JW, Lamendella R, Edge T, Hill S. 2008. Phylogenetic diversity and molecular detection of bacteria in gull feces. Appl. Environ. Microbiol. 74:3969–3976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Frahm E, Obst U. 2003. Application of the fluorogenic probe technique (TaqMan PCR) to the detection of Enterococcus spp. and Escherichia coli in water samples. J. Microbiol. Methods 52:123–131 [DOI] [PubMed] [Google Scholar]
- 34. U.S. EPA Office of Water 2010. Method A: Enterococci in water by TaqMan® quantitative polymerase chain reaction (qPCR) assay. EPA-821-R-10-004. U.S. Environmental Protection Agency, Washington, DC [Google Scholar]
- 35. Arvanitidou M, Katsouyannopoulos V, Tsakris A. 2001. Antibiotic resistance patterns of enterococci isolated from coastal bathing waters. J. Med. Microbiol. 50:1001–1005 [DOI] [PubMed] [Google Scholar]
- 36. Svec P, Sedlacek I. 1999. Occurrence of Enterococcus spp. in waters. Folia Microbiol. (Praha) 44:3–10 [DOI] [PubMed] [Google Scholar]
- 37. Suzuki Y, Kanda N, Furukawa T. 2012. Abundance of Enterococcus species, Enterococcus faecalis and Enterococcus faecium, essential indicators of fecal pollution, in river water. J. Environ. Sci. Health A Tox. Hazard. Subst. Environ. Eng. 47:1500–1505 [DOI] [PubMed] [Google Scholar]
- 38. Svec P, Devriese LA, Sedlacek I, Baele M, Vancanneyt M, Haesebrouck F, Swings J, Doskar J. 2001. Enterococcus haemoperoxidus sp. nov. and Enterococcus moraviensis sp. nov., isolated from water. Int. J. Syst. Evol. Microbiol. 51:1567–1574 [DOI] [PubMed] [Google Scholar]
- 39. Svec P, Vancanneyt M, Devriese LA, Naser SM, Snauwaert C, Lefebvre K, Hoste B, Swings J. 2005. Enterococcus aquimarinus sp. nov., isolated from sea water. Int. J. Syst. Evol. Microbiol. 55:2183–2187 [DOI] [PubMed] [Google Scholar]
- 40. Svec P, Vancanneyt M, Koort J, Naser SM, Hoste B, Vihavainen E, Vandamme P, Swings J, Bjorkroth J. 2005. Enterococcus devriesei sp. nov., associated with animal sources. Int. J. Syst. Evol. Microbiol. 55:2479–2484 [DOI] [PubMed] [Google Scholar]
- 41. Carvalho MdGS, Shewmaker PL, Steigerwalt AG, Morey RE, Sampson AJ, Joyce K, Barrett TJ, Teixeira LM, Facklam RR. 2006. Enterococcus caccae sp. nov., isolated from human stools. Int. J. Syst. Evol. Microbiol. 56:1505–1508 [DOI] [PubMed] [Google Scholar]
- 42. Naser SM, Vancanneyt M, De Graef E, Devriese LA, Snauwaert C, Lefebvre K, Hoste B, Svec P, Decostere A, Haesebrouck F, Swings J. 2005. Enterococcus canintestini sp. nov., from faecal samples of healthy dogs. Int. J. Syst. Evol. Microbiol. 55:2177–2182 [DOI] [PubMed] [Google Scholar]
- 43. Gordon DM, Bauer S, Johnson JR. 2002. The genetic structure of Escherichia coli populations in primary and secondary habitats. Microbiology 148:1513–1522 [DOI] [PubMed] [Google Scholar]