Abstract
The U.S. EPA developed a sample concentration and preparation assay in conjunction with the total culturable virus assay for concentrating and measuring culturable viruses in source and drinking waters as part of the Information Collection Rule (ICR) promulgated in 1996. In an effort to improve upon this method, the U.S. EPA recently developed Method 1615: Measurement of Enterovirus and Norovirus Occurrence in Water by Culture and RT-qPCR. Method 1615 uses a culturable virus assay with reduced equipment and labor costs compared to the costs associated with the ICR virus method and introduces a new molecular assay for the detection of enteroviruses and noroviruses by reverse transcription-quantitative PCR. In this study, we describe the optimization of several new components of the molecular assay and examine virus recovery from ground, reagent-grade, and surface water samples seeded with poliovirus type 3 and murine norovirus. For the culturable virus and molecular assays, mean poliovirus recovery using the complete method was 58% and 20% in groundwater samples, 122% and 39% using low-titer spikes in reagent-grade water, 42% and 48% using high-titer spikes in reagent-grade water, and 11% and 10% in surface water with high turbidity, respectively. Murine norovirus recovery by the molecular assay was 30% in groundwater samples, less than 8% in both low- and high-titer spikes in reagent-grade water, and 6% in surface water with high turbidity. This study demonstrates the effectiveness of Method 1615 for use with groundwater samples and highlights the need for further research into its effectiveness with surface water.
INTRODUCTION
Human enteric viruses are one of the leading causes of nonbacterial gastrointestinal illness. In addition, they are capable of causing respiratory infections, meningitis, conjunctivitis, encephalitis, and paralysis (1–3). The enteroviruses and noroviruses are two types of enteric viruses that replicate within the gastrointestinal tract and are secreted through feces, to be spread by the fecal-oral route (1, 4, 5). Because of their association with feces, these viruses are often found in large numbers in sewage (3, 6). Depending on treatment efficacy, they potentially can be discharged with treated effluent, resulting in possible human exposure through drinking and recreational waters (3, 6). Both enteroviruses and noroviruses have been identified as the causative agents of waterborne outbreaks (5, 7, 8), and the U.S. EPA has listed enteroviruses and noroviruses on their Contaminant Candidate List 3 (CCL3) (http://www.epa.gov/safewater/ccl/ccl3.html) due to their potential public health threat. A rapid, inexpensive, and effective method is needed to measure the occurrences of these viruses in different water matrices so that the actual risk to human health can be better characterized.
In the 1990s, the U.S. EPA issued an Information Collection Rule (ICR) which required that source water be monitored for viruses in U.S. utilities serving over 100,000 customers (9). The rule helped to establish national data on virus levels in source water in the United States in order to evaluate the efficacy of current treatment requirements. For virus monitoring, the ICR virus method was developed, which included a large-volume filtration and concentration component, followed by the detection of infectious viruses by the Buffalo green monkey kidney (BGM) cell line using the total culturable virus assay (TCVA). The results of the ICR indicated that viral contamination of U.S. source waters is widespread. About 80% of drinking water treatment plants utilizing surface water had virus-positive source waters at least once during the 18-month study; overall, 24% of the ICR source water samples were virus positive (10). Since the monitoring period required by the ICR concluded, the ICR virus method has continued to be used, both nationally and internationally for the detection of viruses in water (6, 11–14). The limitations of the ICR virus method, however, are the high cost of sampling due to the type of filter used (Zetapor 1MDS; CUNO, Meriden, CT), the fact that the BGM cell line is selective for only a subset of enteric viruses, and the fact that nonculturable viruses, such as human noroviruses, are not detected since the method lacks a molecular component.
Recent advances in large-volume water sampling have introduced a number of different options for the filtering of viruses. Similar to the 1MDS filter, the NanoCeram (Argonide, Sanford, FL) filter has been shown to be equivalent in performance to the 1MDS (15–17), with the major advantage being that the cost of the filter is about 20% of the cost of the 1MDS filter. In an attempt to offset the cost of the method as well as improve overall virus detection, the U.S. EPA recently developed Method 1615: Measurement of Enterovirus and Norovirus Occurrence in Water by Culture and RT-qPCR (18). The advantages of Method 1615 are that it incorporates the use of the NanoCeram filter and decreases the number of cell culture replicates that are required by the ICR virus method's TCVA, thereby decreasing equipment, reagent, and labor costs. It also incorporates a molecular component which allows the detection of both enteroviruses and noroviruses.
In order to determine the risk of virus exposure through drinking water, a recent study was conducted in 14 small communities in Wisconsin that utilized untreated groundwater for drinking water and which met the requirements of current drinking water regulations (19, 20). Using the same molecular approach as that outlined in Method 1615, that study found viruses present frequently in source and distribution waters from these communities (20–22) and related the presence of virus to adverse health effects (19, 20), particularly when human norovirus genogroup I (GI) and enteroviruses were present. This demonstrates the usefulness of the molecular component of Method 1615.
The U.S. EPA has recently included enterovirus and norovirus among the contaminants for monitoring in the next phase of the Unregulated Contaminant Monitoring Regulation (UCMR) (23). The purpose of the UCMR is to identify and monitor contaminants that are not currently regulated but may pose a public health threat, to determine if they are present at levels which warrant regulation. Under UCMR 3, beginning in January 2013, the U.S. EPA will monitor small public systems utilizing untreated groundwater from vulnerable aquifers for enterovirus and norovirus using Method 1615.
This paper describes the evaluation of components of Method 1615 which have not previously been validated and reports the performance of the method using spiked ground, reagent-grade, and surface water samples. Water samples were spiked with known concentrations of poliovirus type 3 and murine norovirus (MNV), and the method was evaluated for performance by determining virus recovery. Due to the small amounts of stock human norovirus available in our laboratory, we used murine norovirus as a surrogate for human norovirus. Murine norovirus has been shown to be an effective surrogate for human norovirus (24); in addition, this norovirus genotype can be propagated in cell culture, which makes it possible to attain large quantities of virus for spiking experiments.
MATERIALS AND METHODS
Virus stocks and cell lines.
Poliovirus type 3 Sabin was obtained from Mark Borchardt, U.S. Department of Agriculture, Marshfield, WI, and propagated in BGM cells. Murine norovirus was obtained from H. W. Virgin, Washington University, St. Louis, MO, and propagated in mouse macrophage RAW 264.7 cells (ATCC TIB-71; ATCC, Manassas, VA).
Virus stocks were inoculated onto the appropriate cell line and allowed to grow for up to 14 days until cytopathic effects (CPE) were observed. Cultures then underwent three freeze-thaw cycles, low-speed centrifugation at 3,000 × g for 10 min to remove large cell debris, and then high-speed centrifugation at 10,000 × g for 20 min to remove small cell debris. The remaining supernatant containing virus stock was filter sterilized through a 0.22-μm filter (Acrodisc; Pall Corporation, Ann Arbor, MI) to remove bacteria and/or larger organisms and to disperse any viral clumps that may have formed during processing, diluted 1:1 with 1× phosphate-buffered saline (PBS), and divided into 1- to 2-ml aliquots for storage at −70°C for use in spiking experiments. Murine norovirus was quantified by a plaque assay on RAW cells, as described previously (25). Poliovirus was quantified by a TCVA on BGM cells, as described below.
Water samples.
Groundwater samples were obtained from three different water treatment facilities in or around Cincinnati, OH, and from one private well. Reagent-grade water samples were obtained from an ultrapure water system at the U.S. EPA in Cincinnati, OH. Surface water samples were obtained from the Ohio River, Cincinnati, OH.
Sampling procedure.
All water samples were collected by using a five-inch NanoCeram cartridge filter (Argonide, Sanford, FL) (18). A minimum of 1,500 liters was filtered in the field for two replicate groundwater samples at each of three different sampling sites. Two sampling events occurred, on different dates, per groundwater site. Two additional 700-liter groundwater samples were filtered at a private well. At each groundwater sampling site, one 10-liter groundwater sample was collected into a collapsible plastic cubitainer (Cole-Parmer, Vernon Hills, IL) and used for laboratory spiking experiments. In addition, a 1-liter groundwater sample was taken and used for obtaining water sample characteristics. For surface water samples, a minimum of 80 liters of surface water was filtered for two replicate samples and one process blank. Two 10-liter surface water samples were collected into a collapsible plastic cubitainer (Cole-Parmer) and were transported back to the laboratory for spiking experiments. In addition, a 1-liter surface water sample was collected for determining water sample characteristics. For reagent-grade water samples, four 10-liter samples were collected into collapsible plastic cubitainers on separate dates, and three of the four samples were used for spiking experiments, with the fourth sample being a process blank.
Sample spiking experiments, elution, and concentration.
Following the filtration of the groundwater or surface water samples in the field, all filters were transported on ice to the laboratory. Within 24 h of collection, the 10-liter cubitainer samples were spiked with both poliovirus and murine norovirus at concentrations of approximately 3 × 106 most probable number (MPN) and 5 × 106 PFU, respectively. Spiked 10-liter viral suspensions were mixed and then filtered through one of the replicate NanoCeram filters containing the appropriate volume of water previously filtered in the field. For reagent-grade water samples, six 10-liter cubitainer samples were spiked with poliovirus at an MPN of 300 and with 300 PFU of murine norovirus (low titer) and filtered through the NanoCeram filter. In addition, six 10-liter cubitainers were spiked with poliovirus at an MPN of 1,000 and with 1,000 PFU of murine norovirus (high titer) and then filtered. For each separate sampling batch, one blank (unspiked) filter was run with each set of spiked filters to measure any background levels of virus in the water samples.
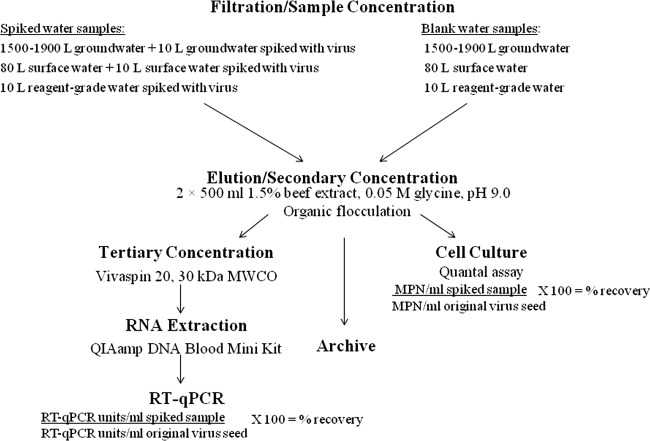
Spiked filters and blanks were eluted and concentrated according to Method 1615 (18). Briefly, elution was performed by adding 500 ml of 1.5% desiccated beef extract (BD Bacto, Franklin Lakes, NJ) containing 0.05 M glycine (pH 9.0) onto the filter in the filter housing and allowing it to soak for 1 min. Using positive pressure, the beef extract was then forced through the filter, and the eluent was collected into a beaker. The filter was again submerged in an additional 500 ml of 1.5% beef extract–0.05 M glycine (pH 9.0) and allowed to soak a second time for 15 min. While the filter was soaking, the pH of the first eluent was dropped to 7 to 7.5 to ensure that no viral inactivation occurred. After the 15-min soak, the remaining beef extract eluent was forced through the filter and added to the first eluent. The pH of the combined eluent was then adjusted to 3.5, and the eluent was allowed to mix slowly for 30 min to allow beef extract proteins to form a floc to which viral particles adsorbed. The sample was then centrifuged for 15 min at 2,500 × g at 4°C to collect the floc. Following centrifugation, the supernatant was discarded, and the resulting pellet containing viral particles was dissolved in 30 ml of 0.15 M sodium phosphate (pH 9.0). The resuspended pellet was again centrifuged at 4,000 × g for 10 min at 4°C to remove any nondissolved materials from the sample. The supernatant was retained, the pH was adjusted to 7 to 7.5, and the supernatant was then sterilized by using a 0.22-μm sterilizing filter (Acrodisc). The final sample was divided into three portions: one for the TCVA, one for tertiary concentration and subsequent molecular analysis, and one for archiving purposes. Figure 1 demonstrates the work flow for sample processing and analysis.
Fig 1.
Flow diagram used in this study for evaluation of Method 1615.
Tertiary concentration.
Several components of Method 1615 were tested separately prior to the evaluation of the complete method in order to identify the most efficient viral concentration and detection assays. These included determining if particulates would have an effect on the sterilizing filter and/or the Vivaspin-20 (Sartorius-Stedim, Aubagne, France) ultrafilter performance and the overall recovery of murine norovirus and poliovirus from Vivaspin-20 ultrafilters. After optimizing these method components, water samples were analyzed for recovery using the complete method.
Vivaspin-20 ultrafilters were used for the tertiary concentration of samples prior to RNA extraction and reverse transcription-quantitative PCR (RT-qPCR). Since the upstream processing of water samples using NanoCeram filters and organic flocculation has the potential to also concentrate particulates from a water sample, it was important to determine if particulates would have a clogging effect on either the sterilizing filter or the Vivaspin-20 ultrafilter. In order to determine if particulates would have an effect, 300 nephelometric turbidity units (NTU) of clay, Tennessee River water sediment, or diatomaceous earth were added to 10 ml of PBS (pH 7 to 7.5) and then processed by filter sterilization through a 0.22-μm filter, followed by Vivaspin concentration according to Method 1615 and as described below.
Following particulate testing, an initial evaluation using Vivaspin-20 ultrafilters with seeded virus compared 100-kDa-molecular-mass-cutoff and 30-kDa-molecular-mass-cutoff ultrafilters to determine which type produced the highest level of recovery, according to the manufacturer's recommendations, with some modifications. Vivaspin-20 filters were soaked overnight with PBSA (PBS with 0.05% bovine serum albumin [BSA]) at 4°C. Prior to the addition of the sample, the PBSA was removed. Poliovirus and murine norovirus stocks were diluted in 0.15 M sodium phosphate (pH 7). Poliovirus was spiked at a low titer of 2.6 × 103 MPN/ml with a murine norovirus spike at 3 × 104 PFU/ml in a total volume of 200 ml. Eight replicates of a 10-ml viral suspension were concentrated by each of the 100,000- and 30,000-molecular-weight-cutoff (MWCO) Vivaspin-20 ultrafilters. Ultrafilters were centrifuged at 3,000 × g for 10 min. Centrifugation was repeated until the sample volume was less than 0.4 ml, whereupon the sample was washed with 1 ml of 0.15 M sodium phosphate (pH 7). This step was repeated a second time, and centrifugation was repeated until the sample volume was less than 0.4 ml. The final sample was transferred from the Vivaspin filter into a clean 1.5-ml microcentrifuge tube, and the final volume was brought to 400 μl with 0.15 M sodium phosphate (pH 7). Using the same concentration steps, an additional Vivaspin comparison was performed in quadruplicate by using poliovirus at a high-titer concentration of 2.6 × 105 MPN/ml, with murine norovirus at the same concentration described above. All final sample concentrates underwent nucleic acid extraction by using a QIAamp DNA Blood minikit (Qiagen, Valencia, CA), as described below, and analyzed by using the GSFTQM primer/probe set (Table 1), as described below.
Table 1.
RT-qPCR primers and probes
| Primer or probea | Sequence (5′→3′)b | Sequence target region (positions)c | Reference |
|---|---|---|---|
| EntFc | CCTCCGGCCCCTGAATG | 444–460 | 26 |
| EntRc | ACCGGATGGCCAATCCAA | 638–621 | |
| EntPc | CGGAACCGACTACTTTGGGTGTCCGT | 532–557 | |
| GSFTQMup | CCCTGAATGCGGCTAAT | 452–468 | This paper |
| GSFTQMlow | TGTCACCATAAGCAGCCA | 594–577 | |
| GSFTQMprober | ACGGACACCCAAAGTAGTCGGTTC | 557–534 | |
| MuNoVF1 | AGATCAGCTTAAGCCCTATTCAGAAC | 3737–3752 | This paper |
| MuNoVR1 | CAAGCTCTCACAAGCCTTCTTAAA | 3884–3861 | |
| MuNoVP1 | TGGCCAGGGCTTCTGT | 3860–3845 | |
| HepF | CGGCCAAAAGGTGGTGGATG | 100–119 | 27 |
| HepR | CGACGAGCCTGACGTCGGG | 285–267 | |
| HepP | AGGTCCCTCTGGCGCTTGTGGCGAG | 172–196 |
The following primers and probes were used: enterovirus forward primers EntF and GSFTQMup, enterovirus reverse primers EntR and GSFTQMlow, enterovirus probes EntP and GSFTQMprober, murine norovirus forward primer MuMoV F1, murine norovirus reverse primer MuMoV R1, murine norovirus probe MuNoV P1, hepatitis G virus forward primer HepF, hepatitis G virus reverse primer HepR, and hepatitis G virus probe HepP. For TaqMan probes, EntP, GSFTQMprober, and HepP are labeled on the 5′ end with 6-carboxyfluorescein (6-FAM) and on the 3′ end with 6-carboxytetramethylrhodamine (TAMRA). The MuNov P1 probe is labeled on the 5′ end with VIC and on the 3′ end with MGB.
Degenerate bases in primers and probes are as follows: N equals a mixture of all four nucleotides, R equals A and G, Y equals T and C, and W equals A and T.
GenBank accession numbers for sequence targets are NC_002058 for enterovirus, AY228235 for murine norovirus, and U44402 for hepatitis G virus primers HepF and HepR; data for the hepatitis G virus probe can be found at http://www.asuragen.com/pdfs/AR_42024.pdf.
For analysis of Method 1615 in its entirety using spiked ground, reagent-grade, and surface water samples, the original sample volume was used to determine how much of the 30-ml concentrated sample (after organic flocculation concentration) should be used in the tertiary concentration, as described by Method 1615 (18). For most samples, this was approximately one-third of the final 30-ml concentrated sample. These samples were tertiary concentrated by using the Vivaspin-20 30,000-MWCO filter, as described above, to a final concentrated volume of 400 μl and analyzed by using the Ent primer/probe set, the MuNoV primer/probe set, and the Hep primer/probe set, as described below.
Nucleic acid extraction.
Initially, several nucleic acid extraction kits and methods were compared. These included NucliSENS (bioMérieux, Durham, NC), the QIAamp MinElute Virus Spin kit with or without carrier RNA (Qiagen), the QIAamp DNA Blood minikit (Qiagen), and heat extraction (28). The manufacturers' instructions were followed for the NucliSENS and MinElute kits. For the QIAamp DNA Blood minikit, buffer AVL (Qiagen) was substituted for buffer AL, as previously described by Lambertini and colleagues (22). Briefly, 200 μl of sample was added to a tube containing 200 μl of buffer AVL with carrier RNA, and the mixture was briefly vortexed before incubation at 56°C for 10 min. Following a brief centrifugation at >5,000 × g, 200 μl of ethanol was added, and the sample was again centrifuged briefly at >5,000 × g. The mixture was then transferred into a QIAamp Minispin column and centrifuged at 6,000 × g for 1 min. Five hundred microliters of buffer AW1 was added, and the mixture was centrifuged at 6,000 × g for 1 min. Five hundred microliters of buffer AW2 was added, and the mixture was centrifuged at 20,000 × g for 3 min. Fifty microliters of buffer AE was added to the column, which was then transferred into a clean tube containing 40 units of RNase inhibitor (Promega, Madison, WI) and incubated at room temperature for 1 min, followed by centrifugation at 6,000 × g for 1 min. An additional elution with 50 μl was performed with buffer AE, and the final sample was collected and stored at −70°C until reverse transcription could be performed. All concentrated ground, reagent-grade, and surface water samples were extracted according to the procedure described above for the QIAamp DNA Blood minikit.
RT-qPCR procedures.
Initially, an independent, in-house RT-qPCR method was employed for preliminary evaluations of several Method 1615 components using the GSFTQM primer/probe set, which targets enteroviruses (Table 1). Reverse transcription was carried out in triplicate by adding 5 μl of sample to a microplate well containing (final concentration) 1× PCR buffer II (Applied Biosystems, Foster City, CA), 1.5 mM MgCl2 (Applied Biosystems), 0.66 mM deoxynucleoside triphosphates (dNTPs) (Promega), and 500 nM lower primer GSFTQMlow (Table 1). Following a 5-min heat release at 99°C, 50 units of murine leukemia virus (MuLV) RT (Applied Biosystems) and 53.3 units of RNase inhibitor (Promega) were added to each well. RT reactions were carried out at 43°C for 60 min, followed by 94°C for 5 min, and then held at 4°C until PCR could be performed (the hold time was less than 24 h). PCRs were carried out by adding 20 μl of a PCR mix that contained (final concentration) 1× PCR buffer II (Applied Biosystems), 10.25 mM MgCl2 (Applied Biosystems), 500 nM GSFTQMup (Table 1), 2.5 units of AmpliTaq Gold (Applied Biosystems), 1 μl/reaction of ROX reference dye (Invitrogen, Grand Island, NY), and 100 nM the GSFTQMprober PCR probe (Table 1). PCR was carried out under cycling conditions of 95°C for 10 min followed by 45 cycles of 94°C for 15 s and 60°C for 1 min.
For analysis of the complete method using ground, reagent-grade, and surface water samples, the Ent primer/probe set, the MuNoV primer/probe set, and the Hep primer/probe set were used (Table 1), which target enterovirus, murine norovirus, and hepatitis G virus, respectively. Reverse transcription was carried out by adding 6.7 μl of each sample to a microplate well containing 10 ng/μl of random primers (Promega), 1 μl of hepatitis G virus Armored RNA (Asuragen, Austin, TX) as an inhibition control, and PCR-grade water to a final volume of 16.5 μl. This random priming approach is the same as that used in Method 1615 and is designed to maximize the range of environmental enteroviruses and noroviruses that can be detected. Following incubation at 99°C for 4 min to release the hepatitis G virus RNA, the samples were quickly cooled, and 16.8 μl of an RT mix containing (final concentration) 10 mM Tris (pH 8.3), 50 mM KCl, 3 mM MgCl2, 0.8 mM dNTPs, 10 mM dithiothreitol (DTT) (Promega), 0.5 units/μl of RNase inhibitor (Promega), and 1.6 units/μl of SuperScript II RT (Promega) was added to each well of a 96-well plate. RT reactions were carried out in triplicate under cycling conditions of 25°C for 15 min, 42°C for 60 min, and 99°C for 5 min, followed by a 4°C hold cycle. Following RT, 6 μl of the RT sample was transferred onto an optical PCR plate containing PCR mix. For enterovirus PCR, the mix contained (final concentration) 10 μl of 2× LightCycler 480 Probes Master Mix (Roche, Indianapolis, IN), 1 μl/reaction of ROX reference dye (Invitrogen), 300 nM primer EntF, 900 nM primer EntR, 100 nM probe EntP, and PCR-grade water to a final volume of 14 μl. For murine norovirus PCR, the mix contained (final concentration) 10 μl of 2× LightCycler 480 Probes Master Mix (Roche), 1 μl/reaction of ROX reference dye (Invitrogen), 100 nM primer MuNoVF1, 100 nM primer MuNoVR1, 100 nM probe MuNoVP1, and PCR-grade water to a final volume of 14 μl. For the hepatitis G virus inhibition control assay, the PCR mix contained (final concentration) 10 μl of 2× LightCycler 480 Probes Master Mix (Roche), 1 μl/reaction of ROX reference dye (Invitrogen), 500 nM primer HepF, 500 nM primer HepR, 100 nM probe HepP, and PCR-grade water to a final volume of 14 μl. PCRs were carried out in triplicate under cycling conditions of 95°C for 10 min followed by 45 cycles of 95°C for 15 s and 60°C for 1 min.
Determination of inhibition using hepatitis G virus Armored RNA.
Methods for the preparation and application of the hepatitis G virus RNA internal inhibition standard have been detailed in Method 1615 and by Gibson et al. (18, 29). In short, buffer AE (Qiagen) was used to prepare 1:5 and 1:25 dilutions of RNA extracts from each test water sample. The inhibition of RT-qPCR was then evaluated for both undiluted and diluted RNA extracts by spiking a known amount of hepatitis G virus Armored RNA into the RT master mix. In addition, a minimum of 10 hepatitis G virus-positive control reaction mixtures containing only hepatitis G virus RNA and no sample were prepared. After heat release, reverse transcription, and amplification using the hepatitis G virus qPCR assay, the relative amounts of hepatitis G virus were measured in all test water samples, sample dilutions, and hepatitis G-positive control reaction mixtures. These amounts were compared and evaluated for differences in threshold cycle (CT) values, that is, the number of temperature cycles at which the target nucleic acid has been sufficiently amplified to reach a defined threshold. A test sample or sample dilution was considered inhibited if the CT of the spiked hepatitis G virus was higher by at least 1 cycle than the mean CT obtained from the hepatitis G virus-positive control reactions. The cDNA from the undiluted test sample, if not inhibited, or from the lowest dilution that did not show inhibition was then used for the quantitation of enteroviruses and MNV by qPCR.
Standard curve approach for quantitation of target viral RNA.
For quantity estimations of poliovirus and murine norovirus by RT-qPCR, a 10-fold dilution series of each virus stock was prepared and extracted as described above. The extracted viral RNA was used for the generation of a standard curve using RT-qPCR and spanned 5 logs. The poliovirus type 3 dilutions included 105 to 101 MPN per RT-qPCR, while the quantity of murine norovirus ranged from 103 to 10−1 PFU per RT-qPCR. A standard curve was produced with every plate of samples analyzed, and a linear regression model was used to evaluate the quality of amplification. The RT-qPCR data were accepted if the results of the linear regression of the standard curve met the criteria defined in Method 1615, which include an amplification efficiency of ≥80% and an R2 value of ≥0.97 (18).
TCVA.
Stock cultures of BGM cells were planted in 2,100-cm2 expanded surface roller bottles (Nalge Nunc International, Rochester, NY) at a concentration of approximately 4 × 105 cells/ml. Cell culture media contained equal parts (50:50) of minimum essential medium Eagle (MEM; Sigma-Aldrich, St. Louis, MO) and Leibovitz's L-15 medium (Sigma-Aldrich) supplemented with 0.67% sodium bicarbonate and 10% calf serum (HyClone, Pittsburg, PA). Cultures for the TCVA were grown on 25-cm2 plastic culture vessels (Greiner, Monroe, NC) in 10 ml of the same media described above at a concentration of approximately 3.5 × 106 cells/ml and were inoculated 4 days after planting.
Rates of recovery of culturable poliovirus from spiked ground, reagent-grade, and surface water samples were determined by using the TCVA described by Method 1615. Prior to all infections, BGM cells were washed with 8 ml of Earle's balanced salts solution. Each sample and viral seed underwent 5-fold dilutions, and each dilution series was divided among 10 replicate BGM cell culture flasks. Three to four dilution series per sample were inoculated. Depending on the original sample volume that was filtered, and as described in Method 1615, approximately 1 ml of inoculum was introduced into each flask, and all flasks were rocked gently for a minimum of 90 min before 10 ml of maintenance medium containing 50:50 MEM and L-15 with 2% serum and antibiotic-antimycotic liquid (Invitrogen) was added. All cultures were incubated for 14 days at 37°C and were checked at least twice weekly for CPE. Those flasks which exhibited 75 to 100% CPE were frozen immediately and marked as positive. Within a dilution series of each sample, the set of flasks containing the lowest dilution that had all cultures positive for CPE, and all subsequent dilutions thereafter, underwent at least one additional passage for the confirmation of CPE. Passage of the samples consisted of freeze-thawing cell culture lysates and filter sterilization of the lysate to ensure that CPE was not caused by bacterial contamination, and then 1 ml of the lysate was inoculated onto fresh BGM cells. The second passage of samples was incubated for an additional 2 weeks; those cultures that were negative upon the first passage but positive upon the second passage were then passed a third time to confirm positive results. The MPN was determined for each sample based on the number of confirmed positive replicates in the 5-fold dilution series for each of the samples; in addition, the MPN was determined for the original poliovirus seed used to spike the 10-liter samples. The MPN of samples was compared to the MPN of the original poliovirus seed in order to determine recovery values. The MPN software used can be found at http://www.epa.gov/nerlcwww/online.html#vis.
Testing of water sample characteristics.
Several biotic and abiotic water quality parameters were examined to characterize the samples. Turbidity was measured by using a LaMotte 2020e turbidity meter (LaMotte, Chesterton, MD), and results are expressed as NTU. Total coliforms and Escherichia coli were measured by using the Colilert test kit (IDEXX, Westbrook, ME) with the Quanti-Tray enumeration procedure, according to the manufacturer's instructions; results are reported as MPN/100 ml. Enterococci were measured by using the Enterolert test kit (IDEXX) with the Quanti-Tray enumeration procedure, according to the manufacturer's instructions; results are reported as MPN/100 ml. The sample pH was measured by using a Corning 440 pH meter (Tewksbury, MA). Total and free chlorine concentrations were measured by using a Hach TNT kit and a DR2800 portable spectrophotometer (Hach, Loveland, CO), according to the manufacturer's instructions; results are reported as mg/liter. Conductivity was measured by using an Oakton CON6 meter (Cole-Parmer), and results are reported as microsiemens (μS). Analyses of total organic carbon and total nitrogen concentrations were performed with a Shimadzu TOC-Vcph/TNM-1 total organic carbon analyzer with a total nitrogen measuring unit (Shimadzu, Columbia, MD), and results are reported as mg/liter for both analytes. For all tests, the calibration of instruments and quality control samples was performed as appropriate.
Statistical evaluation.
All data underwent statistical analyses for a log-normal generalized linear model in SAS, ver. 9 (SAS Institute, Cary, NC). Post hoc multiple comparisons among virus types and matrices were evaluated for statistical significance via the Holms test at a group-wise alpha level of 0.05. Comparisons were limited to only the group comprised of contrasts between either like virus types or like matrices (n = 30).
RESULTS
The initial evaluation of Vivaspin-20 ultrafilters with particulates demonstrated that in all cases where particulates were added, samples were successfully passed through both the sterilizing filters and the Vivaspin-20 ultrafilters, with the particulates collecting primarily on the sterilizing filter. This suggests that samples with high particulate concentrations should not affect the tertiary concentration since the particulates are collected primarily on the sterilizing filter before tertiary concentration (data not shown).
Further evaluation of the recovery of poliovirus and murine norovirus through Vivaspin-20 filters with molecular weight cutoffs of 100,000 and 30,000 resulted in no statistically significant difference in recovery between the two different-MWCO Vivaspin-20 ultrafilters (P > 0.01). For 100,000-MWCO ultrafilters, poliovirus recovery averaged 30% for low-titer spikes and 82% for high-titer spikes, with coefficients of variation (CVs) of 27% and 87%, respectively (Table 2). For 30,000-MWCO ultrafilters, recoveries of poliovirus were 48% for low-titer spikes and 114% for high-titer spikes (Table 2), with CVs of 10% and 36%, respectively. For murine norovirus, recoveries averaged 44% for 30,000-MWCO ultrafilters, with a CV of 15%, and 24% for 100,000-MWCO ultrafilters, with a CV of 18% (Table 2). Because the 30,000-MWCO Vivaspin-20 ultrafilter had better overall recoveries as well as overall lower coefficients of variation across the titers tested, it was selected for use with Method 1615.
Table 2.
Mean percent recoveries of poliovirus and murine norovirus from Vivaspin-20 ultrafilters by RT-qPCR
| Vivaspin type (MWCO) | Poliovirus |
Murine norovirusc (n = 12) |
||||
|---|---|---|---|---|---|---|
| High titera (n = 4) |
Low titerb (n = 8) |
Recovery (%) | CV (%) | |||
| Recovery (%) | CV (%) | Recovery (%) | CV (%) | |||
| 30,000 | 114 | 36 | 48 | 10 | 44 | 15 |
| 100,000 | 82 | 87 | 30 | 27 | 24 | 18 |
The high poliovirus titer is 2.6 × 105 MPN/ml.
The low poliovirus titer is 2.6 × 103 MPN/ml.
The murine norovirus titer is 3 × 104 PFU/ml.
Comparisons of kits and heat release indicated that comparable performances in the extraction of poliovirus RNA could be achieved for all extraction protocols (data not shown).
Two out of seven groundwater samples (28.6%) showed inhibition according to the hepatitis G virus internal inhibition RT-qPCR assay. Inhibition in the sample from the private well was relieved by diluting the RNA extract 25-fold, and a 1:5 dilution was sufficient to alleviate inhibition in a sample from a local water treatment facility. The recoveries of poliovirus from spiked groundwater samples using the complete method averaged 58% by the TCVA, with a range of 8 to 114%, and 20% by the molecular assay, with a range of 9 to 42% (Table 3). For murine norovirus, the recovery from groundwater samples averaged 30%, with a range of 7 to 58% (Table 3). No viruses were detected by the TCVA or molecular assays in blanks for any of the groundwater samples.
Table 3.
Mean percent recoveries of spiked poliovirus and murine norovirus from ground, surface, and reagent-grade water samples
| Assay | Groundwater (n = 7) |
Surface water (n = 2) |
Reagent-grade water |
|||||
|---|---|---|---|---|---|---|---|---|
| Recovery (%) | CV (%) | Recovery (%) | CV (%) | High titera (n = 6) |
Low titerb (n = 6) |
|||
| Recovery (%) | CV (%) | Recovery (%) | CV (%) | |||||
| Poliovirus quantal assay | 58 | 79 | 11 | 47 | 42 | 34 | 122 | 96 |
| Poliovirus RT-qPCR | 20 | 64 | 10 | 6 | 48 | 36 | 39 | 29 |
| Murine norovirus RT-qPCR | 30 | 75 | 6 | 6 | 0.6 | 100 | 8 | 83 |
High-titer spike with poliovirus at a 1,000 MPN and with 1,000 PFU of murine norovirus per 10-liter sample.
Low-titer spike with poliovirus at a 300 MPN and with 300 PFU of murine norovirus per 10-liter sample.
Due to the turbidity of the surface water samples (37 NTU), only 80 liters of water was able to be filtered before filters became clogged. Both surface water samples contained RT-qPCR inhibitors based on the results of the hepatitis G virus inhibition assay. After the RNA extracts were diluted 1:25, inhibition in the surface water samples was mitigated. For the two spiked surface water samples, poliovirus recoveries averaged 11% for the TCVA and 10% for the molecular assay; for murine norovirus, recoveries averaged 6% (Table 3). No culturable viruses were detected in the blank from surface water; however, by the molecular assay, approximately 244 RT-PCR units/liter of enterovirus were found.
As a final step, we determined the recovery of spiked poliovirus and murine norovirus from reagent-grade water. Six 10-liter reagent-grade water samples were spiked with a “low” titer of approximately 300 MPN of poliovirus and 300 PFU of murine norovirus and analyzed by the complete method. A separate set of six samples was tested in the same way with a “high” titer of approximately 1,000 MPN of poliovirus and 1,000 PFU of murine norovirus. For low-titer spikes, the poliovirus recoveries averaged 122% for the TCVA, with a range of 55% to over 100%, and 39% for the molecular assay, with a range of 25 to 54% (Table 3). Murine norovirus recovery from low-titer spikes averaged 8% by the molecular assay, with a range of 2 to 16%. For high-titer spikes, poliovirus recoveries averaged 42% for the TCVA, with a range of 23 to 65%, and 48% for the molecular assay, with a range of 25 to 69% (Table 3). The murine norovirus recovery from high-titer spikes averaged <1%. No culturable viruses were detected in blanks for reagent-grade water samples. There was no statistical significance in recovery between high and low titers of poliovirus by the quantal assay or RT-qPCR in reagent-grade water samples. In addition, of all the different water samples tested, the recovery of murine norovirus from reagent-grade water samples was significantly lower than that from all other water types tested (P < 0.05).
For all ground and surface water samples, water quality data were collected to determine if any correlations existed between water quality parameters and virus recovery. Table 4 shows all water quality data for ground and surface water samples. The pH for all samples ranged from 7 to 8, and free chlorine and total chlorine concentrations were <0.05 mg/liter for all groundwater samples. The level of total coliforms ranged from an MPN of <1 to 90/100 ml in groundwater samples, with the exception of the sample from the private well, which contained 1.1 × 104 MPN/100 ml, and the surface water sample, which contained 1.3 × 103 MPN/100 ml. E. coli and enterococci were detected in the private well sample and a surface water sample at 42 MPN/100 ml and 87 MPN/100 ml for E. coli, respectively, and 16 MPN/100 ml and 12 MPN/100 ml for enterococci, respectively. E. coli and enterococci were detected at an MPN of <1/100 ml in the other groundwater samples. The total organic carbon concentrations ranged from 0.7 to 2.0 mg/liter for all groundwater samples and from 1.2 to 2.4 mg/liter for surface water samples. Turbidity measurements ranged from 0 to 0.6 NTU for groundwater samples and were as high as 37 NTU for the surface water samples, and for all ground and surface water samples, the conductivity ranged from 400 to 1,900 μS/cm, and the total nitrogen concentration ranged from 0.5 to 3.5 mg/liter. No correlation was found between any of the water quality parameters and virus recovery (P > 0.05).
Table 4.
Ground and river water characteristicsa
| Site | pH | Free chlorine concn (mg/liter) | Total chlorine concn (mg/liter) | Concn of total coliforms (MPN/100 ml) | E. coli concn (MPN/100 ml) | Concn of enterococci (MPN/100 ml) | Total organic carbon concn (mg/liter) | Turbidity (NTU) | Conductivity (μS) | Total nitrogen concn (mg/liter) |
|---|---|---|---|---|---|---|---|---|---|---|
| GW1 | ND | ND | ND | 10462 | 41.95 | 16.38 | ND | ND | ND | ND |
| GW2A | 8.1 | <0.05 | <0.05 | <1 | <1 | <1 | 1.89 | 0.28 | 1,079 | 0.52 |
| GW2B | 7.74 | <0.05 | <0.05 | 91.5 | <1 | <1 | 1.6 | 0 | 1,235 | 0.48 |
| GW3A | 7.1 | <0.05 | <0.05 | 11.8 | <1 | <1 | 0.66 | 0.63 | 1,343 | 3.52 |
| GW3B | 7.8 | <0.05 | <0.05 | <1 | <1 | <1 | 1.22 | 0.55 | 1,220 | 3.43 |
| GW4A | 7.44 | <0.05 | <0.05 | 49.4 | <1 | <1 | 1.4 | 0.17 | 1,852 | 2.48 |
| GW4B | 7.72 | <0.05 | <0.05 | <1 | <1 | <1 | 1.18 | 0.028 | 1,901 | 2.47 |
| RW | 7.63 | ND | ND | 1290 | 86.5 | 12 | 2.38 | 36.9 | 375 | 1.19 |
GW, groundwater; RW, river water; ND, not done.
DISCUSSION
This study describes the development and initial evaluation of EPA Method 1615 (18). Like the EPA ICR method for culturable viruses (9), Method 1615 uses a positively charged filter for sample collection and analysis by the TCVA. The advantage of Method 1615 over the ICR method is that it has included the option of a more affordable filter, the NanoCeram filter; in addition, Method 1615 has added tertiary concentration and subsequent molecular detection components to allow the detection of both culturable and nonculturable human enteroviruses and nonculturable human noroviruses; finally, reagent and supply costs have been lowered by decreasing the number of replicates needed for the TCVA.
The QIAamp DNA Blood minikit was selected for inclusion in Method 1615. This kit was previously shown to work effectively on environmental water samples (21, 22), and of the kits tested, it was the most economical, the easiest to use, and the least labor-intensive compared to the other kits.
Recently, Ikner and colleagues employed the Centricon 30,000-MWCO ultrafilter for the secondary concentration of samples (16). In our study, the Vivaspin-20 ultrafilter was employed for tertiary concentrations. Results were very comparable between the ultrafilter employed by Ikner and colleagues and the ultrafilter used in the present study. Ikner et al. reported recoveries of 95% for poliovirus using the Centricon units, and we found recoveries of 114% for poliovirus using the Vivaspin-20 30,000-MWCO ultrafilters. For Method 1615, the Vivaspin-20 ultrafilter is an attractive option for the concentration of viruses prior to RT-qPCR. Like the Centricon units, Vivaspin-20 ultrafilters have a dead-stop volume of 50 μl, which ensures that the entire sample can be concentrated and will not be lost through the ultrafilter. However, the approach used in our study and in Method 1615 allows more of the original sample to be analyzed by RT-qPCR. The Vivaspin-20 ultrafilter is a non-labor-intensive and easily performed tertiary concentration technique resulting in good recovery from seeded samples. The only disadvantage that we observed with the Vivaspin-20 ultrafilters is that for some samples, additional centrifugation was required to achieve the required concentrated sample volume, resulting in a longer processing time. Although we did not encounter a high level of inhibition in our samples, it is important to note that PCR inhibitors potentially can be co-concentrated by the Vivaspin-20 ultrafilters, which highlights the need for kit extraction prior to RT-qPCR. While nucleic acid extraction kits do not necessarily remove all PCR inhibitors, they often remove some of the inhibitory organic materials through the processing steps; in addition, kit extraction concentrates the sample so that more nucleic acid is available per PCR.
The use of the NanoCeram filter for concentrating viruses from water samples has been documented several times recently. Karim and colleagues were able to show that the NanoCeram filter was effective at concentrating enteroviruses (poliovirus type 1, coxsackievirus B5, and echovirus 7) and Norwalk virus, with recoveries of 51% from tap water and 38% from river water for enteroviruses using a plaque assay and 4% from tap water and 12% from river water for Norwalk virus using RT-PCR (17). This is very similar to our results from groundwater (58%), reagent-grade water (42 to 122%), and surface water (11%) for poliovirus using the TCVA and from groundwater (29%) and surface water (9%) for murine norovirus using RT-qPCR.
Recently, Ikner and colleagues were able to demonstrate the effectiveness of the NanoCeram filter in concentrating several virus types from tap water (16). Twenty-liter spiked samples showed an overall recovery rate of 66% for poliovirus, which is comparable to data from both the previous study by Karim et al. (17) and our current study. In the current study and in Method 1615, 1.5% beef extract with 0.05 M glycine (pH 9.0) is employed as the elution buffer. In the studies by both Karim et al. and Ikner and colleagues, the elution solutions used were slightly different. Karim et al. used a nonflocculating beef extract which requires the addition of celite for secondary concentration; Ikner and colleagues employed a 1% sodium polyphosphate elution buffer. Additionally, Karim et al. employed celite for secondary concentration, Ikner et al. used Centricon units for secondary concentration, and we used organic flocculation for secondary concentration. Method 1615 and our current study employed the TCVA, the same approach which was used for the ICR (30), whereas a viral plaque assay was used in the studies by both Karim et al. and Ikner and colleagues. The advantage to the use of the TCVA is that some wild-type viruses will cause visible CPE on cells, but when the cells are covered with an agar overlay medium, as used in a plaque assay, the CPE is not able to be visualized, as oftentimes no plaques form. Thus, the TCVA is more sensitive than the plaque assay (31). Despite the differences in water types and volumes filtered, elution solutions, secondary concentration techniques employed, and cell culture assays used, the recoveries of poliovirus were all comparable between our study, the study by Karim et al., and the study by Ikner et al., highlighting the ability of the NanoCeram filter to efficiently adsorb and desorb poliovirus as well as the stability of the virus in the different processing approaches. This also underscores the need for investigations of potential alternatives that could be included in Method 1615, to allow for alternative elution solutions and secondary concentration procedures that may be tailored to suit the viral target, water matrix, volume, or analytical laboratory's needs.
For poliovirus in groundwater and for the low-titer spikes of poliovirus in reagent-grade water, the recovery rates were higher using the TCVA than those obtained by using RT-qPCR. This could be due to the loss of virus at the Vivaspin-20 concentration and nucleic acid extraction steps. These extra steps are not employed prior to the TCVA and could contribute to the loss of virus, thus resulting in slightly lower percent recoveries obtained by using RT-qPCR. In addition, the amount of original sample that was assayed by RT-qPCR was lower than that assayed by the TCVA. For RT-qPCR, approximately 1/100 of the original sample was assayed. In contrast, approximately one-third of the original sample was assayed by cell culture. The poliovirus recovery from reagent-grade water was greater than 100% and could be due to viral aggregation, as was observed previously in other studies (32, 33). While we tried to eliminate this as a variable when preparing our stocks, at lower titers, it can be more pronounced, thus causing recoveries to appear inflated.
Due to our laboratory's limited supply of human noroviruses, murine norovirus was used as a surrogate for human noroviruses in our evaluation of Method 1615. The advantage to the use of murine norovirus is that high titers can be obtained via cell culture and can be used for spiking experiments; in contrast, the only source of human noroviruses is fecal samples, typically samples obtained through human volunteer studies and/or clinical samples obtained through medical diagnostic laboratories. The disadvantage of the use of murine norovirus is that it may perform differently than human norovirus in Method 1615. Despite the disadvantages, murine norovirus has been demonstrated to be an acceptable surrogate for human norovirus (24). In addition, our mean recovery rate of 29% for murine norovirus in groundwater is similar to that found previously for human noroviruses by Lambertini and colleagues, at 29% (21), and slightly higher than the 18% recovery that was demonstrated previously by Lee and colleagues (34). In addition, the studies by both Karim et al. and Lee and colleagues demonstrated the effectiveness of the NanoCeram filter in recovering human noroviruses from spiked water samples (17, 34).
The percent recovery of murine norovirus from reagent-grade water in our study was significantly low for both low- and high-titer spikes (P < 0.01 in both cases). Gibbons and colleagues previously reported recoveries of >96% for seeded norovirus GII.4 in 40 liters of seawater filtered by the NanoCeram filter (15). This is currently the highest reported percent recovery for noroviruses using the NanoCeram filter and currently the only study to describe the use of the NanoCeram filter for recovering noroviruses from seawater. The high recovery rates that the study by Gibbons et al. reported suggest a possible benefit of salts in aiding norovirus adsorption and elution, as previous studies used tap water, river water, or distilled water (17, 34).
Following the publication of the study by Gibbons et al., Da Silva and colleagues demonstrated various adsorption and aggregation properties of human norovirus virus-like particles (VLPs) (35). Da Silva et al. reported that an increased ionic strength in solutions of NaCl improved the adsorption of both norovirus GI.1 and GII.4 to silica. In addition, the pH had a strong influence on both the adsorption and aggregation of VLPs. Da Silva and colleagues also observed that at pH 9.0, the VLP capsid structure appeared disassembled, suggesting that a high pH may negatively affect these viruses. Since the beef extract solution used in our study has a pH of 9.0, this could negatively affect recovery. While VLPs may respond differently than intact viral particles, these are important observations that should be considered when evaluating norovirus recovery data. Taken together, this adds further evidence to the benefits of salts in the overall adsorption and elution of noroviruses and could help to explain why our recovery was low for murine norovirus from the reagent-grade water samples and higher for groundwater samples (mean conductivity of 1,438 μS).
Lee and colleagues recently reported recoveries of 18% for murine norovirus from distilled water using NanoCeram filters eluted with 1.5% beef extract and 0.05 M glycine (34), which is the same elution buffer used in our study. Our recovery from reagent-grade water was lower, despite the lack of salts in both the study by Lee et al. and our study. This difference in recovery could be due to the fact that we used cartridge filters, while Lee et al. used flat disc filters; in addition, we filtered 10-liter samples, whereas Lee et al. used 1-liter samples. Lee and colleagues also found that the addition of Tween 80 increased the overall recovery of murine norovirus slightly, although not significantly, and they were able to demonstrate that the NanoCeram filter in combination with 1.5% beef extract–0.05 M glycine could recover 27% of seeded norovirus GII.4. This rate could be increased to 85% with the addition of 0.01% Tween to the elution buffer. The low percent recoveries of murine norovirus from reagent-grade water that we report warrant further research efforts, particularly investigations into the addition of salts and/or Tween 80 to enhance adsorption and elution. Despite these low recoveries, Method 1615 recommends the use of reagent-grade water samples for performance evaluation (PE) testing, and PE testing will be used only for analyses of spiked poliovirus samples.
Method 1615 is intended to be used for the detection of enteroviruses and noroviruses from environmental and/or finished drinking water. Our current study examined the recovery of poliovirus and murine norovirus from ground, reagent-grade, and surface water samples. Our experience with a limited number of surface water samples demonstrated that the NanoCeram filter clogged after only 80 liters of water was filtered, when the turbidity was at 37 NTU. Method 1615 suggests the use of a prefilter in such cases or the use of the 1MDS filter, which typically allows more water to be collected at higher turbidities. Further research into the use of the NanoCeram filter in more turbid samples is warranted.
In January 2013, the U.S. EPA will begin monitoring under UCMR 3. Method 1615 will be used in this large national study in order to isolate and identify enteroviruses and/or noroviruses in nondisinfected groundwater sites located in karst aquifers. This study demonstrates the effectiveness of Method 1615 in recovering seeded virus from groundwater samples. By decreasing the cost of sampling and the TCVA, and with the addition of a molecular assay for the detection of enteroviruses and noroviruses, Method 1615 will be an effective tool for gathering important data on the occurrence of enteroviruses and noroviruses. Future research should focus on increasing the rate of recovery of noroviruses from reagent-grade water, increasing the rate of recovery of enteroviruses and noroviruses from surface waters, adding other filtration/concentration approaches, including integrated cell culture (ICC)-PCR approaches, and adding detection assays for other virus types, such as human adenoviruses, for a future revision of Method 1615.
ACKNOWLEDGMENTS
We acknowledge Mary Jean See, Nancy Schable, and Jenifer Jones of Dynamac Corporation for the preparation of BGM cultures, Gretchen Sullivan for assistance in the preparation of stock laboratory reagents, Mohammad Karim for the propagation of murine norovirus stocks, and local private well owners and utilities for allowing us to collect water samples.
Although this work was reviewed by the EPA and approved for publication, it may not necessarily reflect official agency policy. Mention of trade names or commercial products does not constitute endorsement or recommendation for use.
Footnotes
Published ahead of print 19 October 2012
REFERENCES
- 1. Rajtar B, Majek M, Palanski L, Polz-Dacewicz M. 2008. Enteroviruses in water environment—a potential threat to public health. Ann. Agric. Environ. Med. 15:199–203 [PubMed] [Google Scholar]
- 2. Sawyer MH. 2002. Enterovirus infections: diagnosis and treatment. Semin. Pediatr. Infect. Dis. 13:40–47 [DOI] [PubMed] [Google Scholar]
- 3. Sinclair RG, Jones EL, Gerba CP. 2009. Viruses in recreational water-borne disease outbreaks: a review. J. Appl. Microbiol. 107:1769–1780 [DOI] [PubMed] [Google Scholar]
- 4. Glass RI, Parashar UD, Estes MK. 2009. Norovirus gastroenteritis. N. Engl. J. Med. 361:1776–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wyn-Jones AP, Sellwood J. 2001. Enteric viruses in the aquatic environment. J. Appl. Microbiol. 91:945–962 [DOI] [PubMed] [Google Scholar]
- 6. Sedmak G, Bina D, MacDonald J, Couillard L. 2005. Nine-year study of the occurrence of culturable viruses in source water for two drinking water treatment plants and the influent and effluent of a wastewater treatment plant in Milwaukee, Wisconsin (August 1994 through July 2003). Appl. Environ. Microbiol. 71:1042–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Parshionikar SU, Willian-True S, Fout GS, Robbins DE, Seys SA, Cassady JD, Harris R. 2003. Waterborne outbreak of gastroenteritis associated with a norovirus. Appl. Environ. Microbiol. 69:5263–5268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoder J, Roberts V, Craun GF, Hill V, Hicks L, Alexander NT, Radke V, Calderon RL, Hlavsa MC, Beach MJ, Roy SL. 2008. Surveillance for waterborne disease and outbreaks associated with drinking water and water not intended for drinking—United States, 2005-2006. MMWR Surveill. Summ. 57:39–62 [PubMed] [Google Scholar]
- 9. US EPA 1996. Information Collection Rule technical summary, vol EPA 811-F-96–004 US EPA, Washington, DC [Google Scholar]
- 10. Shaw S, Regli S, Chen J. 2002. Virus occurrence and health risks in drinking water, p 437–462 In McLain JL, McGuire MJ, Obolensky A. (ed), Information Collection Rule data analysis. AWWA Research Foundation and American Water Works Association, Denver, CO [Google Scholar]
- 11. Hill VR, Polaczyk AL, Kahler AM, Cromeans TL, Hahn D, Amburgey JE. 2009. Comparison of hollow-fiber ultrafiltration to the USEPA VIRADEL technique and USEPA method 1623. J. Environ. Qual. 38:822–825 [DOI] [PubMed] [Google Scholar]
- 12. Hsu BM, Chen CH, Kung CM, Wan MT, Shen SM. 2007. Evaluation of enterovirus recovery in surface water by different adsorption and elution procedures. Chemosphere 66:964–969 [DOI] [PubMed] [Google Scholar]
- 13. Johnson TB, McKay LD, Layton AC, Jones SW, Johnson GC, Cashdollar JL, Dahling DR, Villegas LF, Fout GS, Williams DE, Sayler G. 2011. Viruses and bacteria in karst and fractured rock aquifers in East Tennessee, USA. Ground Water 49:98–110 [DOI] [PubMed] [Google Scholar]
- 14. Polaczyk AL, Roberts JM, Hill VR. 2007. Evaluation of 1MDS electropositive microfilters for simultaneous recovery of multiple microbe classes from tap water. J. Microbiol. Methods 68:260–266 [DOI] [PubMed] [Google Scholar]
- 15. Gibbons CD, Rodriguez RA, Tallon L, Sobsey MD. 2010. Evaluation of positively charged alumina nanofibre cartridge filters for the primary concentration of noroviruses, adenoviruses and male-specific coliphages from seawater. J. Appl. Microbiol. 109:635–641 [DOI] [PubMed] [Google Scholar]
- 16. Ikner LA, Soto-Beltran M, Bright KR. 2011. New method using a positively charged microporous filter and ultrafiltration for concentration of viruses from tap water. Appl. Environ. Microbiol. 77:3500–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Karim MR, Rhodes ER, Brinkman N, Wymer L, Fout GS. 2009. New electropositive filter for concentrating enteroviruses and noroviruses from large volumes of water. Appl. Environ. Microbiol. 75:2393–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fout GS, Brinkman NE, Cashdollar JL, Griffin SM, McMinn BR, Rhodes ER, Varughese EA, Karim MR, Grimm AC, Spencer SK, Borchardt MA. 2010. Method 1615: measurement of enterovirus and norovirus occurrence in water by culture and RT-qPCR, publication no EPA/600/R-10/181. US Environmental Protection Agency, Cincinnati, OH [Google Scholar]
- 19. Borchardt M, Belongia E, Kieke B, Loge F. 2003. Community-randomized intervention trial with UV disinfection for estimating the risk of pediatric illness from municipal groundwater consumption. US Environmental Protection Agency, Washington, DC [Google Scholar]
- 20. Borchardt MA, Spencer SK, Kieke BJ, Lambertini E, Loge FJ. 2012. Viruses in non-disinfected drinking water from municipal wells and community incidence of acute gastrointestinal illness. Environ. Health Perspect. 120:1272–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lambertini E, Spencer SK, Bertz PD, Loge FJ, Kieke BA, Borchardt MA. 2008. Concentration of enteroviruses, adenoviruses, and noroviruses from drinking water by use of glass wool filters. Appl. Environ. Microbiol. 74:2990–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lambertini E, Spencer SK, Kieke BA, Jr, Loge FJ, Borchardt MA. 2011. Virus contamination from operation and maintenance events in small drinking water distribution systems. J. Water Health 9:799–812 [DOI] [PubMed] [Google Scholar]
- 23. US EPA 2011. Revisions to the Unregulated Contaminant Monitoring Regulation (UCMR 3) for public water systems. EPA OW-2009-0090 US EPA, Washington, DC [Google Scholar]
- 24. Wobus C, Thackray LB, Virgin HW. 2006. Murine norovirus: a model system to study norovirus biology and pathogenesis. J. Virol. 80:5104–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wobus C, Karst SM, Thackray LB, Chang KO, Sosnovtsev SV, Belliot G, Mackenzie JM, Green KY, Virgin HW., IV 2004. Replication of norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2:e432 doi:10.1371/journal.pbio.0020432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. De Leon R, Shieh YSC, Baric R, Sobsey MD. 1990. Detection of enteroviruses and hepatitis A virus in environmental samples by gene probes and polymerase chain reaction, p 833–853 In Water Quality Technology Conference Proceedings AWWA, Denver, CO [Google Scholar]
- 27. Schlueter V, Schmolke S, Stark K, Hess G, Ofenloch-Haehnle B, Engel AM. 1996. Reverse transcription-PCR detection of hepatitis G virus. J. Clin. Microbiol. 34:2660–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fout GS, Martinson BC, Moyer MW, Dahling DR. 2003. A multiplex reverse transcription-PCR method for detection of human enteric viruses in groundwater. Appl. Environ. Microbiol. 69:3158–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gibson KE, Schwab KJ, Spencer SK, Borchardt MA. 2012. Measuring and mitigating inhibition during quantitative real-time PCR analysis of viral nucleic acid extracts from large-volume environmental water samples. Water Res. 46:4281–4291 [DOI] [PubMed] [Google Scholar]
- 30. Fout GS, Schaefer FS, III, Messer JW, Dahling DR. 1996. ICR microbial laboratory manual. US EPA, Washington, DC [Google Scholar]
- 31. Morris R, Waite WM. 1980. Evaluation of procedures for recovery of viruses from water. II. Detection systems. Water Res. 14:795–798 [Google Scholar]
- 32. Floyd R, Sharp DG. 1979. Aggregation of poliovirus and reovirus by dilution in water. Appl. Environ. Microbiol. 38:395–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gassilloud B, Gantzer C. 2005. Adhesion-aggregation and inactivation of poliovirus 1 in groundwater stored in a hydrophobic container. Appl. Environ. Microbiol. 71:912–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee H, Kim M, Paik SY, Lee CH, Jheong WH, Kim J, Ko G. 2011. Evaluation of electropositive filtration for recovering norovirus in water. J. Water Health 9:27–36 [DOI] [PubMed] [Google Scholar]
- 35. Da Silva AK, Kavanagh OV, Estes MK, Elimelech M. 2011. Adsorption and aggregation properties of norovirus GI and GII virus-like particles demonstrate differing responses to solution chemistry. Environ. Sci. Technol. 45:520–526 [DOI] [PMC free article] [PubMed] [Google Scholar]