Abstract
Degradation of terephthalate (TA) through microbial syntrophy under moderately thermophilic (46 to 50°C) methanogenic conditions was characterized by using a metagenomic approach (A. Lykidis et al., ISME J. 5:122–130, 2011). To further study the activities of key microorganisms responsible for the TA degradation, community analysis and shotgun proteomics were used. The results of hierarchical oligonucleotide primer extension analysis of PCR-amplified 16S rRNA genes indicated that Pelotomaculum, Methanosaeta, and Methanolinea were predominant in the TA-degrading biofilms. Metaproteomic analysis identified a total of 482 proteins and revealed a distinctive distribution pattern of microbial functions expressed in situ. The results confirmed that TA was degraded by Pelotomaculum spp. via the proposed decarboxylation and benzoyl-coenzyme A-dependent pathway. The intermediate by-products, including acetate, H2/CO2, and butyrate, were produced to support the growth of methanogens, as well as other microbial populations that could further degrade butyrate. Proteins related to energy production and conservation, and signal transduction mechanisms (that is, chemotaxis, PAS/GGDEF regulators, and stress proteins) were highly expressed, and these mechanisms were important for growth in energy-limited syntrophic ecosystems.
INTRODUCTION
Terephthalate (TA) is produced in large quantities in plastic, textile, and petroleum-related industries, and wastewater generated by the TA manufacturing process is often treated with anaerobic methanogenic processes (1). In these processes, TA is converted to CH4 and CO2 through the microbial syntrophy established among the enriched microbial populations. It is hypothesized that the hydrogen-producing syntrophic bacteria degrade TA to acetate and H2/CO2, which are immediately used by the methanogenic archaea adjacent to final gaseous products (2, 3). To validate the syntrophic interaction, a full-cycle 16S rRNA approach was used to successfully identify the identities of the syntrophic bacteria and methanogenic archaea inside the granules and biofilms of different laboratory-scale TA-degrading methanogenic reactors (3, 4). A recent study further used a metagenomic approach to reveal the diversity and physiological traits of the syntrophs and methanogens involved in the TA degradation under moderately thermophilic conditions (46 to 50°C) (5). All of these studies have provided strong evidence to elucidate the degradation of TA based on the microbial populations and the metabolic pathways involved.
Furthermore, to directly observe the global expression of microbial functions within the TA-degrading consortium, community-level proteomics can be used (6). To do so, total proteins recovered from an environmental sample are first separated and enzymatically fragmented. The resulting peptide mixture is analyzed using advanced liquid chromatography-mass spectrometry (LC-MS) technology, and information related to the key proteins is identified using in silico analysis of the protein sequence database. Thus far, the metaproteomic approach has been successfully applied to analyze the expression of key microbial functions in various environments, including enhanced biological phosphorus removal reactors (7), activated sludge (8), and acid mine drainage sites (9), as well as under soil (10) and marine conditions (11). In the present study, a metaproteomic approach was used to assess the key microbial functions of syntrophs and methanogens involved in TA degradation in an anaerobic fixed-film reactor operated at 50°C. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was used for protein fractionation before a downstream high-throughput detection of proteins using nano-LC coupled to an LTQ-Orbitrap mass spectrometer. This approach was as effective as using two-dimensional gel electrophoresis, which has been extensively used to achieve protein separation (12). By further combining this approach with microbial community analysis and a metagenomic database (5), we were able to closely examine the metabolic pathways used by predominant populations of the TA-degrading syntrophic biofilm.
MATERIALS AND METHODS
Anaerobic fixed-film bioreactor and biofilm samples.
A laboratory-scale anaerobic fixed-film bioreactor (height, 65 cm; diameter, 8 cm; working volume, 2.56 liters) was fabricated with double-layer Pyrex glass and contained 213 ceramic rings (Sera Siporax) (see Fig. S1 in the supplemental material). The bioreactor was inoculated with 250 ml of TA-degrading sludge derived from a previous study (5) and 250 ml of anaerobic sludge obtained from a full-scale upflow anaerobic sludge bed reactor used to treat TA-containing wastewater. The bioreactor was fed with a synthetic medium (pH 7.0) (4) containing TA as the sole substrate and was operated with a hydraulic retention time of 2 to 4 days and an upflow velocity of 7.6 ml/min in the dark. The temperature of the reactor was controlled at 50°C by circulating the heated water through the water jacket. Ceramic rings covered with thick biofilm were collected at days 99, 195, 217, 253, and 464 from the top of the reactor, frozen in liquid nitrogen, and preserved at −20°C before analysis. Dissolved total organic carbon (TOC) concentrations of influent and effluent were routinely monitored using a TOC analyzer (model 1020A; OI Corp., College Station, TX).
DNA extraction and PCR amplification.
The biofilm-covered ceramic rings were grounded in an earthen bowl prechilled with liquid nitrogen, and a TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8]) was added to suspend the biofilm biomass to a concentration of 300 to 600 mg/ml. The cells were subjected to enzymatic and chemical treatments, and the genomic DNAs were purified and precipitated by using phenol-chloroform-isoamyl alcohol and ethanol precipitation procedures (13). 16S rRNA gene fragments of the domains Bacteria and Archaea, were individually obtained by using PCR amplification with the commonly used bacterial (EU27F/U1500R) and archaeal (A109F/U1492R) primer pairs (3, 4), respectively. Approximately 50 ng of genomic DNA was used in a 100-μl PCR mixture and subjected to a thermal program: 95°C for 5 min, 25 cycles of DNA denaturation at 95°C for 45 s, annealing at 52°C for 30 s, extension at 72°C for 1 min, and a final extension at 72°C for 10 min in a thermocycler (Biometra, Germany). The amplicons were then purified using the QIAquick PCR purification kit (Qiagen GmbH, Hilden, Germany), and the concentration was measured and stored at −20°C.
HOPE analysis.
Hierarchical oligonucleotide primer extension (HOPE) assay was conducted to determine the relative abundance of targeted microbial populations in the reactor (14, 15). The reaction in a solution of 5 μl, containing 1.5 μM (each) the unlabeled primers (Table 1), 10 fmol of the purified DNA, and 2.5 μl of the ready-to-use premix from the SNaPshot multiplex kit (Applied Biosystems, USA), was conducted in a thermocycler (Biometra, Germany) for 20 cycles of 96°C for 5 s, 62°C for 10 s, and 72°C for 5 s. The products were treated with shrimp alkaline phosphatase (USB Affymetrix, USA) at 37°C to minimize the influence of unincorporated dye terminators before the analysis of capillary electrophoresis using a ABI Prism 3130 genetic analyzer (Applied Biosystems) as described previously (14). All of the HOPE experiments for each sample were performed in triplicate. The parameters of the HOPE analysis, such as the specificity of group-specific primers, working annealing temperature of the single-base primer extension reaction, and multiplexing analysis, were experimentally determined as described previously (19, 20).
Table 1.
Oligonucleotide primers used in the hierarchical oligonucleotide primer extension assay to determine the relative abundance of microbial populations in the terephthalate-degrading biofilm in this study
| Primer | Target | Sequence (5′–3′)a | Length (nt) |
Extension |
Tube | Source or reference | ||
|---|---|---|---|---|---|---|---|---|
| Binding | Tail | Allele | Color | |||||
| EUB338F | Bacteria | (gtac)7ACTCCTACGGGAGGCAG | 17 | 28 | C | Black | 1,2 | Modified from Amann et al. (16) |
| TA828f | Syntrophorhabdus | GGTGTGGGAGGTGTAATA | 18 | 0 | C | Black | 1 | This study |
| DFM230r | Pelotomaculum | GCTAATGGGACGCGGACC | 18 | 0 | C | Black | 1 | Modified from Loy et al. (17) |
| OP5-80f | Caldisericum | TCTCTGATAAGGGTGCTGGC | 20 | 0 | G | Blue | 1 | This study |
| Syb459f | Syntrophobacter | GGAGGAATATGCTCTGTG | 18 | 0 | A | Green | 2 | This study |
| Syn742r | Syntrophus | CGCGTCTCAGCGTCAGTATAGGA | 23 | 0 | C | Black | 2 | This study |
| OP8-466r | Candidate division OP8 | GAGGGTACCGTCAGTCCCT | 19 | 0 | T | Red | 2 | This study |
| MCD823f | Methylocaldum | AACTAGCCGTTGGGCACAAT | 20 | 0 | T | Red | 2 | This study |
| TA758f | Candidate division WWE1 | (atgc)2AACTGCGAAGGTGTGGGGAT | 20 | 8 | C | Black | 2 | This study |
| ARC911fm | Archaea | (cgta)6TAAAGGAATTGGCGCGG | 17 | 24 | G | Blue | 3 | Modified from Narihiro et al. (18) |
| MX802f | Methanosaeta | GTCCTAGCCGTAAACGTAA | 19 | 0 | C | Black | 3 | This study |
| ML398fm | Methanolinea | CCCGAGTGCCCGTAAATTC | 19 | 0 | G | Blue | 3 | This study |
Lowercase letters represent tail sequences.
The oligonucleotide primers used in the HOPE assay were designed using the ARB package with an aligned 16S rRNA sequence database (SSU reference version 96) downloaded from SILVA (http://www.arb-silva.de/). The bacterial primers were designed to target the domain Bacteria (EUB338F), Pelotomaculum group (DFM230r), Syntrophorhabdus group (TA828f), Syntrophus group (Syn742r), Syntrophobacter group (Syb459f), uncultured Caldiserica group (OP5-80f), Methylocaldum group (MCD823f), uncultured OP8 group (OP8-466r), and uncultured WWE1 group (TA785f). The archaeal primers ARC911fm, MX802f, and ML398fm were designed to target the domain Archaea, Methanosaeta, and Methanolinea groups, respectively (Table 1 and see Fig. S2 in the supplemental material). The group-specific primers were designed based on key criteria, including primer properties (i.e., direction, extended nucleotides, length, GC content, and melting temperature), mismatched bases, and mismatched positions (19). The specificity of each primer was validated in silico using the Probe Match tool provided by the Ribosomal Database Project (http://rdp.cme.msu.edu/probematch/search.jsp) and was experimentally confirmed using perfect-match and mismatched reference DNAs at various annealing temperatures (55 to 65°C) (19, 20). Thus, the relative abundances of targeted microbial groups within the Bacteria and Archaea domains could be analyzed using three multiplexing tubes for the HOPE reactions (Table 1).
Recovery of total protein from biofilm.
The biofilm samples were frozen in liquid nitrogen and grounded as described previously. The powder was mixed with 1 ml of a lysis buffer (CelLytic B; Sigma), 25 μl of dithiothreitol (DTT; 1 M, pH 5.2), and 10 μl of a protease inhibitor (Sigma) and was subsequently subjected to five cycles of freeze (−80°C)-thaw (60°C) treatment. The total protein was purified using phenol extraction (21) and stored in acetone at −20°C overnight. The protein was precipitated through centrifugation, and resuspended in a buffer containing 7 M urea, 2 M thiourea, and 4% (wt/vol) CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}. The concentration was quantified by using a 2D-Quant kit (GE-Amersham Biosciences).
Protein fractionation using SDS-PAGE.
The protein mixture was fractionated using an SDS-PAGE method (22). Approximately 100 μg of the total protein extract was mixed with a buffer (5:1 [vol/vol]) containing 0.35 M Tris-HCl (pH 6.8), 10% SDS, 30% glycerol, 9.3% DTT, and 0.2% bromophenol blue and was then denatured in boiling water for 5 min. Next, the samples were dispensed into three separate wells (30 μg each) in a polyacrylamide gel (4% acrylamide stacking gel, 12% acrylamide gel) and electrophoresed at 12°C in Protean II xi 2-D Cell (Bio-Rad) at 200 V for 1 h, 400 V for 1 h, and 600 V for 15 to 30 min. The banding patterns were visualized by using silver staining (23). The image of the stained gel was recorded using the U:Genius system (Syngene). The triplicate strips of the stained gel were excised into 17 sections for subsequent proteomic analysis.
In-gel digestion and peptide extraction.
Prior to in-gel protein digestion, the gel slices were washed with sterilized MiniQ water three times and destained in a solution containing 15 mM potassium ferricyanide and 50 mM sodium thiosulfate for 30 min. Next, the gel slices were dehydrated with acetonitrile and air dried. The protein molecules in the gels were digested enzymatically into short peptide fragments with trypsin (Trypsin Gold, mass spectrometry grade; Promega) in a buffer solution containing 40 mM ammonium hydrogen carbonate and 10% acetonitrile at 37°C for 16 h. After centrifugation at 12,000 × g for 10 min, the supernatant was transferred to new tubes, whereas the gel particles were incubated with 50 μl of an extraction solution containing 50% acetonitrile and 5% trifluoroacetic acid (TFA), and centrifuged at 12,000 × g for 10 min. The peptides generated in the supernatant were recovered and combined together. The combined supernatant was evaporated in a centrifugal evaporator (DNA 120 Savant SpeedVac concentrator; Thermo Scientific, Rochester, NY), and the proteolytic peptides were dried and stored at −20°C before analysis.
Nano-HPLC-MS/MS analysis.
The proteolytic peptides were first desalted by using ZipTip C18 pipette tips (Millipore) and eluted with 5 μl of a solvent containing 0.1% TFA and 70% acetonitrile. Next, 1 μl of a purified peptide mixture was analyzed by using an LTQ Orbitrap XL (Thermo Fisher Scientific, San Jose, CA) coupled with a Nanoflow LC system (Agilent Technologies, Santa Clara, CA). A total of 8 μl from the desalted digest solution was loaded onto an Agilent Zorbax XDB C18 precolumn (0.3 by 5 mm, 5 μm), followed by separation using a C18 column (inner diameter, 75 μm by 15 cm, 3.5 μm; Zorbax 300SB-C18; Agilent). Buffer A (0.1% formic acid) and buffer B (0.1% formic acid in 98% acetonitrile) solutions were prepared for LC separation. An elution program was applied by using 5% of buffer B for 5 min, followed by a linear gradient from 5 to 35% of buffer B for 60 min and then 35% to 100% of buffer B for 5 min at a flow rate of 300 nl/min. The peptides were analyzed in the positive ion mode by electrospray ionization (spray voltage = 1.8 kV). The MS was operated in a data-dependent mode, in which one full scan with m/z 300 to 2,000 in the Orbitrap (R = 60,000 at m/z 400) was performed using a rate of 30 ms/scan. The five most intense peaks were selected for fragmentation with a normalized collision energy value of 35% in the LTQ, and a repeat duration of 3 min was applied to exclude the same m/z ions from the reselection for fragmentation.
Data processing and protein annotation.
The obtained spectra were processed using the software Proteome Discoverer v1.0 (Thermo Fisher Scientific) and the integrated microbial genomes and metagenomes (IMG/M) sequence database (http://img.jgi.doe.gov/cgi-bin/m/main.cgi) for TA-degrading biofilm (24). To facilitate bioinformatics analysis, multiple raw files of MS/MS spectra generated from analyses of each protein fraction were merged into a single contiguous input file. The combined data files were searched by using the protein databases and the SEQUEST algorithm using the following parameters: a precursor mass within ±5 ppm of the theoretical mass, a fragment ion tolerance of <0.8 Da, and a maximum of two missed cleavages allowed. The cross correction values (Xcorrs) versus charges > 1.5 (1+), 2.0(2+), and 2.5(3+), only counting rank 1 peptide and searching for the top protein were selected for filtering the output data set. The protein identity fulfilling the previously mentioned criteria was considered correct identification. For positive identification, a matching of at least two peptides was required for each protein (9).
The resulting data sets generated by the duplicate analyses were manually combined to compose an identification list. Annotation was performed using the stand-alone BLAST program combined with the Conserved Domain Database (CDD) and the Clusters of Orthologous Group (COG) database at the National Center for Biotechnology Information (25, 26), as well as the Pfam and TIGRfam databases (27, 28), to predict the protein functions. The reactions in which the proteins were likely involved in the metabolic pathways were inferred using the KEGG automatic annotation server (29). The results of identification and annotation were processed using Microsoft Office Excel 2007.
RESULTS AND DISCUSSION
Reactor performance.
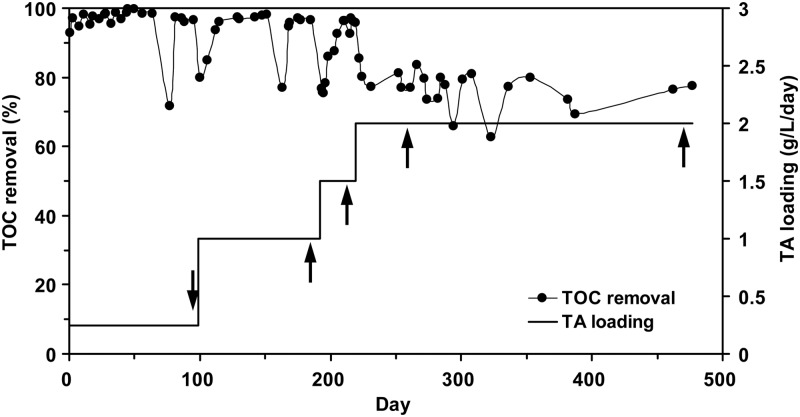
The anaerobic fixed-film reactor system was successfully operated with TA as the carbon and energy sources for ∼500 days. Figure 1 shows that the TA loadings were increased from 0.25 to 2.0 g of TA/liter/day by gradually increasing the TA concentrations from 1.0 to 4.0 g/liter. High removal efficiencies of dissolved TOC were achieved at a low TA loading rate of 0.25 g of TA/liter/day without any lag time. The removal efficiency was reduced when the TA loadings were increased each time. Eventually, the TOC removal efficiencies leveled off at ca. 70 to 80% at a TA loading rate of 2.0 g of TA/liter/day. Throughout the operation, biofilm samples on the surface of the media were periodically obtained and used for microbial community and proteome analyses.
Fig 1.
Operational performance of laboratory-scale anaerobic fixed-film bioreactor fed with synthetic metium that contained TA as the sole substrate at 50°C. The arrows showed the sampling time of biofilm samples.
Microbial population structure and dynamics.
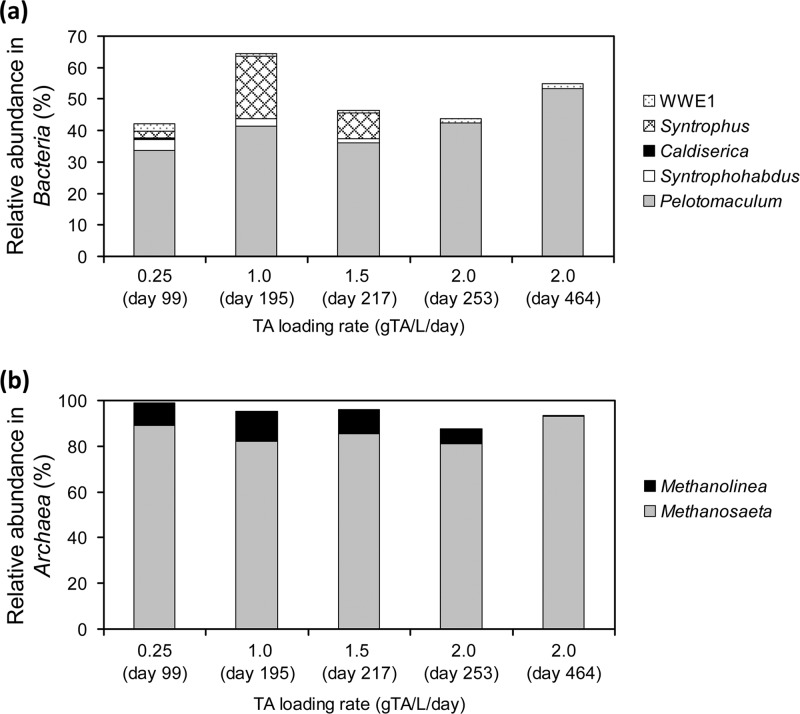
The microbial composition in the TA-degrading biofilm sampled at day 99 was analyzed by using the 16S rRNA gene clone library. The predominant microbial populations were found to include Pelotomaculum spp., the Caldiserica group (formerly known as candidate division OP5), Syntrophorhabdus spp., Methanosaeta spp., and Methanolinea spp. (see Fig. S2 in the supplemental material). Using HOPE, the population dynamics of key microbial populations found in the clone library (see Fig. S2 in the supplemental material) was determined in samples taken on days 99, 195, 217, 253, and 464 under various TA loading rates. Figure 2a shows that Pelotomaculum spp. with a relative abundance of 33.7 to 53.4% (standard deviations, 1.05 to 4.97%) represented the most predominant bacterial population in the TA-degrading biofilm during the operation. The Syntrophorhabdus-related group accounted for 3.4% ± 2.2% of the total bacterial population at a TA loading rate of 0.25 g of TA/liter/day, and this gradually decreased to undetectable levels (<0.1%) when the TA loading rates were increased to 2.0 g of TA/liter/day. The uncultured WWE1 group detected by the primer TA758f remained at low abundances between 0.8% ± 0.02% and 2.6% ± 0.13%. The Caldiserica group, which was highly abundant in the bacterial clone library (see Fig. S2 in the supplemental material), accounted for <0.6% of the total bacterial populations. This difference was likely due to the biases associated with PCR and cloning procedures (30), as well as the specificity of the HOPE primers designed to target members of the Caldiserica group. In contrast, the Syntrophus-related group was not detected in the cloning library but represented 19.8% ± 0.3% of the total bacterial populations at 1.0 g of TA/liter/day. The abundance of the Syntrophus-related group became undetectable at a loading of 2.0 g of TA/liter/day. Members associated with Syntrophobacter spp., Methylocaldum spp., and uncultured OP8 were not detected in the biofilm samples throughout the operation. Figure 2b indicates that Methanosaeta spp. (81.0 to 92.9% [0.25 to 3.53%]) and Methanolinea spp. (0.7 to 13.1% [0.14 to 1.86%]) accounted for almost all of the archaeal populations in the biofilm. The relative abundance of the Methanolinea group was observed to decline gradually with an increase in the TA loadings.
Fig 2.
Relative abundances of specific bacterial (a) and archaeal (b) populations in the syntrophic TA-degrading biofilms sampled at various TA loading rates. The relative abundances were analyzed with the group- and domain-specific primers in multiplexing hierarchical oligonucleotide primer extension (HOPE) analyses.
Proteome of TA-degrading community.
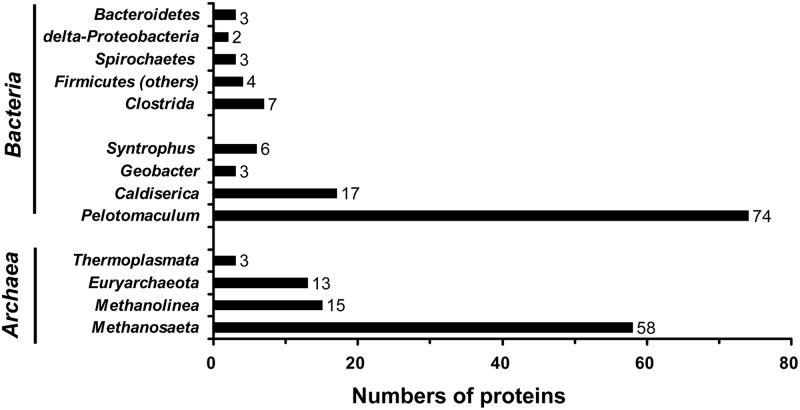
The total proteins extracted from TA-degrading biofilm sampled at day 464 were fractionated using one-dimensional gel electrophoresis (see Fig. S3 in the supplemental material) and were then analyzed by using a shotgun community proteomic approach. In total, 5,560 peptide fragments were detected from two independent experiments. These were assigned to 482 different proteins (54.8% showed positive identification); among them, 208 protein sequences were encoded by the genes located on the contigs or scaffolds of the known microbial populations (Fig. 3). The Pelotomaculum bin (74 proteins) and Methanosaeta bin (58 proteins) contained the highest numbers of identifiable proteins, followed by the uncultured Caldiserica group (17 proteins), Methanolinea (15 proteins), Syntrophus (6 proteins), and Geobacter (3 proteins). The identities of the remaining 35 proteins were distributed to five bacterial populations and two archaeal populations. The phylogeny distribution of microbial composition based on the proteomic analysis was highly similar to that based on the 16S rRNA gene clone library.
Fig 3.
Numbers of proteins annotated with the known microbial populations identified in the proteomic analysis with the metagenomic database of syntrophic TA-degrading biofilm.
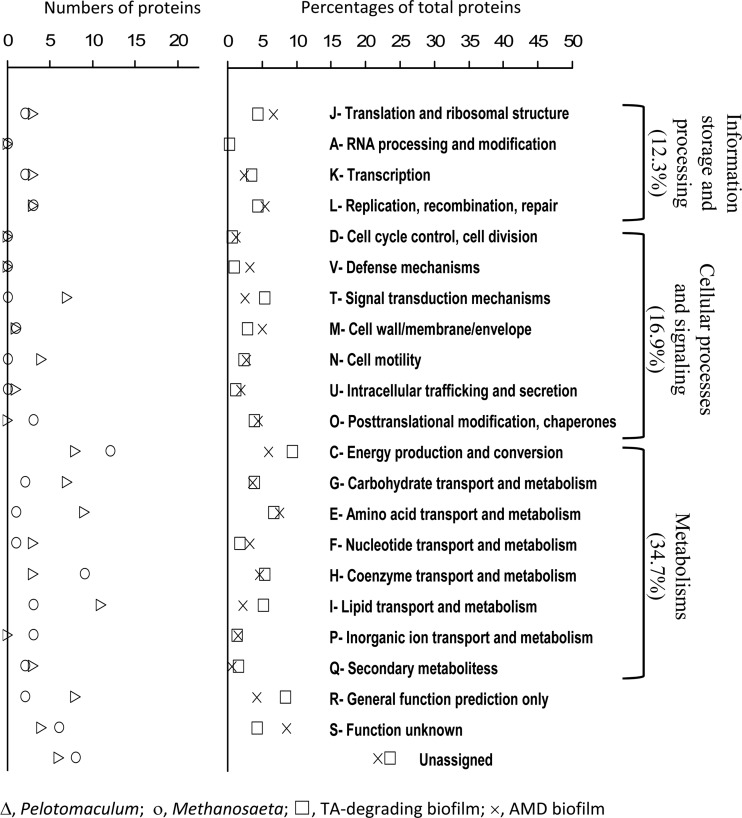
The identifiable proteins were annotated and assigned to 281 nonrepeated COGs in 21 COG categories (Fig. 4). Among them, 12.3% were related to information storage and processing (categories J to L), 16.9% were related to cellular processes and signaling (categories D to O), and 34.7% were related to metabolisms (categories C to Q). Categories C, E, H, I, and T each contained >5% of the identified proteins, and the highest number of proteins (9.3%) was observed in category C (i.e., energy production and conservation). Approximately 36.1% of the total identified proteins could not be assigned to those COG functions. As shown in Fig. 4, the distribution was comparable to that of the natural biofilms of acid mine drainage (pH ∼0.8, 42°C. and metal-rich) (9). However, higher percentages of proteins related to the energy production and conservation (higher by 3.4%), lipid transport and metabolism (higher by 2.9%), and signal transduction (higher by 2.8%) could be observed in the TA-degrading syntrophic community. These differences in the protein expressions reflected ecological differences of two biofilms such as nutrient conditions (autotrophic versus heterotrophic), redox states (aerobic versus anaerobic), and environmental stress (pH 0.8 versus 7.0 and with versus without metal toxicity).
Fig 4.
Global functional distributions of Pelotomaculum, Methanosaeta, and whole biofilm community actually expressed in the syntrophic TA-degrading biofilm based on the functional classification of the Cluster of Orthologous Groups (COG). The distribution of the proteins in the biofilm of acid mine drainage (AMD) reported in Ram et al. (9) was included for comparison.
The distribution of those 74 and 58 identifiable proteins related to Pelotomaculum spp. and Methanosaeta spp., respectively, was also compared (Fig. 4). Their distribution in the COG categories was similar for proteins associated with information storage and processing, and for cellular processing and signaling (except for categories T, N, and O). Concurrently, differences in the number of identified proteins related to metabolisms were clearly observed. Pelotomaculum spp. appeared to express more proteins associated with signal transduction (T), cell motility (N), carbohydrates (G), amino acids (E), and lipid transport and metabolism (I), whereas Methanosaeta spp. had more proteins involved in coenzymes transport and metabolism (H). These differences are likely related to the unique microbial functions performed by syntrophs, which degraded TA to acetate and H2/CO2, and acetoclastic methanogens, which produced CH4.
Proteins related to the degradation of TA to CH4.
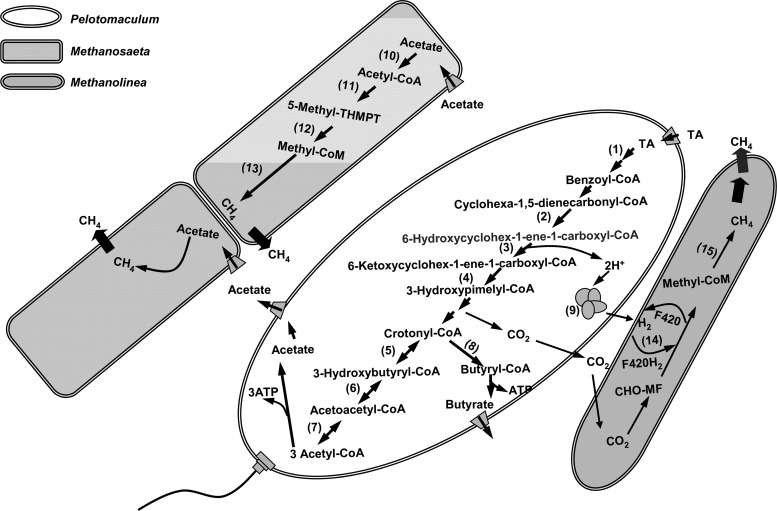
Metagenomic analysis strongly suggested that, through syntrophic methanogenic interactions, Pelotomaculum spp. could degrade TA to acetate and H2/CO2, which were then converted to CH4 by the methanogens (5). The proposed degradation pathway (Fig. 5) was further supported by the results of community proteomic analysis. In the Pelotomaculum bin, 11 proteins were identified to be involved in the catabolism of TA to acetyl coenzyme A (acetyl-CoA) (Table 2). The UbiD family decarboxylase (tadcc27178), which was predicted to be responsible for the decarboxylation of TA in the first step of the proposed pathway (5), was detected in duplicate analyses. Likewise, cyclohexa-1,5-dienecarbonyl-CoA hydratase (tadcc30311), 6-hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase (tadcc30312), and 6-oxo-cyclohex-1-ene-carbonyl-CoA hydrolase (tadcc30310) in turn, which were predicted to be responsible for the catalytic reactions of the aromatic metabolites into 3-hydroxypimelyl-CoA, were identified. In the subsequent metabolism of 3-hydroxypimelyl-CoA to acetyl-CoA through β-oxidation, crotonase (tadcc30320), β-hydroxybutyryl-CoA dehydrogenase (tadcc30319), and acetyl-CoA acetyltransferase (tadcc30308 and tadcc30318) were identified.
Fig 5.
Metabolic pathways for TA degradation in the TA-degrading Pelotomaculum spp., and for methane formation in acetoclastic Methanosaeta spp. and hydrogenotrophic Methanolinea spp. (Modified from Lykidis et al. [5].) The proteins detected by the proteomic analysis are indicated (by numbers in parentheses in the figure) as follows: 1, UbiD family decarboxylases (tadcc27178); 2, cyclohexa-1,5-dienecarbonyl-CoA hydratase (tadcc30311); 3, 6-hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase (tadcc30312); 4, 6-oxo-cyclohex-1-ene-carbonyl-CoA hydrolase (tadcc30310); 5, crotonase (tadcc30320); 6, β-hydroxybutyryl-CoA dehydrogenase (tadcc30319); 7, acetyl-CoA acetyltransferase (tadcc30308 and tadcc30318); 8, butyryl-CoA dehydrogenase (tadcc30313); 9, Fe-only hydrogenase (tadcc12813); 10, acetyl-CoA synthetase (tadcc27522, tadcc27524, and tadcc32283); 11, acetyl-CoA decarbonylase/synthase (tadcc17916 and tadcc8403); 12, tetrahydromethanopterin S-methyltransferase (tadcc28098 and tadcc28101); 13, methyl-CoM reductase alpha subunit (tadcc11668 and tadcc5364), beta subunit (tadcc6056, tadcc6057, and tadcc5367), and gamma subunit (tadcc6054 and tadcc5365); 14, F420-reducing hydrogenase (tadcc3700); and 15, methyl-CoM reductase beta subunit (tadcc3596) and gamma subunit (tadcc3599).
Table 2.
Identified protein entries related to terephthalate degradation, methanogenesis, signal transduction, and motility of Pelotomaculum, Methanosaeta, and Methanolinea in terephthalate-degrading biofilm at 50°C
| Gene ID | Protein annotation | Putative mechanism involvementa | Sequence coverageb (%) | Peptide hitc (no.) | Microorganismd |
|---|---|---|---|---|---|
| tadcc27178 | UbiD family decarboxylases | TA degradation | (5.8, 38.7) | (7, 273) | Pelotomaculum |
| tadcc30311 | Cyclohexa-1,5-dienecarbonyl-CoA hydratase | TA degradation | 46.0 | 69 | Pelotomaculum |
| tadcc30312 | 6-Hydroxycyclohex-1-ene-1-carboxyl-CoA dehydrogenase | TA degradation | 14.7 | 44 | Pelotomaculum |
| tadcc30310 | 6-Oxo-cyclohex-1-ene-carbonyl-CoA hydrolase | TA degradation | (2.1, 27.7) | (6, 187) | Pelotomaculum |
| tadcc30320 | Crotonase | TA degradation | 56.5 | 115 | Pelotomaculum |
| tadcc30319 | β-Hydroxybutyryl-CoA dehydrogenase | TA degradation | 11.0 | 10 | Pelotomaculum |
| tadcc30308 | Acetyl-CoA acetyltransferase | TA degradation | (6.5, 35.7) | (14, 86) | Pelotomaculum |
| tadcc30318 | Acetyl-CoA acetyltransferase | TA degradation | 41.4 | 55 | Pelotomaculum |
| tadcc30313 | Butyryl-CoA dehydrogenase | TA degradation | 38.4 | 102 | Pelotomaculum |
| tadcc12813 | Fe-only hydrogenase | TA degradation | 3.5 | 2 | Pelotomaculum |
| tadcc23108 | Zinc-finger protein HypA/HybF | TA degradation | 8.0 | 2 | Pelotomaculum |
| tadcc16835 | PAS-containing transcriptional regulator | Signaling | 3.38 | 2 | Pelotomaculum |
| tadcc24013 | Diguanylate cyclase (GGDEF) domain | Signaling | (6.1, 6.1) | (1, 2) | Pelotomaculum |
| tadcc35368 | PAS/PAC sensor signal transduction histidine kinase | Signaling | 2.6 | 1 | Pelotomaculum |
| tadcc7371 | Methyl-accepting chemotaxis protein | Signaling/motility | 13.2 | 61 | Pelotomaculum |
| tadcc23087 | Methyl-accepting chemotaxis protein | Signaling/motility | 6.91 | 4 | Pelotomaculum |
| tadcc12422 | Chemotaxis response regulator containing a CheY-like receiver domain and a methylesterase domain | Signaling/motility | 8.7 | 1 | Pelotomaculum |
| tadcc5742 | Universal stress protein UspA and related nucleotide-binding proteins | Signaling | 9.5 | 1 | Pelotomaculum |
| tadcc18253 | Flagellin and related hook-associated proteins | Motility | 12.6 | 1 | Pelotomaculum |
| tadcc27522 | Acetyl-coenzyme A synthetase | HAc methanogenesis | 8.2 | 29 | Methanosaeta |
| tadcc27524 | Acetyl-coenzyme A synthetase | HAc methanogenesis | 4.4 | 12 | Methanosaeta |
| tadcc32283 | Acetyl-coenzyme A synthetase | HAc methanogenesis | 6.4 | 10 | Methanosaeta |
| tadcc17916 | Acetyl-CoA decarbonylase/synthase alpha subunit | HAc methanogenesis | 6.3 | 24 | Methanosaeta |
| tadcc8403 | Acetyl-CoA decarbonylase/synthase beta subunit | HAc methanogenesis | 6.2 | 68 | Methanosaeta |
| tadcc35500 | Formylmethanofuran dehydrogenase, subunit A | Methanogenesis | 3.9 | 1 | Methanosaeta |
| tadcc1199 | Formylmethanofuran dehydrogenase subunit B | Methanogenesis | 10.29 | 1 | Methanosaeta |
| tadcc28098 | Tetrahydromethanopterin S-methyltransferase, subunit A | Methanogenesis | (6.6, 11.5) | (6, 13) | Methanosaeta |
| tadcc28101 | Tetrahydromethanopterin S-methyltransferase, subunit H | Methanogenesis | 5.8 | 6 | Methanosaeta |
| tadcc11668 | Methyl-coenzyme M reductase, alpha subunit | Methanogenesis | (14.2, 17.4) | (5, 51) | Methanosaeta |
| tadcc5364 | Methyl-coenzyme M reductase, alpha subunit | Methanogenesis | (6.6, 15.0) | (17, 54) | Methanosaeta |
| tadcc5367 | Methyl-coenzyme M reductase, beta subunit | Methanogenesis | 23.5 | 36 | Methanosaeta |
| tadcc6056 | Methyl-coenzyme M reductase, beta subunit | Methanogenesis | (26.5, 26.5) | (6, 204) | Methanosaeta |
| tadcc6057 | Methyl-coenzyme M reductase, beta subunit | Methanogenesis | (7.0, 24.4) | (9, 203) | Methanosaeta |
| tadcc6054 | Methyl-coenzyme M reductase, gamma subunit | Methanogenesis | 32.9 | 61 | Methanosaeta |
| tadcc5365 | Methyl-coenzyme M reductase, gamma subunit | Methanogenesis | 11.1 | 628 | Methanosaeta |
| tadcc3596 | Methyl-coenzyme M reductase, beta subunit | Methanogenesis | 5.03 | 99 | Methanolinea |
| tadcc3599 | Methyl-coenzyme M reductase, gamma subunit | Methanogenesis | (7.1, 7.1) | (3, 8) | Methanolinea |
| tadcc3700 | F420-reducing hydrogenase, gamma subunit | Methanogenesis | 13.5 | 18 | Methanolinea |
| tadcc3028 | Universal stress protein UspA and related nucleotide-binding proteins | Signaling | 5.7 | 64 | Methanolinea |
| tadcc3181 | Chemotaxis protein histidine kinase and related kinases | Signaling/motility | 4.0 | 1 | Methanolinea |
| tadcc39810 | Osmosensitive K+ channel histidine kinase | Signaling | 13.8 | 1 | Methanolinea |
TA degradation, anaerobic degradation pathway of terephthalate; HAc methanogenesis, acetoclastic methanogenesis pathway.
That is, the percentage of the protein sequence covered by identified peptides. The numbers of duplicated results are indicated in parentheses.
That is, the number of peptide sequences matched to the protein group. Numbers of duplicated results are indicated in parentheses.
The microbial origins detected with the IMG/M metagenomic database of syntrophic terephthalate-degrading biofilm.
For hydrogen production, two Fe-only hydrogenases encoded by the gene segments tadcc25255 and tadcc12813 were previously observed with the Pelotomaculum bin using metagenomics, and one (tadcc12813) of them was detected in the present study. To support the expression of hydrogenase in Pelotomaculum spp., the zinc-finger protein HypA/HybF (tadcc23108) (Table 2), which participates in the regulation of the hydrogenase expression, was also detected.
Both Methanosaeta spp. and Methanolinea spp. in the TA-degrading biofilm were identified to be responsible for CH4 production (5). In the Methanosaeta bin, three and seven proteins respectively related to AMP-forming acetyl-CoA synthetases (tadcc27522, tadcc27524, tadcc32283) and methyl coenzyme M (methyl-CoM) reductase (α, β, and γ subunits) were identified (Table 2 and Fig. 5). These enzymes are known to mediate the biochemical reactions in the first and final steps of the acetoclastic methanogenesis (29). In addition, the α and β subunits (tadcc17916 and tadcc8403) of acetyl-CoA decarbonylase/synthase and the A and H subunits (tadcc28098 and tadcc28101) of tetrahydromethanopterin S-methyltransferase were detected. For the Methanolinea spp., three proteins related to methyl-CoM reductase (β and γ subunits) (tadcc3596 and tadcc3599) and the F420-reducing hydrogenase (γ subunit) (tadcc3700), which could reduce the low-potential two-electron acceptor coenzyme F420 using H2 (32), were identified. Thus, the proteomic data revealed the activities of Methanosaeta and Methanolinea populations in metabolizing the methanogenic precursors, acetate and H2/CO2, which could be derived from the TA catabolism.
Proteins involved in the metabolism of butyrate.
In addition to the proteins from the Pelotomaculum, Methanosaeta, and Methanolinea bins, 17 proteins were classified into the Caldiserica bin (Fig. 3). Combined with the microbial community analysis, this observation suggested that the Caldiserica group was an active member in the TA-degrading syntrophic biofilm. Based on previous metagenomic analysis, Caldiserica spp. were predicted to perform CO2 fixation, H2 consumption, and butyrate production (5). The butyrate production by Caldiserica spp. was supported by the detection of butyrate kinase encoded by tadcc17547.
Our results further suggested that butyrate could be produced from various routes during the TA metabolism. First, crotonyl-CoA might serve as the interchange hub for the formation of acetyl-CoA (acetate) and/or butyryl-CoA (butyrate). Alternatively, the acetyl-CoA could be synthesized to crotonyl-CoA by the amphibolic activities of acetyl-CoA acetyltransferase, 3-hydroxyacyl-CoA dehydrogenase, and crotonase in sequence in an anabolic process (Fig. 5). Finally, a butyryl-CoA dehydrogenase encoded by the gene tadcc30313 was identified (Table 2), suggesting that butyrate-CoA could also be produced.
The production of butyrate was also detected during the cultivation of P. terephthalicum JT, where crotonate was used as the growth substrate (33). Actually, the fatty acids in the effluent from the reactor was analyzed, but only acetate (ca. 4.2 to 7.8 mg/liter) could be detected. The butyrate should be present at very low concentrations due to low hydrogen partial pressures in the well-established syntrophic consortia. However, a detectable level of butyrate was observed with the TA-enriching consortia when acetate and H2 were accumulated (3). Therefore, the produced butyrate might serve as a key metabolite to support the growth of other microbial populations present at low abundances, possibly through secondary syntrophy, to maintain the process stability.
Proteins involving in cellular regulation.
It was reported that Pelotomaculum thermopropionicum can potentially regulate its cellular activities with the PAS- and GGDEF-related two-component signal transduction systems (34) that are associated with environmental sensing and motility (31, 35, 36). In the present study, several essential proteins involved in the signal transduction and cell motility were identified. These proteins included, for example, the diguanylate cyclase (GGDEF) domain (tadcc24013), a PAS/PAC sensor signal transduction histidine kinase (tadcc35368), and a PAS-containing transcriptional regulator (tadcc16835). These proteins were all known as two-component signal transduction systems for the regulation of cellular processes. Furthermore, a chemotaxis response regulator containing a CheY-like receiver domain and a methylesterase domain (tadcc12422), a methyl-accepting chemotaxis protein (tadcc23087 and tadcc7371), and flagellin and related hook-associated proteins (tadcc18253) (Table 2) were also detected. These observations suggested that Pelotomaculum spp. could effectively regulate the relevant catabolism, cell motility, and chemotaxis in response to environmental conditions and global cellular situations in the syntrophic biofilm (34).
Several peptide sequences were also matched to signaling proteins such as chemotaxis-related histidine kinases (tadcc3181 and tadcc39810) and the universal stress protein UspA (tadcc3028) from Methanolinea spp. (Table 2). UspA has been reported as a small cytoplasmic protein induced to enhance the rate of cell survival when the cell is subjected to starvation or prolonged exposure to stress agents (37). Similar to Pelotomaculum spp., the cellular activities of Methanolinea spp. were highly regulated in syntrophic ecosystems where substrates and nutrients are quite limited, and the energy conservation efficiency is extremely low (34). In contrast, Methanosaeta spp. appeared to exhibit a different growth strategy, because proteins relevant to the signal transduction were not observed.
In the present study, metaproteomics could provide useful insight into the community structure and functions of TA-degrading syntrophic biofilm and could elucidate distinct microbial syntrophy ecology for anaerobic TA degradation.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate the LC-MS/MS technical assistance provided by the Instrument Development Center, NCKU, Taiwan, Republic of China, as well as the assistance of Y.-A. Tsai and M.-H. Hsu.
This study was supported by the Taiwan National Science Council (97-2221-E006-038-MY3 and 100-2628-E006-026-MY3).
Footnotes
Published ahead of print 12 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02327-12.
REFERENCES
- 1. Kleerebezem R, Lettinga G. 2000. High-rate anaerobic treatment of purified terephthalic acid wastewater. Water Sci. Technol. 42:259–268 [Google Scholar]
- 2. Kleerebezem R, Hulshoff Pol LW, Lettinga G. 1999. Anaerobic degradation of phthalate isomers by methanogenic consortia. Appl. Environ. Microbiol. 65:1152–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wu JH, Liu WT, Tseng IC, Cheng SS. 2001. Characterization of microbial consortia in a terephthalate-degrading anaerobic granular sludge system. Microbiology 147:373–382 [DOI] [PubMed] [Google Scholar]
- 4. Chen CL, Macarie H, Ramirez I, Olmos A, Ong SL, Monroy O, Liu WT. 2004. Microbial community structure in a thermophilic anaerobic hybrid reactor degrading terephthalate. Microbiology 150:3429–3440 [DOI] [PubMed] [Google Scholar]
- 5. Lykidis A, Chen CL, Tringe SG, McHardy AC, Copeland A, Kyrpides NC, Hugenholtz P, Macarie H, Olmos A, Monroy O, Liu WT. 2011. Multiple syntrophic interactions in a terephthalate-degrading methanogenic consortium. ISME J. 5:122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. VerBerkmoes NC, Denef VJ, Hettich RL, Banfield JF. 2009. Functional analysis of natural microbial consortia using community proteomics. Nat. Rev. Microbiol. 7:196–205 [DOI] [PubMed] [Google Scholar]
- 7. Wilmes P, Andersson AF, Lefsrud MG, Wexler M, Shah M, Zhang B, Hettich RL, Bond PL, VerBerkmoes NC, Banfield JF. 2008. Community proteogenomics highlights microbial strain-variant protein expression within activated sludge performing enhanced biological phosphorus removal. ISME J. 2:853–864 [DOI] [PubMed] [Google Scholar]
- 8. Wilmes P, Wexler M, Bond PL. 2008. Metaproteomics provides functional insight into activated sludge wastewater treatment. PLoS One 3:e1778 doi:10.1371/journal.pone.0001778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ram RJ, VerBerkmoes NC, Thelen MP, Tyson GW, Baker BJ, Blake RC, Shah M, Hettich RL, Banfield JF. 2005. Community proteomics of a natural microbial biofilm. Science 308:1915–1920 [PubMed] [Google Scholar]
- 10. Williams MA, Taylor EB, Mula HP. 2010. Metaproteomic characterization of a soil microbial community following carbon amendment. Soil Biol. Biochem. 42:1148–1156 [Google Scholar]
- 11. Powell MJ, Sutton JN, Del Castillo CE, Timperman AI. 2005. Marine proteomics: generation of sequence tags for dissolved proteins in seawater using tandem mass spectrometry. Mar. Chem. 95:183–198 [Google Scholar]
- 12. Wilmes P, Bond PL. 2004. The application of two-dimensional polyacrylamide gel electrophoresis and downstream analyses to a mixed community of prokaryotic microorganisms. Environ. Microbiol. 6:911–920 [DOI] [PubMed] [Google Scholar]
- 13. Liu WT, Marsh TL, Cheng H, Forney LJ. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hong PY, Wu JH, Liu WT. 2009. A high-throughput and quantitative hierarchical oligonucleotide primer extension (HOPE)-based approach to identify sources of faecal contamination in water bodies. Environ. Microbiol. 11:1672–1681 [DOI] [PubMed] [Google Scholar]
- 15. Wu JH, Hsu MH, Hung CH, Tseng IC, Lin TF. 2010. Development of a hierarchical oligonucleotide primer extension assay for the qualitative and quantitative analysis of Cylindrospermopsis raciborskii subspecies in freshwater. Microbes Environ. 25:103–110 [DOI] [PubMed] [Google Scholar]
- 16. Amann RI, Binder BJ, Olson RJ, Chisholm SW, Devereux R, Stahl DA. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Loy A, Lehner A, Lee N, Adamczyk J, Meier H, Ernst J, Schleifer KH, Wagner M. 2002. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Appl. Environ. Microbiol. 68:5064–5081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Narihiro T, Terada T, Ohashi A, Wu JH, Liu WT, Araki N, Kamagata Y, Nakamura K, Sekiguchi Y. 2009. Quantitative detection of culturable methanogenic archaea abundance in anaerobic treatment systems using the sequence-specific rRNA cleavage method. ISME J. 3:522–535 [DOI] [PubMed] [Google Scholar]
- 19. Wu JH, Hong PY, Liu WT. 2009. Quantitative effects of position and type of single mismatch on single base primer extension. J. Microbiol. Methods 77:267–275 [DOI] [PubMed] [Google Scholar]
- 20. Wu JH, Liu WT. 2007. Quantitative multiplexing analysis of PCR-amplified rRNA genes by hierarchical oligonucleotide primer extension reaction. Nucleic Acids Res. 35:e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuhn R, Benndorf D, Rapp E, Reichl U, Palese LL, Pollice A. 2011. Metaproteome analysis of sewage sludge from membrane bioreactors. Proteomics 11:2738–2744 [DOI] [PubMed] [Google Scholar]
- 22. Cleveland DW, Fischer SG, Kirschner MW, Laemmli UK. 1977. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J. Biol. Chem. 252:1102–1106 [PubMed] [Google Scholar]
- 23. Heukeshoven J, Dernick R. 1985. Simplified method for silver staining of proteins in polyacrylamide gels and the mechanism of silver staining. Electrophoresis 6:103–112 [Google Scholar]
- 24. Markowitz VM, Szeto E, Palaniappan K, Grechkin Y, Chu K, Chen IMA, Dubchak I, Anderson I, Lykidis A, Mavromatis K, Ivanova NN, Kyrpides NC. 2008. The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 36:D528–D533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192–D196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA. 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, Studholme DJ, Yeats C, Eddy SR. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138–D141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Haft DH, Selengut JD, White O. 2003. The TIGRFAMs database of protein families. Nucleic Acids Res. 31:371–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35:W182–W185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Forney LJ, Zhou X, Brown CJ. 2004. Molecular microbial ecology: land of the one-eyed king. Curr. Opin. Microbiol. 7:210–220 [DOI] [PubMed] [Google Scholar]
- 31. Ausmees N, Mayer R, Weinhouse H, Volman G, Amikam D, Benziman M, Lindberg M. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163–167 [DOI] [PubMed] [Google Scholar]
- 32. Alex LA, Reeve JN, Orme-Johnson WH, Walsh CT. 1990. Cloning, sequence determination, and expression of the genes encoding the subunits of the nickel-containing 8-hydroxy-5-deazaflavin reducing hydrogenase from Methanobacterium thermoautotrophicum ΔH. Biochemistry 29:7237–7244 [DOI] [PubMed] [Google Scholar]
- 33. Qiu YL, Sekiguchi Y, Hanada S, Imachi H, Tseng IC, Cheng SS, Ohashi A, Harada H, Kamagata Y. 2006. Pelotomaculum terephthalicum sp. nov. and Pelotomaculum isophthalicum sp. nov.: two anaerobic bacteria that degrade phthalate isomers in syntrophic association with hydrogenotrophic methanogens. Arch. Microbiol. 185:172–182 [DOI] [PubMed] [Google Scholar]
- 34. Kosaka T, Kato S, Shimoyama T, Ishii S, Abe T, Watanabe K. 2008. The genome of Pelotomaculum thermopropionicum reveals niche-associated evolution in anaerobic microbiota. Genome Res. 18:442–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simm R, Morr M, Kader A, Nimtz M, Romling U. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123–1134 [DOI] [PubMed] [Google Scholar]
- 36. Taylor BL, Zhulin IB. 1999. PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63:479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Freestone P, Nystrom T, Trinei M, Norris V. 1997. The universal stress protein, UspA, of Escherichia coli is phosphorylated in response to stasis. J. Mol. Biol. 274:318–324 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.