Abstract
Chitin amendment is a promising soil management strategy that may enhance the suppressiveness of soil toward plant pathogens. However, we understand very little of the effects of added chitin, including the putative successions that take place in the degradative process. We performed an experiment in moderately acid soil in which the level of chitin, next to the pH, was altered. Examination of chitinase activities revealed fast responses to the added crude chitin, with peaks of enzymatic activity occurring on day 7. PCR-denaturing gradient gel electrophoresis (DGGE)-based analyses of 16S rRNA and chiA genes showed structural changes of the phylogenetically and functionally based bacterial communities following chitin addition and pH alteration. Pyrosequencing analysis indicated (i) that the diversity of chiA gene types in soil is enormous and (i) that different chiA gene types are selected by the addition of chitin at different prevailing soil pH values. Interestingly, a major role of Gram-negative bacteria versus a minor one of Actinobacteria in the immediate response to the added chitin (based on 16S rRNA gene abundance and chiA gene types) was indicated. The results of this study enhance our understanding of the response of the soil bacterial communities to chitin and are of use for both the understanding of soil suppressiveness and the possible mining of soil for novel enzymes.
INTRODUCTION
The suppressiveness of soils toward plant pathogens can be enhanced by adding polymers such as chitin to them (http://www.wageningenur.nl/en/location/PPO-Vredepeel-1.htm). There is currently great interest in such applications. In addition to enhancing suppression, chitinolytic bacteria can be successfully used as biological control agents against particular fungal or nematodal plant diseases (1–4). Such bacteria might be involved as natural agents in the suppression of plant pathogens. Concurrently, such endeavors drive research on other potential applications of the relevant chitin-degrading enzymes involved.
However, as a result of soil bacterial communities being highly diverse in soil (5–7), the ecology of the processes driven by them is still poorly understood. Hence, we do not quite understand how such communities respond, in terms of the succession of groups and activities and prominence of the enzymes these express, to chitin amendments. Moreover, the genetic diversity and potential of the relevant enzymes has remained largely unknown, which is mainly due to the difficulties associated with culturing the majority of bacteria under standard laboratory conditions (8–10).
Current DNA-based technologies allow opening the black box of soil functional and phylogenetic diversity. Moreover, an increasing research interest focuses on genes that encode biotechnologically applicable enzymes which are capable of degrading a natural polymer such as chitin. Chitin is clearly spread among many soil organisms, as it is a major component of the cell walls of fungi, in addition to the exoskeletons of invertebrates. Structurally, it is composed of a chain of β-1,4-glucosidic bonds linking N-acetyl-d-glucosamine moieties. The lack of evidence for the accumulation of chitin in soil suggests that it is normally degraded at a substantial rate (11). Thus, the existence of a great reservoir of degrading enzymes is indicated. In general, bacteria are hypothesized to be among the main degraders of chitin in natural ecosystems, using it as a source of nutrition and energy (11). Although exact estimates are lacking, many soil bacteria are thought to possess enzymes involved in chitin degradation. Data from aquatic bacteria indicate that between one and a few percent of these are able to degrade chitin (12, 13). Involvement of chitin-degradative enzymes in the parasitism of other (micro)organisms has also been reported (14).
All known chitinases belong to glycosyl hydrolase families 18, 19, and 20 (15, 16). Bacterial chitin degradation involves, in the initial step, cleavage of the β-(1→4) bond by exochitinases, which are assigned mostly to family 18 of the glycosyl hydrolases. This protein family is subdivided in groups A, B, and C on the basis of the amino acid sequences of the respective members. As the majority of the characterized bacterial chitinases have been assigned to group A, it has been assumed that this group is the most abundant one in the environment (17). These chitinases (ChiA) are important given their capacity to produce short oligosaccharide chains and chitin derivatives which are ecologically relevant as substrates. In addition, ChiA enzymes, in particular, those that work well at raised pH, find application in agriculture, industry, and medicine (11, 18–23). Currently, limited examples of bacterial chitinases are commercially available. The relevant enzymes, originating from Bacillus, Serratia, and Streptomyces spp., have limited optimal activities, mainly at acid to neutral pH. Given the interest in chitin degradation, in particular, as related to plant pathogen suppression, as well as the potential exploitation of chitinases, the objective of this study was to examine the impact of chitin amendment of soil, at two different pHs, on the diversity and abundance of the soil bacterial community. We included a high-pH treatment, as bacterial chitinases active under alkaline conditions are not yet available. We placed a special focus on changes in the family 18 gene chiA. A short-term (60-day) soil microcosm experiment was set up, which allowed the emergence of different bacterial communities enriched for chitin degraders. Changes in the structures and diversities of the bacterial communities were analyzed based on the 16S rRNA and chiA genes. In the light of reports on the importance of actinobacteria in soil chitinolytic processes (17, 24, 25), actinobacterium-specific analyses were also performed. Finally, deep sequencing was applied to foster our understanding of the changes of chiA gene diversity and abundance as driven by the experimental factors applied.
MATERIALS AND METHODS
Extraction of chitin from shrimp shell waste.
Shrimp shell waste obtained from Heiploeg (Zoutkamp, The Netherlands) was first intensively washed with demineralized water. Chitin extraction was performed according to a protocol modified from that of Xu et al. (26). Proteins were removed by being briefly soaked twice in 0.25 M NaOH, followed by overnight incubation in 0.25 M NaOH at room temperature. Samples were then soaked (30 min) in 0.12 M HCl followed by rinsing with deionized water until neutral pH was reached and drying overnight in a 60°C oven. The dried material was ground and sieved through a 2-mm-pore-size mesh. The product contained chitin at over ∼90% on a dry weight basis, the remainder consisting of mostly pigments, proteins, and ash.
Soil microcosms.
Soil samples were collected in June 2010 at the experimental farm De Vredepeel in The Netherlands. The soil was characterized as sandy, with pH 5.7 and 2.2% organic matter. Triplicate 4-kg soil samples were removed from subplots in the top 10 cm of the soil. Soil was homogenized by passage through a 2-mm-pore-size mesh sieve. The moisture was adjusted to 65% of the water-holding capacity. Three replicates of microcosms containing 220 g of soil were prepared in 720-ml jars for four different sets of treatments (12 jars in total). Two sets contained the soil of the original pH 5.7 measured in deionized water (1:2 soil:water ratio), whereas the pH of the other two sets was changed to 8.7 by adding 1 M sodium carbonate to each. One set of microcosms of each pH value was supplemented with 1.8% of the preprepared ground shrimp shell waste. The microcosms were incubated at room temperature. After 0, 1, 3, 7, 15, 30, and 60 days of incubation (T0 through T60, respectively), 5-g portions of soil were removed from each microcosm, immediately frozen, and stored at −80°C for further analyses.
Chitinase activity enzymatic assay.
Chitinase activities were measured fluorimetrically on the basis of enzymatic hydrolysis of the substrates 4-methylumbelliferyl N-acetyl-β-d-glucosaminide (a substrate for β-N-acetylglucosaminidase), 4-methylumbelliferyl N,N′-diacetyl-β-d-chitobioside (a substrate for chitobiosidase), and 4-methylumbelliferyl-β-d-N,N′,N″-triacetylchitotriose (a substrate for endochitinase) with production of 4-methylumbelliferone using a chitinase assay kit (Sigma, Saint Louis, MO). Half a gram from each of the three replicates (at each time point) was collected and vortex mixed (full speed; 2 min) with 1 ml of sterile water. Samples were centrifuged for 5 min at 13,000 rpm. A 10-μl volume of supernatant was used for the enzymatic assay. The enzymatic activity was calculated according to the manufacturer's specifications. One unit of chitinase activity was defined as the release of 1 μmol of 4-methylumbelliferone from the appropriate substrate per min at pH 5.0 and 37°C. Fluorescence was measured at an excitation of 360 nm and emission of 450 nm for 1 s on a microplate reader (Synergy Mx Monochromator-Based Multi-Mode; BioTek Instruments Inc., Winooski, VT).
Nucleic acid extractions.
DNA was isolated from all soil samples (300 mg wet weight) using a PowerSoil DNA isolation kit according to the manufacturer's specifications (MoBio Laboratories, Carlsbad, CA).
PCR-DGGE.
ChiA gene amplicons were obtained as previously described (25). Actinobacterial and bacterial 16S rRNA gene amplicons were obtained as previously described (27). All PCR mixtures contained 0.2 μM each primer and 1.25 U GoTaq polymerase (Promega, Madison, WI). Amplifications were carried out on a Veriti 96-well thermal cycler (Applied Biosystems, Bleiswijk, The Netherlands). Denaturing gradient gel electrophoresis (DGGE) profiles were obtained using 16 h of electrophoresis at 100 V, 0.5× Tris-acetate-EDTA (TAE) buffer at 60°C, and an Ingeny Phor-U system (Ingeny International, Goes, Netherlands). After electrophoresis, polyacrylamide gels were stained with SYBR gold (Invitrogen, Breda, The Netherlands) and visualized on a UV transilluminator. Three replicates from each of the treatments and each time point were analyzed. DGGE profiles were compared using GelCompar II software (version 5.6; Applied Maths, Sint-Martens-Latem, Belgium).
Quantitative PCR.
Quantitative PCR assays were performed using Maxima SYBR green mix (Fermentas, ThermoFisher Scientific) on an Applied Biosystems 7300 real-time PCR system. Primers and conditions as previously described (28) were applied to quantify total bacterial and actinobacterial 16S rRNA copy numbers. chiA gene copy numbers were quantified as previously described (29). For standards, chiA (approximately 450 bp) and full-length 16S rRNA gene products obtained after amplification of pure template DNA isolated from a Streptomyces griseus strain were used. Tenfold dilutions of the standard concentrations were adjusted to 101 to 108 (estimated) gene targets per reaction. We thus calculated the gene copy numbers for all three of the aforementioned systems. We interpreted these with the cautionary note in mind that such data are intrinsically biased in the light of known and well-accepted (but unsolved) questions about DNA extraction efficiencies, numbers of genes per genome, and potentially differential PCR rates.
Pyrosequencing analysis.
Partial chiA gene amplifications (T0, T3, and T7 samples) were carried out in 50-μl volumes containing 5 μl of 10× PCR buffer, 0.2 mM deoxynucleoside triphosphate (dNTP) mix, 3.75 mM MgCl2, 2% dimethyl sulfoxide (DMSO), 20 μm of 10-bp barcoded GA1F and GA1R primers (25), and 1.5 U of GoTaq Flexi DNA polymerase (Promega, Madison, WI). All amplifications were performed on a Veriti 96-well thermal cycler (Applied Biosystems-Life Technologies Europe BV, Bleiswijk, The Netherlands). The amplification conditions were preceded by an initial denaturation step (95°C for 5 min) and followed by 35 cycles consisting of denaturation at 94°C for 1 min, annealing at 60°C, and elongation at 72°C for 1 min, followed by a final elongation step for 10 min at 72°C. Amplicon mixtures were subjected to pyrosequencing using a Genome Sequencer FLX system (Roche Diagnostics GmbH/454-Life Science Corporation, Branford, CT). Sequences were deposited in the Sequence Read Archive (SRA).
Mothur (30) was utilized to analyze the reads. The sequences were evaluated for quality and the presence of ambiguous bases and homopolymers and were trimmed to remove primers and barcodes. All sequences which did not pass quality control requirements (mean quality of reads > 25, no ambiguous bases allowed, homopolymers < 8 bases), as well as sequences without identifiable primers and barcode, were removed. Potential chimeras were removed using ChimeraSlayer implemented in Mothur. The remaining reads (50% on average) were translated into amino acid sequences. All sequences containing internal stop codons and unidentified amino acids due to sequencing errors were removed. Each translated sequence was used as a Blast-P query against a 1,754 sequence database obtained from CAZy (31) using a 10−20 E cutoff value. Sequences which did not pass this filter were removed. Due to the protein-based comparisons, we refer to “-type” or “-like” sequences rather than to defined affiliations. Amino acid sequences were aligned using ARB software (32), together with the corresponding region of the reference sequences. Dissimilarity matrices were calculated from the pairwise alignment of amino acid sequences using PHYLIP 3.67 software (33). The obtained matrix was fed to Mothur for complete linkage clustering. Qualified sequences were assigned to operational taxonomic units (OTUs) based on a 20% dissimilarity cutoff. This 20% cutoff was chosen based on slope stabilization by plotting the number of unique OTUs at different OTU cutoff values.
Statistics.
The Shannon index of bacterial diversity was calculated as H = −ΣPilogPi based on the relative band intensities (Pi) as formulated by Eichner (34). Bacterial and actinobacterial abundances were analyzed by one-way analysis of variance (ANOVA) (STATISTICA 8; StatSoft Inc., Tulsa, OK) to determine the significance of the differences between treatments. The analysis of coverage was based on post hoc analysis using Tukey's honestly significant difference (HSD) test.
Stepwise multiple regression calculations were conducted by the REG procedure (SAS version 8.02; SAS Institute Inc., Cary, NC) to determine to what extent variations in biological or chemical parameters could explain the variations in the enzymatic activities. Regression analyses of normalized data were conducted. The parameters were as follows: log[actinobacterial 16S rRNA gene copy number](Actino16SrRNA), sqrt[log(16S rRNA gene copy number)/log(16S rRNA gene copy number)max](16SrRNA). Actinobacterial, bacterial, and chiA gene diversity and richness as determined by PCR-DGGE (Actino16SrRNA Shannon, 16SrRNA Shannon, Actino16SrRNA Chao1, and 16SrRNAChao1) were included after the following transformation: log[chiA gene abundance] (chiA) and diversity (chiA and chiA Shannon, respectively), log[pH] (pH), sampling time point (incubation), chitin addition (chitin). Variables in the regression models were significant at the 0.15 level. Models were restricted to a maximum of two parameters.
Relative abundance data were fitted to the power function (35), using nonlinear regression (Gauss-Newton method; SAS version 8.02). In the model, Cr = a rm; where Cr is the relative abundance at rank r (the most abundant rank is given a value of 1, the second most abundant a value of 2, etc.), a is an empirical type- and location-specific constant, and m is the shape parameter (36). The significance and fit were assessed using the F value of the nonlinear regression and the nonlinear coefficient of determination (pseudo-R2) for each rank/abundance curve, respectively. Differences in the relative abundances were analyzed by two-sided t tests.
Finally, Jaccard similarity matrices calculated based on the PCR-DGGE patterns were used in principal coordinate analysis (PCoA) (Canoco 4.55; Microcomputer Power).
Nucleotide sequence accession number.
Sequences were deposited in the Sequence Read Archive (SRA) under the number SRA052538.
RESULTS
Effects of chitin addition and pH on the chitinolytic activity in soil.
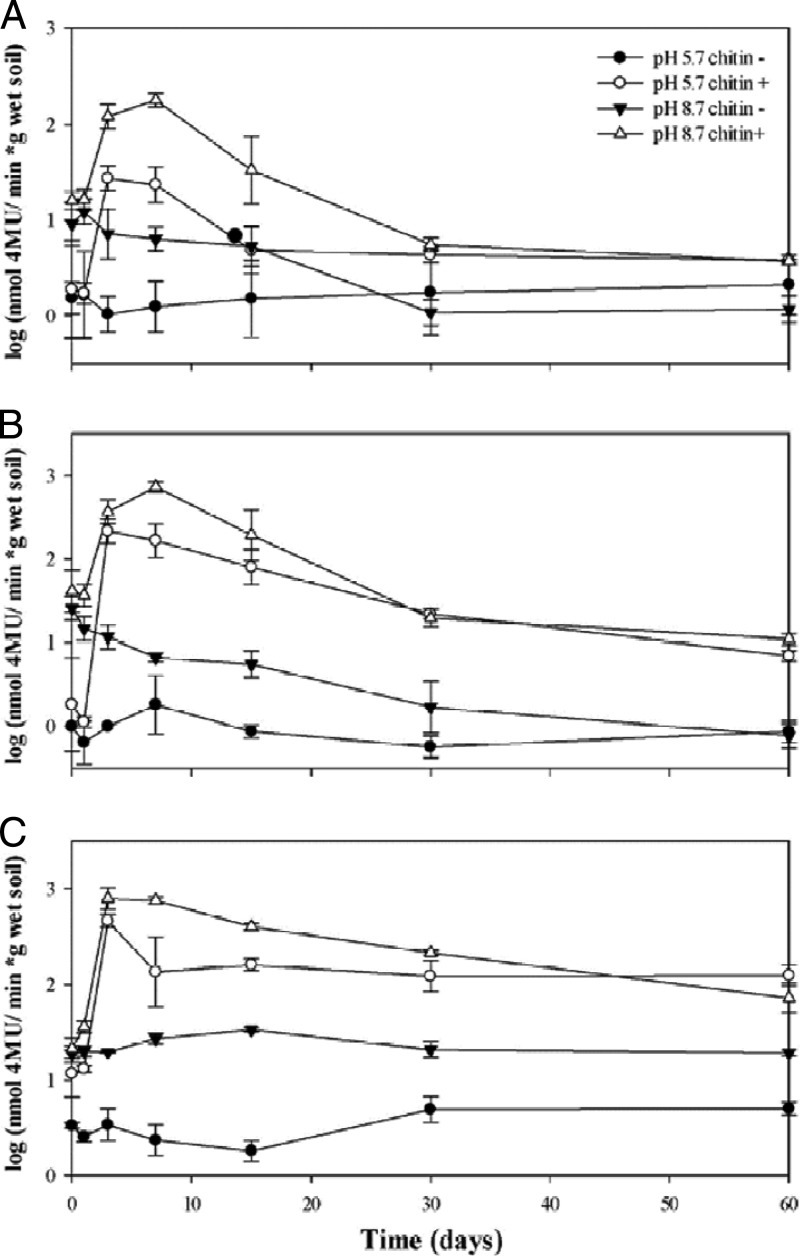
As expected, in all microcosms without added chitin, the levels of chitinolytic activity were low throughout the course of the experiment (Fig. 1). First, there were no significant differences (P > 0.05) between the values over time in the control (“native pH, no chitin”) microcosms. In the “altered pH” microcosm without added chitin, there was a progressive decrease of chitobiosidase activity (Fig. 1B) between the beginning and the end of the experiment (T0, T1, T3, T7, and T15 versus T30 and T60; P < 0.001).
Fig 1.
Changes in endochitinase (A), chitobiosidase (B), and β-N-acetylglucosaminidase (C) activities measured during 60 days of microcosm incubation. Triplicate microcosms were supplemented with chitin obtained from shrimp waste or left unsupplemented at each of the time points for soils with native pH (5.7) and increased pH (8.7). Error bars represent standard errors (SEs) of the mean results of three replicate experiments.
Overall, the highest chitinolytic activities were observed in the chitin-amended microcosms at pH 8.7 at T3 and T7 for all three measured activities, with rapid decreases being recorded at T15. From this time onward, the initial levels, in particular those of the endochitinase and chitobiosidase activities, were reached (P > 0.05). In the native-pH microcosm with added chitin, values were raised as well. There were no differences in the endochitinase activities over time (P > 0.05; Fig. 1C). A comparison of the native-pH and pH 8.7 microcosms with or without added chitin revealed that the measured activities were lower in the former than in the corresponding latter systems throughout the duration of the experiment.
Effect of chitin addition and pH on the soil bacterial community—abundance and community structure.
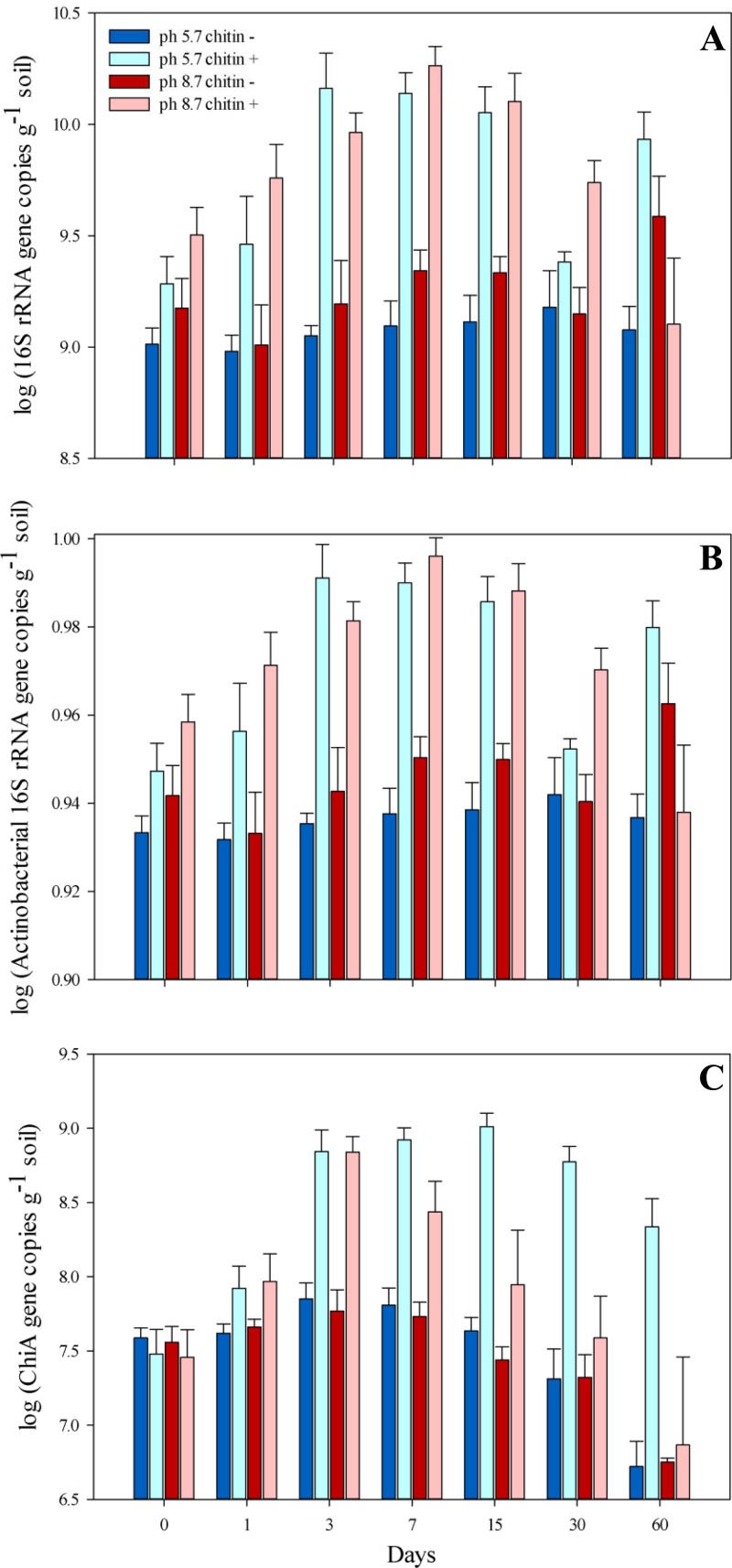
The bacterial abundances in the non-chitin-amended soils (pHs 5.7 and 8.7) were statistically similar (P > 0.05) (Fig. 2A) during the whole experiment. In the chitin-amended soil at native pH, a rapid increase of the 16S rRNA gene copy abundance was observed from T0 to T3 (from 2 × 109 to 1.5 × 1010 gene copies per g of soil; P = 1.8 × 10−4) and the numbers remained high until T15 (1.4 × 1010 gene copies per g of soil). At T30, the initial copy number was reached again. In contrast, in chitin-amended pH 8.7 soil, the 16S rRNA gene copy numbers increased steadily from T1 to T15, at which time they reached the maximal level of 1.3 × 1010 copies per g dry soil.
Fig 2.
Abundance of total bacterial (A) and actinobacterial (B) 16S rRNA and chiA gene copy numbers (C) measured at seven time points during the microcosm experiment. Days of sampling are indicated below the columns. Error bars represent SEs of the mean results of three replicate experiments.
Although fluctuations in the actinobacterial 16S rRNA gene copy numbers were observed in the soil at pH 8.7 (Fig. 2B), there was no clear trend which could explain the changes as the result of a response to chitin addition. In the control soil microcosms, the abundances remained at similar, stable levels (Fig. 2B). After addition of chitin, the actinobacterial abundances increased slightly but significantly from T0 to T1 (from 3.8 × 108 to 1.05 × 109 copies per g of soil; P = 7.1 × 10−4) and stayed at this increased level until the end of the experiment. However, the relative abundance decreased, suggesting that Actinobacteria were not the prime responders to the chitin addition.
As expected, the chiA gene copy numbers did not change in the control soils (no chitin addition) throughout the experiment. Also, the change of pH from 5.7 to 8.7 did not significantly influence the chiA gene copy numbers (T0), which remained at levels that were similar for the high-pH and the native-pH soils (Fig. 2C). In contrast, the chitin-treated soils, at both native and high pHs, revealed strong effects of added chitin on chiA gene abundance. As from T1, this abundance was greatly increased compared to that in the corresponding control soil (Fig. 2C). Thus, due to the treatment, the chiA gene abundance had increased from 3.0 × 107 to 7.0 × 108 gene copies g−1 soil in pH 8.7 soil (ANOVA; log-transformed data, P = 1.5 × 10−3) and from 3.2 × 107 to 7.2 × 108 gene copies g−1 soil (ANOVA; log-transformed data, P = 1.7 × 10−4) in native-pH soil at T3. The abundance remained at a similar high level in the native-pH chitin-amended soil until T30. Interestingly, in the pH 8.7 chitin-amended soil, in which the maximal chiA gene abundance had already been reached at T3, the chiA gene abundance gradually decreased, reaching a level at T15 that was comparable to the initial one (Fig. 2C) and significantly different from that at T3 (P = 4.4 × 10−2).
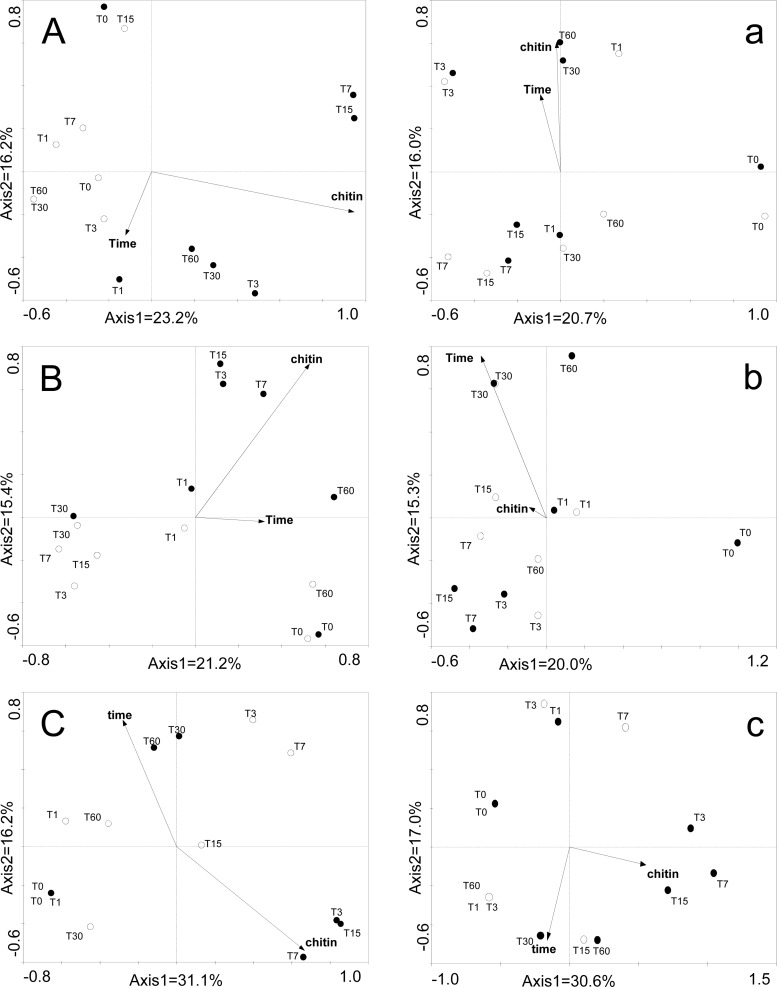
The differences among replicates of the bacterial and actinobacterial as well as chiA-based PCR-DGGE profiles were negligible (see Fig. S1 in the supplemental material); hence, we used single profiles in our clustering analyses. The bacterial profiles were affected as a result of the shifted pH as well as chitin addition. Moreover, per treatment, they shifted over the time of the experiment (Fig. 3A and a). In the native pH soil, a strong effect of added chitin was observed, as is evident from the shifts of the community compositions at T7 and T15 (Fig. 3A). In contrast, in the pH 8.7 soil, the detectable chitin addition effect was weaker. Similar observations were made with respect to the actinobacterial PCR-DGGE profiles. Here, there was a clear separation of the community profiles in the native-pH soil supplemented with chitin (Fig. 3B), whereas the community structures in the pH 8.7 soils were stronger influenced by incubation time (Fig. 3b).
Fig 3.
DGGE patterns from principal coordinate analysis based on Jaccard similarity. (A, a) 16S rRNA. (B, b) Actinobacterial 16S rRNA. (C, c) chiA gene. Uppercase letters represent comparisons of samples at pH 5.7. Lowercase letters represent comparisons of samples at pH 8.7. Empty circles represent samples without chitin and filled circles samples with chitin addition. Positions of circles represent the mean positions of the replicates. The DGGE photos can be found in the supplemental material.
The chiA gene-based PCR-DGGE analyses revealed progressive and major changes in the profiles as a result of the addition of chitin (T3 through T15 compared to T0, for both native-pH and pH 8.7 soils). Moreover, extended incubation times (30 to 60 days) had a strong influence on the chiA gene profiles, as the similarities between the profiles at T30 and T60 and the remaining ones were the lowest (Fig. 3C and c). Based on these observations, expecting major differences in the bacterial chitinolytic communities at T7 in contrast to longer incubation times, the T0, T3, and T7 samples were thus selected for deep pyrosequencing of the chiA gene.
Analysis of the relationship between enzymatic activity, bacterial community, and treatment.
Multiple regression analyses were performed in order to identify the factors that explained most of the variations in the enzymatic activities measured in the microcosms (Table 1). Bacterial abundance (16S rRNA gene copy numbers) was the best predictor of chitinolytic activities for both β-N-acetylglucosaminidase activities (P < 0.0001) and chitobiosidase activities (P < 0.0001), explaining 67% and 54% of the variations, respectively. In both cases, increases in bacterial abundances correlated well with increases in enzymatic activities. The remaining variation was explained by pH, which significantly affected both measured enzymatic activities (P < 0.0001), with increases of activity at higher pH values.
Table 1.
Best regression models for enzymatic activity in microcosm experiment
| Enzyme | Modela | R2 | P |
|---|---|---|---|
| β-N-Acetylglucosaminidase | −17.2 (±1.96)*** + 11.57 (±2.49)*** × 16S rDNA + 0.24 (±0.02)*** × pH | 0.75 | 0.001 |
| Chitobiosidase | −22.73 (±4.29)*** + 12.36 (±5.1)*** × 16S rDNA + 0.36 (±0.05)*** × pH | 0.63 | 0.001 |
***, P < 0.0001. All models have to be restricted to a maximum of only two parameters (see Materials and Methods).
RADs.
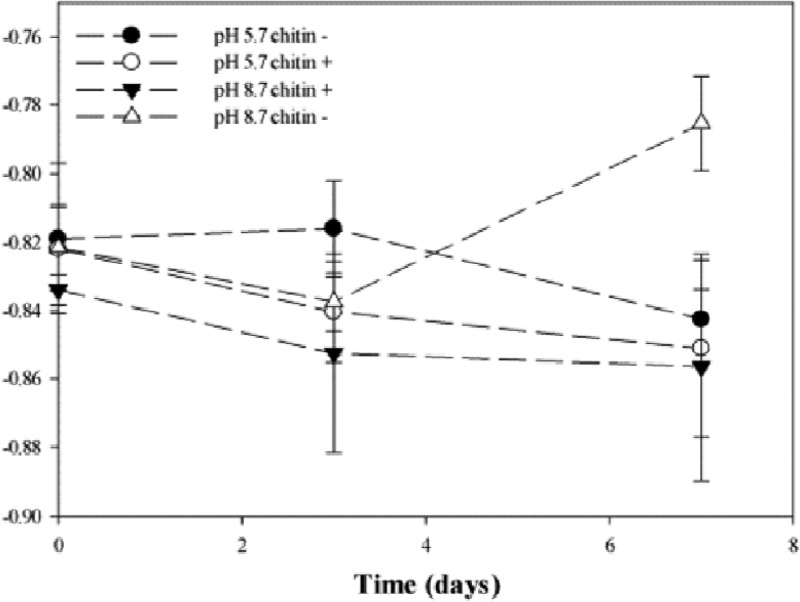
In order to better understand underexamined processes in microbial communities, we focused on rank-abundance distributions (RADs). We examined the effects of chitin and pH on the bacterial community structures (evenness) over time, and the evenness of the RADs of OTUs obtained from the pyrosequencing analysis was examined. The shape (slope) of a distribution curve is associated with evenness; lower values are observed if more of the OTUs have similar numbers of individuals (higher evenness). The shape parameter, m, was plotted over time for each of the treatments (Fig. 4). Although slight shifts in the evenness of the microbial communities (m values ranging from −0.82 to −0.85) between chitin-untreated and chitin-treated soils were observed at native pH, these changes were not significant (P > 0.05). In the chitin-treated pH 8.7 soils, the general trend was different, as a significant shift toward higher evenness (the m value increased from −0.84 to −0.80; P = 0.031) was found after day 7 of incubation.
Fig 4.
Changes in the shape parameter, m, which characterizes the evenness of the rank-abundance distributions (RADs) over time.
Analysis of the diversity of the chiA gene across treatments.
Given the fast activity and bacterial responses to the chitin amendment and pH change, our interest in the immediate responses to the treatment was raised. Thus, the samples taken at T0, T3, and T7 were selected for deep pyrosequencing of the chiA gene. Sequencing was performed for all four treatments, each in three replicate experiments. In total, 592,924 sequence reads of, on average, 450 bp were obtained; of those, 322,167 reads passed the filtration settings. For each replicate per treatment, an average 8,949 reads were thus obtained, giving 26,793 reads per treatment (Table 2). We defined chiA-based OTUs on the basis of an 80% similarity criterion. OTU-based analyses were performed on normalized numbers of sequences (3,854 sequences). Good's coverage estimator (Table 2) then revealed that the main fraction of family 18 chiA sequence types (OTUs) was detected across all samples. Based on the Chao1 and abundance-based coverage estimator (ACE) richness estimators, the chiA gene richness decreased over time in the pH 8.7 soil, especially after chitin addition (Chao1 T0 versus T3; P = 0.018), suggesting a doubly selective effect of pH and chitin. The T7 samples of this treatment yielded the lowest predicted richness values (ACE, 582 ± 59; Chao1, 418 ± 9). Similarly, the chiA gene diversity, expressed as the Shannon index, decreased over time in the pH 8.7 soil supplemented with chitin (T0 versus T3, P = 0.035; T0 versus T7, P < 0.001) and in comparison to the control (T3, P = 0.004; T7, P = 0.001), with the lowest value at T3 (1.95 ± 0.08; Table 2). In both native-pH microcosms, the decrease of richness was observed from T0 to T3, with an increase at T7 (Table 2).
Table 2.
Observed richness and diversity estimates based on 80% OTU clusters
| Sample | Avg score ± SE |
Coverageb | ||||
|---|---|---|---|---|---|---|
| No. of sequences | Richness estimatord |
Shannon diversity indexd | ||||
| Sobsa | ACE | Chao1 | ||||
| T0 pH 8.7 chitinc | 5,825 ± 1,946 | 478 ± 5 | 1,202 ± 8 | 888 ± 25 | 4.96 ± 0.01 | 0.88 |
| T0 pH 8.7 | 6,735 ± 197 | 508 ± 12 | 1,278 ± 61 | 932 ± 26 | 5.09 ± 0.02 | 0.88 |
| T0 pH 5.7 | 5,506 ± 593 | 476 ± 13 | 1,216 ± 25 | 876 ± 15 | 4.87 ± 0.1 | 0.88 |
| T0 pH 5.7 chitin | 5,176 ± 425 | 453 ± 14 | 930 ± 128 | 782 ± 37 | 4.83 ± 0.07 | 0.88 |
| T3 pH 8.7 chitin | 11,799 ± 1,530 | 177 ± 15 | 580 ± 125 | 421 ± 67 | 1.95 ± 0.08 | 0.96 |
| T3 pH 8.7 | 9,505 ± 214 | 339 ± 6 | 907 ± 74 | 705 ± 73 | 3.45 ± 0.05 | 0.94 |
| T3 pH 5.7 | 6,315 ± 375 | 428 ± 15 | 1,032 ± 86 | 767 ± 51 | 4.74 ± 0.02 | 0.92 |
| T3 pH 5.7 chitin | 11,151 ± 1,049 | 243 ± 9 | 625 ± 25 | 465 ± 12 | 2.33 ± 0.09 | 0.92 |
| T7 pH 8.7 chitinc | 13,091 ± 564 | 227 ± 2 | 582 ± 59 | 418 ± 9 | 2.42 ± 0.02 | 0.95 |
| T7 pH 8.7 | 10,199 ± 548 | 369 ± 23 | 983 ± 92 | 739 ± 74 | 3.76 ± 0.16 | 0.90 |
| T7 pH 5.7 | 7,586 ± 452 | 458 ± 7 | 1,175 ± 86 | 855 ± 35 | 4.78 ± 0.07 | 0.91 |
| T7 pH 5.7 chitin | 19,631 ± 8,849 | 267 ± 14 | 728 ± 34 | 511 ± 33 | 2.78 ± 0.18 | 0.99 |
Sobs, observed richness.
Data represent Good's coverage, calculated as the number of OTUs that were sampled once/the total number of individuals in the sample.
Data represent averages of the results of two replicate experiments.
Data were calculated based on 3,854 sequences per replicate obtained by a random resampling approach.
Representatives of each OTU were then used for a BLAST-P query against the database. The number of OTUs affiliated with each of the best-hit chitinases and the identities between best hit and representatives of OTUs are given in Table S1 in the supplemental material.
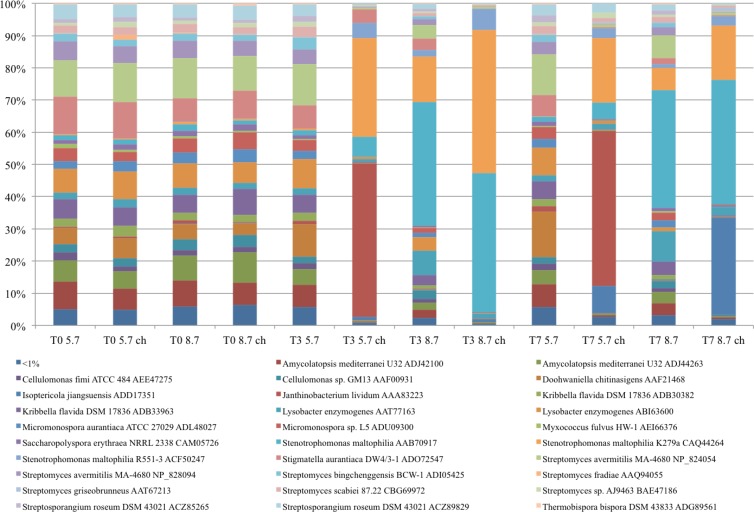
The BLAST-P results revealed high similarities between the chiA gene profiles in all four treatments at T0 (Fig. 5). Hence, there were no immediate changes related to the chitin addition and pH modulation that were applied. Furthermore, the chiA gene diversity and composition remained stable in the native control soil over the T7 experimental period. Addition of chitin lowered the observed diversities in both native-pH and pH 8.7 soils. Also, strong chiA gene diversity shifts concomitant with the pH were observed (See Table S2 in the supplemental material). Stenotrophomonas maltophilia-related chiA sequences dominated in the pH 8.7 soil (both with and without chitin) at T3 (P < 0.001) (Fig. 5; see also Table S2 in the supplemental material), with increased dominance in the chitin-treated soil. Although still present at T7, the relative abundance of the S. maltophilia-related sequences had decreased. After T7, Isoptericola jiangsuensis-type gene sequences dominated the chiA gene pools in the chitin-treated pH 8.7 soils (increase from <1% to 30%; P < 0.001) (Fig. 5; see also Table S2 in the supplemental material). Thus, this type was the second dominant chiA gene type after that of S. maltophilia. Also, in native-pH soil treated with chitin, there was an increase of such sequences but not to the same extent as in the pH 8.7 soil (from <1% to 8%; P < 0.001) (Fig. 5; see also Table S2 in the supplemental material). The native-pH soil treated with chitin was already dominated by Janthinobacterium lividum-type chiA sequences at T3 (48%; P < 0.001) (Fig. 5; see also Table S2 in the supplemental material).
Fig 5.
Comparison of levels of chiA gene diversity at different sampling points. The stacked-column graph represents the relative distributions of chiA genes affiliated with different bacterial species based on BLAST-P analysis. All calculations were performed on normalized data. Average relative abundance data from three replicates were calculated as the ratio between the sequence type abundance and the total number of sequences in the group. Sequence types represented by less than 1% of the sequences are grouped and presented on the graph as the “<1%” group.
The same trends in the chiA gene diversity as described above became visible at the phylum level (see Fig. S2 in the supplemental material). Irrespective of time-, chitin-, and/or pH-related differences, Gamma- and Betaproteobacteria-like chiA gene sequences became predominant after the addition of chitin to the soil used. There were clear differences between the native-pH and pH 8.7 soils in the gene sequence composition. In pH 8.7 soils with and without added chitin, Gammaproteobacteria-like chiA sequences dominated, with higher percentages in the chitin-treated versus untreated soil at T3 (96% versus 66%, respectively). At T7, these percentages had decreased and were comparable between the two systems (61% versus 55%). In the native-pH soil treated with chitin, Betaproteobacteria-like chiA sequences dominated (an increase from 6.6% at T0 to 48% at T3 and T7). However, there was also an increase in the percentage of Gammaproteobacteria-like chiA gene sequences. The relative abundance of sequences with similarity to chiA genes from Actinobacteria also shifted, as it appeared that both the chitin treatment and pH increase had a negative effect on the relative numbers of sequences assigned to this phylum.
DISCUSSION
In general terms, microbially driven chitin degradation is well documented at the overall process level. Chitin degradation in soil has been shown to be a relatively fast process (60% mass loss of buried chitin in soil nets after 180 days [37], 40% mass loss after 5 months [24]). However, in spite of previous important work (2, 25, 37), there still is a paucity of knowledge on the actual successions that take place, in the face of chitin supplementation, during chitin degradation in soil among the chitinolytic and other microbial communities. For instance, no deep-sequencing studies have been conducted that would provide a more comprehensive insight into the effects of chitin and pH on the chiA gene diversity in soil. Here, we combined enzymatic assays with bacterial abundance and diversity assessments and deep chiA gene sequencing in order to shed light on such effects.
Chitin addition to soil was shown to stimulate the overall community chitinase activities, which is likely to reflect the activity and growth of a chitinolytic microbiota. In the process, it is likely that the microbial growth following chitin amendment was affected and possibly limited by other prevailing conditions, such as the availabilities of other compounds that became limiting (e.g., phosphorus and/or iron) or predatory forces exerted by protozoa. It is also important that the chitinase activity assays measure the total enzymatic activity of the soil community and are not specific for bacterially driven chitin degradation. In natural or agricultural soils, chitin polymers are mainly of fungal origin, fungi also being among major degraders of this compound. Thus, it is expected that the measured enzymatic activity also reflects fungus-based chitin degradation (38). Moreover, there is no clear distinction between endo- and exochitinolytic activities (39). However, our results showed the best predictors of chitinolytic activities for both β-N-acetylglucosaminidase and chitobiosidase activities were correlated with bacterial abundance and soil pH (explaining 89% and 77% of the variations, respectively). Indeed, for both enzymes, the increases in bacterial abundances were related to increases in enzyme activities. Hallmann et al. (40) observed that bacteria respond much faster than fungi to chitin amendment of soil.
Species (OTU) richness is considered to reflect the stability of an environment (41, 42). Indeed, in our study, the number of chiA-based OTUs was highest in the control soil microcosm. As expected, OTU richness decreased after chitin addition and/or pH changes, probably as a result of the necessary adaptation of the soil microbiota to the new conditions. The evenness of the species distribution allows an evaluation of the overall biological activity of a habitat, as high biological activity often coincides with low evenness. This suggests that the conditions created in soil by chitin addition as well as pH alteration induced changes in local conditions, allowing selection of particular bacterial groups. Such differential responses thus allow a better understanding, and possibly mining, of the chitinolytic activities from such different locally responsive communities.
Actinobacteria, which are widely distributed in soil, have been pinpointed as important decomposers of complex organic molecules in the soil environment. They are also often taken as representative models of chitin degradation in soil (17, 24, 25). Actinobacteria are indeed commonly found in soil screens for chitinolytic activities by culture-based approaches (37). However, in our study, their numbers were negatively correlated with the soil chitin level. This was observed for both the actinobacterial 16S rRNA gene quantification and the actinobacterium-related chiA gene sequences. One explanation could be that these organisms exhibit slower responses to chitin addition to soil than those of other groups. Moreover, they may show better culturability than other bacterial groups, thus showing up abundantly on plates, including those supplemented with chitin. Indirectly, this is consistent with “the great plate count anomaly” (43), corroborating the fraction of known culturable bacteria that is known to possess chitinolytic activity (44). However, being aware of molecular-technique biases (e.g., accounting for genome copy numbers, DNA extraction efficiency, or the presence of inhibitors in the DNA extracts influencing PCR efficiency), we are not able to measure absolute gene/bacterium abundances.
To analyze the changes in richness and diversity of the chiA gene in our system, we employed analyses using chiA OTUs (defined at the amino acid sequence level using an 80% similarity cutoff). The initial BLAST-P analyses of our sequences gave an impression of rather low chiA gene richness in our systems. However, our analyses may have been biased by the depth with which the available reference sequences represent the genes found in ecosystems. The existing database may actually be rather limited, and so our data set adds a large amount of novelty to the database.
Furthermore, although we tentatively assigned affiliated species to the partial chiA sequences obtained, reliable identification of the chiA gene-carrying bacterial species is clearly difficult on this basis. The reason for this cautionary note is that 16S rRNA gene-based phylogeny is often not consistent with chiA gene-based phylogeny. Cottrell et al. (45) revealed a closer relationship of chiA genes of the gammaproteobacterium Vibrio with those of Alphaproteobacteria than with those of bacteria belonging to the Gammaproteobacteria. Moreover, Xiao et al. (46) showed that the chiA genes obtained from the taxonomically divergent Janthinobacterium lividum, Cytophaga sp., and Stenotrophomonas sp. shared surprisingly high (95%) identity. Similarly, Ramaiah et al. (47) showed that the chiA gene of Vibrio sheri did not group with that of other Vibrio types. Horizontal gene transfer is suggested to be responsible for these observations of unexpected high sequence similarities between similar gene types in unrelated taxa. In contrast to these findings, Metcalfe et al. (17) found a perfect match between chiA and 16S rRNA gene sequence-based phylogenies in streptomycetes as well as in Stenotrophomonas maltophilia. Thus, given these contrasting findings, more data comparing chiA and 16S rRNA gene sequence phylogenies on the basis of the same organisms are needed to allow a definition of the relationship between these two genes. With respect to this, we have referred to “-type” or “-like” sequences rather than to defined affiliations.
Our data demonstrated, irrespective of time-, chitin-, and pH-related differences, that Gamma- and Betaproteobacteria-like chiA gene sequences became predominant after the addition of chitin to the soil used. An abundance of Gammaproteobacteria (studies based on the 16S rRNA gene marker) in chitin-amended soil has recently been reported (44), with dominance of Cellvibrio species (37). However, although C. gilvus-like sequences were also identified in the current study, they covered only about 0.01% of the total chiA gene diversity and their relative abundance was not affected by chitin addition.
The main group of sequences found in our study, mainly as responders to chitin addition, was related to sequences from a limited number of Gram-negative organisms. Interestingly, by inference, organisms potentially involved in plant pathogen suppression were thus stimulated. One major gene type was affiliated with the chiA gene of S. maltophilia. Stenothrophomonas and Lysobacter, which were also identified in our study, are rather opportunistic organisms which can respond extremely quickly to emerging ecological opportunities in soil. The former genus is under investigation as a candidate for the biological control of, primarily, plant-pathogenic nematodes (48–51). S. maltophilia strains have an extraordinarily high hydrolytic enzyme potential (52). In addition to chitinases, they produce diverse proteases, glucanases, DNases, RNases, lipases, and laccases. Both chitinolytic and proteolytic activities are thought to contribute to their biocontrol capacity. S. maltophilia as well as Lysobacter spp. occur widely in soils (53). The second major chiA gene type found was related to a Janthinobacterium lividum chiA gene. Janthinobacterium species are common in soil (54) and aquatic (55) environments. J. lividum was recently found to inhibit fungal pathogens of amphibians (56–58), again highlighting a potential role in pathogen suppression. The third major chiA gene type was related to Isoptericola jiangsuensis chiA. Two chitinases, i.e., Is-chiA (with chitobiosidase activity) and Is-chiB (showing endochitinase activity), have been recently identified and characterized in I. jiangsuensis. Interestingly, the optimal pH of Is-chiB was observed to be around 9.0 and the enzyme retained 80% of its activity during 1 h of incubation in buffers from pH 7.0 to 10.0, with the highest stability at pH 8.5. The enzyme does not reveal a chitin-binding domain; however, this did not affect its activity toward a soluble substrate. The genus Isoptericola comprises six identified species; however, only I. jiangsuensis was shown to have chitinolytic activity (59, 60). Strikingly, we found that, in spite of their established role in chitin degradation, actinobacteria were not among the primary responders to the added chitin. This finding can be explained by the fact that we placed a focus on the immediate responders to the substrate, which may well have been mainly the r-responding Gram-negative bacteria described. In contrast, K-responding actinobacteria may be among the late responders, which in this study fell beyond the chosen focus on the immediate responders.
In summary, this study showed that modulation of chitin availability and pH in an agricultural soil induces shifts in the chitin-degrading microbial communities, which has potential relevance for the pathogen-suppressive nature of soil. The soil used, Vredepeel, had been treated with chitin in the field, and indeed its suppressiveness toward particular pathogenic nematodes has been raised (61). Our study lays the basis for further work on the specific activities exerted by the substrate-responsive communities. In particular, their exact role in the suppressiveness needs elucidation, as well as the specificity of the enzymes they produce for biotechnological purposes.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the EU Metaexplore project (KBBE-222625).
We acknowledge Reinier van der Flier of the Heiploeg Group for providing samples, Richard Finkers (Plant Breeding, WUR) for help with computing, and Waleed Abu Al-Soud (University of Copenhagen) for his support.
Footnotes
Published ahead of print 26 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.02546-12.
REFERENCES
- 1. Downing K, Thomson JA. 2000. Introduction of the Serratia marcescens chiA gene into an endophytic Pseudomonas fluorescens for the biocontrol of phytopathogenic fungi. Can. J. Microbiol. 46:363–369 [DOI] [PubMed] [Google Scholar]
- 2. Hjort K, Bergstrom M, Adesina MF, Jansson JK, Smalla K, Sjoling S. 2010. Chitinase genes revealed and compared in bacterial isolates, DNA extracts and a metagenomic library from a phytopathogen-suppressive soil. FEMS Microbiol. Ecol. 71:197–207 [DOI] [PubMed] [Google Scholar]
- 3. Kobayashi DY, Reedy RM, Bick J, Oudemans PV. 2002. Characterization of a chitinase gene from Stenotrophomonas maltophilia strain 34S1 and its involvement in biological control. Appl. Environ. Microbiol. 68:1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kotan R, Dikbas N, Bostan H. 2009. Biological control of post harvest disease caused by Aspergillus flavus on stored lemon fruits. Afr. J. Biotechnol. 8:209–214 [Google Scholar]
- 5. Curtis TP, Sloan WT, Scannell JW. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. U. S. A. 99:10494–10499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gans J, Wolinsky M, Dunbar J. 2005. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 309:1387–1390 [DOI] [PubMed] [Google Scholar]
- 7. Torsvik V, Sorheim R, Goksoyr J. 1996. Total bacterial diversity in soil and sediment communities—a review. J. Ind. Microbiol. 17:170–178 [Google Scholar]
- 8. Curtis TP, Sloan WT. 2004. Prokaryotic diversity and its limits: microbial community structure in nature and implications for microbial ecology. Curr. Opin. Microbiol. 7:221–226 [DOI] [PubMed] [Google Scholar]
- 9. Schloss PD, Handelsman J. 2004. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68:686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Torsvik V, Ovreas L. 2002. Microbial diversity and function in soil: from genes to ecosystems. Curr. Opin. Microbiol. 5:240–245 [DOI] [PubMed] [Google Scholar]
- 11. Gooday GW. 1990. The ecology of chitin degradation. Adv. Microb. Ecol. 11:387–430 [Google Scholar]
- 12. Beier S, Jones CM, Mohit V, Hallin S, Bertilsson S. 2011. Global phylogeography of chitinase genes in aquatic metagenomes. Appl. Environ. Microbiol. 77:1101–1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cottrell MT, Moore JA, Kirchman DL. 1999. Chitinases from uncultured marine microorganisms. Appl. Environ. Microbiol. 65:2553–2557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Patil RS, Ghormade V, Deshpande MV. 2000. Chitinolytic enzymes: an exploration. Enzyme Microb. Technol. 26:473–483 [DOI] [PubMed] [Google Scholar]
- 15. Henrissat B, Bairoch A. 1993. New families in the classification of glycosyl hydrolases based on amino-acid-sequence similarities. Biochem. J. 293:781–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Henrissat B. 1991. A classification of glycosyl hydrolases based on amino-acid-sequence similarities. Biochem. J. 280:309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Metcalfe AC, Krsek M, Gooday GW, Prosser JI, Wellington EMH. 2002. Molecular analysis of a bacterial chitinolytic community in an upland pasture. Appl. Environ. Microbiol. 68:5042–5050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dutta PK, Dutta J, Chattopadhyaya MC, Tripathi VS. 2004. Chitin and chitosan: novel biomaterials waiting for future developments. J. Polym. Mater. 21:321–324 [Google Scholar]
- 19. Dutta PK, Ravikumar MN, Dutta J. 2002. Chitin and chitosan for versatile applications. J. Macromol. Sci. Polym. Rev. C 42:307–354 [Google Scholar]
- 20. Felse PA, Panda T. 1999. Studies on applications of chitin and its derivatives. Bioprocess Eng. 20:505–512 [Google Scholar]
- 21. Krajewska B. 2004. Application of chitin- and chitosan-based materials for enzyme immobilizations: a review. Enzyme Microb. Technol. 35:126–139 [Google Scholar]
- 22. Olander LP, Vitousek PM. 2000. Regulation of soil phosphatase and chitinase activity by N and P availability. Biogeochemistry 49:175–190 [Google Scholar]
- 23. Qiu YZ, Zhang N, Kang Q, An YH, Wen XJ. 2009. Chemically modified light-curable chitosans with enhanced potential for bone tissue repair. J. Biomed. Mater. Res. A 89:772–779 [DOI] [PubMed] [Google Scholar]
- 24. Krsek M, Wellington EMH. 2001. Assessment of chitin decomposer diversity within an upland grassland. Antonie Van Leeuwenhoek 79:261–267 [DOI] [PubMed] [Google Scholar]
- 25. Williamson N, Brian P, Wellington EMH. 2000. Molecular detection of bacterial and streptomycete chitinases in the environment. Antonie Van Leeuwenhoek 78:315–321 [DOI] [PubMed] [Google Scholar]
- 26. Xu Y, Gallert C, Winter J. 2008. Chitin purification from shrimp wastes by microbial deproteination and decalcification. Appl. Microbiol. Biotechnol. 79:687–697 [DOI] [PubMed] [Google Scholar]
- 27. Heuer H, Krsek M, Baker P, Smalla K, Wellington EMH. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233–3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fierer N, Jackson JA, Vilgalys R, Jackson RB. 2005. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 71:4117–4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yergeau E, Kang S, He Z, Zhou J, Kowalchuk GA. 2007. Functional microarray analysis of nitrogen and carbon cycling genes across an Antarctic latitudinal transect. ISME J. 1:163–179 [DOI] [PubMed] [Google Scholar]
- 30. Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233–D238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Felsenstein J. 2005. PHYLIP (Phylogeny Inference Package) version 3.6. Department of Genome Sciences, University of Washington, Seattle, WA [Google Scholar]
- 34. Eichner CA, Erb RW, Timmis KN, Wagner-Dobler I. 1999. Thermal gradient gel electrophoresis analysis of bioprotection from pollutant shocks in the activated sludge microbial community. Appl. Environ. Microbiol. 65:102–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mitzenmacher M. 2004. A brief history of generative models for power law and lognormal distributions. Internet Math. 1:226–251 [Google Scholar]
- 36. van der Gast C, Duane A, Lilley A. 2008. Temporal scaling of bacterial taxa is influenced by both stochastic and deterministic ecological factors. Environ. Microbiol. 10:1411–1418 [DOI] [PubMed] [Google Scholar]
- 37. Sato K, Azama Y, Nogawa M, Taguchi G, Shimosaka M. 2010. Analysis of a change in bacterial community in different environments with addition of chitin or chitosan. J. Biosci. Bioeng. 109:472–478 [DOI] [PubMed] [Google Scholar]
- 38. Kellner H, Vandenbol M. 2010. Fungi unearthed: transcripts encoding lignocellulolytic and chitinolytic enzymes in forest soil. PLoS One 5:e10971 doi:10.1371/journal.pone.0010971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. LeCleir GR, Buchan A, Maurer J, Moran MA, Hollibaugh JT. 2007. Comparison of chitinolytic enzymes from an alkaline, hypersaline lake and an estuary. Environ. Microbiol. 9:197–205 [DOI] [PubMed] [Google Scholar]
- 40. Hallmann J, Rodriguez-Kabana R, Kloepper JW. 1999. Chitin-mediated changes in bacterial communities of the soil, rhizosphere and within roots of cotton in relation to nematode control. Soil Biol. Biochem. 31:551–560 [Google Scholar]
- 41. Ives AR, Klug JL, Gross K. 2000. Stability and species richness in complex communities. Ecol. Lett. 3:399–411 [Google Scholar]
- 42. Li W, Stevens MHH. 2010. How enrichment, ecosystem size, and their effects on species richness co-determine the stability of microcosm communities. Oikos 119:686–695 [Google Scholar]
- 43. Staley JT, Konopka A. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321–346 [DOI] [PubMed] [Google Scholar]
- 44. Das SN, Sarma P, Neeraja C, Malati N, Podile AR. 2010. Members of Gammaproteobacteria and Bacilli represent the culturable diversity of chitinolytic bacteria in chitin-enriched soils. World J. Microbiol. Biotechnol. 26:1875–1881 [Google Scholar]
- 45. Cottrell MT, Wood DN, Yu LY, Kirchman DL. 2000. Selected chitinase genes in cultured and uncultured marine bacteria in the α- and γ-subclasses of the Proteobacteria. Appl. Environ. Microbiol. 66:1195–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiao X, Yin X, Lin J, Sun L, You Z, Wang P, Wang F. 2005. Chitinase genes in lake sediments of Ardley Island, Antarctica. Appl. Environ. Microbiol. 71:7904–7909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ramaiah N, Hill RT, Chun J, Ravel J, Matte MH, Straube WL, Colwell RR. 2000. Use of a chiA probe for detection of chitinase genes in bacteria from the Chesapeake Bay. FEMS Microbiol. Ecol. 34:63–71 [DOI] [PubMed] [Google Scholar]
- 48. Dunne C, Crowley JJ, Moenne-Loccoz Y, Dowling DN, de Bruijn FJ, O'Gara F. 1997. Biological control of Pythium ultimum by Stenotrophomonas maltophilia W81 is mediated by an extracellular proteolytic activity. Microbiology 143:3921–3931 [DOI] [PubMed] [Google Scholar]
- 49. Dunne C, Moenne-Loccoz Y, de Bruijn FJ, O'Gara F. 2000. Overproduction of an inducible extracellular serine protease improves biological control of Pythium ultimum by Stenotrophomonas maltophilia strain W81. Microbiology 146(Pt 8):2069–2078 [DOI] [PubMed] [Google Scholar]
- 50. Kobayashi DY, Reedy RM, Palumbo JD, Zhou JM, Yuen GY. 2005. A clp gene homologue belonging to the crp gene family globally regulates lytic enzyme production, antimicrobial activity, and biological control activity expressed by Lysobacter enzymogenes strain C3. Appl. Environ. Microbiol. 71:261–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Messiha NAS, van Diepeningen AD, Farag NS, Abdallah SA, Janse JD, van Bruggen AHC. 2007. Stenotrophomonas maltophilia: a new potential biocontrol agent of Ralstonia solanacearum, causal agent of potato brown rot. Eur. J. Plant Pathol. 118:211–225 [Google Scholar]
- 52. Ryan RP, Monchy S, Cardinale M, Taghavi S, Crossman L, Avison MB, Berg G, van der Lelie D, Dow JM. 2009. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7:514–525 [DOI] [PubMed] [Google Scholar]
- 53. Hayward AC, Fegan N, Fegan M, Stirling GR. 2010. Stenotrophomonas and Lysobacter: ubiquitous plant-associated Gamma-Proteobacteria of developing significance in applied microbiology. J. Appl. Microbiol. 108:756–770 [DOI] [PubMed] [Google Scholar]
- 54. Gleave AP, Taylor RK, Morris BAM, Greenwood DR. 1995. Cloning and sequencing of a gene encoding the 69-kDa extracellular chitinase of Janthinobacterium lividum. FEMS Microbiol. Lett. 131:279–288 [DOI] [PubMed] [Google Scholar]
- 55. Saeger JL, Hale AB. 1993. Genetic variation within a lotic population of Janthinobacterium lividum. Appl. Environ. Microbiol. 59:2214–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Becker MH, Harris RN. 2010. Cutaneous bacteria of the redback salamander prevent morbidity associated with a lethal disease. PLoS One 5:e10957 doi:10.1371/journal.pone.0010957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Brucker RM, Harris RN, Schwantes CR, Gallaher TN, Flaherty DC, Lam BA, Minbiole KPC. 2008. Amphibian chemical defense: antifungal metabolites of the microsymbiont Janthinobacterium lividum on the salamander Plethodon cinereus. J. Chem. Ecol. 34:1422–1429 [DOI] [PubMed] [Google Scholar]
- 58. Harris RN, Brucker RM, Walke JB, Becker MH, Schwantes CR, Flaherty DC, Lam BA, Woodhams DC, Briggs CJ, Vredenburg VT, Minbiole KP. 2009. Skin microbes on frogs prevent morbidity and mortality caused by a lethal skin fungus. ISME J. 3:818–824 [DOI] [PubMed] [Google Scholar]
- 59. Wu Y, Li WJ, Tian W, Zhang LP, Xu L, Shen QR, Shen B. 2010. Isoptericola jiangsuensis sp. nov., a chitin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 60:904–908 [DOI] [PubMed] [Google Scholar]
- 60. Wu Y, Liu F, Liu YC, Zhang ZH, Zhou TT, Liu X, Shen QR, Shen B. 2011. Identification of chitinases Is-chiA and Is-chiB from Isoptericola jiangsuensis CLG and their characterization. Appl. Microbiol. Biotechnol. 89:705–713 [DOI] [PubMed] [Google Scholar]
- 61. Molendijk LPG. 1999. Control strategy for nematodes works ([example of] Vredepeel). PAV-Bull. Akkerbouw 1999:4–8 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.