Abstract
The human short PLUNC1 (SPLUNC1) protein has been identified as a component of the pulmonary antimicrobial response based on its structural similarity to the bactericidal/permeability-increasing (BPI) protein. Using a genetically modified mouse model, we recently verified the antimicrobial activity of SPLUNC1 against Pseudomonas aeruginosa in vivo. To further define the mechanism of epithelial SPLUNC1-mediated antibacterial action, we carried out studies to determine how SPLUNC1 protects the host from acute respiratory infections. P. aeruginosa treated with recombinant human SPLUNC1 protein showed decreased growth in vitro. This antibacterial activity was due to growth inhibition, as a consequence of a SPLUNC1-induced increase in bacterial cell permeability. Removal of SPLUNC1 allowed the recovery of P. aeruginosa and suggested no permanent cell injury or direct killing of bacteria. Further investigation showed coating of bacterial cells by SPLUNC1. We suggest that this “bacterial cell coating” is necessary for the bacteriostatic function of SPLUNC1. Additionally, we demonstrated a novel role for SPLUNC1 as a chemoattractant that facilitated migration of macrophages and neutrophils. Taking the findings together, we propose synergistic roles for human SPLUNC1 as an antibacterial agent with bacteriostatic and chemotactic activities.
INTRODUCTION
Humans are constantly exposed to different types of bacteria; however, respiratory infections are relatively rare. Various proteins play important roles in innate immunity to eliminate invading microbes or to keep them under control so that the adaptive immune response can subsequently clear the remaining pathogenic microbes. Conversely, dysfunction of innate immune responses may contribute to infection and inflammatory disorders. The human SPLUNC1 (Short Palate, Lung, Nasal Epithelium Clone 1) protein has been identified as a component of innate immunity because of its structural similarity to BPI and its abundant expression throughout the oral cavity and respiratory tract of humans and many other mammals (1–7). Since the upper respiratory tract, including the nasal and oral cavities, is the major route of entry of most pathogens into the body, we carried out the present study to determine the mechanisms by which the SPLUNC1 protein controls microbial growth.
SPLUNC1 is also referred to as SPURT, LUNX, NASG, or BPIFA1 and belongs to the BPI-fold-containing (BPIF) protein family based on its structural similarity to BPI (2, 3, 8, 9). SPLUNC1 encodes a secreted protein found abundantly in human sputum and tracheobronchial secretions, as well as in saliva (10), nasal lavage fluid (5), and apical secretions from cultured human tracheobronchial epithelial cells (3, 11). Normally, SPLUNC1 proteins are present in healthy individuals at relatively low concentrations (∼10 to 250 μg/ml in airway epithelial secretion [12]) and increase significantly upon infection by pathogenic microbes (11). Recent studies from our laboratory and those of others have demonstrated its in vivo function against several pathogenic bacteria, including Pseudomonas aeruginosa and Mycoplasma pneumoniae (13–16). However, the detailed mechanism of SPLUNC1-mediated antibacterial activity has not yet been conclusively demonstrated.
P. aeruginosa, a Gram-negative bacterium, was used as model organism in this study because this opportunistic pathogen is often associated with hospital-acquired infections and also represents a great threat to immunocompromised people. P. aeruginosa is widespread in nature, inhabiting soil, water, plants, and animals (including humans). It is the most common pathogen isolated from patients who have been hospitalized longer than 1 week and is a frequent cause of nosocomial infections such as pneumonia, urinary tract infections, and bacteremia. It also causes secondary infections in immune-deficient individuals with tuberculosis, cancer, and, particularly, cystic fibrosis (17–20). Although we have clearly demonstrated that mice overexpressing SPLUNC1 exhibit enhanced protection against P. aeruginosa (15), in vitro studies of the antimicrobial activity of SPLUNC1 were inconclusive (21, 22). It has been suggested that SPLUNC1 may be required for regulation of certain physical properties, such as airway surface liquid volume (23) and surface tension, that may interfere with the biofilm formation of the bacteria (12, 16). In this study, we extended our previous in vivo studies of SPLUNC1 antimicrobial activity to elucidate the mechanisms of SPLUNC1-mediated activity against P. aeruginosa using several in vitro assays. We report that SPLUNC1 plays a dual role as an antibacterial agent by directly increasing bacterial cell permeability and as a chemoattractant signal that recruits macrophages and neutrophils to the site of infection.
MATERIALS AND METHODS
Bacterial strains, culture media, and chemicals.
Pseudomonas aeruginosa PAO1 containing green fluorescent protein (GFP) plasmid (pMF230; carbenicillin resistant) was kindly provided by Garth Ehrlich of Allegheny Singer Research Institute, Allegheny General Hospital, Pittsburgh, PA. Trypticase soy broth (TSB; Becton, Dickinson and Company) or agar (TSA; Becton, Dickinson and Company) supplemented with 400 μg/ml carbenicillin was used to grow these bacteria.
Purification of human recombinant SPLUNC1 protein.
The human SPLUNC1 gene (NCBI reference sequence NM_130852.2) was cloned into p2N vector without the secretion signal sequence (nucleotides 1 to 127, amino acids 1 to 19) for expression in Escherichia coli. The His tag sequence was attached at the N-terminal end of SPLUNC1 to facilitate protein purification. A standard DNA extraction method using a Qiagen miniprep kit (Qiagen) was used to extract DNA from E. coli. Cloned DNA was verified through PCR amplification using forward (5′-CGAAGAGGCCAAGAAACAG-3′) and reverse (5′-GTCAAGCTCCTTGACCAAATCC-3′) primers before sequencing. DNA sequencing was performed by the Genomics and Proteomics Core Laboratories at the University of Pittsburgh, Pittsburgh, PA. SPLUNC1 protein was expressed in E. coli, and cell lysates were purified using nickel and ion exchange columns. The expression of SPLUNC1 was confirmed by Western blotting with both anti-His and anti-SPLUNC1 antibodies (R&D, Minneapolis, MN).
Determination of SPLUNC1 antimicrobial activity.
The antimicrobial activity of SPLUNC1 protein was tested by incubating Pseudomonas aeruginosa-pMF230 with recombinant human SPLUNC1 protein (1 to 5 μg/ml) for 2 h, a typical time for a bactericidal assay in Gram-negative bacteria (24). Pseudomonas aeruginosa-pMF230 was grown in TSB (pH 7.2) supplemented with 400 μg/ml carbenicillin at 37°C. The bacterial culture was adjusted to an optical density at 600 nm (OD600) of ∼0.225 (∼8 × 107 CFU/ml), and ∼106 cells were added to a flat-bottom 96-well plate (Greiner Bio-One, Germany). SPLUNC1 (1 μg and 5 μg) was added to determine the antibacterial activity. Samples were collected every 30 min for bacterial plating and determining the number of CFU per milliliter. A similar antibacterial activity assay was performed with “protein-carrying buffer” and was used as a negative control. Since P. aeruginosa-pMF230 contains GFP, GFP fluorescence measurements were also used as an alternative approach to quantitatively monitor growth of P. aeruginosa-pMF230 (data not shown). To determine the long-term antibacterial activity of SPLUNC1, P. aeruginosa-pMF230 was grown and the OD600 was adjusted to ∼0.201 (∼4 × 106 CFU/ml) to avoid the overgrowth of P. aeruginosa-pMF230. Bacteria at that concentration were grown with or without SPLUNC1 (1 μg) for 24 h, and the effect of SPLUNC1 on bacterial growth was determined by counting the number of CFU after growing serial dilutions on TSB agar plates. All experiments were performed at least 3 times, and data were presented as means ± standard deviations (SD).
Detection of lipopolysaccharide (LPS) contamination in the recombinant SPLUNC1 protein.
The presence of LPS in SPLUNC1 recombinant protein was determined with a HEK-Blue LPS detection kit (InvivoGen, San Diego, CA) according to the manufacturer's protocol. Quantitation of residual LPS in the sample was performed with a ToxinSensor Chromogenic LAL endotoxin assay kit (GenScript, Piscataway, NJ) using the manufacturer's protocol.
Detection of SPLUNC1 binding to LPS.
A modified, enzyme-linked immunosorbent assay (ELISA)-based LPS binding method was used to detect interaction between LPS and SPLUNC1 (25). Briefly, a 96-well plate was coated overnight with purified LPS (500 ng/well) from Pseudomonas aeruginosa 10 (Sigma, St. Louis, MO) or E. coli K-12 (Life Technologies, Grand Island, NY). Wells were washed and blocked with 1% bovine serum albumin (BSA)–phosphate-buffered saline (PBS) for 1 h; then, 2-fold dilutions of purified SPLUNC1 (500 ng) were added to each well in duplicate. PBS was used as a control for this experiment. An antibody specific to human SPLUNC1 (R&D, Minneapolis, MN) was used to detect the LPS-bound SPLUNC1. After the secondary antibody reaction performed using horseradish peroxidase (HRP)-conjugated anti-goat IgG, enzyme activity was detected using a TMB Ultra 1-step assay (Pierce Biotechnology, Inc., Rockford, IL). Absorbance was detected in a BioTek spectrophotometer (BioTek, Winooski, VT). This experiment was performed three times, and the data were presented as means ± SD.
Removal of SPLUNC1 from Pseudomonas aeruginosa-pMF230 and growth recovery assay.
P. aeruginosa-pMF230 was grown as described above for 4 h at 37°C in a black, flat-bottom 96-well plate with 5 μg SPLUNC1. Bacterial cells were briefly centrifuged to remove the culture supernatant. P. aeruginosa-pMF230 cells were washed with either 0.1% Triton X-100 or PBS to remove the attached SPLUNC1 from bacterial cells. Fresh TSB medium (∼100 μl) was added immediately to the washed bacterial cells, which were then grown for 6 h at 37°C. P. aeruginosa-pMF230 treated with SPLUNC1 without any washing was used as a control. GFP fluorescence readings were recorded at 0 h, 3 h, and 6 h to show the bacterial growth recovery. This experiment was performed 3 times, and data were presented as means ± SD.
Outer membrane assay of P. aeruginosa-pMF230 permeabilization by SPLUNC1 protein.
The outer membrane permeabilization ability of SPLUNC1 was determined by a 1-N-phenyl-napthylamine (NPN; Molecular Probes, Eugene, OR) assay (26). P. aeruginosa-pMF230 was grown with 3 μg (the lowest concentration that exhibited measurable antibacterial activity in 2 h; data not shown) of SPLUNC1. Bacterial cells were washed and resuspended in 5 mM HEPES buffer (pH 7.4) and were used for the permeabilization assay. The uptake of the fluorescent (blue color) hydrophobic probe NPN was measured as described by Loh et al. (26). Fluorescence of NPN was measured by a fluorescent spectrophotometer (BioTek, Winooski, VT) at wavelengths of 350 nm (excitation) and 420 nm (emission). The classic permeabilizing agent EDTA (2 mM) was used as a positive control, and buffer alone was used as a negative control. This experiment was performed three times, and data were presented as means ± SD. The same experiments were performed with ethidium bromide or propidium iodide, and the data were presented as means ± SD of the results of the three experiments.
Confocal microscopy to detect the coating of SPLUNC1 on P. aeruginosa-pMF230.
P. aeruginosa-pMF230 was grown for 2 h from diluted overnight culture. Bacterial cells were mixed with SPLUNC1 and incubated at 37°C for 15 min. The mixture was added onto clean glass slides and allowed to bind onto glass for 15 min. The control slide had P. aeruginosa-pMF230 only (without SPLUNC1). Slides were fixed in 4% paraformaldehyde for 10 min at room temperature. After three washes in PBS, slides were incubated with a SPLUNC1-specific antibody and detected with a Cy5-conjugated secondary antibody. Images were captured in a Fluoview 1000 confocal microscope equipped with a 60× (1.4 numerical aperture [NA]) oil immersion optic (Olympus, Center Valley, PA).
In vitro cell migration assay.
Murine RAW246.7 cells (ATCC TIB-71; ATCC, Manassas, VA) and human neutrophils differentiated from HL-60 cells (ATCC CCL-240; ATCC, Manassas, VA) were used in the cell migration assay. This assay was performed in a Boyden chamber with polycarbonate membranes and a pore size of 8 μm or 3 μm (Costar, Corning, NY) for murine RAW246.7 cells or human neutrophils, respectively, according to a method described by Yona et al. (27). SPLUNC1 (1 μg) mixed in Dulbecco's modified Eagle's medium (DMEM) or RPMI medium was added into the lower compartment of the chamber. A 100-ng volume of monocyte chemoattractant protein-1 (MCP-1) (R&D) (a potent chemoattractant for macrophages) and 1 μg of N-formyl-methionyl-leucyl-phenylalanine (fMLP) (Sigma) (a known chemoattractant for neutrophil) were added in separate wells as positive controls. Buffer alone was used as a negative control. The RAW246.7 cells (∼2.5 × 105/ml) or neutrophils (∼5 × 106/ml) were added in the upper compartment of the chambers, on top of the membrane. The incubations were carried out at 37°C in a 5% CO2 atmosphere for 2 h and 4 h. EDTA (10% [0.5 M]) was added to the bottom chamber to release the migrated cells at 4°C for 20 min before the cells were counted using a Cellometer Vision system (Nexcelom Bioscience, Lawrence, MA). The data were reported as the means ± SD of three representative cell counts.
Statistical analysis.
For all experiments, results were expressed as the means ± SD of the results of at least 3 independent experiments. One-way or two-way analysis of variance (ANOVA) was used to calculate the statistical significance of the differences between the treatment group results. A P value of <0.05 was considered to be statistically significant.
RESULTS
Characterization of recombinant human SPLUNC1.
Sequencing results confirmed the successful cloning of SPLUNC1 into a p2N vector without any changes in nucleotide sequence. Western blotting of this purified protein showed strong signals corresponding to the SPLUNC1-specific antibody and suggested that the recombinant SPLUNC1 protein was pure and ∼25 kDa in size. Both Lowry tests and ELISAs were performed to determine the protein concentrations. LPS is a cell membrane component of Gram-negative bacteria such as E. coli. During lysis of bacterial cells, LPS may be released from the outer membrane into lysate and contaminate the sample. Since a recombinant SPLUNC1 expression purification was performed in E. coli, a possible LPS contamination in the SPLUNC1 protein sample was analyzed prior to the LPS binding assay with the HEK-Blue LPS assay kit. This assay showed minimal presence of LPS in the SPLUNC1 preparation. Further quantification with a ToxinSensor Chromogenic Limulus Amebocyte lysate (LAL) endotoxin kit showed negligible LPS contamination in SPLUNC1 (<0.03 endotoxin units [EU] of LPS/1 μg SPLUNC1).
Antibacterial activity of SPLUNC1 on P. aeruginosa-pMF230.
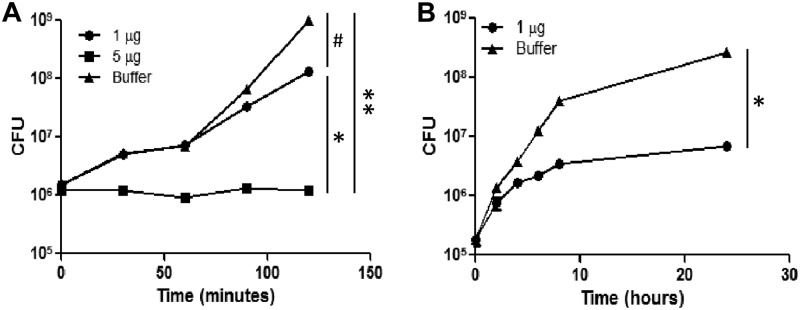
The antibacterial function of SPLUNC1 was tested in vitro on P. aeruginosa-pMF230. The number of bacterial CFU per milliliter was slightly decreased (∼1 log decrease in growth) compared to the number seen with the control (Fig. 1A) with 1 μg recombinant protein. However, a significant inhibition of bacterial growth was observed with 5 μg at all time points (Fig. 1A). This clearly indicated a dose-dependent antibacterial function of SPLUNC1 on P. aeruginosa-pMF230. An alternative approach to monitor the growth inhibition by measuring GFP fluorescence intensity also showed similar trends in the bacterial growth inhibition and confirmed the antibacterial function of this protein (data not shown). To determine if SPLUNC1 could maintain its antibacterial activity for a longer period of time, we monitored the bacterial growth with 1 μg SPLUNC1 for 24 h. SPLUNC1 displayed significant growth inhibition of P. aeruginosa-pMF230 during the 24-h incubation period when monitored by bacterial CFU count (Fig. 1B).
Fig 1.
Activity of SPLUNC1 against Pseudomonas aeruginosa-pMF230. (A) P. aeruginosa-pMF230 was grown with or without SPLUNC1. Bacterial CFU with 1 μg or 5 μg of SPLUNC1 or in the presence of buffer, as a negative control, were measured. The growth reduction of P. aeruginosa-pMF230 was reproducible in repeated experiments. (B) P. aeruginosa-pMF230 was grown with or without SPLUNC1 for 24 h. Bacterial CFU with 1 μg of SPLUNC1 or in the presence of buffer, as a negative control, were measured. All experiments were performed at least 3 times, and data are presented as means ± SD. Both graphs showed statistical significance (*, P < 0.005; **, P < 0.01) of the differences between the SPLUNC1 protein and buffer control results. #, P < 0.01 (1 μg versus 5 μg for the SPLUNC1-treated groups).
Binding of SPLUNC1 to bacterial LPS.
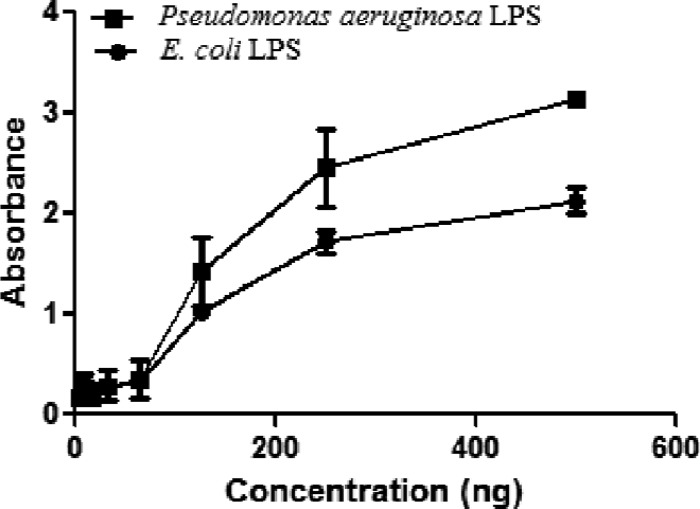
The LPS binding property may enhance interaction between the Gram-negative bacteria and SPLUNC1 during infection. In order to determine how SPLUNC1 interacts with bacterial LPS, we used the ELISA-based LPS binding assay (25). Results obtained indicated strong LPS binding with SPLUNC1 (Fig. 2). Our LPS detection assay showed LPS contamination at a negligible level with SPLUNC1 (<0.03 EU of LPS/1 μg of SPLUNC1). The very minute quantity of LPS that existed in the SPLUNC1 was unlikely to affect the results of the LPS binding assay. Buffer alone (PBS) showed no LPS binding, and LPS (from P. aeruginosa 10 or E. coli K-12), used as a positive control, showed strong binding when detected with a LPS-specific antibody (data not shown).
Fig 2.
LPS binding property of SPLUNC1. Binding of LPS from P. aeruginosa 10 or LPS from E. coli K-12 with different concentrations of SPLUNC1 is shown. All experiments were performed at least 3 times, and data are presented as means ± SD.
SPLUNC1 displays a bacteriostatic property in preventing bacterial growth via affecting cell permeability.
When SPLUNC1-treated P. aeruginosa-pMF230 was observed under a time-lapse microscope, bacterial cells appeared nonmotile in the presence of SPLUNC1. It also showed no change in bacterial cell shape or size, which suggested an inhibition of cellular processes, especially regarding motility (data not shown). This result suggested that SPLUNC1 may control bacterial number by inhibiting bacterial growth (bacteriostatic) rather than by killing bacteria directly (bactericidal).
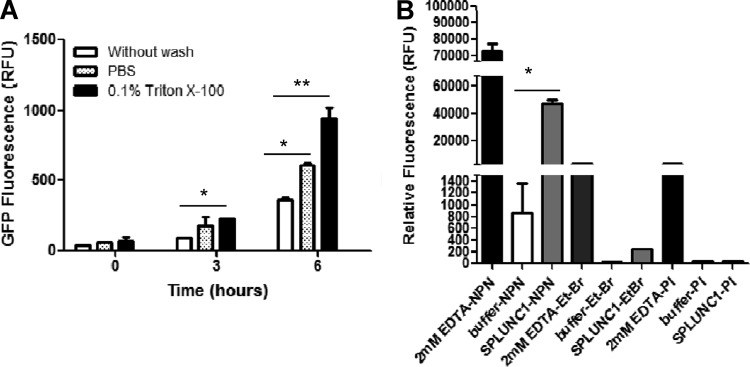
We performed pulse-chase experiments to determine if SPLUNC1 is bacteriostatic or bactericidal against P. aeruginosa-pMF230. Bacteria were first treated with 5 μg of SPLUNC1 and grown for 4 h, washed with PBS or 0.1% Triton X-100, and then grown for an additional 6 h in fresh TSB medium to examine the bacterial growth recovery. P. aeruginosa-pMF230 showed significant recovery after the removal of SPLUNC1 with 0.1% Triton X-100 or PBS washes. Removal of SPLUNC1 with 0.1% Triton X-100 or PBS washes resulted in the significant recovery in bacterial growth compared with the bacteria without any wash (Fig. 3A; P < 0.001 [**] and P < 0.05 [*], respectively).
Fig 3.
Bacteriostatic activity of SPLUNC1 against Pseudomonas aeruginosa-pMF230 via increased cell permeability. (A) The antibacterial activity of SPLUNC1 is bacteriostatic. Pseudomonas aeruginosa-pMF230 was grown with 5 mg SPLUNC1 for 4 h and then briefly washed with PBS or 0.1% Triton X-100. P. aeruginosa-pMF230 was grown for 6 h in fresh medium, and the GFP fluorescence was measured at 0 h, 3 h, and 6 h. Bacteria grown without washes were used as a control. Experiments were performed 3 times, and data are presented as means ± SD. The data showed statistical significance (*, P < 0.05; **, P < 0.001) in comparisons of PBS and Triton X-100 treatments with the no-wash control. (B) A P. aeruginosa-pMF230 cell permeability assay was performed by growing the bacterial cell in the presence of 3 mg of SPLUNC1 (gray bars). EDTA (2 mM) was added to 1-N-phenyl-naphthylamine (NPN), ethidium bromide (Et-Br), or propidium iodide (PI) as positive controls (black bars), and the fluorescence of these positive controls was measured. Bacteria were grown alone (buffer; white bars) as a negative control. Experiments were repeated 3 times, and data are presented as means ± SD. The graph showed a statistically significant difference (**, P < 0.001) between SPLUNC1 proteins and the no-protein (buffer) control when treated with NPN.
To further understand SPLUNC1-mediated bacteriostatic function, we determined if SPLUNC1 changed bacterial cell wall permeability. 1-N-Phenyl-naphthylamine (NPN) emits fluorescence weakly in an aqueous environment but strongly in the hydrophobic interior of cell membranes. Upon destabilization of the bacterial outer membrane by antimicrobial agents, the dye enters the damaged membrane, where it emits stronger fluorescence. Uptake of the NPN blue fluorescent probe (molecular weight, 219.28) by P. aeruginosa-pMF230 after 1 h of preincubation with SPLUNC1 correlated with a considerable increase in fluorescence compared to the buffer control (Fig. 3B). A relatively small amount of NPN fluorescence was detected in the buffer control (∼1% of the amount seen with the positive EDTA control and ∼1.6% of that seen with treatment with SPLUNC1), likely due to the fact that the fluorescence of this compound is higher in a lipid environment (intracellular) than in an aqueous environment (extracellular) (28). A similar permeability-increasing effect by SPLUNC1 on bacterial cell walls was also seen with ethidium bromide dye (molecular weight, 394.31), although at a lower intensity than that seen with NPN.
Interestingly, the permeability assay performed with propidium iodide (molecular weight, 668.4) failed to show any increased dye intensity after SPLUNC1 was added (Fig. 3B). These observations strongly suggest that the increase of cell permeability in P. aeruginosa-pMF230 was due to SPLUNC1-mediated formation of small pores that are large enough to be permeable with NPN and, to a lesser extent, ethidium bromide but not with propidium iodide.
Coating of P. aeruginosa-pMF230 with SPLUNC1.
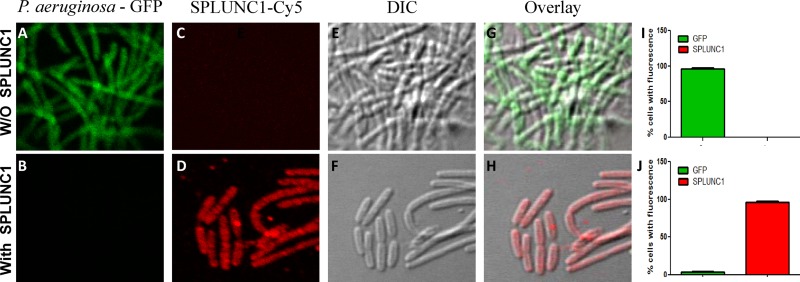
SPLUNC1-mediated bacteriostatic activity and bacterial growth inhibition may require direct binding of SPLUNC1 to P. aeruginosa to initiate changes in cell permeability. To determine if SPLUNC1 binds to bacteria, GFP-labeled P. aeruginosa-pMF230 cells were treated with SPLUNC1, and the SPLUNC1 was detected by immunofluorescence (IF) microscopy. P. aeruginosa-pMF230 exhibited green fluorescence (constitutive GFP expression; Fig. 4A), and SPLUNC1 was stained red (Cy5-conjugated secondary antibody; Fig. 4D). The coating of SPLUNC1 on the bacterial cells, which inhibited bacterial growth, resulted in a loss of bacterial green fluorescence (Fig. 4B), and the bacteria appeared red due to the fluorescent staining of SPLUNC1 (Fig. 4H). There were a small number of bacterial cells, however, that escaped the coating of SPLUNC1, probably due to the inclusion of washing steps during the staining procedure. Fluorescent quantitation showed coating with SPLUNC1 in most of the P. aeruginosa-pMF230 cells (Fig. 4J), and very limited GFP fluorescence indicated the bacterial inactivation caused by the SPLUNC1 treatment.
Fig 4.
Binding of SPLUNC1 to Pseudomonas aeruginosa-pMF230. Immunostaining experiments were performed without (panels A, C, E, and G) and with (panels B, D, F, and H) SPLUNC1. P. aeruginosa-pMF230 is shown in green (GFP) and SPLUNC1 in red (Cy5). (I and J) Quantitation of double fluorescence. Fluorescence was calculated using the percentages of total cells counted in 15 different randomly selected fields, and data are presented as means ± SD.
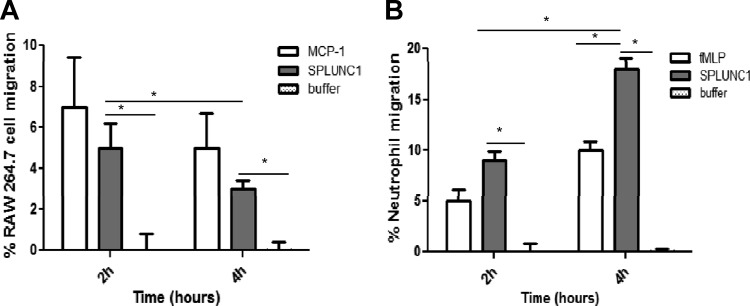
SPLUNC1 stimulates macrophage and neutrophil migration.
Since recruited inflammatory cells also contribute to innate immunity through the phagocytosis of bacteria, we examined if secreted SPLUNC1 in the airway plays a role in the migration of phagocytic cells. Recombinant SPLUNC1 protein (1 μg) showed significant effects on the migration of RAW264.7 cells and neutrophils (Fig. 5). It appeared that SPLUNC1 showed slightly less recruitment to macrophages, as exemplified by the migration of RAW264.7 cells compared to that of the neutrophils (Fig. 5) after 2 h of incubation. Macrophages showed decreased migration after 4 h, but we observed a significant increase in neutrophil migration in the same 4-h time interval.
Fig 5.
Chemotaxis of macrophage and neutrophils by SPLUNC1. The assay was performed in a Boyden chamber, where 1 μg of SPLUNC1 mixed in growth medium was added in the lower compartment of the chambers and RAW264.7 cells or HL-60 induced neutrophils were added in the top chambers. Cell migration was measured after 2-h and 4-h incubation periods. (A) Percentage of RAW264.7 cell migration in the presence of SPLUNC1. A 100-ng volume of MCP-1 was used as a positive control, and buffer alone was used as a negative control. Data are presented as the means ± SD of three representative cell counts. (B) Percentage of neutrophil migration in the presence of SPLUNC1. A 1-μg volume of fMLP was used as a positive control and buffer alone as a negative control. Data are presented as the means ± SD of three representative cell counts. Both graphs showed statistically significance (*, P < 0.05) of the differences between the SPLUNC1 protein and buffer control results.
DISCUSSION
The human innate immune system keeps the microbial population in the internal environment under control. There are many players in this system, and each has its unique way to eliminate pathogenic microbes. Since oral and nasal areas are the main portals of microbial entry into the human body, these areas generally require maximum protection against invading pathogens. Recent investigations by our group and others have shown that SPLUNC1 is abundant in these areas and that its level increases considerably upon infection (1, 5, 11, 14, 15, 29). Our previous published in vivo study demonstrated that SPLUNC1 plays a major role in bacterial clearance (15) and that the increased expression of SPLUNC1 in mouse airways significantly enhanced survival of Pseudomonas-induced severe respiratory infection. Other investigators showed in vitro findings on the antibacterial action of SPLUNC1 on either the planktonic or biofilm forms (12, 22) of bacterial growth. However, the mechanism of the antimicrobial function of SPLUNC1 remains unexplained. Results from our current in vitro studies have shed light on the antibacterial mechanisms of SPLUNC1 during infection.
The SPLUNC1 gene has a sequence similarity to BPI, another antibacterial protein in the primary granules of polymorphonuclear leukocytes (PMN). Since airway epithelial cells do not usually express BPI, SPLUNC1 may have a biological activity similar to that of BPI but represents the epithelial cell-originated antibacterial response of the BPI-PLUNC (BPIF) protein family. The biological role of BPI has been studied extensively, and BPI has been shown to have functions in killing bacteria and in neutralizing the effects of bacterial LPS (30). Similar antibacterial activity was reported for SPLUNC1 in in vivo experiments, but the mechanism of antimicrobial function remains unclear (15). In this study, we successfully expressed and purified recombinant human SPLUNC1 in order to understand the biological role associated with this protein. The recombinant SPLUNC1 was biologically active and showed antibacterial activity when used against P. aeruginosa-pMF230. Our LPS binding assay indicated strong LPS binding activity with SPLUNC1 (Fig. 2).
SPLUNC1 showed antibacterial activity, although differences in dosages were required to achieve complete growth inhibition. P. aeruginosa-pMF230 showed a lower growth rate instead of a decline in the cell numbers after addition of exogenous SPLUNC1, which suggested that SPLUNC1 does not possess direct bacterial killing functions. Removal of bound SPLUNC1 with 0.1% Triton X-100 or PBS (Fig. 3A) reinstated normal bacterial growth, confirming that SPLUNC1-mediated antibacterial activity was due to the bacterial growth inhibition (bacteriostatic) and not to bacterial killing (bactericidal).
Our analysis suggested that the concentration ratio between bacteria and SPLUNC1 is critical for total growth inhibition (as indicated by the dosage versus antibacterial activity results in Fig. 1). Furthermore, we demonstrated that the growth inhibition was due to the formation of discrete pores in the outer membrane and that this may represent a key feature in the SPLUNC1-mediated antibacterial mechanism (Fig. 3B). A similar antibacterial characterization has been reported with two other immune proteins, attacin and collectin (31, 32). These proteins can also restrict the growth of bacteria through membrane permeabilization without directly killing bacteria. The McCormack laboratory demonstrated that the bacterial membrane integrity of E. coli K-12 could be compromised by surfactant protein D (SP-D) but also that the increased permeability did not lead to reduced bacterial viability. They used the thiol-specific fluorophore ThioGlo1 and the fluorescent probe STYO 9 to determine the release of thiol-containing proteins after the treatment and membrane permeabilization (32). Additionally, limited migration of ethidium bromide and no observed migration of propidium iodide dye in P. aeruginosa-pMF230 treated with SPLUNC1 suggested the formation of a relatively small pore size during growth inhibition. This estimated pore size was sufficient to reduce growth of the bacteria but not large enough to cause bacterial lysis. A similar finding (creation of a small pore and growth inhibition) was reported by Dowling et al. (33) when they treated bacteria (E. coli) with rotavirus VP5 protein.
Our immunostaining data clearly demonstrated the coating of P. aeruginosa-pMF230 by SPLUNC1. We did not observe any colocalization or equal intensities of the two fluorescence signals after the treatment with SPLUNC1 because the GFP was not functionally expressed in the inhibited bacterial cells (a similar observation in GFP fluorescent reduction/elimination in inhibited cells was reported by Kamau et al. [34]).
Since growth inhibition alone is not sufficient for complete bacterial removal, we looked for an additional role for SPLUNC1. We demonstrated that SPLUNC1 acts as an antimicrobial agent by coating the bacteria, resulting in the growth inhibition of bacteria. This activity keeps the number of bacterial cells under control and may allow time for other immune components to act for a complete removal of the pathogenic microorganism. Results from our cell migration assay clearly demonstrated that bacterium-bound SPLUNC1 can enhance the migration of macrophages and neutrophils to the site of infection. This may happen in real-life situations, and it could represent an additional, indirect role of SPLUNC1 in bacterial clearance.
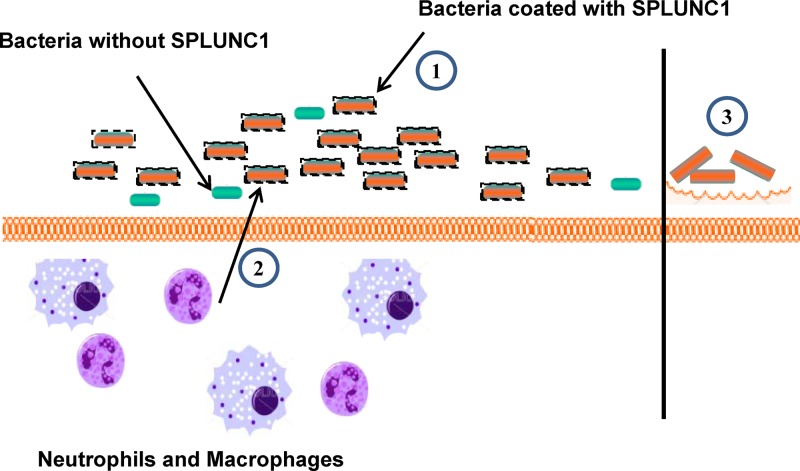
Based on our observation, we proposed a “SPLUNC1-mediated antimicrobial model” to describe the multiple functions of SPLUNC1 during infection as illustrated in Fig. 6. The main function of this protein appears to be to bind to microbes and arrest/inhibit microbial growth, thereby keeping the number of both endogenous and invading bacteria relatively low with its bacteriostatic effect. In the meantime, SPLUNC1 enhances the migration of phagocytic cells, which is important for the final elimination of microbes. Based on its abundant expression in upper respiratory tract, it is likely that SPLUNC1 also plays a similar antimicrobial role during the normal “noninfected” homeostasis stages to maintain a fairly sterile environment in airways to combat overgrowth of indigenous bacteria.
Fig 6.
Proposed model for the synergistic function of SPLUNC1. We have proposed that the SPLUNC1 secretion increases severalfold upon infection to keep up with the ratio between the protein and microbes. SPLUNC1 binds to bacteria upon contact and inhibits bacterial growth through its bacteriostatic activity by increasing the bacterial cell permeability, keeping the number of existing bacteria from significantly increasing (1). At the same time, SPLUNC1 recruits phagocytic cells to the site of infection to remove the controlled bacterial population (2). SPLUNC1 was previously shown to have surfactant properties, which were suggested to inhibit the biofilm form of bacterial growth (3). This cumulative function of SPLUNC1 controls the bacterial load and shows antimicrobial activity during bacterial infection.
Taken together, our data suggest the following: (i) the SPLUNC1 concentration increases during bacterial infection to coat the increased number of bacterial cells; (ii) SPLUNC1-mediated antibacterial properties likely act through a bacteriostatic effect to suppress the growth of bacteria, and this suppression is through the formation of small pores on the bacterial outer membrane; and (iii) SPLUNC1 enhances the migration of macrophages and neutrophils after its binding to bacteria for subsequent, successful removal of bacteria from sites of infection. We propose that these are the cumulative activities (direct and indirect) of SPLUNC1-mediated antimicrobial functions in controlling the bacterial load in the oral and nasopharyngeal regions. Further demonstrations of an association between the SPLUNC1 structural integrity and these multiple functions and characterizations of the potential motifs critical to the bacterial binding and growth inhibition, phagocyte migration, and LPS binding should provide insights into how SPLUNC1 interacts with bacteria and may provide an alternative approach for treating respiratory infection.
ACKNOWLEDGMENTS
This work was supported by grants from NIH/NHLBI (HL091938).
We thank Garth Ehrlich, Fen Z. Hu, and Azad Ahmed of the Allegheny Singer Research Institute, Allegheny General Hospital, Pittsburgh, PA, for providing Pseudomonas aeruginosa containing pMF230 (GFP) and allowing access to the confocal microscope facility. Special thanks to Bruce Pitt, Bruce McClane, Jihong Li, Kelly Brant, and K. Balasubramanian of the University of Pittsburgh and Susan Reynolds at National Jewish Health for their helpful suggestions. We also thank Marissa Di for her editorial assistance with respect to the manuscript.
Footnotes
Published ahead of print 6 November 2012
REFERENCES
- 1. Bingle CD, Craven CJ. 2002. PLUNC: a novel family of candidate host defence proteins expressed in the upper airways and nasopharynx. Hum. Mol. Genet. 11:937–943 [DOI] [PubMed] [Google Scholar]
- 2. Bingle CD, LeClair EE, Havard S, Bingle L, Gillingham P, Craven CJ. 2004. Phylogenetic and evolutionary analysis of the PLUNC gene family. Protein Sci. 13:422–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Di YP, Harper R, Zhao Y, Pahlavan N, Finkbeiner W, Wu R. 2003. Molecular cloning and characterization of spurt, a human novel gene that is retinoic acid-inducible and encodes a secretory protein specific in upper respiratory tracts. J. Biol. Chem. 278:1165–1173 [DOI] [PubMed] [Google Scholar]
- 4. Ghafouri B, Irander K, Lindbom J, Tagesson C, Lindahl M. 2006. Comparative proteomics of nasal fluid in seasonal allergic rhinitis. J. Proteome Res. 5:330–338 [DOI] [PubMed] [Google Scholar]
- 5. Ghafouri B, Kihlstrom E, Stahlbom B, Tagesson C, Lindahl M. 2003. PLUNC (palate, lung and nasal epithelial clone) proteins in human nasal lavage fluid. Biochem. Soc. Trans. 31:810–814 [DOI] [PubMed] [Google Scholar]
- 6. Ghafouri B, Kihlstrom E, Tagesson C, Lindahl M. 2004. PLUNC in human nasal lavage fluid: multiple isoforms that bind to lipopolysaccharide. Biochim. Biophys. Acta 1699:57–63 [DOI] [PubMed] [Google Scholar]
- 7. Ghafouri B, Stahlbom B, Tagesson C, Lindahl M. 2002. Newly identified proteins in human nasal lavage fluid from non-smokers and smokers using two-dimensional gel electrophoresis and peptide mass fingerprinting. Proteomics 2:112–120 [PubMed] [Google Scholar]
- 8. Bingle D, Craven CJ. 2003. Comparative analysis of the PLUNC (palate, lung and nasal epithelium clone) protein families. Biochem. Soc. Trans. 31:806–809 [DOI] [PubMed] [Google Scholar]
- 9. Di YP. 2011. Functional roles of SPLUNC1 in the innate immune response against Gram-negative bacteria. Biochem. Soc. Trans. 39:1051–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vitorino R, Lobo MJ, Ferrer-Correira AJ, Dubin JR, Tomer KB, Domingues PM, Amodo FM. 2004. Identification of human whole saliva protein components using proteomics. Proteomics 4:1109–1115 [DOI] [PubMed] [Google Scholar]
- 11. Campos MA, Abreu AR, Nlend MC, Cobas MA, Conner GE, Whitney PL. 2004. Purification and characterization of PLUNC from human tracheobronchial secretions. Am. J. Respir. Cell Mol. Biol. 30:184–192 [DOI] [PubMed] [Google Scholar]
- 12. Gakhar L, Bartlett JA, Penterman J, Mizrachi D, Singh PK, Mallampalli RK, Ramaswamy S, McCray PBJ. 2010. PLUNC is a novel airway surfactant protein with anti-biofilm activity. PLoS One 5:e9098 doi:10.1371/journal.pone.0009098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chu HW, Thaikoottathil J, Rino JG, Zhang G, Wu Q, Moss T, Refaeli Y, Bowler R, Wenzel SE, Chen Z, Zdunek J, Breed R, Young R, Allaire E, Martin RJ. 2007. Function and regulation of SPLUNC1 protein in Mycoplasma infection and allergic inflammation. J. Immunol. 179:3995–4002 [DOI] [PubMed] [Google Scholar]
- 14. Gally F, Di YP, Smith SK, Minor MN, Liu Y, Bratton DL, Frasch SC, Michels NM, Case SR, Chu HW. 2011. SPLUNC1 promotes lung innate defense against Mycoplasma pneumoniae infection in mice. Am. J. Pathol. 178:2159–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lukinskiene L, Liu Y, Reynolds SD, Steele C, Stripp BR, Leikauf GD, Kolls JK, Di YP. 2011. Antimicrobial activity of PLUNC protects against Pseudomonas aeruginosa infection. J. Immunol. 187:382–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McGillivary G, Bakaletz LO. 2010. The multifunctional host defense peptide SPLUNC1 is critical for homeostasis of the mammalian upper airway. PLoS One 5:e13224 doi:10.1371/journal.pone.0013224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bou R, Lorente L, Aguilar A, Perpiñán J, Ramos P, Peris M, Gonzalez D. 2009. Hospital economic impact of an outbreak of Pseudomonas aeruginosa infections. J. Hosp. Infect. 71:138–142 [DOI] [PubMed] [Google Scholar]
- 18. Driscoll JA, Brody SL, Kollef MH. 2007. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67:351–368 [DOI] [PubMed] [Google Scholar]
- 19. Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129–1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McCann S, Byrne JL, Rovira M, Shaw P, Ribaud P, Sica S, Volin L, Olavarria E, Mackinnon S, Trabasso P, VanLint MT, Ljungman P, Ward K, Browne P, Gratwohl A, Widmer AF, Cordonnier C. 2004. Outbreaks of infectious diseases in stem cell transplant units: a silent cause of death for patients and transplant programs. Bone Marrow Transplant. 33:519–529 [DOI] [PubMed] [Google Scholar]
- 21. Bartlett JA, Hicks BJ, Schlomann JM, Ramachandran S, Nauseef WM, Cray PBJ. 2008. PLUNC is a secreted product of neutrophil granules. J. Leukoc. Biol. 83:1201–1206 [DOI] [PubMed] [Google Scholar]
- 22. Zhou HD, Li XL, Li GY, Zhou M, Liu HY, Yang YX, Deng Y, Ma J, Sheng SR. 2008. Effect of SPLUNC1 protein on the Pseudomonas aeruginosa and Epstein-Barr virus. Mol. Cell. Biochem. 309:191–197 [DOI] [PubMed] [Google Scholar]
- 23. Garcia-Caballero A, Rasmussen JE, Gaillard E, Watson MJ, Olsen JC, Donaldson SH, Stutts MJ, Tarran R. 2009. SPLUNC1 regulates airway surface liquid volume by protecting ENaC from proteolytic cleavage. Proc. Natl. Acad. Sci. U. S. A. 106:11412–11417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Taylor PW. 1983. Bactericidal and bacteriolytic activity of serum against gram-negative bacteria. Microbiol. Rev. 47:46–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bantroch S, Bühler T, Lam JS. 1994. Appropriate coating methods and other conditions for enzyme-linked immunosorbent assay of smooth, rough, and neutral lipopolysaccharides of Pseudomonas aeruginosa. Clin. Diagn. Lab. Immunol. 1:55–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Loh B, Grant C, Hancock RE. 1984. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 26:546–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yona S, Hayhoe R, Avraham-Davidi I. 2010. Monocyte and neutrophil isolation and migration assays. Curr. Protoc. Immunol. 88:14.15.1–14.15.14 [DOI] [PubMed] [Google Scholar]
- 28. Hancock REW, Wong PGW. 1984. Compounds which increase the permeability of the Pseudomonas aeruginosa outer membrane. Antimicrob. Agents Chemother. 26:48–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chu HW, Gally F, Thaikoottathil J, Janssen-Heininger YM, Wu Q, Zhang G, Reisdorph N, Case S, Minor M, Smith S, Jiang D, Michels N, Simon G, Martin RJ. 2010. SPLUNC1 regulation in airway epithelial cells: role of Toll-like receptor 2 signaling. Respir. Res. 11:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Elsbach P. 1998. The bactericidal/permeability-increasing protein (BPI) in antibacterial host defense. J. Leukoc. Biol. 64:14–18 [DOI] [PubMed] [Google Scholar]
- 31. Carlsson A, Nyström T, de Cock H, Bennich H. 1998. Attacin—an insect immune protein—binds LPS and triggers the specific inhibition of bacterial outer-membrane protein synthesis. Microbiology 144(Pt 8):2179–2188 [DOI] [PubMed] [Google Scholar]
- 32. Wu H, Kuzmenko A, Wan S, Schaffer L, Weiss A, Fisher JH, Kim KS, McCormack FX. 2003. Surfactant proteins A and D inhibit the growth of Gram-negative bacteria by increasing membrane permeability. J. Clin. Invest. 111:1589–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dowling W, Denisova E, LaMonica R, Mackow ER. 2000. Selective membrane permeabilization by the rotavirus VP5* protein is abrogated by mutations in an internal hydrophobic domain. J. Virol. 74:6368–6376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kamau SW, Grimm F, Hehl AB. 2001. Expression of green fluorescent protein as a marker for effects of antileishmanial compounds in vitro. Antimicrob. Agents Chemother. 45:3654–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]