Abstract
Anaplasma phagocytophilum, a member of the family Anaplasmataceae, is the tick-transmitted obligate intracellular bacterium that causes human granulocytic anaplasmosis. The life cycle of A. phagocytophilum is biphasic, transitioning between the noninfectious reticulate cell (RC) and infectious dense-cored (DC) forms. We analyzed the bacterium's DC surface proteome by selective biotinylation of surface proteins, NeutrAvidin affinity purification, and mass spectrometry. Transcriptional profiling of selected outer membrane protein candidates over the course of infection revealed that aph_0248 (designated asp14 [14-kDa A. phagocytophilum surface protein]) expression was upregulated the most during A. phagocytophilum cellular invasion. asp14 transcription was induced during transmission feeding of A. phagocytophilum-infected ticks on mice and was upregulated when the bacterium engaged its receptor, P-selectin glycoprotein ligand 1. Asp14 localized to the A. phagocytophilum surface and was expressed during in vivo infection. Treating DC organisms with Asp14 antiserum or preincubating mammalian host cells with glutathione S-transferase (GST)–Asp14 significantly inhibited infection of host cells. Moreover, preincubating host cells with GST-tagged forms of both Asp14 and outer membrane protein A, another A. phagocytophilum invasin, pronouncedly reduced infection relative to treatment with either protein alone. The Asp14 domain that is sufficient for cellular adherence and invasion lies within the C-terminal 12 to 24 amino acids and is conserved among other Anaplasma and Ehrlichia species. These results identify Asp14 as an A. phagocytophilum surface protein that is critical for infection, delineate its invasion domain, and demonstrate the potential of targeting Asp14 in concert with OmpA for protecting against infection by A. phagocytophilum and other Anaplasmataceae pathogens.
INTRODUCTION
Members of the family Anaplasmataceae are major causes of tick-borne illness in humans and animals (1, 2). These obligate intracellular bacteria infect cells of hematopoietic origin or endothelial cells to reside in host cell-derived vacuoles. Anaplasma phagocytophilum, an Anaplasmataceae member, is the causative agent of granulocytic anaplasmosis in humans (human granulocytic anaplasmosis [HGA]) and animals. A. phagocytophilum is transmitted primarily by Ixodes sp. ticks (3). However, it can also be transmitted perinatally (4, 5) or by blood transfusion (6–9), and it has been suggested to be transmitted nosocomially (10, 11). HGA is an emerging infection in the United States, Europe, and Asia (3, 12). Since becoming a reportable disease in the United States in 1999 (13), the number of clinically documented HGA cases has risen annually, with an increase of >50% between 2000 and 2007 (3, 12–14). HGA is a febrile infection associated with laboratory findings that can include leukopenia, thrombocytopenia, elevated serum transaminase levels, and increased susceptibility to potentially fatal opportunistic infections. The hallmark of HGA is A. phagocytophilum colonization of neutrophils (3).
A. phagocytophilum undergoes a biphasic developmental cycle, transitioning between an infectious dense-cored (DC) form and a noninfectious, replicative reticulate cell (RC) form. DC bacteria are able to invade and infect promyelocytic and endothelial cell lines (15–22). Within 4 h of adherence, DC organisms enter host cell-derived vacuoles (23–26). Between 4 and 8 h after adherence, DC bacteria transition to the RC form and initiate replication to produce bacterium-filled vacuoles called morulae. The majority of RCs develop into DCs between 28 and 32 h. DC organisms are released between 32 and 36 h, initiating new waves of infections (26). The DC form presents an ideal subject for identifying outer membrane proteins (OMPs) that promote infection.
The ability of intracellular bacteria to colonize eukaryotic host cells depends on interactions between bacterial invasins and host cell receptors. P-selectin glycoprotein ligand 1 (PSGL-1) is the only confirmed A. phagocytophilum receptor (27, 28). We recently identified A. phagocytophilum outer membrane protein A (OmpA) as an invasin that recognizes the α2,3-sialic acid determinant of the sialyl Lewis X (sLex) tetrasaccharide, which caps PSGL-1 on myeloid cells and decorates unknown glycoprotein receptors on endothelial cell surfaces. Using glutathione S-transferase (GST)–OmpA as an agonist reduces A. phagocytophilum infection of host cells by approximately 50% (29). Moreover, the OmpA invasin domain is conserved among Anaplasma and Ehrlichia species (29), the latter of which are other Anaplasmataceae members that cause potentially deadly tick-borne diseases (30). These findings collectively serve as both a promising advance toward the development of standardized vaccines that protect against infection by Anaplasmataceae pathogens and a reminder that additional bacterial ligand-receptor interactions promote cellular invasion via redundant and/or complementary routes (2, 27, 28, 31–35). Identifying additional A. phagocytophilum invasins that are conserved among the Anaplasmataceae is desirable because targeting them in concert with OmpA has the potential to more completely inhibit infection by these pathogens.
MATERIALS AND METHODS
Cell lines and cultivation of uninfected and A. phagocytophilum-infected HL-60 cells.
Uninfected HL-60 cells (CCL-240; American Type Culture Collection [ATCC], Manassas, VA) and HL-60 cells infected with the A. phagocytophilum NCH-1 strain or a transgenic HGE1 strain expressing green fluorescent protein (GFP) (36) were cultivated as described previously (33). Spectinomycin (100 μg/ml; Sigma-Aldrich, St. Louis, MO) was added to HL-60 cultures harboring transgenic HGE1 bacteria. Uninfected and A. phagocytophilum-infected RF/6A rhesus monkey choroidal endothelial cells (ATCC CRL-1780) and Ixodes scapularis embryo-derived ISE6 cells were cultivated as described previously (16).
A. phagocytophilum DC organism surface biotinylation and affinity purification.
A. phagocytophilum DC organisms were selectively enriched from 109 infected (≥90%) HL-60 cells by sonication and differential centrifugation as previously described (17). Electron microscopic examination of sonicated samples confirmed the presence of DC but not RC bacteria. To purify DC organisms away from the majority of contaminating host and RC organism cellular debris, the sonicate was fractionated using discontinuous Renografin (diatrizoate sodium; Bracco Diagnostics, Princeton, NJ) density gradient centrifugation (37). For biotinylation of surface proteins, purified DC organisms were resuspended in 1 ml of phosphate-buffered saline (PBS) (pH 8.0) containing 1 mM MgCl2 and 10 mM sulfo-NHS-SS-biotin (Pierce, Rockland, IL) and incubated for 30 min at room temperature. Free sulfo-NHS-SS-biotin was quenched by washing the sample with 50 mM Tris (pH 8.0), followed by two washes with PBS. Biotinylated bacteria were solubilized in radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 1 mM sodium orthovanadate, 1 mM sodium fluoride, and Complete EDTA-free protease inhibitor cocktail [Roche, Indianapolis, IN]) on ice for 1 h. Every 20 min during the 1-h incubation, the sample was subjected to eight 8-s bursts on ice interspersed with 8-s rest periods, using a Misonix S4000 ultrasonic processor (Misonix, Farmingdale, NY) on an amplitude setting of 30. Insoluble material was removed by spinning at 10,000 × g for 10 min at 4°C. To purify biotinylated proteins, the clarified lysate was mixed with high-capacity NeutrAvidin agarose beads (Pierce) by end-over-end rotation overnight at 4°C. The gel slurry was pelleted by centrifugation at 1,000 × g for 1 min. After removal of the supernatant, the beads were resuspended in 8 ml PBS and parceled into 10 800-μl aliquots, each of which was added to a spin column optimized for affinity purification (Pierce). The columns were washed three times with PBS and centrifuged at 1,000 × g to remove any nonbiotinylated proteins. The captured biotinylated proteins were eluted from the beads by end-over-end rotation with 150 mM dithiothreitol (DTT) in 0.25% sodium deoxycholate for 2 h at room temperature. The agarose beads were centrifuged at 1,000 × g for 2 min, and the supernatant containing the biotinylated proteins was saved. The Bradford assay was used to determine the protein concentration of the eluate (38). To ensure that this procedure had enriched DC bacterial surface proteins, an aliquot of the affinity-purified sample was resolved by SDS-PAGE alongside an A. phagocytophilum whole-cell lysate, NeutrAvidin beads plus unlabeled DC whole-cell lysate, and NeutrAvidin beads alone, followed by silver staining (39).
Two-dimensional (2D) LC-MS/MS proteome analysis.
Unless otherwise stated, all buffers were made with liquid chromatography-mass spectrometry (LC-MS)-grade solvents (Fisher Chemical, Fairlawn, NJ). Samples were processed for proteomic analysis (40, 41). Following biotinylation enrichment of A. phagocytophilum surface proteins, 300 μg of protein mass in 400 μl of lysis buffer was concentrated and exchanged into 25 μl of ammonium bicarbonate buffer (ABC; 50 mM NH4CO3–0.05% C24H39O4Na) by use of a Centriprep YM-10 filter unit (Millipore, Billerica, MA). DTT was added to achieve a final concentration of 20 mM, and disulfide bonds were reduced at 90°C for 30 min. After cooling to room temperature, cysteine alkylation was performed on the sample by incubation with freshly prepared iodoacetamide (32 mM) for 30 min at room temperature in the dark. Trypsin Gold (100 ng/μl; Promega, Madison, WI) was added to a final enzyme/protein ratio of 1:100, and the sample was incubated at 37°C overnight. The digested sample was dried within a speed vacuum and stored dry at −20°C.
The digested sample was reconstituted in 60 μl of 100 mM ammonium formate (pH 10) for multidimensional peptide separation and mass spectrometry analysis on a 2D-nanoAcquity chromatography system online with a Synapt quadrupole–time-of-flight tandem mass spectrometer (Waters) (41). Two replicate injections were analyzed for the sample. Resulting data were processed using ProteinLynx Global (PLGS) software, v2.4 (Waters), as described elsewhere (41, 42). Data were then searched against an A. phagocytophilum-specific FASTA database (RefSeq and UniProt sources; downloaded February 2010) and its reversed sequences as a decoy database. Search parameters required a minimum precursor ion intensity of 500 counts, two or more peptide sequences per protein, and a minimum of seven matching fragment ions. Trypsin selectivity was specified, allowing for 1 missed cleavage event and variable methionine oxidation. Using a decoy database method, the score threshold was calculated at the 5% false discovery rate. Confidence in protein identification was also increased for those that were identified against both the RefSeq and UniProt A. phagocytophilum databases.
In silico analyses.
SignalP 3.0 (43) was used to predict if candidates carried signal peptide sequences. TMPred (www.ch.embnet.org/software/TMPRED_form.html) was used to predict if candidates carried transmembrane domains. Alignments of Asp14 and its homologs from other Anaplasma and Ehrlichia species were generated using Clustal W (44).
Differential gene expression studies.
Synchronous infections of HL-60 cells with A. phagocytophilum DC organisms were established (26). Microscopic examination of aliquots recovered at 24 h confirmed that ≥60% of HL-60 cells were infected. The infection time course proceeded for 36 h at 37°C in a humidified atmosphere of 5% CO2. The length of the time course allowed for the bacteria to complete their biphasic developmental cycle and initiate a second round of infection (26). Every 4 h, aliquots were processed for RNA isolation. Reverse transcriptase quantitative PCR (RT-qPCR) was performed (41) using the gene-specific primers listed in Table 1. Relative transcript levels for each target were normalized to the transcript levels of the A. phagocytophilum 16S rRNA gene (aph_1000), using the 2−ΔΔCT method (45). To facilitate identification of genes that were upregulated in the infectious DC form compared to the noninfectious RC form, normalized transcript levels for each gene at each time point were calculated as fold changes in expression relative to expression at 16 h, a time point at which the A. phagocytophilum population consists exclusively of RC organisms (26). To monitor asp14 expression during the tick blood meal, A. phagocytophilum-infected I. scapularis nymphs were fed on C3H/HeJ mice, followed by RNA extraction from salivary glands of transmission-fed, unfed, and uninfected control nymphs (46), and RT-qPCR was performed using asp14-specific primers.
Table 1.
Oligonucleotides used in this study
| Designation | Sequence (5′ to 3′)a | Targeted nucleotides |
|---|---|---|
| Ap 16S-527F | TGTAGGCGGTTCGGTAAGTTAAAG | 527–550 (+ strand) |
| Ap 16S-753R | GCACTCATCGTTTACAGCGTG | 753–773 (− strand) |
| aph_0240-731F | CCATTGCTCATCATAGCTGAAGAC | 731–754 (+ strand) |
| aph_0240-927R | CATCTTAACAGCAAGCTCGTCATTT | 927–951 (− strand) |
| aph_0248-012F | AGCTCCTTGGAAGAGCATTTCGG | 12–34 (+ strand) |
| aph_0248-211R | TCTGCCCGACAGTTCCAGTATCG | 189–211 (− strand) |
| aph_0248-001F-ENTR | CACCATGATACCATTAGCTCCTTGGAAGAGC | 1–27 (+ strand) |
| aph_0248-181F-ENTR | CACCCAGCAAGACGATACTGGAACTG | 181–202 (+ strand) |
| aph_0248-192R | CTAATCGTCTTGCTGCATACTAGACGC | 169–192 (− strand) |
| aph_0248-228R | CTAACCCTCGCCGGACTCTATCT | 209–228 (− strand) |
| aph_0248-264R | CTACTGCTGAACTTGCTCGTCAGACAAC | 240–264 (− strand) |
| aph_0248-300R | CTAATCATCTTTGAACTCTTCCCGAATGCTATTCATC | 264–300 (− strand) |
| aph_0248-336R | CTATTCAAGCTTGAGAATCCTTCTTTTAATAGCCC | 305–336 (− strand) |
| aph_0248-375R | TTAGCTTTCTTTAGGAGTATTGGCACCGTAA | 345–375 (− strand) |
| aph_0346-229F | AGATACGATGACATGAGGGATTTGA | 229–252 (+ strand) |
| aph_0346-401R | TTCTTCACTGGAGCACCAAAGTATC | 401–425 (− strand) |
| aph_0404-258F | GCATCTAGATTGGAGCGAAGATT | 258–280 (+ strand) |
| aph_0404-407R | CTGTTTATGAAGGCCTTTCCAACTT | 407–432 (− strand) |
| aph_0405-409F | GAGAGCAGGGTTTTTGCTGATACT | 409–432 (+ strand) |
| aph_0405-580R | GAAAAGGTACACCCTTCGCATACTT | 580–605 (− strand) |
| aph_0441-341F | GTGTAATGTACAATGCTGGTGAGC | 341–365 (+ strand) |
| aph_0441-491R | GATTCGTCAACCCTTTCTTCTTT | 491–514 (− strand) |
| aph_0625-444F | GATGTTGGGGCTTATGGAGTTGAAGG | 444–469 (+ strand) |
| aph_0625-0621R | AACTGAACCACACGTCGCAGC | 601–621 (− strand) |
| aph_0874-250F | GGTAGATGAGGAGGCATTAAATCA | 250–273 (+ strand) |
| aph_0874-497R | GTAGACAGGACGCGATAACTAGC | 497–519 (− strand) |
| aph_1032-910Fb | CAAGGCGGAAGTATACGTATTGAA | 910–933 (+ strand) |
| aph_1032-1096Rb | CAGGCTTATCCAACGCTACCTCTAT | 1096–1120 (− strand) |
| aph_1049-124F | ACTGTGGTTGAACAAGCGCCG | 124–144 (+ strand) |
| aph_1049-288F | TTGCGATACCATACCAAGTTCCGC | 265–288 (− strand) |
| aph_1170-53F | GTGATGCCAAAGGGATTGCACC | 53–75 (+ strand) |
| aph_1170-233R | GTGCCACGCATATCCGAGTTG | 233–254 (− strand) |
| aph_1210-024F | CCTGATGGCAACGATGTTTGCC | 24–45 (+ strand) |
| aph_1210-172R | ATCTTCCCTGTGTTATCTCCTCCGG | 148–172 (− strand) |
| aph_1359-387F | TTTCTTTAGTGAGCAGCACGGGG | 387–409 (+ strand) |
| aph_1359-573R | AAACCCTGCACATACATAAGGCCTC | 549–573 (− strand) |
| msp2 (P44)- 130Fc | GGTTTGGATTACAGTCCAGCGTTTAG | 130–156 (+ strand) |
| msp2 (P44)-278Rc | GGATCAGGTGTGTTCCAGTCAAACT | 278–302 (− strand) |
Underlined nucleotides correspond to a Gateway entry vector-compatible sequence. Underlined and italicized nucleotides correspond to an added stop codon.
aph_0278 and aph_1032, both of which encode EF-Tu, are 100% identical at the nucleotide level.
The msp2 (P44) primer set targets nucleotides that encode the N-terminal conserved region of all Msp2 (P44) paralogs. The designated nucleotide positions correspond to those of aph_1221, which encodes Msp2 (P44)-18.
Recombinant protein and antiserum production.
A. phagocytophilum genes of interest were amplified using the primers listed in Table 1 and Platinum Pfx DNA polymerase (Invitrogen). Amplicons were cloned into pENTR/TEV/D-TOPO (Invitrogen) (18) to yield pENTR candidate gene entry plasmids harboring the genes of interest. Plasmid inserts were sequenced to verify their integrity. Recombination of the candidate gene insert downstream of and in frame with the gene encoding GST was achieved using the pDest-15 vector (Invitrogen) (18). GST-Asp14, GST–Omp-1A, and GST were affinity purified and used to generate murine polyclonal antisera (41).
Western blotting and spinning disk confocal microscopy.
Antisera generated in this study and previous studies targeted Asp14, outer membrane protein 1A (Omp-1A), APH_0032, Asp55 (55-kDa A. phagocytophilum surface protein), major surface protein 2 (Msp2 [P44]) (17, 47–49), and VirB9 (50). Western blot analyses were performed as described previously (41). A. phagocytophilum-infected HL-60 cells were processed and analyzed via indirect immunofluorescence using spinning disk confocal microscopy (37, 51).
Surface trypsin digestion of intact A. phagocytophilum DC organisms.
Intact A. phagocytophilum DC organisms were subjected to surface trypsinolysis (29, 52). Lysates of trypsin- and PBS-treated DC bacteria were resolved by SDS-PAGE, transferred to a nitrocellulose membrane, and screened with antibodies targeting Asp14, Asp55 (47), Omp-1A, and APH_0032 (17).
Flow cytometry.
Host cell-free GFP-expressing transgenic HGE1 organisms were prepared as described previously (26), followed by resuspension in PBS containing preimmune mouse serum, serum from a mouse that had been infected experimentally with A. phagocytophilum (53), mouse anti-Asp14, or a secondary antibody control (rabbit anti-mouse IgG conjugated to Alexa Fluor 594; BD Biosystems). Antibody incubations and wash steps were performed as described previously (29, 32).
Asp14 antibody inhibition of A. phagocytophilum infection.
DC organisms were incubated with heat-inactivated mouse polyclonal antiserum raised against GST or GST-Asp14 (1.5 mg/ml), after which bacterial adhesion to and infection of HL-60 cells were monitored (29).
Binding of GST-Asp14 to mammalian host cells and competitive inhibition of A. phagocytophilum infection.
For recombinant protein binding studies, RF/6A or HL-60 cells were incubated with 4 μM GST, GST-tagged Asp14 or fragments thereof, or GST-tagged APH_1387 amino acids 112 to 559 (GST-APH_1387112–579) at 37°C for 1 h. In some experiments, HL-60 cells were incubated with 4 μM (each) GST-Asp14 and GST-OmpA or 8 μM GST as a control. Binding of recombinant proteins to host cells and competitive inhibition of A. phagocytophilum infection were assayed (29).
Statistical analyses.
Statistical analyses were performed using the Prism 5.0 software package (Graphpad, San Diego, CA). If one-way analysis of variance (ANOVA) indicated a group difference (P < 0.05), then Dunnett's post hoc test was used to test for a significant difference among groups. In some instances, the Student t test was used to assess statistical significance. Statistical significance was set at P values of <0.05.
RESULTS
Proteomic analysis of affinity-purified A. phagocytophilum DC surface proteins identifies novel outer membrane protein candidates.
To identify DC surface proteins, DC organisms were surface labeled with biotin and solubilized, and biotinylated proteins were captured by NeutrAvidin affinity chromatography. Comparison of the banding patterns of the input lysate and eluate revealed enrichment for many proteins (Fig. 1). With the exception of 44-kDa and 70-kDa proteins, both of which were recovered in low abundances, nonbiotinylated DC whole-cell lysate proteins did not bind to NeutrAvidin beads (Fig. 1). Eluted biotinylated proteins were subjected to 2D LC-MS/MS analysis. Resulting data were searched against A. phagocytophilum-specific FASTA databases (RefSeq and UniProt sources). Table 2 summarizes a total of 56 identified A. phagocytophilum proteins, 47 of which were identified in both the RefSeq and UniProt sources. All proteins for which at least 2 peptides were identified from either the RefSeq or UniProt source and scored above a 5% false discovery rate cutoff are listed. Nine proteins had previously been delineated as being surface localized (47), thereby validating the efficacy of our approach. Ten paralogs of the Msp2 (P44) family (54) were identified, 8 of which yielded the highest PLGS scores.
Fig 1.
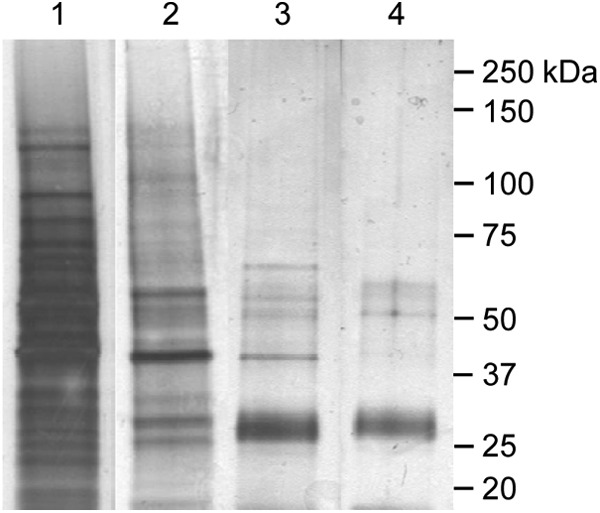
NeutrAvidin affinity purification of biotinylated A. phagocytophilum surface proteins. DC bacteria were purified to remove the majority of contaminating host cellular debris. DC surface proteins labeled with sulfo-NHS-SS-biotin were recovered by NeutrAvidin affinity chromatography. Aliquots of input host cell-free DC lysate (lane 1), affinity-captured DC surface proteins (lane 2), NeutrAvidin beads plus unlabeled DC whole-cell lysate (lane 3), and NeutrAvidin beads alone (lane 4) were resolved by SDS-PAGE followed by silver staining.
Table 2.
A. phagocytophilum DC proteins recovered post-surface labeling and -affinity chromatography and analyzed by 2D nano-LC-MS/MS protein analysis
| Locusc | JCVI description | Molecular mass (Da) | pI | RefSeq dataa |
UniProt datab |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Score | No. of peptides | Coverage (%) | Amount (fmol)d | Score | No. of peptides | Coverage (%) | Amount (fmol)d | ||||
| APH_1221 | P44 18ES outer membrane protein expression locus with P44-18 | 45,799 | 5.6 | 20,608.4 | 131 | 78.0 | 269.0 | 20,363.3 | 133 | 78.0 | 222.1 |
| APH_1287 | P44 32 outer membrane protein | 44,350 | 5.4 | 19,848.5 | 137 | 73.9 | 343.3 | 19,451.2 | 137 | 75.1 | 375.9 |
| APH_1229 | P44 2b outer membrane protein | 44,884 | 5.2 | 18,321.9 | 138 | 81.4 | 29.1 | 17,898.4 | 135 | 76.5 | 31.7 |
| APH_1169 | P44 19 outer membrane protein | 33,033 | 5.3 | 18,185.8 | 62 | 82.3 | 524.8 | 17,902.6 | 65 | 82.3 | 633.6 |
| APH_1269 | P44 16 outer membrane protein | 45,261 | 5.6 | 16,839.7 | 114 | 69.0 | 314.4 | 16,779.1 | 116 | 69.0 | 550.3 |
| APH_1275 | P44 16b outer membrane protein | 45,194 | 5.9 | 16,695.3 | 122 | 78.0 | 44.4 | 16,427.1 | 122 | 78.0 | 37.5 |
| APH_1215 | P44 14 outer membrane protein | 46,133 | 5.4 | 13,580.3 | 129 | 77.1 | 788.0 | 13,490.5 | 123 | 76.7 | 664.6 |
| APH_0172 | P44 outer membrane protein C-terminal fragment | 7,236 | 4.4 | 11,994.3 | 18 | 94.0 | 0 | 11,807.8 | 18 | 94.0 | 0 |
| APH_1235 | Hypothetical protein | 14,762 | 5.3 | 4,190.9 | 33 | 91.8 | 189.4 | 4,998.2 | 30 | 97.0 | 189.4 |
| APH_0240 | Chaperonin GroEL | 58,263 | 5.0 | 1,436.7 | 69 | 68.7 | 76.9 | 1,403.2 | 64 | 71.6 | 76.9 |
| APH_0494 | FoF1 ATP synthase subunit beta | 51,478 | 4.8 | 641.4 | 32 | 58.9 | 40.8 | 628.2 | 30 | 70.1 | 40.8 |
| APH_0405 | Asp62 outer membrane protein | 57,538 | 9.5 | 489.5 | 27 | 45.5 | 84.9 | 471.9 | 21 | 38.6 | 106.4 |
| APH_1087 | Putative competence lipoprotein ComL | 26,084 | 4.8 | 458.2 | 10 | 36.9 | 32.9 | 519.9 | 10 | 36.9 | 32.9 |
| APH_1032 | Elongation factor Tu | 42,831 | 5.1 | 415.5 | 19 | 44.8 | 0.0 | 398.1 | 19 | 35.1 | 51.1 |
| APH_1190 | Putative ATP synthase Fo B subunit | 18,837 | 5.9 | 415.5 | 2 | 14.4 | 31.7 | 458.5 | 10 | 47.9 | 31.7 |
| APH_0404 | Asp55 outer membrane protein | 63,644 | 8.9 | 413.1 | 21 | 26.8 | 49.3 | 413.9 | 22 | 25.8 | 49.3 |
| APH_0397 | 30S ribosomal protein S2 | 32,118 | 9.2 | 406.4 | 12 | 32.8 | 66.8 | 392.5 | 12 | 36.1 | 66.8 |
| APH_0036 | Co chaperone GrpE | 22,646 | 5.8 | 394.7 | 4 | 33.2 | 0.0 | 372.7 | 4 | 33.2 | 0 |
| APH_1404 | Type IV secretion system protein VirB10 | 46,871 | 4.7 | 388.9 | 8 | 22.8 | 34.5 | 379.4 | 7 | 21.7 | 34.5 |
| APH_0346 | Chaperone protein DnaK | 69,676 | 4.9 | 381.2 | 25 | 34.4 | 177.7 | 380.1 | 24 | 36.4 | 177.7 |
| APH_0248 | Hypothetical protein (Asp14) | 13,824 | 4.9 | 359.0 | 10 | 58.1 | 0 | ||||
| APH_1049 | Major surface protein 5 | 23,341 | 4.7 | 353.7 | 4 | 22.5 | 170.6 | 339.9 | 3 | 22.5 | 170.6 |
| APH_1334 | FoF1 ATP synthase subunit alpha | 54,068 | 5.3 | 312.1 | 30 | 34.8 | 180.0 | 270.5 | 23 | 28.5 | 0 |
| APH_0051 | Iron binding protein | 37,317 | 5.2 | 252.9 | 4 | 14.6 | 0 | 318.8 | 5 | 17.9 | 109.1 |
| APH_0853 | Hypothetical protein | 10,833 | 9.3 | 249.9 | 4 | 62.9 | 0 | 162.7 | 1 | 15.5 | 0 |
| APH_0625 | Immunogenic protein; membrane transporter | 34,653 | 5.9 | 229.0 | 6 | 28.6 | 0 | 207.9 | 5 | 28.6 | 0 |
| APH_1050 | Putative phosphate ABC transporter; periplasmic phosphate binding protein | 37,567 | 5.6 | 221.0 | 3 | 16.5 | 0 | 192.1 | 1 | 2.7 | 0 |
| APH_1246 | Glutamine synthetase type I | 52,383 | 6.0 | 216.0 | 9 | 10.2 | 0 | 228.0 | 10 | 10.2 | 0 |
| APH_1232 | Citrate synthase I | 45,591 | 5.8 | 213.8 | 5 | 19.7 | 0 | 151.0 | 2 | 3.6 | 0 |
| APH_0600 | Thiamine biosynthesis protein ThiC | 61,522 | 6.0 | 203.3 | 4 | 11.0 | 0 | 206.0 | 4 | 13.5 | 0 |
| APH_0059 | Phenylalanyl tRNA synthetase alpha subunit | 39,277 | 6.5 | 197.0 | 7 | 14.0 | 0 | 180.0 | 8 | 11.4 | 0 |
| APH_0555 | Cysteinyl tRNA synthetase | 51,774 | 5.8 | 192.8 | 5 | 18.6 | 0 | 197.2 | 4 | 16.0 | 0 |
| APH_0794 | Hypothetical protein | 27,119 | 7.1 | 183.9 | 2 | 8.4 | 0 | 164.8 | 1 | 4.2 | 0 |
| APH_0740 | AnkA | 131,081 | 6.1 | 182.8 | 11 | 7.2 | 0 | 189.2 | 13 | 8.2 | 0 |
| APH_1258 | Fructose bisphosphate aldolase | 32,685 | 6.7 | 182.0 | 5 | 9.2 | 0 | 193.7 | 4 | 9.2 | 0 |
| APH_1025 | 50S ribosomal protein L7 L12 | 14,122 | 4.8 | 181.5 | 2 | 23.9 | 0 | ||||
| APH_1292 | Cell division protein FtsZ | 41,975 | 5.0 | 181.3 | 3 | 13.3 | 0 | 205.0 | 3 | 10.5 | 0 |
| APH_1210 | OMP85 family outer membrane protein | 85,652 | 8.5 | 173.9 | 7 | 8.3 | 0 | 165.5 | 6 | 5.7 | 0 |
| APH_0283 | 50S ribosomal protein L2 | 29,772 | 11.5 | 169.5 | 3 | 8.3 | 0 | 154.1 | 2 | 6.2 | 0 |
| APH_0893 | Heat shock protein 90 | 71,123 | 4.9 | 167.9 | 6 | 12.7 | 0 | 173.7 | 9 | 17.0 | 0 |
| APH_0111 | Uridylate kinase | 26,347 | 6.9 | 164.4 | 2 | 13.1 | 0 | 176.4 | 3 | 18.0 | 0 |
| APH_0608 | PpiC parvulin rotamase family protein | 67,363 | 4.9 | 161.4 | 10 | 13.1 | 0 | 144.2 | 8 | 9.0 | 0 |
| APH_1359 | Major outer membrane protein Omp-1A | 31,617 | 9.0 | 157.8 | 2 | 5.5 | 0 | 142.4 | 2 | 5.5 | 0 |
| APH_1084 | Cytochrome c oxidase subunit II | 29,873 | 6.1 | 155.0 | 3 | 13.0 | 0 | ||||
| APH_0422 | Acetylglutamate kinase | 35,726 | 4.6 | 151.9 | 2 | 7.0 | 0 | ||||
| APH_0971 | Putative trigger factor | 49,358 | 4.8 | 140.8 | 3 | 13.0 | 0 | 138.3 | 2 | 10.0 | 0 |
| APH_0038 | CTP synthetase | 59,416 | 5.5 | 139.6 | 2 | 5.9 | 0 | 136.9 | 2 | 5.9 | 0 |
| APH_1355 | P44 79 outer membrane protein | 50,321 | 8.7 | 139.0 | 2 | 3.9 | 0 | 147.7 | 2 | 4.6 | 0 |
| APH_0669 | Bifunctional proline dehydrogenase-pyrroline 5 carboxylate dehydrogenase | 114,508 | 5.1 | 139.0 | 4 | 6.9 | 0 | 159.1 | 5 | 7.6 | 0 |
| APH_0450 | ATP-dependent Clp protease; ATP-binding subunit ClpA | 86,715 | 6.2 | 138.0 | 2 | 1.6 | 0 | ||||
| APH_0231 | Leucyl aminopeptidase | 54,611 | 5.5 | 128.8 | 3 | 11.4 | 0 | ||||
| APH_0874 | Hypothetical protein | 115,420 | 6.6 | 123.2 | 5 | 2.9 | 0 | ||||
| APH_1017 | Outer membrane protein of Msp2 family | 46,971 | 8.4 | 131.9 | 2 | 3.6 | 0 | ||||
| APH_1339 | Conserved domain protein | 47,356 | 7.3 | 128.6 | 2 | 5.1 | 0 | ||||
| APH_0168 | Heme exporter protein CcmC | 26,310 | 9.5 | 126.7 | 4 | 6.9 | 0 | ||||
| APH_0502 | tRNA pseudouridine synthase A | 28,012 | 8.8 | 131.9 | 2 | 3.6 | 0 | ||||
Refseq data for A. phagocytophilum (downloaded February 2010).
UniProt data for A. phagocytophilum (downloaded February 2010).
Proteins that were previously confirmed to be on the A. phagocytophilum surface and/or were recovered by surface biotinylation and affinity chromatography in the study by Ge and Rikihisa (47) are denoted in bold.
Peptides that are considered in-source fragments were given a 0-fmol value, as their quantification was confounded by the signal lost within the mass spectrometer.
Selection of A. phagocytophilum OMP candidates.
Several candidates were selected for differential gene expression analysis. Asp14, APH_0625, and APH_0874 were chosen because they were uncharacterized (hitherto hypothetical) proteins. APH_0441, APH_1049 (Msp5), APH_1170, APH_1210 (Omp85), and APH_1359 (Omp-1A) were selected because they are confirmed Anaplasma sp. proteins that are uncharacterized and/or have yet to be studied for differential gene expression (47). APH_0240 (the chaperonin GroEL), APH_0346 (DnaK), and APH_1032 (the elongation factor Tu) were chosen because these housekeeping proteins have been shown to moonlight as surface proteins in A. phagocytophilum and other bacteria. Moreover, these proteins have been implicated as adhesins in other bacteria (47, 55–64). Of the 11 candidates, Omp-1A, Omp85, Msp5, and APH_0441 carry predicted N-terminal signal peptide sequences. All except for Asp14 have one or more predicted transmembrane domains.
Differential transcription profiling of OMP candidate genes.
To gain insight into the transcription of the 11 genes of interest during A. phagocytophilum infection, we synchronously infected HL-60 cells with DC organisms and allowed the infection to proceed until the biphasic developmental cycle was complete and a second round of infection had begun (26). Based on when during infection they were expressed, genes of interest were classified as early (0 to 12 h)-, mid (12 to 24 h)-, or late (24 to 36 h)-stage genes (Fig. 2A). The early stage correlates with DC adhesion and invasion, DC-to-RC differentiation, and initiation of RC replication. Early-stage gene transcription increased at 4 h and peaked at 8 h or 12 h, except for that of asp14 and aph_0346, both of which peaked at 4 h (Fig. 2B). Expression of all early-stage genes increased again between 28 and 36 h, which corresponds to the period during which A. phagocytophilum RC organisms differentiate to DC organisms and initiate the second round of infection (26). Midstage gene expression, which coincides with a period of extensive A. phagocytophilum replication (26), peaked at 16 h (Fig. 2C). Late-stage genes were upregulated between 24 and 36 h (Fig. 2D), a period that correlates with the conversion of RC to DC organisms, DC exit, and initiation of the second round of infection (26). Transcript levels of asp14, aph_0346, and aph_0874 were more abundant in DC bacteria used as the inoculum than at 16 h postinfection, when the bacterial population consisted exclusively of RC organisms (26) (Fig. 2).
Fig 2.
Differential transcription profiling of OMP candidate genes throughout the A. phagocytophilum infection cycle. DC organisms were used to synchronously infect HL-60 cells, and the infection proceeded for 36 h, a period that allowed the bacteria to complete their biphasic developmental cycle and to reinitiate infection. Total RNAs were isolated from the DC inoculum and from infected host cells at several postinfection time points. RT-qPCR was performed using gene-specific primers. Relative transcript levels for each target were normalized to A. phagocytophilum 16S rRNA gene transcript levels by using the 2−ΔΔCT method. To determine the relative transcription of OMP candidate genes between RC and DC organisms, normalized transcript levels of each gene at each time point were calculated as fold changes in expression relative to expression at 16 h (encircled in the experimental timeline in panel A), a time point at which the A. phagocytophilum population consists exclusively of RC organisms. (A) Diagram of the experimental design, highlighting the time points at which RNA was isolated, the A. phagocytophilum biphasic developmental and infection stages, and the expression categories into which the genes of interest were classified based on their expression profiles. (B to D) RT-qPCR results for OMP candidate-encoding genes of interest, grouped into early-stage (B), midstage (C), and late-stage (D) genes according to when during the course of infection they are most highly expressed. (E) RT-qPCR results for control genes. The data in panels B to E are the means and standard deviations (SD) of results for triplicate samples and are representative of two independent experiments that yielded similar results.
msp2 (P44), asp62, and asp55 encode confirmed A. phagocytophilum surface proteins, and because the last two genes constitute an operon (3, 26, 47), they were analyzed as controls. The transcriptional profiles of asp55 and asp62 were highly similar (Fig. 2E), which reinforces the accuracy of the expression data obtained for all genes. Coincident with the kinetics of the biphasic infection cycle (26), msp2 (P44) transcription steadily increased from 4 to 28 h, after which it pronouncedly declined by 32 h (Fig. 2E). Of the genes analyzed, asp14 was the most abundantly expressed at 4 h (Fig. 2B). We rationalized that since asp14 is transcriptionally upregulated during the initial 4 h that coincide with both A. phagocytophilum invasion and transcriptional upregulation of the ompA-encoded invasin (29), it may contribute to establishing infection. Accordingly, we focused on Asp14 for the remainder of this study.
A. phagocytophilum upregulates asp14 expression upon binding to PSGL-1 and during tick transmission feeding.
Next, we more closely examined asp14 expression during two critical stages of the A. phagocytophilum life cycle—cellular invasion and tick transmission feeding. Expression of asp14 was significantly upregulated at all time points relative to carryover mRNA levels present in DC organisms during A. phagocytophilum invasion of HL-60 cells, exhibiting a maximal increase at 2 h (Fig. 3A). Transcription of asp14 exhibited little change over the course of invasion of RF/6A endothelial cells (Fig. 3B).
Fig 3.
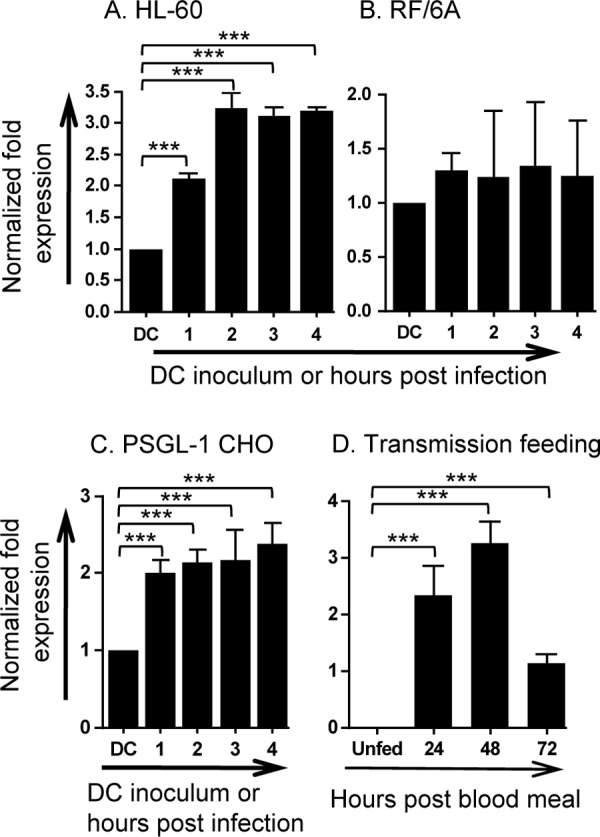
A. phagocytophilum-host cell interactions and transmission feeding of infected ticks upregulate asp14 expression. (A to C) A. phagocytophilum DC organisms were incubated with HL-60 (A), RF/6A (B), and PSGL-1 CHO (C) cells for 4 h, a period that is required for bacterial adherence and for ≥90% of bound bacteria to invade host cells. A. phagocytophilum cannot invade PSGL-1 CHO cells. Total RNAs were isolated from the DC inoculum and from host cells after 1, 2, 3, and 4 h of bacterial addition. (D) A. phagocytophilum-infected I. scapularis nymphs were allowed to feed on mice for 72 h. Total RNAs were isolated from the salivary glands of uninfected and transmission-fed ticks that had been removed at 24, 48, and 72 h postattachment. Total RNA was isolated from combined salivary glands and midguts from unfed ticks. (A to D) RT-qPCR was performed using gene-specific primers. Relative transcript levels for asp14 were normalized to A. phagocytophilum 16S rRNA gene transcript levels. The normalized values in panels A to C are presented relative to asp14 transcript levels of the DC inoculum. Data are the means and standard deviations of results for triplicate samples and are representative of two independent experiments that yielded similar results. Statistically significant (***, P < 0.001) values are indicated.
Since A. phagocytophilum infection of HL-60 cells and neutrophils is predicated, at least in part, on binding to PSGL-1 (28), we examined whether DC binding to PSGL-1 upregulated asp14 expression. PSGL-1 CHO cells (Chinese hamster ovary [CHO] cells transfected to express PSGL-1) are excellent models for studying A. phagocytophilum--PSGL-1 interactions (26, 29, 31, 35, 65, 66). A. phagocytophilum binding to PSGL-1 CHO cells occurs exclusively through recognition of PSGL-1, and the bacterium cannot bind to untransfected CHO cells (31, 35, 67). DC binding to PSGL-1 CHO cells significantly upregulated asp14 transcription, with the most pronounced increase relative to carryover mRNA levels present in DC organisms occurring within the first hour (Fig. 3C). A. phagocytophilum genes that are induced during transmission feeding of infected ticks on mammals conceivably aid in establishing infection. asp14 transcripts were not detected in unfed infected I. scapularis nymphs but were significantly induced within the first 24 h of I. scapularis nymphs feeding on naïve mice and continued to be expressed throughout the blood meal (Fig. 3D).
Asp14 exhibits a propensity to multimerize and is expressed by A. phagocytophilum during infection of humans or mice.
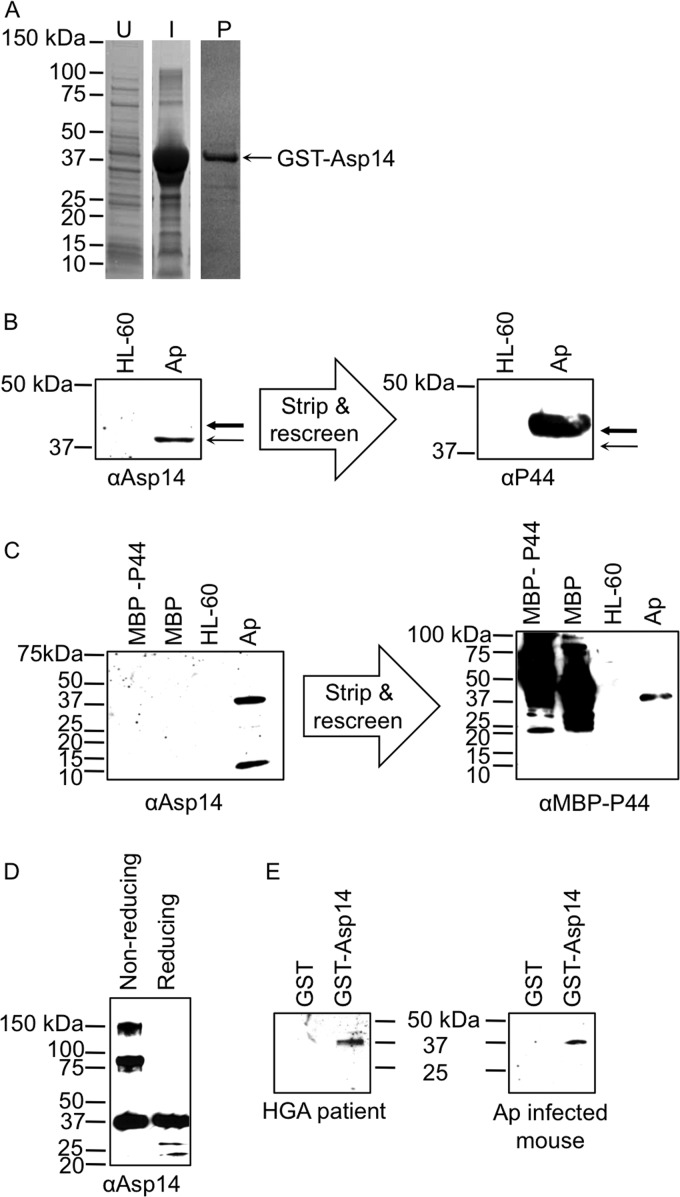
The asp14 coding region (encoding 13.8 kDa) was cloned and expressed in Escherichia coli as an N-terminally GST-tagged fusion protein (GST-Asp14) (Fig. 4A). Purified GST-Asp14 appeared as a 39.8-kDa band upon SDS-PAGE. Purified GST-Asp14 was used to immunize mice. In addition to the anticipated 13.8-kDa band, anti-Asp14 detected a band of approximately 42 kDa in an A. phagocytophilum lysate but not an uninfected HL-60 cell lysate (Fig. 4B and C). Anti-Asp14 occasionally detected another band, of approximately 28 kDa, on blots of A. phagocytophilum lysates (data not shown). Even though the 42-kDa band is close in size to that of Msp2 (P44), anti-Asp14 failed to recognize A. phagocytophilum-derived and maltose binding protein (MBP)-tagged Msp2 (P44) (Fig. 4B and C) (68). An amino acid sequence alignment revealed that Asp14 exhibits no homology to Msp2 (P44) paralogs (data not shown). GST-Asp14 multimerized when it was fractionated by nonreducing SDS-PAGE (Fig. 4D). Thus, the 28- and 42-kDa bands in the A. phagocytophilum lysate recognized by anti-Asp14 are presumably multimeric complexes that consist exclusively of or contain Asp14. HGA patient serum and serum from an A. phagocytophilum-infected mouse each recognized GST-Asp14 (Fig. 4E), indicating that Asp14 is expressed by A. phagocytophilum and elicits a humoral immune response during infection of humans or mice. Two additional HGA patient serum samples recognized GST-Asp14 (data not shown).
Fig 4.
A. phagocytophilum (Ap) expresses Asp14 during in vitro and in vivo infections. (A) Whole-cell lysates of E. coli (U), E. coli induced (I) to express GST-Asp14, and GST-Asp14 purified (P) by glutathione Sepharose affinity chromatography were separated by SDS-PAGE and stained with Coomassie blue. (B) Western blot analyses in which mouse anti-Asp14 (αAsp14; raised against GST-Asp14) was used to screen whole-cell lysates of uninfected HL-60 cells and A. phagocytophilum organisms. The blot was stripped and rescreened with anti-Msp2 (P44) (αP44). The thin and thick arrows denote Asp14 and Msp2 (P44), respectively. (C) Western-blotted MBP-P44, MBP, and whole-cell lysates of uninfected HL-60 cells and A. phagocytophilum organisms were screened with anti-Asp14. The blot was stripped and rescreened with anti-MBP-P44. (D) GST-Asp14 was resolved by SDS-PAGE under nonreducing and reducing conditions, Western blotted, and screened with anti-Asp14. (E) Western-blotted GST-Asp14 and GST were screened with sera from an HGA patient and an experimentally infected mouse.
Asp14 is located in the A. phagocytophilum outer membrane and colocalizes with confirmed outer membrane proteins.
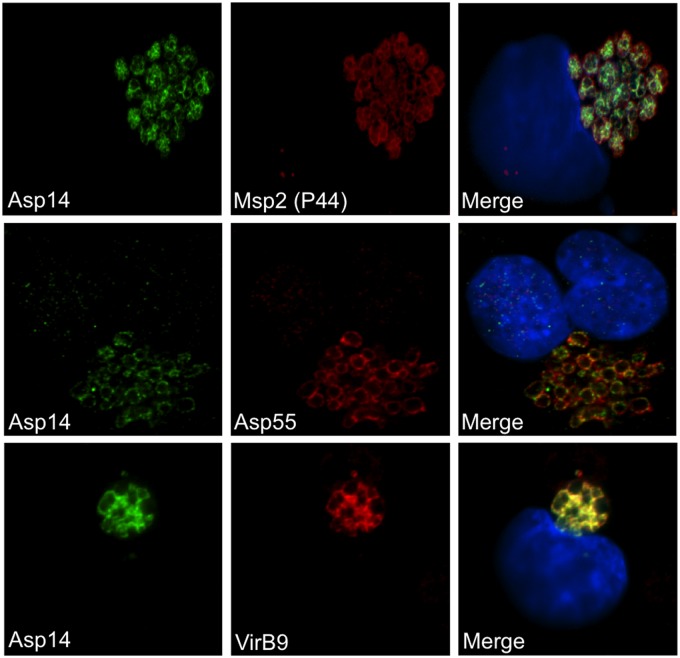
To determine if Asp14 colocalizes with confirmed outer membrane proteins, we screened infected RF/6A cells by confocal microscopy, using Asp14 antiserum in conjunction with antiserum targeting Msp2 (P44), Asp55, or VirB9 (47, 50, 69). Recognition of intravacuolar A. phagocytophilum bacteria with each antiserum generated ring-like staining patterns on the peripheries of the organisms. Asp14 and VirB9 signals colocalized (Fig. 5). Asp55 antiserum is specific for a surface-exposed peptide epitope (47). The Asp55 signal colocalized with the Asp14 signal in punctate patterns on the peripheries of the bacteria. The Msp2 (P44) signal colocalized with and also extended beyond the Asp14 signal. Screening of A. phagocytophilum-infected HL-60, RF/6A, and ISE6 (I. scapularis embryonic cell line) cells with Asp14 and Msp2 (P44) antibodies at multiple time points throughout infection revealed that 100.0% of morulae in each cell line were Asp14 positive (data not shown).
Fig 5.
Asp14 colocalizes with confirmed outer membrane proteins. A. phagocytophilum-infected RF/6A cells were fixed and viewed by confocal microscopy to determine immunoreactivity with Asp14 antiserum in conjunction with antiserum against Msp2 (P44), Asp55, or VirB9, each of which is a confirmed outer membrane protein. Host cell nuclei were stained with DAPI (4′,6-diamidino-2-phenylindole).
Surface localization of Asp14.
We next verified that Asp14 is a surface protein. Intact DC bacteria were incubated with trypsin, followed by Western blotting with Asp14 antiserum to determine if immunoaccessible domains are on the A. phagocytophilum surface. Positive-control antisera targeted Omp-1A or a surface-exposed Asp55 epitope (47). Negative-control antiserum targeted the A. phagocytophilum-occupied vacuolar membrane protein APH_0032 (17). Following surface trypsinolysis, the signal from Asp14, Asp55, or Omp-1A was completely eliminated, or very little signal was detected (Fig. 6A). There was no difference in APH_0032 signal intensity between treated and vehicle control-treated organisms. Live transgenic DC organisms expressing GFP (33, 36) were screened by flow cytometry with anti-Asp14 or serum from an A. phagocytophilum-infected mouse obtained at the peak of bacteremia to assess the presentation of Asp14 on the bacterial surface (53). We had verified that the infected mouse serum detected Msp2 (P44) and other A. phagocytophilum antigens by Western blotting and immunofluorescence assays (data not shown). This positive-control serum recognized 1.5- ± 0.3-fold more GFP-positive bacteria than preimmune mouse serum (Fig. 6B), thereby demonstrating that antibodies that recognize Msp2 (P44) and other A. phagocytophilum antigens bind to the surfaces of intact A. phagocytophilum organisms. Asp14 antiserum yielded a similar result, as it bound to 3.6- ± 0.9-fold more transgenic bacteria than preimmune serum, indicating that Asp14 or a portion thereof is exposed on the A. phagocytophilum surface.
Fig 6.
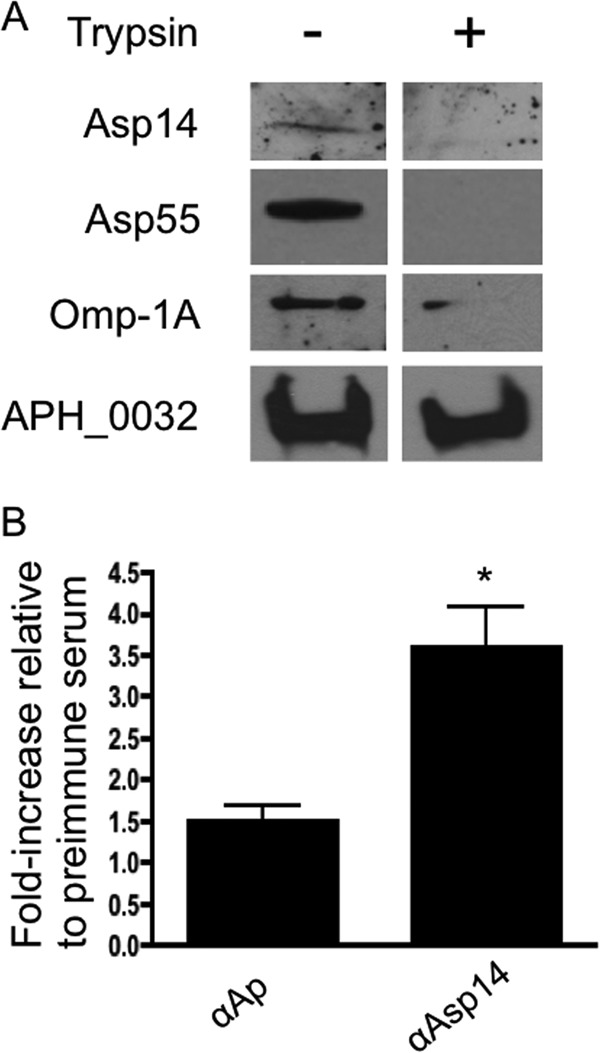
Asp14 is presented on the A. phagocytophilum surface. (A) Intact DC bacteria were incubated with trypsin or vehicle control, lysed in RIPA buffer, fractionated by SDS-PAGE, and immunoblotted. Western blots were screened with antiserum targeting Asp14, Asp55, Omp-1A, or APH_0032. Data are representative of at least two experiments with similar results. (B) Live transgenic A. phagocytophilum DC organisms expressing GFP were incubated with preimmune mouse serum, mouse anti-Asp14, or serum recovered from an A. phagocytophilum-infected mouse. Primary antibodies were detected with anti-mouse IgG conjugated to Alexa Fluor 647. Flow cytometry was used to determine the percentage of Alexa Fluor 647- and GFP-positive DC organisms per sample. The fold increase in the percentage of Alexa Fluor 647-positive, GFP-positive DC organisms for each sample relative to preimmune serum is provided. Results presented are the means ± SD for three experiments. Statistically significant (*, P < 0.05) values are indicated.
Pretreating A. phagocytophilum with anti-Asp14 inhibits infection of HL-60 cells.
Since Asp14 is a surface protein, we examined whether incubating A. phagocytophilum DC organisms with heat-inactivated Asp14 antiserum prior to adding them to HL-60 cells would inhibit bacterial binding or infection. Anti-Asp14 had no effect on A. phagocytophilum adhesion, but it reduced infection by approximately 33% and lowered the mean number of morulae per cell by approximately 54% (Fig. 7A to D). Inhibition was specific to Asp14 antiserum, as GST antiserum did not alter bacterial binding or infection.
Fig 7.
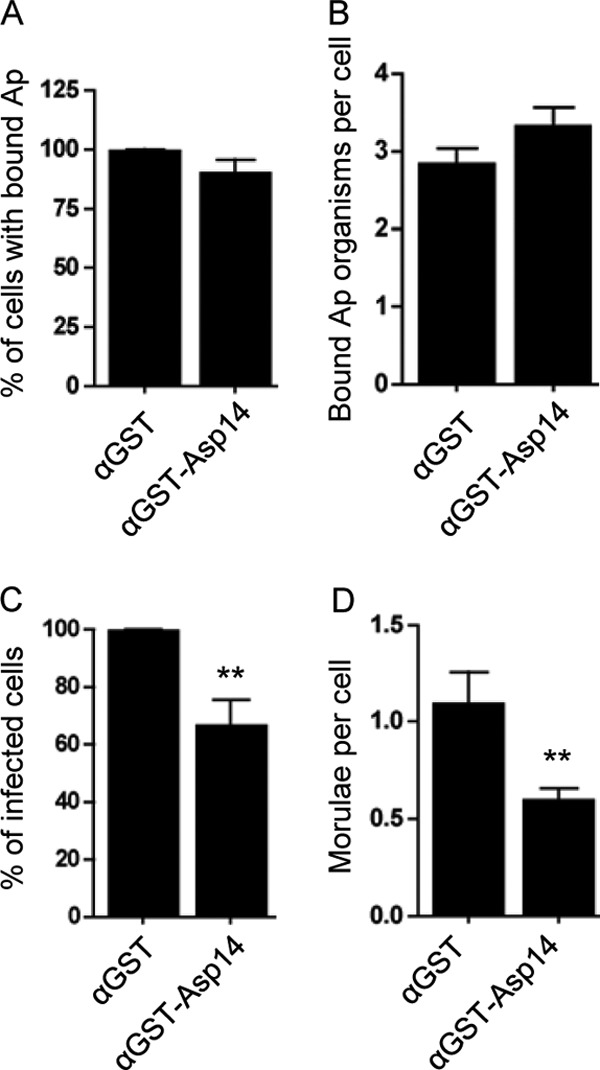
Pretreatment of A. phagocytophilum (Ap) with anti-Asp14 partially abrogates infection of but not cellular adherence to HL-60 cells. Host cell-free A. phagocytophilum DC organisms were incubated with mouse polyclonal antiserum raised against GST-Asp14 or GST alone. Next, the treated bacteria were incubated with HL-60 cells for 60 min. After removal of unbound bacteria, the infection of HL-60 cells was allowed to proceed for 48 h, during which A. phagocytophilum binding and infection were assessed using an antibody targeting A. phagocytophilum Msp2 (P44) and by confocal microscopy. (A) Percentages of HL-60 cells with bound A. phagocytophilum organisms. (B) Mean number (±SD) of bound bacteria per cell. (C) Percentage of infected HL-60 cells following incubation with A. phagocytophilum organisms in the presence of anti-Asp14 or anti-GST. (D) Mean number (±SD) of morulae per cell. Results in each panel are the means ± SD for three independent experiments. Statistically significant (**, P < 0.005) values are indicated.
The Asp14 C-terminal region binds mammalian host cells.
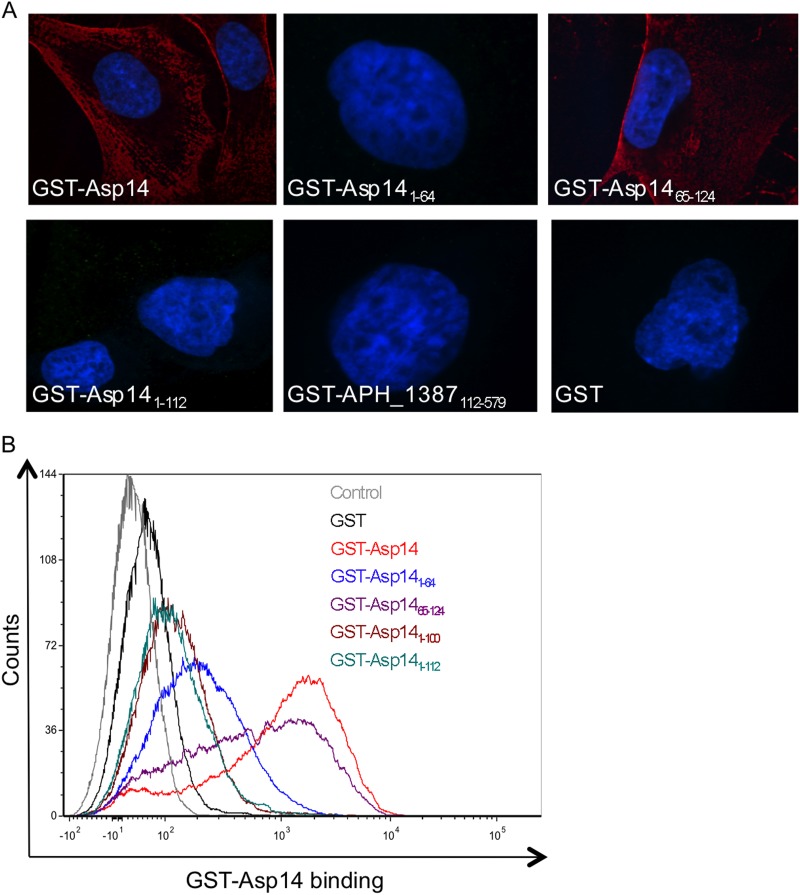
Since Asp14 is an exposed outer membrane protein and anti-Asp14 reduces A. phagocytophilum infection, we rationalized that Asp14 may interact with mammalian host cell surfaces to promote infection. To test this possibility and to identify the Asp14 region that is sufficient for optimal adherence, we examined if GST-tagged Asp14 or portions thereof bind to RF/6A cells. GST alone and GST-tagged APH_1387 amino acids 112 to 579 (GST-APH_1387112–579) were used as negative controls. APH_1387 is an A. phagocytophilum protein that localizes to the pathogen's vacuolar membrane and does not associate with the host cell surface (18). GST-Asp14 but neither GST nor GST-APH_1387112–579 bound to RF/6A cells (Fig. 8). The binding domain is carried on the Asp14 C-terminal half, as GST-Asp1465–124 but not GST-Asp141–64 exhibited binding. GST-Asp141–100 and GST-Asp141–112 were unable to bind RF/6A cells (Fig. 8). Thus, Asp14 residues 101 to 124 contain the minimal region that is sufficient to facilitate adhesion to mammalian cell surfaces.
Fig 8.
The Asp14 C-terminal region binds mammalian host cells. RF/6A cells were incubated with GST-Asp14, GST-Asp141–64, GST-Asp1465–124, GST-Asp141–100, GST-Asp141–112, GST-APH_1387112–579 (negative control; does not associate with mammalian cell membranes), or GST alone for 60 min, followed by removal of unbound protein. (A) Confocal microscopy analyses of GST fusion proteins bound to RF/6A cells. The host cells were fixed and successively incubated with anti-GST antibody and anti-mouse IgG conjugated to Alexa Fluor 594. Gel mounting medium containing DAPI was added. Representative merged fluorescent images are shown. Results are representative of two to four independent experiments. (B) Flow cytometric analysis of GST fusion protein binding to RF/6A cells. The host cells were successively incubated with GST antibody and Alexa Fluor 488-conjugated anti-mouse IgG and then analyzed by flow cytometry.
GST-Asp14 requires Asp14 residues 101 to 124 to competitively inhibit A. phagocytophilum infection of mammalian host cells.
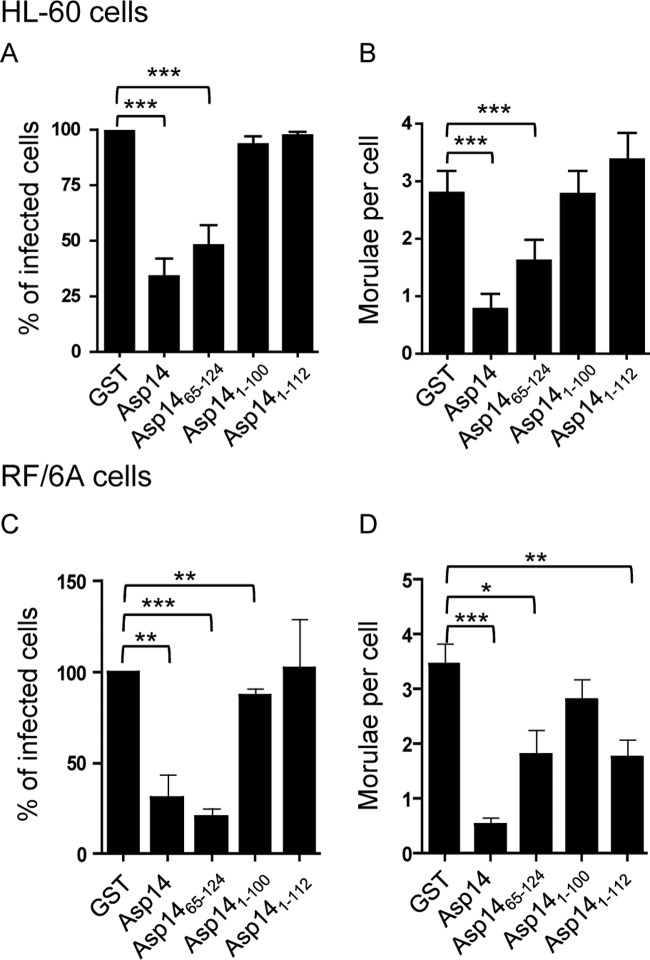
We next determined if GST-tagged Asp14 or fragments thereof could inhibit A. phagocytophilum infection. GST-Asp14 and GST-Asp1465–124 each significantly reduced infection of HL-60 and RF/6A cells relative to that with GST alone (Fig. 9). GST-Asp141–100 and GST-Asp141–112 had no effect on infection of HL-60 cells (Fig. 9A and B). GST-Asp141–112 did not lower the percentage of infected RF/6A cells but reduced the mean number of morulae per RF/6A cell comparably to the case with GST-Asp1465–124 (Fig. 9C and D). Pretreating host cells with GST-Asp14 fusion proteins prior to incubation with bacteria failed to inhibit A. phagocytophilum binding (data not shown). Thus, A. phagocytophilum binding to mammalian host cells is Asp14 independent, but Asp14 is important for bacterial invasion.
Fig 9.
GST-Asp14 requires Asp14 residues 101 to 124 to competitively inhibit A. phagocytophilum infection of mammalian host cells. HL-60 (A and B) and RF/6A (C and D) cells were incubated with A. phagocytophilum (Ap) DC organisms in the presence of GST alone, GST-Asp14, GST-Asp1465–124, GST-Asp141–100, or GST-Asp141–112 for 1 h. After removal of unbound organisms, host cells were incubated for 24 h (A and B) or 48 h (C and D) and subsequently examined by confocal microscopy to assess the percentage of infected cells (A and C) or the mean number (±SD) of morulae per cell (B and D). Results shown are relative to GST-treated host cells and are the means ± SD for 3 experiments. Statistically significant (*, P < 0.05; **, P < 0.005; ***, P < 0.001) values are indicated.
The Asp14 C terminus is positively charged, and residues 101 to 115 constitute a conserved domain among homologs from Anaplasma and Ehrlichia species.
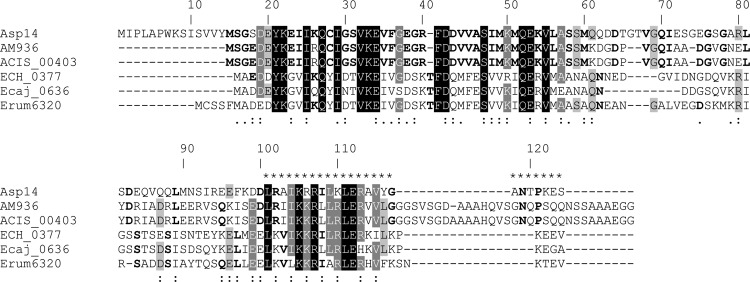
We rationalized that a domain that lies within Asp14 amino acids 101 to 124 is involved in mediating interactions with host cells that promote A. phagocytophilum infection. To determine if this or any other Asp14 region is conserved among Anaplasmataceae members, we aligned the primary amino acid sequences of Asp14 and its homologs from two Anaplasma marginale strains and three monocytotropic Ehrlichia species. Doing so identified two conserved regions, the first of which corresponds to Asp14 amino acids 19 to 61 (Fig. 10). The second conserved region aligns with Asp14 residues 101 to 115. The consensus sequence for this region among the Anaplasma and Ehrlichia sp. Asp14 homologs is L[RK]aIKKR[IL]LRLERxV, where “a” and “x” represent a nonpolar amino acid and any amino acid, respectively. Beginning at tyrosine 116, the Asp14 C terminus bears no sequence homology to its A. marginale and Ehrlichia counterparts. The Asp14 C terminus (amino acids 101 to 124) has a charge of +4.91, despite the entire protein sequence having a charge of −3.10 (Table 3). A similar trend is observed when the charges of the Asp14 homologs' C termini and entire protein sequences are examined.
Fig 10.
Alignment of A. phagocytophilum Asp14 with its homologs from other Anaplasma and Ehrlichia species. Clustal W alignment was performed with A. phagocytophilum Asp14 and its homologs from an A. marginale St. Maries strain (AM936), an A. marginale subspecies centrale Israel strain (ACIS_00403), an Ehrlichia chaffeensis Arkansas strain (ECH_0377), an Ehrlichia canis Jake strain (Ecaj_0636), and an Ehrlichia ruminantium Welgevonden strain (Erum6320). The A. phagocytophilum Asp14 amino acid numbers are listed above the aligned sequences. White text with black shading denotes amino acids that are identical among all six sequences. White text with dark gray shading denotes amino acids that are identical among five of six sequences. Light gray shading denotes amino acids that are identical among four of six sequences. Amino acids in bold are those that are identical among three of six sequences. If a given amino acid is identical among the three Anaplasma sequences and the amino acid at the same position is identical among the three Ehrlichia sequences but is different from the Anaplasma sequences, then the amino acid for only the Anaplasma sequences is shown in bold. A. phagocytophilum amino acids that contain the invasin domain are denoted by asterisks above them. Amino acids that are highly similar and weakly similar are denoted by colons and dots, respectively, below them.
Table 3.
Charges of Asp14 and Asp14 homolog full-length sequences and C-terminal regions
| Proteina | Residues in full-length sequence | Charge | Residues in C-terminal region | Charge |
|---|---|---|---|---|
| Asp14 | 1–124 | −3.10 | 101–124 | +4.91 |
| AM936 | 1–130 | −4.93 | 84–130 | +3.08 |
| ACIS_00403 | 1–131 | −4.93 | 84–131 | +3.08 |
| ECH_0377 | 1–104 | −3.07 | 83–104 | +5.91 |
| Ecaj_0636 | 1–98 | −1.91 | 77–98 | +6.07 |
| Erum6320 | 1–110 | −0.94 | 88–110 | +6.08 |
Annotated genome designations correspond to Asp14 homologs from an A. marginale St. Maries strain (AM936), an A. marginale subspecies centrale Israel strain (ACIS_00486), an E. chaffeensis Arkansas strain (ECH_0462), an E. canis Jake strain (Ecaj_0563), and an E. ruminantium Welgevonden strain (Erum_5620).
GST-Asp14 and GST-OmpA together more pronouncedly inhibit A. phagocytophilum infection of HL-60 cells than either protein alone.
We examined whether we could improve upon the protection against A. phagocytophilum infection afforded by GST-Asp14 (Fig. 9) or GST-OmpA (29) by pretreating HL-60 cells with both recombinant proteins. Consistent with previous results, 35.5% ± 7.4% of GST-OmpA-treated and 53.2% ± 11.8% of GST-Asp14-treated HL-60 cells became infected (Fig. 11A). However, HL-60 cells that had been preincubated with both GST-Asp14 and GST-OmpA were better protected against A. phagocytophilum infection, as only 9.9% ± 9.4% of these cells developed morulae. To prove that the synergistic reduction in infection was specific to the combinatorial effect of GST-Asp14 and GST-OmpA and not simply due to the presence of excess recombinant protein, we treated HL-60 cells with GST-Asp14 and GST-OmpA, GST-Asp141–100 (does not block infection) (Fig. 9) and GST-OmpA, or GST-Asp14 and GST-OmpA75–205 (does not block infection) (29). HL-60 cells treated with GST-Asp141–100 and GST-OmpA or GST-Asp14 and GST-OmpA75–205 exhibited reductions in infection and bacterial load comparable to those of cells treated with GST-Asp14 or GST-OmpA alone (Fig. 11A and B) (29). HL-60 cells treated with GST-Asp14 and GST-OmpA exhibited an approximately 4.5-fold reduction in the percentage of infected cells relative to cells treated with either GST-Asp141–100 and GST-OmpA or GST-Asp14 and GST-OmpA75–205 (Fig. 11A).
Fig 11.
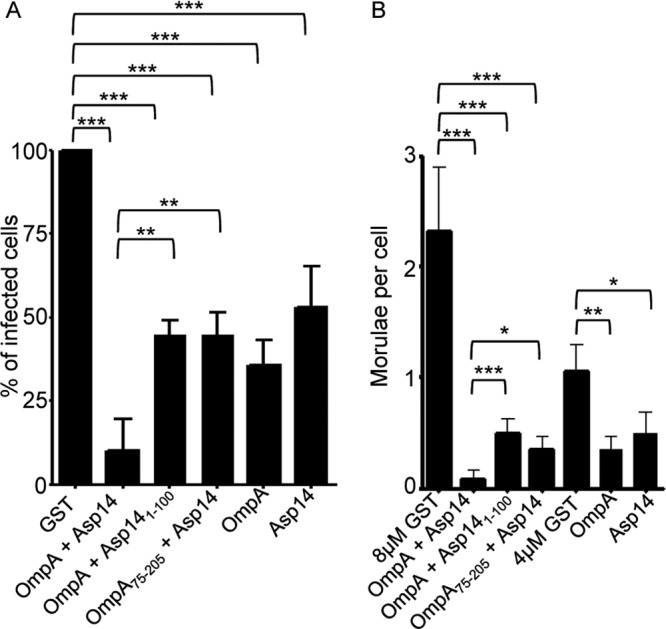
GST-Asp14 and GST-OmpA cooperatively block A. phagocytophilum infection of HL-60 cells. HL-60 cells were incubated with A. phagocytophilum (Ap) DC organisms in the presence of GST alone, GST-Asp14, GST-OmpA, GST-Asp14 and GST-OmpA, GST-Asp141–100 and GST-OmpA, or GST-Asp14 and GST-OmpA75–205 for 1 h. After removal of unbound organisms, host cells were incubated for 24 h and subsequently examined by confocal microscopy to assess the percentage of infected cells (A) or the mean number (±SD) of morulae per cell (B). Results shown are relative to GST-treated host cells and are the means ± SD for three to six experiments. Cells treated with either 4 μM GST-Asp14 or GST-OmpA were compared to cells treated with 4 μM GST, while cells treated with 4 μM GST-Asp14 and 4 μM GST-OmpA were compared to cells treated with 8 μM GST. Statistically significant (*, P < 0.05; **, P < 0.005; ***, P < 0.001) values are indicated.
DISCUSSION
Over the past nearly 2 decades, Anaplasma and Ehrlichia species have emerged as major causes of tick-transmitted diseases of humans and animals worldwide and are significant causes of human morbidity in the United States (1, 12, 30, 70, 71). Identifying targets that are conserved among these bacteria would be advantageous from both a cross-protective and a cost-effective standpoint because of their potential to prevent infection by multiple Anaplasmataceae members rather than a single species. Evidence suggests that cellular invasion by each species is likely multifactorial, involving interactions between numerous bacterial invasins and host cell surface determinants that evoke entry through redundant or complementary pathways. Spotted fever group Rickettsia species, which are in the order Rickettsiales with Anaplasma and Ehrlichia, utilize multiple entry routes (72, 73). A. phagocytophilum infection of human neutrophils and HL-60 cells largely depends on recognition of three determinants of sLex-capped PSGL-1—α2,3-sialic acid and α1,3-fucose of sLex and the PSGL-1 N-terminal peptide (27, 28, 35)—but also involves unidentified sLex- and PSGL-1-independent receptors (32–34). Given that endothelial cells do not express PSGL-1 (74) and that the same region of Asp14 mediates invasion of RF/6A and HL-60 cells, we presume that the Asp14 C terminus does not directly bind the PSGL-1 N-terminal peptide to promote bacterial entry. The invasion domains of both Asp14 and OmpA are conserved among Anaplasma and Ehrlichia species (29). Using GST-OmpA or GST-Asp14 alone competitively inhibits A. phagocytophilum infection of host cells by 50 to 70% (29). However, using GST-OmpA and GST-Asp14 together more pronouncedly inhibits infection than using either protein alone, indicating that Asp14 promotes entry by a complementary pathway that is independent of invasion mediated by the OmpA–α2,3-sialic acid interaction. It will be important to investigate the potential of targeting Asp14 and OmpA together for protection against in vivo infection by A. phagocytophilum, A. marginale, and Ehrlichia species.
A. phagocytophilum invasion of mammalian host cells is predicated on the recognition of host cell surfaces by bacterial outer membrane proteins, whose identities have long remained elusive. We identified Asp14 as an A. phagocytophilum surface protein that interacts with mammalian host cell surfaces via its C-terminal invasin domain to promote infection. The invasin domain lies within amino acids 101 to 124. Asp14 residues 101 to 115 are highly conserved among homologs of other Anaplasma and Ehrlichia species, implying that this region may exert a functionally conserved role. Yet deleting amino acids 113 to 124, which are unique to Asp14, eliminates the ability of GST-Asp14 to bind to and competitively inhibit A. phagocytophilum infection of mammalian host cells. One possible explanation for this result is that the invasin domain lies exclusively within residues 113 to 124 and that residues 101 to 112, although conserved among Anaplasma and Ehrlichia species, are not requisite for cellular invasion. A second rationalization is that the invasin domain includes amino acids 113 to 115 of the highly conserved region and that deleting them eliminates the ability of Asp14 to interact with host cells. A third possibility is that the net positive charge generated by residues 101 to 124 is important for overcoming the charge repulsion barrier that occurs between negatively charged bacterial and host cell surfaces (75). Support for the third premise comes from the fact that a net positive charge is maintained for the most C-terminal portion of each Anaplasma and Ehrlichia Asp14 homolog, despite the lack of primary amino acid sequence conservation. Another highly conserved domain of the Anaplasma and Ehrlichia homologs is formed by Asp14 amino acids 19 to 61. However, this region likely plays no role in cellular invasion, as GST-Asp141–64 binds poorly to host cells.
A. phagocytophilum binding to sLex-capped PSGL-1 upregulates asp14 expression. Myeloid but not endothelial cells express PSGL-1 (74), which likely accounts for the increase in asp14 transcription upon A. phagocytophilum binding to HL-60 cells but not RF/6A cells. The greater abundance of Asp14 that presumably results likely enhances cellular invasion of myeloid cells. Indeed, the increase in asp14 mRNA is coincident with the 4-hour period that A. phagocytophilum requires to efficiently invade HL-60 cells (23–25). Asp14 is not transcribed in unfed A. phagocytophilum-infected I. scapularis nymphs but is transcriptionally induced during transmission feeding on mice, an observation that is consistent with a recent proteomic profiling study of A. phagocytophilum proteins induced during I. scapularis transmission feeding (46). Thus, while Asp14 is dispensable for A. phagocytophilum survival in ticks, it may be part of the bacterium's armamentarium required for establishing infection in mammals. Support for this argument comes from our and others' reports that transcription of A. phagocytophilum ompA and the Borrelia sp. virulence factor ospC (encoding outer surface protein C), the latter of which is essential for mammalian infection, is induced during ixodid tick transmission feeding (29, 76, 77). Furthermore, A. phagocytophilum expresses Asp14 during infection of humans and mice. A. phagocytophilum expresses Asp14 throughout its development in ISE6 cells, which suggests that environmental factors present in the tick vector but absent in tick cell culture may regulate Asp14 expression.
Asp14 is presented on the A. phagocytophilum outer membrane and colocalizes with confirmed outer membrane proteins. Yet Asp14 is not predicted to carry a traditional signal sequence or a transmembrane domain. How Asp14 associates with the bacterial outer membrane is unknown, and this question is further complicated by its lack of structural similarity to any known crystal structure (data not shown). It may be an atypical transmembrane protein or a peripheral membrane protein that is anchored to the A. phagocytophilum surface via a posttranslational modification. Both endogenous and recombinant forms of Asp14 display propensities to multimerize. Asp14 may form homomeric complexes with itself in the A. phagocytophilum outer membrane or may form heteromeric complexes with other OMPs. Asp14 and Msp2 (P44) colocalize on the A. phagocytophilum outer membrane. Msp2 (P44) has been suggested to form heteromeric complexes that facilitate interactions with host cells (78).
Prior studies have comprehensively delineated the A. phagocytophilum proteome in infected HL-60 cells or infected, transmission-feeding I. scapularis ticks or have used surface biotinylation and affinity proteomics to identify select A. phagocytophilum surface proteins. These studies were performed on mixed populations of DC and RC bacteria (46, 47, 79). Our study is the first to analyze the A. phagocytophilum DC surface proteome, which facilitated the discovery of the importance of Asp14 to infection. It should be noted, however, that not all DC OMPs were identified. Affinity proteomics using amine-labeling reagents is a valuable approach, but its ability to label surface proteins is limited by whether or not the proteins have free amines and, if so, whether they are accessible. The OmpA predicted N-terminal ectodomain carries five lysines (29), yet affinity proteomics failed to detect OmpA. OmpA and other OMPs may be folded such that their free amines are inaccessible for biotinylation. Several recovered proteins are predicted to be cytoplasmic. While many of these so-called cytoplasmic proteins were recovered in another surface biotinylation and affinity proteomic study of A. phagocytophilum or have homologs in other bacteria that localize to and function on bacterial surfaces (47, 55–64), it remains to be verified if all recovered proteins in this study are in fact surface localized. Though we initially identified Asp14 on the A. phagocytophilum DC surface, indirect immunofluorescence analyses revealed that the bacterium constitutively expresses Asp14 throughout its development in HL-60, RF/6A, and ISE6 cells. Thus, the RC form also expresses Asp14.
Of the profiled OMP candidates, we selected Asp14 because it, like OmpA (29), was transcriptionally upregulated during cellular invasion. An equally plausible approach would be to focus on late-stage genes, such as APH_0874 and APH_1170, because they may be important for RC-to-DC transition, adhesion, invasion, and/or establishing infection. A precedent for this rationalization is provided by APH_1235, which is a late-stage gene that is pronouncedly upregulated at the DC stage and is critical for propagating infection in HL-60 cell culture (41, 46).
In summary, this work advances the knowledge of A. phagocytophilum pathogenesis by identifying Asp14 as a novel surface protein that is critical for infection of mammalian host cells and mapping its invasion domain-containing region. The conservation of the Asp14 and OmpA invasion domains among Anaplasma and Ehrlichia species, combined with the observation that treating host cells with recombinant forms of both proteins protects against A. phagocytophilum cellular invasion in vitro, serves as a harbinger that targeting both invasins in vivo may protect against infection by multiple Anaplasmataceae pathogens.
ACKNOWLEDGMENTS
We thank Ulrike Munderloh of the University of Minnesota for providing transgenic A. phagocytophilum expressing GFP, J. Stephen Dumler of The Johns Hopkins University for providing MAb 20B4, Durland Fish of Yale University for providing A. phagocytophilum-infected ticks, and Yasuko Rikihisa of The Ohio State University for donating antisera against Asp55 and VirB9.
This study was supported by NIH grants R01 AI072683, R01 AI072683-04S1, and R21 AI090170 to J.A.C. and R01 AI141440 to E.F. The VCU Flow Cytometry and Imaging Shared Resource Facility is supported in part by funding from NIH-NCI Cancer Center support grant 5 P30 CA016059.
Footnotes
Published ahead of print 15 October 2012
REFERENCES
- 1. Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, Rikihisa Y, Rurangirwa FR. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145–2165 [DOI] [PubMed] [Google Scholar]
- 2. Palmer GH, Noh SM. 2012. Rickettsial entry into host cells: finding the keys to unlock the doors. Infect. Immun. 80:3746–3747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rikihisa Y. 2011. Mechanisms of obligatory intracellular infection with Anaplasma phagocytophilum. Clin. Microbiol. Rev. 24:469–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dhand A, Nadelman RB, Aguero-Rosenfeld M, Haddad FA, Stokes DP, Horowitz HW. 2007. Human granulocytic anaplasmosis during pregnancy: case series and literature review. Clin. Infect. Dis. 45:589–593 [DOI] [PubMed] [Google Scholar]
- 5. Horowitz HW, Kilchevsky E, Haber S, Aguero-Rosenfeld M, Kranwinkel R, James EK, Wong SJ, Chu F, Liveris D, Schwartz I. 1998. Perinatal transmission of the agent of human granulocytic ehrlichiosis. N. Engl. J. Med. 339:375–378 [DOI] [PubMed] [Google Scholar]
- 6. Alhumaidan H, Westley B, Esteva C, Berardi V, Young C, Sweeney J. 7 May 2012. Transfusion-transmitted anaplasmosis from leukoreduced red blood cells. Transfusion doi:10.1111/j.1537-2995.2012.03685.x [DOI] [PubMed] [Google Scholar]
- 7. Annen K, Friedman K, Eshoa C, Horowitz M, Gottschall J, Straus T. 2012. Two cases of transfusion-transmitted Anaplasma phagocytophilum. Am. J. Clin. Pathol. 137:562–565 [DOI] [PubMed] [Google Scholar]
- 8. CDC 2008. Anaplasma phagocytophilum transmitted through blood transfusion—Minnesota, 2007. MMWR Morb. Mortal. Wkly. Rep. 57:1145–1148 [PubMed] [Google Scholar]
- 9. Jereb M, Pecaver B, Tomazic J, Muzlovic I, Avsic-Zupanc T, Premru-Srsen T, Levicnik-Stezinar S, Karner P, Strle F. 2012. Severe human granulocytic anaplasmosis transmitted by blood transfusion. Emerg. Infect. Dis. 18:1354–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Krause PJ, Wormser GP. 2008. Nosocomial transmission of human granulocytic anaplasmosis? JAMA 300:2308–2309 [DOI] [PubMed] [Google Scholar]
- 11. Zhang L, Liu Y, Ni D, Li Q, Yu Y, Yu XJ, Wan K, Li D, Liang G, Jiang X, Jing H, Run J, Luan M, Fu X, Zhang J, Yang W, Wang Y, Dumler JS, Feng Z, Ren J, Xu J. 2008. Nosocomial transmission of human granulocytic anaplasmosis in China. JAMA 300:2263–2270 [DOI] [PubMed] [Google Scholar]
- 12. Thomas RJ, Dumler JS, Carlyon JA. 2009. Current management of human granulocytic anaplasmosis, human monocytic ehrlichiosis and Ehrlichia ewingii ehrlichiosis. Expert Rev. Anti Infect. Ther. 7:709–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hall-Baker PA, Nieves E, Jajosky R, Adams DA, Sharp P, Anderson WJ, Aponte JJ, Aranas AE, Katz SB, Wodajo MS, Onweh DH, Baillie J, Park M. 2010. Summary of notifiable diseases: United States, 2008. MMWR Morb. Mortal. Wkly. Rep. 57:1–94 [Google Scholar]
- 14. Dahlgren FS, Mandel EJ, Krebs JW, Massung RF, McQuiston JH. 2011. Increasing incidence of Ehrlichia chaffeensis and Anaplasma phagocytophilum in the United States, 2000–2007. Am. J. Trop. Med. Hyg. 85:124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Herron MJ, Ericson ME, Kurtti TJ, Munderloh UG. 2005. The interactions of Anaplasma phagocytophilum, endothelial cells, and human neutrophils. Ann. N. Y. Acad. Sci. 1063:374–382 [DOI] [PubMed] [Google Scholar]
- 16. Huang B, Ojogun N, Ragland SA, Carlyon JA. 2011. Monoubiquitinated proteins decorate the Anaplasma phagocytophilum-occupied vacuolar membrane. FEMS Immunol. Med. Microbiol. 64:32–41 [DOI] [PubMed] [Google Scholar]
- 17. Huang B, Troese MJ, Howe D, Ye S, Sims JT, Heinzen RA, Borjesson DL, Carlyon JA. 2010. Anaplasma phagocytophilum APH_0032 is expressed late during infection and localizes to the pathogen-occupied vacuolar membrane. Microb. Pathog. 49:273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang B, Troese MJ, Ye S, Sims JT, Galloway NL, Borjesson DL, Carlyon JA. 2010. Anaplasma phagocytophilum APH_1387 is expressed throughout bacterial intracellular development and localizes to the pathogen-occupied vacuolar membrane. Infect. Immun. 78:1864–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Munderloh UG, Lynch MJ, Herron MJ, Palmer AT, Kurtti TJ, Nelson RD, Goodman JL. 2004. Infection of endothelial cells with Anaplasma marginale and A. phagocytophilum. Vet. Microbiol. 101:53–64 [DOI] [PubMed] [Google Scholar]
- 20. Sukumaran B, Carlyon JA, Cai JL, Berliner N, Fikrig E. 2005. Early transcriptional response of human neutrophils to Anaplasma phagocytophilum infection. Infect. Immun. 73:8089–8099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sukumaran B, Mastronunzio JE, Narasimhan S, Fankhauser S, Uchil PD, Levy R, Graham M, Colpitts TM, Lesser CF, Fikrig E. 2011. Anaplasma phagocytophilum AptA modulates Erk1/2 signalling. Cell. Microbiol. 13:47–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiong Q, Rikihisa Y. 2011. The prenylation inhibitor manumycin A reduces the viability of Anaplasma phagocytophilum. J. Med. Microbiol. 60:744–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Borjesson DL, Kobayashi SD, Whitney AR, Voyich JM, Argue CM, Deleo FR. 2005. Insights into pathogen immune evasion mechanisms: Anaplasma phagocytophilum fails to induce an apoptosis differentiation program in human neutrophils. J. Immunol. 174:6364–6372 [DOI] [PubMed] [Google Scholar]
- 24. Carlyon JA, Abdel-Latif D, Pypaert M, Lacy P, Fikrig E. 2004. Anaplasma phagocytophilum utilizes multiple host evasion mechanisms to thwart NADPH oxidase-mediated killing during neutrophil infection. Infect. Immun. 72:4772–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. IJdo JW, Mueller AC. 2004. Neutrophil NADPH oxidase is reduced at the Anaplasma phagocytophilum phagosome. Infect. Immun. 72:5392–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Troese MJ, Carlyon JA. 2009. Anaplasma phagocytophilum dense-cored organisms mediate cellular adherence through recognition of human P-selectin glycoprotein ligand 1. Infect. Immun. 77:4018–4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Goodman JL, Nelson CM, Klein MB, Hayes SF, Weston BW. 1999. Leukocyte infection by the granulocytic ehrlichiosis agent is linked to expression of a selectin ligand. J. Clin. Invest. 103:407–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Herron MJ, Nelson CM, Larson J, Snapp KR, Kansas GS, Goodman JL. 2000. Intracellular parasitism by the human granulocytic ehrlichiosis bacterium through the P-selectin ligand, PSGL-1. Science 288:1653–1656 [DOI] [PubMed] [Google Scholar]
- 29. Ojogun N, Kahlon A, Ragland SA, Troese MJ, Mastronunzio JE, Walker NJ, Viebrock L, Thomas RJ, Borjesson DL, Fikrig E, Carlyon JA. 2012. Anaplasma phagocytophilum outer membrane protein A interacts with sialylated glycoproteins to promote infection of mammalian host cells. Infect. Immun. 80:3748–3760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mansueto P, Vitale G, Cascio A, Seidita A, Pepe I, Carroccio A, di Rosa S, Rini GB, Cillari E, Walker DH. 2012. New insight into immunity and immunopathology of rickettsial diseases. Clin. Dev. Immunol. 2012:967852 doi:10.1155/2012/967852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Carlyon JA, Akkoyunlu M, Xia L, Yago T, Wang T, Cummings RD, McEver RP, Fikrig E. 2003. Murine neutrophils require alpha1,3-fucosylation but not PSGL-1 for productive infection with Anaplasma phagocytophilum. Blood 102:3387–3395 [DOI] [PubMed] [Google Scholar]
- 32. Reneer DV, Kearns SA, Yago T, Sims J, Cummings RD, McEver RP, Carlyon JA. 2006. Characterization of a sialic acid- and P-selectin glycoprotein ligand-1-independent adhesin activity in the granulocytotropic bacterium Anaplasma phagocytophilum. Cell. Microbiol. 8:1972–1984 [DOI] [PubMed] [Google Scholar]
- 33. Reneer DV, Troese MJ, Huang B, Kearns SA, Carlyon JA. 2008. Anaplasma phagocytophilum PSGL-1-independent infection does not require Syk and leads to less-efficient AnkA delivery. Cell. Microbiol. 10:1827–1838 [DOI] [PubMed] [Google Scholar]
- 34. Sarkar M, Reneer DV, Carlyon JA. 2007. Sialyl-Lewis X-independent infection of human myeloid cells by Anaplasma phagocytophilum strains HZ and HGE1. Infect. Immun. 75:5720–5725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yago T, Leppanen A, Carlyon JA, Akkoyunlu M, Karmakar S, Fikrig E, Cummings RD, McEver RP. 2003. Structurally distinct requirements for binding of P-selectin glycoprotein ligand-1 and sialyl Lewis X to Anaplasma phagocytophilum and P-selectin. J. Biol. Chem. 278:37987–37997 [DOI] [PubMed] [Google Scholar]
- 36. Felsheim RF, Herron MJ, Nelson CM, Burkhardt NY, Barbet AF, Kurtti TJ, Munderloh UG. 2006. Transformation of Anaplasma phagocytophilum. BMC Biotechnol. 6:42 doi:10.1186/1472-6750-6-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Carlyon JA. 2005. Laboratory maintenance of Anaplasma phagocytophilum. Curr. Protoc. Microbiol. 2005:Unit 3A.2. doi:10.1002/9780471729259.mc03a02s00 [DOI] [PubMed] [Google Scholar]
- 38. Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248–254 [DOI] [PubMed] [Google Scholar]
- 39. Sarkar M, Troese MJ, Kearns SA, Yang T, Reneer DV, Carlyon JA. 2008. Anaplasma phagocytophilum MSP2(P44)-18 predominates and is modified into multiple isoforms in human myeloid cells. Infect. Immun. 76:2090–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ottens AK, Bustamante L, Golden EC, Yao C, Hayes RL, Wang KK, Tortella FC, Dave JR. 2010. Neuroproteomics: a biochemical means to discriminate the extent and modality of brain injury. J. Neurotrauma 27:1837–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Troese MJ, Kahlon A, Ragland SA, Ottens AK, Ojogun N, Nelson KT, Walker NJ, Borjesson DL, Carlyon JA. 2011. Proteomic analysis of Anaplasma phagocytophilum during infection of human myeloid cells identifies a protein that is pronouncedly upregulated on the infectious dense-cored cell. Infect. Immun. 79:4696–4707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li GZ, Vissers JP, Silva JC, Golick D, Gorenstein MV, Geromanos SJ. 2009. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9:1696–1719 [DOI] [PubMed] [Google Scholar]
- 43. Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 44. Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- 46. Mastronunzio JE, Kurscheid S, Fikrig E. 2012. Postgenomic analyses reveal development of infectious Anaplasma phagocytophilum during transmission from ticks to mice. J. Bacteriol. 194:2238–2247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ge Y, Rikihisa Y. 2007. Identification of novel surface proteins of Anaplasma phagocytophilum by affinity purification and proteomics. J. Bacteriol. 189:7819–7828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park J, Choi KS, Dumler JS. 2003. Major surface protein 2 of Anaplasma phagocytophilum facilitates adherence to granulocytes. Infect. Immun. 71:4018–4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Scorpio DG, Caspersen K, Ogata H, Park J, Dumler JS. 2004. Restricted changes in major surface protein-2 (msp2) transcription after prolonged in vitro passage of Anaplasma phagocytophilum. BMC Microbiol. 4:1 doi:10.1186/1471-2180-4-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Niu H, Rikihisa Y, Yamaguchi M, Ohashi N. 2006. Differential expression of VirB9 and VirB6 during the life cycle of Anaplasma phagocytophilum in human leucocytes is associated with differential binding and avoidance of lysosome pathway. Cell. Microbiol. 8:523–534 [DOI] [PubMed] [Google Scholar]
- 51. Huang B, Hubber A, McDonough JA, Roy CR, Scidmore MA, Carlyon JA. 2010. The Anaplasma phagocytophilum-occupied vacuole selectively recruits Rab-GTPases that are predominantly associated with recycling endosomes. Cell. Microbiol. 12:1292–1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang Y, Berg EA, Feng X, Shen L, Smith T, Costello CE, Zhang YX. 2006. Identification of surface-exposed components of MOMP of Chlamydia trachomatis serovar F. Protein Sci. 15:122–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wang T, Malawista SE, Pal U, Grey M, Meek J, Akkoyunlu M, Thomas V, Fikrig E. 2002. Superoxide anion production during Anaplasma phagocytophila infection. J. Infect. Dis. 186:274–280 [DOI] [PubMed] [Google Scholar]
- 54. Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen J, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21 doi:10.1371/journal.pgen.0020021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Balasubramanian S, Kannan TR, Baseman JB. 2008. The surface-exposed carboxyl region of Mycoplasma pneumoniae elongation factor Tu interacts with fibronectin. Infect. Immun. 76:3116–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Barel M, Hovanessian AG, Meibom K, Briand JP, Dupuis M, Charbit A. 2008. A novel receptor-ligand pathway for entry of Francisella tularensis in monocyte-like THP-1 cells: interaction between surface nucleolin and bacterial elongation factor Tu. BMC Microbiol. 8:145 doi:10.1186/1471-2180-8-145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bergonzelli GE, Granato D, Pridmore RD, Marvin-Guy LF, Donnicola D, Corthesy-Theulaz IE. 2006. GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect. Immun. 74:425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Candela M, Centanni M, Fiori J, Biagi E, Turroni S, Orrico C, Bergmann S, Hammerschmidt S, Brigidi P. 2010. DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology 156:1609–1618 [DOI] [PubMed] [Google Scholar]
- 59. Frisk A, Ison CA, Lagergard T. 1998. GroEL heat shock protein of Haemophilus ducreyi: association with cell surface and capacity to bind to eukaryotic cells. Infect. Immun. 66:1252–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Garduno RA, Garduno E, Hoffman PS. 1998. Surface-associated hsp60 chaperonin of Legionella pneumophila mediates invasion in a HeLa cell model. Infect. Immun. 66:4602–4610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Granato D, Bergonzelli GE, Pridmore RD, Marvin L, Rouvet M, Corthesy-Theulaz IE. 2004. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 72:2160–2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hickey TB, Thorson LM, Speert DP, Daffe M, Stokes RW. 2009. Mycobacterium tuberculosis Cpn60.2 and DnaK are located on the bacterial surface, where Cpn60.2 facilitates efficient bacterial association with macrophages. Infect. Immun. 77:3389–3401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Macellaro A, Tujulin E, Hjalmarsson K, Norlander L. 1998. Identification of a 71-kilodalton surface-associated Hsp70 homologue in Coxiella burnetii. Infect. Immun. 66:5882–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tsugawa H, Ito H, Ohshima M, Okawa Y. 2007. Cell adherence-promoted activity of Plesiomonas shigelloides groEL. J. Med. Microbiol. 56:23–29 [DOI] [PubMed] [Google Scholar]
- 65. Li F, Wilkins PP, Crawley S, Weinstein J, Cummings RD, McEver RP. 1996. Post-translational modifications of recombinant P-selectin glycoprotein ligand-1 required for binding to P- and E-selectin. J. Biol. Chem. 271:3255–3264 [PubMed] [Google Scholar]
- 66. Xia L, Ramachandran V, McDaniel JM, Nguyen KN, Cummings RD, McEver RP. 2003. N-terminal residues in murine P-selectin glycoprotein ligand-1 required for binding to murine P-selectin. Blood 101:552–559 [DOI] [PubMed] [Google Scholar]
- 67. Troese MJ, Sarkar M, Galloway NL, Thomas RJ, Kearns SA, Reneer DV, Yang T, Carlyon JA. 2009. Differential expression and glycosylation of Anaplasma phagocytophilum major surface protein 2 paralogs during cultivation in sialyl Lewis X-deficient host cells. Infect. Immun. 77:1746–1756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. IJdo JW, Wu C, Magnarelli LA, Fikrig E. 1999. Serodiagnosis of human granulocytic ehrlichiosis by a recombinant HGE-44-based enzyme-linked immunosorbent assay. J. Clin. Microbiol. 37:3540–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fronzes R, Christie PJ, Waksman G. 2009. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7:703–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rikihisa Y. 2010. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat. Rev. Microbiol. 8:328–339 [DOI] [PubMed] [Google Scholar]
- 71. Suarez CE, Noh S. 2011. Emerging perspectives in the research of bovine babesiosis and anaplasmosis. Vet. Parasitol. 180:109–125 [DOI] [PubMed] [Google Scholar]
- 72. Chan YG, Riley SP, Martinez JJ. 2010. Adherence to and invasion of host cells by spotted fever group Rickettsia species. Front. Microbiol. 1:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Riley SP, Goh KC, Hermanas TM, Cardwell MM, Chan YG, Martinez JJ. 2010. The Rickettsia conorii autotransporter protein Sca1 promotes adherence to nonphagocytic mammalian cells. Infect. Immun. 78:1895–1904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Laszik Z, Jansen PJ, Cummings RD, Tedder TF, McEver RP, Moore KL. 1996. P-selectin glycoprotein ligand-1 is broadly expressed in cells of myeloid, lymphoid, and dendritic lineage and in some nonhematopoietic cells. Blood 88:3010–3021 [PubMed] [Google Scholar]
- 75. Ofek I, Hasty DL, Doyle RJ. 2003. Basic concepts in bacterial adhesion, p 1–17 In Bacterial adhesion to animal cells and tissues. ASM Press, Washington, DC [Google Scholar]
- 76. Grimm D, Tilly K, Byram R, Stewart PE, Krum JG, Bueschel DM, Schwan TG, Policastro PF, Elias AF, Rosa PA. 2004. Outer-surface protein C of the Lyme disease spirochete: a protein induced in ticks for infection of mammals. Proc. Natl. Acad. Sci. U. S. A. 101:3142–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Tilly K, Krum JG, Bestor A, Jewett MW, Grimm D, Bueschel D, Byram R, Dorward D, Vanraden MJ, Stewart P, Rosa P. 2006. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun. 74:3554–3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Park J, Kim KJ, Grab DJ, Dumler JS. 2003. Anaplasma phagocytophilum major surface protein-2 (Msp2) forms multimeric complexes in the bacterial membrane. FEMS Microbiol. Lett. 227:243–247 [DOI] [PubMed] [Google Scholar]
- 79. Lin M, Kikuchi T, Brewer HM, Norbeck AD, Rikihisa Y. 2011. Global proteomic analysis of two tick-borne emerging zoonotic agents: Anaplasma phagocytophilum and Ehrlichia chaffeensis. Front. Microbiol. 2:24. [DOI] [PMC free article] [PubMed] [Google Scholar]