Abstract
Genome-wide expression analyses have provided clues on how Salmonella proliferates inside cultured macrophages and epithelial cells. However, in vivo studies show that Salmonella does not replicate massively within host cells, leaving the underlying mechanisms of such growth control largely undefined. In vitro infection models based on fibroblasts or dendritic cells reveal limited proliferation of the pathogen, but it is presently unknown whether these phenomena reflect events occurring in vivo. Fibroblasts are distinctive, since they represent a nonphagocytic cell type in which S. enterica serovar Typhimurium actively attenuates intracellular growth. Here, we show in the mouse model that S. Typhimurium restrains intracellular growth within nonphagocytic cells positioned in the intestinal lamina propria. This response requires a functional PhoP-PhoQ system and is reproduced in primary fibroblasts isolated from the mouse intestine. The fibroblast infection model was exploited to generate transcriptome data, which revealed that ∼2% (98 genes) of the S. Typhimurium genome is differentially expressed in nongrowing intracellular bacteria. Changes include metabolic reprogramming to microaerophilic conditions, induction of virulence plasmid genes, upregulation of the pathogenicity islands SPI-1 and SPI-2, and shutdown of flagella production and chemotaxis. Comparison of relative protein levels of several PhoP-PhoQ-regulated functions (PagN, PagP, and VirK) in nongrowing intracellular bacteria and extracellular bacteria exposed to diverse PhoP-PhoQ-inducing signals denoted a regulation responding to acidic pH. These data demonstrate that S. Typhimurium restrains intracellular growth in vivo and support a model in which dormant intracellular bacteria could sense vacuolar acidification to stimulate the PhoP-PhoQ system for preventing intracellular overgrowth.
INTRODUCTION
Salmonella enterica serovars are food-borne bacterial pathogens that cause gastroenteritis and systemic disease (typhoid fever) in humans and livestock (1–3). Salmonellae invade a variety of eukaryotic cell types and have been extensively studied in animal models (2, 4, 5) and in vitro in models involving cultured mammalian cell lines (6). Master elements of Salmonella pathogenicity include two type III secretion systems encoded in the Salmonella pathogenicity islands 1 and 2 (SPI-1 and SPI-2), which secrete proteins promoting invasion and survival/proliferation inside the host cell. Other widely studied Salmonella functions linked to virulence include regulatory proteins, such as the two-component system PhoP-PhoQ, which orchestrates adaptation of the pathogen to the intracellular environment of the infected cell (7, 8).
Despite the bulk of information collected on pathogen functions required for virulence, it remains largely unknown how this pathogen regulates virulence functions in distinct host cell types. Host cells targeted by this pathogen in vivo include epithelial cells, macrophages, neutrophils, and dendritic cells (9, 10). However, Salmonella preferentially resides within macrophages in both acute and chronic infections (11–14). A feature that distinguishes the behavior of intracellular bacteria in vivo is their limited capacity to proliferate inside host cells, reaching progenies of only 3 to 4 individuals per infected cell (12–15). The most widely accepted model indicates that S. Typhimurium colonizes the animal by increasing the number of infection foci rather than increasing the number of intracellular bacteria per cell. Repetitive cycles of limited proliferation rounds inside macrophages, exit from the infected cells and infection of neighbor cells may account for the increase of infection foci (16). Of interest, recent studies in cultured macrophages and epithelial cells reported a marked heterogeneity in the population of intracellular Salmonella, with some bacteria undergoing active replication while others remain in a nongrowing state for long periods of time (17, 18). This heterogeneity, inherent to many natural processes (19), is also known in other pathogens exposed to antimicrobials (20) and in the expression by Salmonella of certain virulence determinants such as the pathogenicity island 1, SPI-1 (20, 21).
The visualization of low numbers of intracellular Salmonella per infected cell in chronically infected mice (11) suggests that, similar to what has been described for other intracellular pathogens, such as Mycobacterium tuberculosis (22), strategies limiting intracellular growth (driven by the host and/or by the pathogen) may operate in chronic and asymptomatic infections (15, 23–25). Serovar Typhi establishes this condition in about 5% of humans recovering from typhoid fever, and nontyphoidal serovars causing infections in humans can also persist asymptomatically in livestock and domestic fowl (23). Despite the relevance of these phenomena, it is still unknown whether Salmonella restricts intracellular growth in vivo. The existing information relates to anatomical sites where bacteria are visualized during chronic infections and pathogen functions influencing such a state (26, 27). Macrophages present in the mesenteric lymph nodes (11), hemophagocytic macrophages (28), and epithelial cells of the gallbladder (29) are proposed to act as serovar Typhimurium reservoirs during chronic infections. Microarray-based negative genetic selections revealed that SPI-1, SPI-2, prophages, fimbrial operons, and genes regulated by the PhoP-PhoQ two-component regulatory system contribute to long-lasting colonization of the mouse spleen (30). Functions encoded in the genomic island CS54 as ShdA and RatB are also required for efficient fecal shedding in mice (26). Other functions linked to persistence include the secretion system ZirT/ZirS, highly expressed in bacteria shed in fecal pellets (31), and SciS, proposed to attenuate Salmonella growth inside macrophages and to downregulate virulence in vivo (32). Together, these studies support the idea that intracellular Salmonella is capable of limiting intracellular growth in vivo.
Our previous studies revealed that S. Typhimurium does not proliferate inside cultured fibroblasts (33, 34). The pathogen contributes to this condition, since bacterial overgrowth is observed upon inactivation of bacterial regulators, such as PhoP-PhoQ, the sigma factor RpoS, or the plasmid-encoded regulator SpvR (33). In addition to growth restraint, nonreplicating intracellular bacteria ensure viability using SPI-2 and the sigma factor RpoE (33). Here, we undertook a study in mice to investigate whether S. Typhimurium has the capacity to attenuate intracellular growth in vivo. We obtained evidence for attenuation of S. Typhimurium proliferation in nonphagocytic cells located in the intestinal lamina propria which was dependent on a functional PhoP-PhoQ system. The establishment of such a intracellular nonproliferative state was also demonstrated in primary fibroblasts isolated from intestinal tissue. On the basis of this information, genome profiling was defined in nonreplicating intracellular bacteria using an in vitro model of persistence. These data allowed us to further characterize the mode in which the PhoP-PhoQ system is activated in intracellular dormant bacteria.
MATERIALS AND METHODS
Bacterial strains, culture media, and growth conditions.
The S. enterica serovar Typhimurium strains used in this study are shown in Table 1. All strains derive from SV5015, a His+ derivative of the mouse-virulent strain SL1344 (36). Bacteria were grown in Luria broth (LB) at 37°C. When appropriate, kanamycin (30 μg/ml) or ampicillin (50 μg/ml) was added to the growth media. For the transcriptomic analyses (see below), bacteria were grown in LB medium at 37°C with aeration (180 rpm) to exponential phase (optical density at 600 nm [OD600] of ∼0.2) or to stationary phase (final OD600 of ∼3.0) and maintained in the latter condition for an additional 12 h. To infect BALB/c mice and eukaryotic cells (see below), bacteria were grown at 37°C in standing nonaerated cultures obtained upon inoculation of 2 ml of LB medium with a bacterial colony and subsequent overnight incubation (final OD600 of ∼1.0). To analyze gene regulation mediated by PhoP-PhoQ, bacteria were grown in N minimal medium (39) containing 38 mM glycerol as the carbon source and supplemented with 10 mM or 8 μM MgCl2 as described previously (40). For SPI-2-inducing conditions, the PCN minimal medium adjusted to a pH of 5.8 was used as described previously (41).
Table 1.
S. enterica serovar Typhimurium strains used in this studya
| Strain | Relevant genotype | Source or reference |
|---|---|---|
| SL1344 | hisG64, rpsL, mouse virulent isolate | 36 |
| SV4056 | SL1344 phoP7953::Tn10 | 33 |
| SV4386 | SL1344 ssaC (spiA)::KIXX | 33 |
| MD1682 | SL1344 glpK::3×FLAG-Kn | 38 |
| SV5015 | Wild type, SL1344 His+ | 37 |
| MD1120 | phoP7953::Tn10 | 37 |
| MD1178 | pagC::3×FLAG-Kn | This study |
| MD1182 | pagC::3×FLAG-Kn phoP7953::Tn10 | This study |
| MD1942 | mgtC::3×FLAG-Kn | This study |
| MD1951 | mgtC::3×FLAG-Kn phoP7953::Tn10 | This study |
| MD2926 | glpK::3×FLAG-Kn | This study |
| MD2927 | glpK::3×FLAG-Kn phoP7953::Tn10 | This study |
| MD3702 | virK::3×FLAG-Kn | This study |
| MD3729 | virK::3×FLAG-Kn phoP7953::Tn10 | This study |
| MD3703 | pagN::3×FLAG-Kn | This study |
| MD3726 | pagN::3×FLAG-Kn phoP7953::Tn10 | This study |
| MD3727 | pagP::3×FLAG-Kn | This study |
| MD3723 | pagP::3×FLAG-Kn phoP7953::Tn10 | This study |
Unless otherwise indicated, all strains are isogenic to the wild-type strain SV5015 (SL1344 His+).
Construction of chromosomal epitope-tagged genes.
The strains carrying chromosomal 3× FLAG epitope-tagged genes were constructed using the method described by Uzzau et al. (42). Plasmids and oligonucleotides used for this procedure are listed in Table S8 in the supplemental material. Correct insertion of the epitope at the 3′ end of the targeted gene was verified in all cases by PCR and sequencing.
Fibroblast cells.
NRK-49F normal rat kidney fibroblasts (ATCC CRL-1570) were used throughout the study. These fibroblasts were propagated in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) containing 5% (vol/vol) fetal bovine serum (FBS) and 4 mM l-glutamine. Mouse intestinal fibroblasts were isolated from six C57BL/10 female mice of 12 weeks of age by following the method of Strong et al. described for human intestinal fibroblasts (43). These primary fibroblasts were propagated in the presence of antibiotics for the first three passages and then in antibiotic-free medium to avoid interference with the bacterial invasion and proliferation assays. The infection experiments were performed between passages 5 and 9.
Intracellular bacterial proliferation assays in primary intestinal fibroblasts and NRK-49F rat fibroblasts.
Mouse primary intestinal fibroblasts and NRK-49F normal rat kidney fibroblasts were infected with bacteria for 20 min using a multiplicity of infection (MOI) of 10:1 (bacteria to eukaryotic cells) as previously described (44). After extensive washing, infected cells were incubated in fresh tissue culture medium containing 100 μg/ml gentamicin for the first 2 h postinfection and 10 μg/ml for the remainder of the experiment. Infected cells were lysed at the desired postinfection times in phosphate-buffered saline (PBS), pH 7.4, 1% Triton X-100. The number of viable intracellular bacteria was determined by plating. To inhibit vacuolar acidification, 100 nM bafilomycin (BAF) was added to the infected fibroblasts in the fresh tissue culture medium containing gentamicin to avoid any effect in bacterial entry. BAF was maintained during the incubation periods with high (100 μg/ml) and low (10 μg/ml) doses of gentamicin.
Bacterial infection of BALB/c mice and immunohistochemistry.
Wild-type and phoP mutant bacteria grown overnight in LB medium at 37°C in nonshaking conditions were collected by centrifugation (5,000 × g, 10 min, 4°C), washed twice in cold sterile PBS, pH 7.4, and suspended at a density of ∼8 × 1010 CFU/ml. Serial dilutions were used to infect orally (25 μl) groups of 8-week-old female BALB/c mice as described previously (45). The protocols used in these studies were approved by the Comité Ético de Experimentación of the Consejo Superior de Investigaciones Científicas (CSIC). The infectious dose was in the range of 108 to 109 viable bacteria per mouse, as confirmed by plating and counting of CFU. At distinct postinfection times, 6 and 24 h, a laparotomy was performed to localize the small intestine. Ileum was extracted aseptically and fixed in 4% paraformaldehyde (PFA) during 1 h at room temperature. After three washes with PBS, pH 7.4, buffer, the ileum was incubated at 4°C overnight in a 20% sucrose-PBS, pH 7.4, solution. Tissue blocks of ca. 1 cm3 were embedded in Jung tissue freezing medium (Leica) and rapidly deep-frozen in cold acetone (−50 to −60°C). Fifteen-μm-thick sections were obtained by cryotomy and mounted in glass slides pretreated with SuperFrost-Plus (Menzel-Glaser). Blocks and slices were kept at −80°C until further processing. For immunohistochemistry, glass slides containing the tissue section were thawed overnight at room temperature to increase tissue adhesiveness. Sections were further incubated in cold acetone (−20°C) during 10 min, the acetone was evaporated at room temperature for 15 min, and finally the sections were rehydrated in PBS, pH 7.4, for 15 min. Prior to immunostaining, sections were incubated for 1 h at room temperature in blocking solution (10% fetal bovine serum [FBS], 0.2% saponin). Primary and secondary antibodies were diluted as appropriate in 2% FBS, 0.2% saponin. Sections were incubated with the primary antibodies for 48 h at 4°C, followed by three washes with PBS, pH 7.4, and further incubation with secondary fluorochrome-conjugated antibodies for 1 h. Nuclei were stained with the cyanine fluorochrome To-Pro3 (Invitrogen) at a 1:200 dilution during 20 min at room temperature. Sections were blotted to remove excess PBS buffer. A drop of inclusion medium containing polyvinyl alcohol and DABCO (Fluka) was added, and the sections were covered with 24- by 60-mm glass coverslips (Menzel-Glaser). Samples were visualized in a Zeiss Axiovert 200 fluorescence microscope equipped with a confocal Radiance 2100 unit (Bio-Rad). LaserSharp 2000 software was used to capture the image, and LaserPix and Adobe Photoshop were used for image processing.
Large-scale infection of fibroblasts to obtain RNA and protein from intracellular bacteria.
NRK-49F normal rat fibroblasts were seeded in BioDish-XL 500-cm2 plates (reference 351040; BD Biosciences) at a density of 2 × 107 cells per dish and infected at an MOI of 10:1 (bacteria:fibroblast) and 40 to 60% confluence. After 20 min, the infected cells were washed five times with prewarmed Hank's balance salt solution (HBSS). These cells were incubated until 1 h postinfection in fresh culture medium containing 100 μg/ml of gentamicin. The culture medium was then replaced with new fresh medium containing 10 μg/ml gentamicin until the desirable postinfection time (1, 2, 6, 8, or 24 h). Infected fibroblasts were processed as described for the Salmonella-macrophage infection model (46), with slight modifications. Briefly, infected fibroblasts were washed five times with cold PBS, pH 7.4, and lysed (at 30 ml per plate) in a solution containing 0.4% SDS, 1% acidic phenol, and 19% ethanol in water. After 30 min of incubation at 4°C, intracellular bacteria were collected by centrifugation (27,500 × g, 4°C, 30 min) and washed three times with 1 ml of a 1% acidic phenol, 19% ethanol solution. For each infection time point at which RNA or protein was extracted, four BioDish-XL 500-cm2 plates usually were pooled. For microarray hybridizations, RNA of nongrowing intracellular bacteria was extracted and pooled from a minimum of 20 independent experiments with four BioDish-XL 500-cm2 plates each. For protein extraction, intracellular bacteria were washed twice with cold PBS, pH 7.4, recovered by centrifugation (15,000 × g, 4°C, 10 min), and processed as described previously (37, 38). RNA and protein extraction in extracellular bacteria was performed as described previously (37, 38, 46) by following the same treatment as that for intracellular bacteria with a solution containing 1% acidic phenol, 19% ethanol, and 0.4% SDS.
Genome expression analyses and RT-qPCR.
Total RNA purified from intracellular and extracellular bacteria were processed as previously described to generate the corresponding cDNAs (47). The Salgenomics microarray used for these studies has been described previously and contains 70-mer antisense oligonucleotides specific to 4,369 open reading frames (ORFs), 21 rRNAs, 86 tRNAs, and 47 sRNAs identified in the genome of S. Typhimurium strain SL1344 (47). The hybridization conditions, data acquisition, normalization, and statistical analyses have been described elsewhere (37, 47). Validation assays were performed by quantitative reverse transcription-PCR (RT-qPCR) as described previously (37), using ompA as an internal control.
Antibodies and immunofluorescence microscopy.
The following primary antibodies were used for Western assays and immunofluorescence microscopy studies: rabbit polyclonal anti-Salmonella flagellin (FliC/FljB) (48); rabbit polyclonal KH1331 anti-TlpA (gift from Reini Hurme, Karolinska Institutet, Stockholm, Sweden); mouse monoclonal anti-FLAG epitope (clone M2; Sigma); rabbit polyclonal anti-S. Typhimurium lipopolysaccharide (LPS), group B, factors 1:4:5:12 (Difco Laboratories); mouse monoclonal antibacterial RNA polymerase sigma S subunit, RpoS (clone 1RS1; Santa Cruz Biotechnology); rabbit polyclonal anti-OmpA (gift of H. Schwarz, Tübingen, Germany); rabbit polyclonal anti-calnexin (Stressgen); rat monoclonal anti-CD18 (clone M18/2; Developmental Studies Hybridoma Bank [DSHB], IA); rat monoclonal anti-CD45 (clone 30-F11; BD PharMingen); and mouse monoclonal anti-alpha actin of smooth muscle (α-SMA) conjugated to Cy3 (clone 1A4; Sigma). For immunofluorescence microscopy, the following secondary antibodies were used at a 1:500 dilution: goat polyclonal anti-rabbit IgG conjugated to Alexa 488 (Molecular Probes), goat polyclonal anti-rat IgG conjugated to Alexa 594 (Molecular Probes), and goat polyclonal anti-mouse IgG conjugated to Alexa 594 (Molecular Probes). Goat polyclonal anti-mouse IgG conjugated to horseradish peroxidase (HRP; Bio-Rad) was used as secondary antibody at a 1:5,000 dilution for Western assays. Polyclonal rabbit HRP-conjugated anti-GroEL (Sigma) was also used. Infected NRK-49F fibroblasts and mouse intestinal primary fibroblasts were fixed and processed for immunofluorescence microscopy as previously described (44). Cells were examined in a Leica fluorescence inverted microscope (DMI6000B).
Statistical analysis.
Data were analyzed with GraphPad Prism 5.0 software (GraphPad Inc., San Diego, CA) using Student's t test. Differences in values with P < 0.05 were considered significant.
Accession numbers.
The characteristics and configuration of the Salgenomics microarray were deposited in the MIAME database (http://www.ebi.ac.uk/miamexpress) under accession number A-MEXP-846. Gene expression data were deposited in the Array Express database (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-MEXP-1774 (intracellular phoP transcriptome), E-MEXP-1775 (intracellular wild-type transcriptome), and E-MEXP-1776 (extracellular wild type, stationary phase).
RESULTS
S. Typhimurium attenuates growth in nonphagocytic cells located in the lamina propria of intestinal villi.
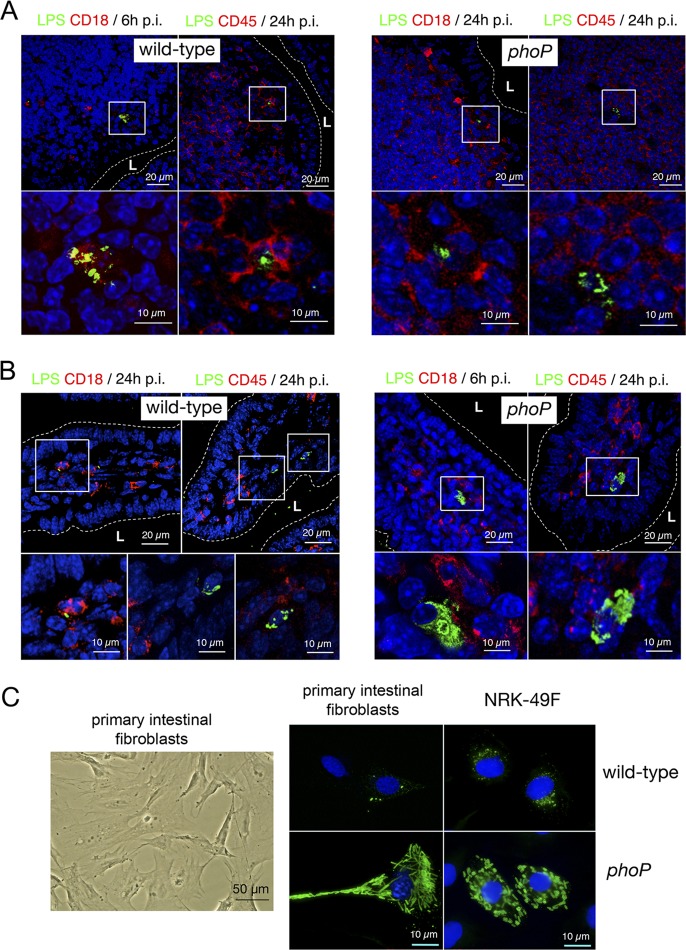
S. Typhimurium uses the PhoP-PhoQ system to attenuate growth inside cultured fibroblasts (33, 34). To determine whether the pathogen also triggers an attenuation response in vivo, BALB/c mice were challenged orally with wild-type and phoP isogenic bacteria. Host cell types harboring bacteria were searched by microscopy in intestinal tissue sections. Due to the lack of cell markers suitable for distinguishing by immunohistochemistry and in an exclusive manner the fibroblast populations present in the intestinal tissue (49), we examined the distribution of CD18 and CD45, two panleukocyte cell markers. CD18 is the β2 chain of the leukocyte-specific integrins LFA-1, Mac-1, gp150, and gp95 (50). CD45, also known as leukocyte common antigen, is a transmembrane glycoprotein present exclusively in nucleated cells of hematopoietic origin (51). More than 200 sections of the intestinal tissue were examined at the microscope for each postinfection time and bacterial strain (i.e., wild-type and phoP mutant strains). In the Peyer's patches, wild-type and phoP mutant bacteria were visualized inside CD18+ CD45+ cells located underneath the intestinal epithelium (Fig. 1A). A similar inspection in the intestinal villi resulted in the visualization of CD18+ CD45+ and CD18− CD45− cells positioned in the lamina propria (Fig. 1B). Surprisingly, the nonphagocytic CD18− CD45−-negative cells contained a higher intracellular bacterial load only in the phoP mutant strain-infected mice (Fig. 1B). Such a difference was consistently observed in all tissue sections in which infected cells were present in the lamina propria of the villi. Thus, of a total of 14 CD18− CD45− cells visualized in the villi of mice challenged with wild-type bacteria, none of them exhibited massive amounts of bacteria. In contrast, all the 17 CD18− CD45− cells observed in the villi of mice challenged with the phoP mutant harbored large quantities of intracellular bacteria. Additional assays proved that the stromal cells in which the phoP mutant overgrows were negative for smooth muscle actin (SMA) (see Fig. S1 in the supplemental material). SMA is a marker present in smooth muscle cells, myofibroblasts, and interstitial cells of Cajal (ICC) but is absent from leukocytes and interstitial stromal fibroblasts (49, 52). To assess the possibility that the CD45− CD18− cells in which S. Typhimurium restricts growth correspond to interstitial fibroblasts, we isolated primary fibroblasts from the intestinal lamina propria of the ileum. When propagated in vitro, intestinal primary fibroblasts exhibited uniform morphology (Fig. 1C) and were invaded at similar rates by wild-type and phoP mutant strain bacteria (data not shown). In contrast to wild-type bacteria, the phoP mutant strain proliferated extensively within these intestinal primary fibroblasts in a fashion reminiscent of that exhibited by the same mutant in CD18− CD45− nonphagocytic cells of the lamina propria (Fig. 1B). A similar behavior was also observed for the phoP mutant upon invasion of cultured NRK-49F rat fibroblasts (Fig. 1C) in which this intracellular growth-attenuating response was uncovered (33). To our knowledge, these data provided the first in vivo evidence that S. Typhimurium can restrain intracellular growth in the host.
Fig 1.
S. Typhimurium attenuates growth inside nonphagocytic cells positioned in the lamina propria of intestinal villi. (A) Tissue sections of the intestinal ileum corresponding to Peyer's patch areas were labeled with antibodies recognizing S. Typhimurium lipopolysaccharide (LPS) and the panphagocytic marker CD18 or CD45. To-pro3 was used to stain nuclei. Samples were collected at 6 or 24 h postchallenge of BALB/c mice with the SV5015 (wild-type) and MD1120 (phoP mutant) strains. Areas in boxes in upper panels are magnified in the lower panels. (B) Tissue sections showing bacterium-containing cells in the lamina propria of intestinal villi. Samples were collected at 6 or 24 h postinfection as described for panel A and were labeled with antibodies against S. Typhimurium LPS, CD18, or CD45. Note the presence of nonphagocytic stromal cells containing large numbers of intracellular phoP mutant bacteria. Areas in boxes in upper panels are magnified in the lower panels. (C) Morphology of primary intestinal fibroblasts isolated from intestinal tissue. These primary fibroblasts were infected with the SV5015 (wild-type) or MD1120 (phoP mutant) strain. In parallel, NRK-49F fibroblasts were also infected with the same strains. Bacteria were detected with anti-S. Typhimurium LPS antibodies, and nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI). Note the similar bacterial phenotypes in both types of fibroblasts. L, intestinal lumen.
Genome-wide expression profiling of nongrowing intracellular Salmonella located inside fibroblasts.
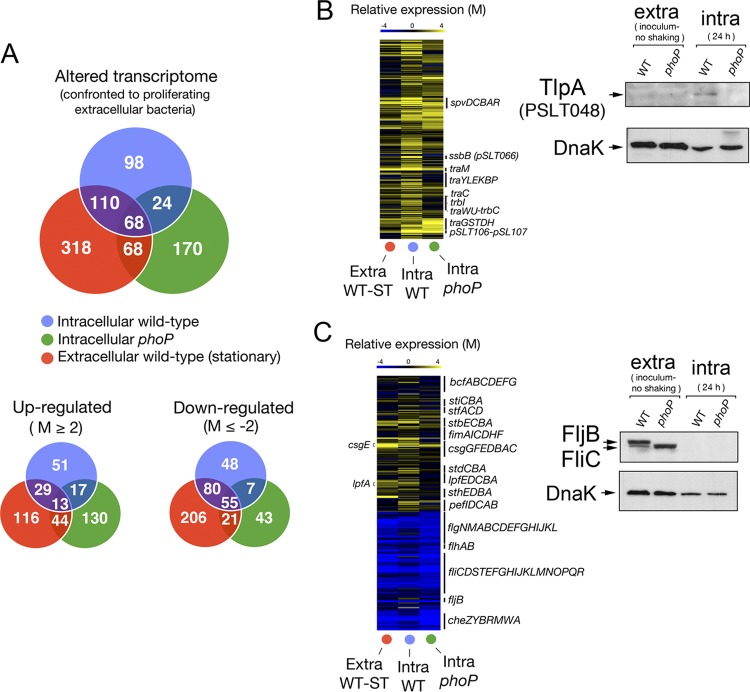
The marked parallelism found in the behavior of intracellular bacteria in vivo and in cultured fibroblasts (Fig. 1B and C) led us to exploit the in vitro model to dissect the growth-attenuating response at the transcriptome level. Total RNA was extracted from nongrowing wild-type bacteria collected at 24 h postinfection of NRK-49F fibroblasts. RNA extraction from intracellular bacteria located inside fibroblasts required optimization of the protocol previously described for the Salmonella-macrophage infection model (46). Given the low number of nongrowing wild-type bacteria residing inside fibroblasts (average of ∼2 to 3 bacteria per infected cell), we infected ca. 109 fibroblasts to obtain the minimal amount of RNA required for hybridization purposes. Transcriptomic data were obtained using the Salgenomics 70-mer oligonucleotide microarray platform (37, 47), which covers the genome of S. Typhimurium strain SL1344. Only relative expression changes greater than 4-fold were considered significant. Total RNA was also purified from extracellular wild-type and phoP mutant strain bacteria in an active (exponential) phase of growth and for wild-type bacteria in stationary phase. The latter condition was included to differentiate genes genuinely expressed in nongrowing conditions inside the fibroblast. The initial assays revealed that the transcriptomes of wild-type and phoP mutant bacteria were rather similar in extracellular conditions (LB medium), with only three genes, phoP, STM0939 (ybjD), and STM0940 (ybjX), exhibiting expression differences greater than 4-fold (see Table S1 in the supplemental material). The expression profile of extracellular wild-type bacteria growing exponentially in LB was used as a comparator for nongrowing wild-type bacteria in intracellular (inside fibroblasts) and extracellular (stationary-phase) environments (Fig. 2A; also see Table S2). The expression profile of the phoP mutant in exponential phase in LB medium was also compared to that of the same mutant in the intracellular (overgrowing) condition (Fig. 2A; also see Table S2). A total of 98 genes (ca. 2% of the genome) showed differential regulation in nongrowing wild-type bacteria located inside the fibroblast (Fig. 2A; also see Table S3). Of these, 51 genes were upregulated (see Table S4) and 48 downregulated (Fig. 2A; also see Table S5). Some fimbria-related genes, such as stbB, stbC, lpfA, and fimF, were expressed at higher levels in nongrowing intracellular wild-type bacteria than in the overgrowing phoP mutant (Fig. 2B; also see Table S2). Similarly, most of the pSLT virulence plasmid genes were expressed at higher levels in nongrowing intracellular wild-type bacteria (Fig. 2B; also see Table S2). Upregulation of virulence plasmid functions was confirmed at the protein level for PSLT048 (TlpA), a protein regulated by PhoP-PhoQ that was detected only in nongrowing intracellular wild-type bacteria (Fig. 2B). Chemotaxis and flagellar genes were also strongly downregulated in intracellular bacteria (see Table S5), which agreed with the absence of flagellin noted by Western assays in wild-type and phoP mutant bacteria located inside the fibroblast (Fig. 2C). This response seems to make sense for bacteria persisting within a tightly apposed membrane-bound vacuole and, therefore, not requiring motility. This observation contrasts with the upregulation of flagellin occurring at late infection times in Salmonella proliferating within epithelial cells (18, 53). Nongrowing intracellular bacteria also upregulated metabolic functions responding to low-oxygen conditions. Examples are dmsB and STM1499, which encode subunits of the anaerobic dimethyl sulfoxide reductase, and glpB, encoding a subunit of the anaerobic glycerol-3-phosphate dehydrogenase (see Table S2). Genes encoding functions related to the utilization of propanediol (pduT) or ethanolamine (eutG and eutS) and certain heat shock proteins (ibpB) were also exclusively upregulated in nonproliferating wild-type bacteria (see Table S2). Conversely, gntT, a gene encoding a high-affinity gluconate permease and previously reported to be induced by S. Typhimurium inside macrophages (46), was strongly downregulated by nongrowing bacteria inside fibroblasts (see Table S2). Overall, these data indicated that the nonproliferative lifestyle of S. Typhimurium involves a transcriptional profile distinct from those reported for macrophages and epithelial cells. Some features of this unique lifestyle include metabolic reprogramming to microaerophilic conditions and gene expression changes that can be tentatively interpreted as energetic restraint.
Fig 2.
Genome-wide expression analyses reveal unique signatures in nongrowing intracellular wild-type S. Typhimurium. (A) Number of genes displaying expression changes higher than 4-fold (log2 = M value of ≤−2 or ≥2) in intracellular nongrowing bacteria (wild-type strain SV5015) or intracellular proliferating bacteria (phoP mutant strain MD1120) at 24 h postinfection of NRK-49F fibroblasts. A third sample, corresponding to extracellular bacteria grown overnight to stationary phase in LB medium in shaking conditions, was included for comparison. The three samples are referenced to the expression pattern displayed by actively growing extracellular bacteria grown in LB medium to an OD of 0.2 (exponential phase) (see the text for details). The number of genes that displayed up- and downregulation compared to bacteria grown to exponential phase are also indicated. (B) Heat map showing the upregulation in intracellular bacteria of genes mapping in the virulence plasmid pSLT. None that, relative to extracellular bacteria, pSLT plasmid genes are upregulated to a greater extent in nongrowing intracellular wild-type bacteria than in intracellular phoP mutant bacteria (also see Table S2 in the supplemental material). A representative case confirming this difference is shown for the TlpA (PSLT048) plasmid protein. (C) Heat map showing the expression changes of gene clusters encoding distinct fimbriae or flagellar proteins. Note the downregulation in the expression of flagellar and chemotaxis genes in intracellular wild-type and mutant bacteria (both wild-type and phoP mutant strains) (see Table S2). A Western assay demonstrating the marked drop in flagellin (FliC/FljB) relative levels in intracellular bacteria is shown.
Characterization of the Salmonella PhoP-PhoQ regulon in nongrowing intracellular bacteria.
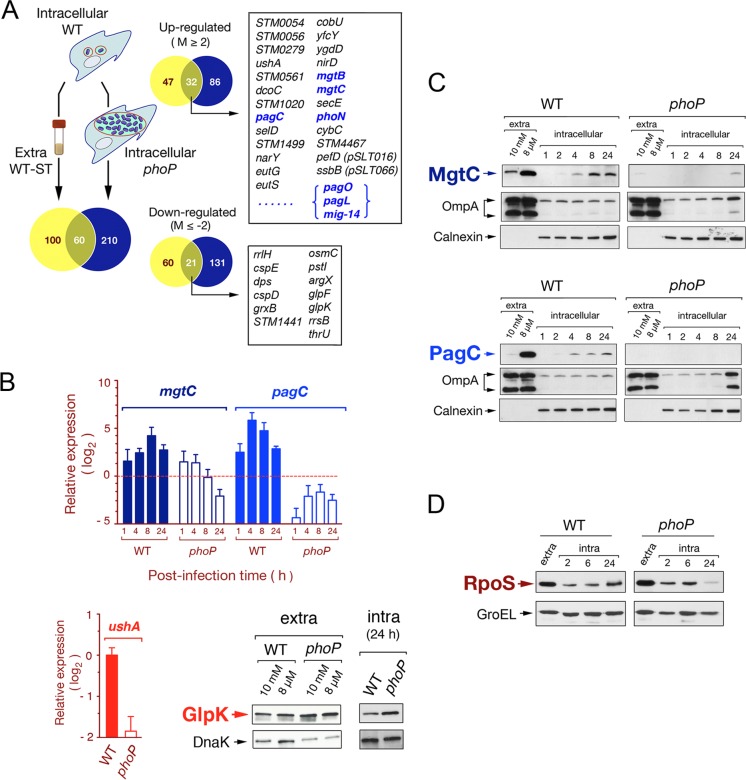
As expected, some known PhoPQ-regulated genes, such as pagC and mgtC, were upregulated by intracellular wild-type bacteria in the nongrowing dormant state, while such upregulation was not observed in overgrowing phoP mutant bacteria (see Table S2 in the supplemental material). Based on these observations, we reasoned that comparison of the expression profiles of nongrowing wild-type bacteria (intracellular and extracellular, stationary phase) to that of overgrowing intracellular phoP mutant bacteria could reveal features of the nonproliferative intracellular lifestyle (Fig. 3A). A total of 160 genes were found to be differentially expressed in nongrowing intracellular bacteria compared to extracellular stationary-phase bacteria, therefore they were considered to respond genuinely to intracellular cues (Fig. 3A; also see Table S6). On the other hand, nongrowing wild-type bacteria differed in the expression of 270 genes compared to overgrowing intracellular phoP mutant bacteria (Fig. 3A; also see Table S7). Of these 270 genes, 60 responded in a PhoP-PhoQ-dependent manner to both traits, the intracellular environment and a nongrowing condition (Fig. 3A). Interestingly, some of these genes were not previously ascribed to the PhoP-PhoQ regulon in extracellular growing conditions (Fig. 3A). To validate these observations, we quantified the relative transcript levels of mgtC and pagC in intracellular and extracellular bacteria. Another gene, ushA, not previously assigned to the PhoP-PhoQ regulon and displaying altered expression exclusively in nongrowing intracellular bacteria (Fig. 3A; also see Table S2), was also included in the analysis. ushA encodes a putative UDP-sugar hydrolase/5′-nucleotidase that is present in S. enterica and Escherichia coli (54). RT-qPCR assays confirmed that the expression of mgtC, pagC, and ushA in nonproliferating intracellular bacteria was PhoP-PhoQ dependent (Fig. 3B). Interestingly, the ushA allele harbored by the strain used in our study (SV5015; a His+ derivate of SL1344) encodes a protein not containing the S139Y missense mutation reported to abrogate the activity of this enzyme in other S. Typhimurium strains, such as LT2 (54) (see Fig. S2). In addition to ushA, we also validated glpK, a gene encoding a putative glycerol-kinase that displayed negative regulation by the PhoP-PhoQ system in intracellular bacteria (see Tables S2 and S7). Using a glpK::3×FLAG-tagged strain from a previous proteomic study (38), we constructed a derivate phoP glpK::3×FLAG isogenic strain to determine relative levels of GlpK in intracellular and extracellular bacteria. In concordance with the transcriptomic data, GlpK levels were found to be higher in the phoP mutant than in wild-type bacteria (Fig. 3B). The transcriptomes obtained with RNA extracted from wild-type and phoP mutant strains therefore provide a valuable source to identify new genes hitherto not assigned to the PhoP-PhoQ regulon.
Fig 3.
Characterization of the S. Typhimurium PhoP-PhoQ regulon in dormant nongrowing intracellular bacteria. (A) Venn diagram showing the overlapping among transcriptomes obtained from intracellular nongrowing bacteria (wild-type strain SV5015), intracellular proliferating bacteria (phoP mutant strain MD1120), and extracellular wild-type bacteria grown to stationary phase in LB medium. Each of these transcriptomes refers to the expression of extracellular bacteria grown to exponential phase (see Table S2 in the supplemental material). Numbers of genes differing in expression among transcriptomes by more than 4-fold are indicated. Highlighted in boxes are some of the genes displaying differential expression among transcriptomes (intra-WT versus extra-WT and intra-WT versus intra-phoP), which respond to both the intracellular environment and the functional status of the PhoP-PhoQ system. Genes previously reported to be regulated by PhoP-PhoQ are indicated in blue (see Tables S6 and S7 for details). (B) Validation data obtained by RT-qPCR for two PhoP-PhoQ-regulated genes, mgtC and pagC. Two other genes hitherto not assigned to the PhoP-PhoQ regulon, ushA and glpK, were also validated at the transcript and protein levels, respectively. mgtC and pagC data refer to different postinfection times (1, 4, 8, and 24 h) upon invasion of NRK-49F fibroblasts and are relative to expression levels detected in extracellular bacteria growing to exponential phase. ushA data are relative to the expression levels registered at 24 h postinfection in nongrowing intracellular bacteria. In the RT-qPCR assays, expression values were normalized to those obtained for the ompA gene used as an internal control. (C) Relative levels of the MgtC-3×FLAG- and PagC-3×FLAG-tagged proteins detected in intracellular nongrowing wild-type bacteria and in the overgrowing phoP mutant at the indicated postinfection times. As a control of canonical PhoP-PhoQ regulation, these two proteins were also monitored in extracellular bacteria grown in inducing (8 μM Mg2+) or repressing (10 mM Mg2+) conditions. (D) Levels of the alternative sigma factor RpoS detected in intracellular bacteria at different postinfection times upon entry into NRK-49F fibroblasts. DnaK, OmpA (bacterial proteins), and calnexin (eukaryotic protein) were used as loading controls.
Kinetics of induction of the PhoP-PhoQ system in intracellular nongrowing bacteria.
Using recombinant S. Typhimurium strains harboring a phoP::GFP transcriptional fusion, Martin-Orozco et al. showed in cultured macrophages that phoP expression is upregulated in intracellular bacteria from 30 min postentry (55). At this time, GFP-derived fluorescence was detected in ∼50% of the infected macrophages. These kinetics were more rapid than those observed in extracellular bacteria exposed to PhoP-PhoQ-inducing signals at a low magnesium concentration, which required about 1 h (55). Based on this, we investigated whether activation of the PhoP-PhoQ system inside fibroblasts shares similarities with macrophages. To that aim, we engineered strains carrying mgtC::3×FLAG and pagC::3×FLAG alleles tagged in their 3′ ends and in their respective chromosomal locations. Under these conditions, the regulatory scheme remains unaltered. Protein extracts were prepared from extracellular bacteria grown in inducing (8 μM Mg2+) and repressing (10 mM Mg2+) conditions for the PhoP-PhoQ system. Relative protein levels were compared to those detected in intracellular bacteria collected at early and late postinfection times (1 and 24 h). Both proteins, PagC and MgtC, were detected in the nongrowing intracellular wild type from 2 h postinfection, with a progressive increase in protein levels over time (Fig. 3C). As expected, PagC and MgtC levels were negligible at all postinfection times tested in the overgrowing phoP mutant collected from fibroblasts (Fig. 3C). Taken together, our data show that in fibroblasts the PhoP-PhoQ system is activated in intracellular S. Typhimurium in the absence of any noticeable increase in bacterial growth, most probably between 1 and 2 h postinfection.
We next focused on reinforcing the idea that intracellular S. Typhimurium adapts to a nongrowing state inside the fibroblasts. We hypothesized that the alternative sigma factor RpoS, required for adaptation of bacteria to nonproliferating (stationary phase) conditions (56) and regulated posttranscriptionally by PhoP-PhoQ in Salmonella (57), could be produced in larger amounts by nongrowing wild-type bacteria. Our early studies also revealed that S. Typhimurium rpoS mutants overgrow inside fibroblasts (33). Western assays showed that nongrowing intracellular wild-type bacteria contained larger amounts of RpoS than the overgrowing phoP mutant bacteria (Fig. 3D). This difference was more evident at 24 h postinfection, when wild-type bacteria may require more RpoS to face stresses linked to long-lasting residence in the infected cell. These data support the existence of a positive regulation of the PhoP-PhoQ system over RpoS in bacteria persisting inside the fibroblast.
Induction of PhoP-PhoQ in nonproliferating intracellular S. Typhimurium located inside fibroblasts responds to vacuolar acidification.
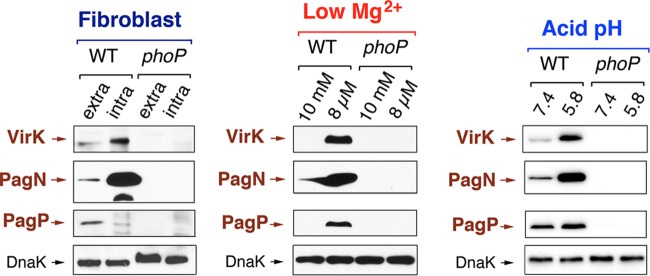
The S. Typhimurium regulatory system PhoP-PhoQ has been shown to respond in vitro to diverse signals, including Mg2+ limitation (58, 59), antimicrobial peptides (60, 61), and acidic pH (62). The extent to which these signals activate PhoP-PhoQ in bacteria located in the phagosome is still a matter of debate (8, 63). Given that PhoP-PhoQ induction could be easily monitored at the protein level in nongrowing intracellular bacteria (Fig. 3), we sought to determine in fibroblast the signals sensed by this system by comparing distinct inducing conditions in extra- and intracellular bacteria. For this purpose, we generated additional epitope-tagged strains in three genes positively regulated by PhoP-PhoQ, namely, virK, pagN, and pagP. The relative levels of 3× FLAG-tagged VirK, PagN, and PagP proteins were quantified in intracellular bacteria at 24 h postinfection of NRK-49F and compared to extracellular bacteria grown in 8 μM or 10 mM Mg2+ concentration. We also included in the analysis bacteria grown in PCN minimal medium containing 1 mM Mg2+ and adjusted to two distinct pH values, 5.8 or 7.4. The induction pattern observed in nongrowing intracellular bacteria indicated that PagN was strongly induced inside the fibroblast as well as VirK, although in the latter case the induction was less pronounced (Fig. 4). Interestingly, the relative level of PagP did not increase in intracellular bacteria, although its expression was clearly PhoP-PhoQ dependent (Fig. 4). The expression pattern of VirK, PagN, and PagP in intracellular bacteria was reproduced in extracellular bacteria grown in acidified (pH 5.8) PCN medium but not in a low (8 μM) Mg2+ concentration. Thus, VirK and PagN were induced in both conditions while PagP responded exclusively to Mg2+ limitation (Fig. 4). Taken together, these observations suggest that induction of PhoP-PhoQ by nongrowing dormant intracellular S. Typhimurium residing in vacuoles of fibroblasts occurs upon sensing of acidic pH.
Fig 4.
Regulation exerted by the PhoP-PhoQ system in nongrowing dormant intracellular bacteria matches the regulatory pattern observed in extracellular bacteria incubated in acidified growth medium. Western assays showing the relative levels of three distinct 3× FLAG-tagged proteins regulated by the PhoP-PhoQ system (VirK, PagN, and PagP) in extracellular and intracellular bacteria. Induction in intracellular bacteria was monitored by analysis of protein levels in extracellular bacteria used to infect the NRK-49F fibroblasts (inoculum, nonshaking growth conditions) and intracellular bacteria collected at 24 h postinfection. These samples are marked as extra and intra, respectively. The positive regulation of these three proteins by PhoP-PhoQ was tested in low Mg2+ concentrations and acid pH using the N and PCN media, respectively (see Materials and Methods). Shown are the levels of VirK, PagN, and PagP detected in extracellular bacteria grown in inducing (either 8 μM Mg2+ or pH 5.8) or repressing (either 10 mM Mg2+ or pH 7.4) conditions. Note that the response observed in acidified PCN medium matches, to a large extent, that observed in intracellular bacteria. However, the marked increase of PagP levels observed in 8 μM Mg2+ is not observed in nonproliferating intracellular bacteria. Loading controls based on DnaK are shown for the pagN::3×FLAG-tagged strains with equivalent results obtained for the other sets of strains shown.
An acidified vacuole directs survival of nongrowing intracellular S. Typhimurium inside the fibroblast.
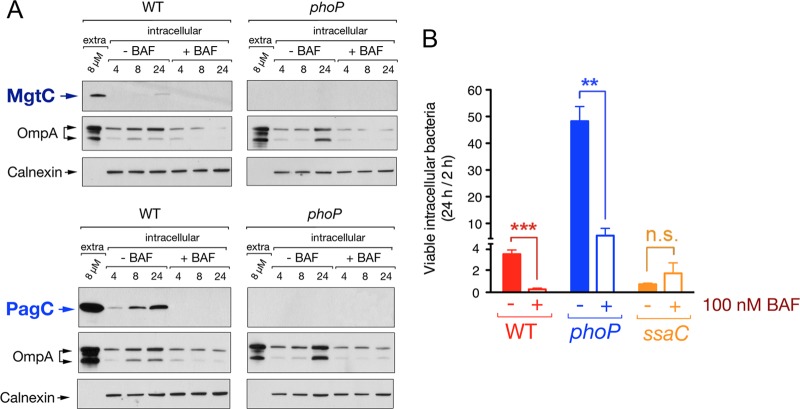
Since our data supported the idea of PhoP-PhoQ being induced upon bacterial sensing of intravacuolar acidic pH (Fig. 4), we reasoned that loss of such acidification could have consequences in PhoP-PhoQ function and fitness of intracellular bacteria. Indeed, early studies reported a key role of acid pH for PhoP-PhoQ induction in S. Typhimurium located inside macrophages (35, 55). To test whether a similar phenomenon occurs in fibroblasts, we monitored MgtC and PagC relative levels in S. Typhimurium residing within NRK-49F fibroblasts that were treated with the intravacuolar acidification inhibitor bafilomycin A1 (64) after bacterial entry. Control experiments showed that no cytotoxicity occurred in the fibroblasts at the concentration of the drug used (100 nM). Dissipation of intravacuolar acidification resulted in lower production of MgtC or PagC by nongrowing intracellular bacteria, confirming the important role played by acid pH in PhoP-PhoQ induction (Fig. 5A). This effect was especially evident in the case of PagC, which was produced by nongrowing intracellular bacteria at relatively higher levels than MgtC in the untreated fibroblasts (Fig. 5A). Previous studies in macrophages showed that S. Typhimurium viability relies on the maintenance of an intravacuolar pH (65). Counting of viable intracellular bacteria at 2 and 24 h postinfection of fibroblasts also revealed that inhibition of intravacuolar acidification results in loss of viability of nonproliferating intracellular bacteria (Fig. 5B). Dissipation of intravacuolar acidification also decreased the growth rate of the phoP mutant in the fibroblast (Fig. 5B). Interestingly, bafilomycin A1 did not affect survival of an SPI-2 mutant (sseC) (Fig. 5B) which is known to lose viability inside normal untreated fibroblasts (33). Taken together, these data indicate that PhoP-PhoQ induction, SPI-2 activation, and maintenance of survival by nongrowing intracellular S. Typhimurium are interconnected phenomena requiring vacuolar acidification.
Fig 5.
Activity of the PhoP-PhoQ system in nonproliferating intracellular S. Typhimurium responds to intravacuolar acidic pH. (A) Effect of the dissipation of intravacuolar acidification on the induction of the PhoP-PhoQ system. Shown are the relative levels of the 3× FLAG-tagged proteins MgtC and PagC produced by intracellular bacteria isolated from NRK-49F fibroblasts that were left untreated or were treated with 100 nM bafilomycin (BAF), an inhibitor of vacuolar acidification. OmpA (bacterial protein) and calnexin (eukaryotic protein) were used for loading controls. (B) Effect of loss of vacuolar acidification on the viability of intracellular bacteria. Shown are the ratios of viable intracellular bacteria enumerated at 24 h versus 2 h. Data are the means and standard deviations from three independent experiments. **, P = 0.001 to 0.01; ***, P < 0.001; n.s., not significant by a Student t test.
DISCUSSION
This study reports the first genome-wide expression analysis performed in intracellular S. Typhimurium while persisting in a nongrowing state within the infected host cell. The occurrence of negative regulation of Salmonella intracellular proliferation due to the action of pathogen functions was envisioned in our early studies on fibroblasts, which unraveled the requirement of the PhoP-PhoQ system to restrict bacterial growth (33, 34). Other authors reported mutants exhibiting increased loads of intracellular bacteria in macrophages (32). However, none of these studies investigated the physiology of nonproliferating intracellular bacteria and the basis of growth restraint. In addition, the tissue(s) and cell type(s) in which Salmonella may activate in vivo these intracellular responses were unknown. Microscopy analyses shown here unequivocally demonstrate that nonphagocytic cells positioned in the lamina propria of intestinal villi harbor bacteria that attenuate intracellular growth in a PhoP-PhoQ-dependent manner. Aside from phagocytic cells such as neutrophils, T and B lymphocytes, monocytes, and dendritic cells, fibroblasts are the only cells known to populate the lamina propria of intestinal villi. The notion that Salmonella is capable of infecting fibroblasts in this location is supported by the remarkable identity in the phenotypes exhibited by wild-type and phoP mutant bacteria in primary fibroblasts isolated from the lamina propria (Fig. 1). Recent studies of the streptomycin mouse model of S. Typhimurium diarrhea revealed that the pathogen targets epithelial cells and lamina propria phagocytes (66). Our assays were, however, performed in a typhoid infection model, for which only scarce information exists on the early events occurring in the intestine and the intestinal cell types colonized by S. Typhimurium. In the latter model, parallel routes involving traffic and dissemination of the pathogen through the lymphatic system after being ingested by dendritic cells or CD18+ phagocytes (monocytes or DCs) seem to occur (10). However, a detailed microscopy analysis at the level of host cell populations containing the pathogen is not available yet. Although the lack of highly specific fibroblast markers suitable for immunohistochemistry makes this type of study difficult, our findings provide the first in vivo evidence of an S. Typhimurium response directed to restrain growth within the infected cell.
The transcriptome profile obtained from cultured fibroblasts shared some features with genome-wide expression data reported for S. Typhimurium proliferating inside macrophages and epithelial cells (46, 53, 67). Examples included stress-related functions of the family of phage-shock proteins (Psp) that respond to impaired membrane function (68) and functions regulated positively by PhoP-PhoQ, such as those of MgtB, MgtC, PagC, Mig-3, and PhoN. An intriguing observation was the late expression (24 h postinfection) of SPI-1 genes by dormant nonproliferating intracellular bacteria (see Tables S2 and S3 in the supplemental material). This finding is in line with the upregulation of SPI-1 observed in intracellular bacteria located inside cultured epithelial cells (18, 53). However, SPI-1 upregulation was claimed to favor subsequent invasion events as bacteria are extruded from infected epithelial cells having a high bacterial load, a phenomenon that is not seen in fibroblasts. It is worth noting that SPI-1 genes were shown to be important for persistence of S. Typhimurium in vivo in a mouse chronic infection model (30), so it is possible that S. Typhimurium controls intracellular growth using yet-unknown mechanisms dependent on SPI-1. Future work should address this appealing hypothesis.
Genome expression data also shed light on new features that differentiate the unique lifestyle of nonproliferating S. Typhimurium persisting inside fibroblasts. Thus, unlike bacteria residing within macrophages, dormant intracellular S. Typhimurium does not upregulate expression of the gluconate transporter gene gntT, which was proposed to be important for nutrient acquisition (46). Moreover, nongrowing intracellular bacteria completely repress expression of flagellin inside fibroblasts, a phenomenon that contrasts with the synthesis of flagellin reported to occur in bacteria proliferating inside epithelial cells or macrophages (18, 53, 69). Since it is an energy-costly process, repressing the synthesis of flagella not needed for dormant bacteria living enclosed in a tightly apposed vacuole may facilitate metabolic reprogramming and long-lasting intracellular residence of the pathogen. Of interest, intracellular S. Typhimurium is known to inject flagellin into the macrophage cytosol, which alerts immune recognition systems based on cytosolic receptors (69). The persistence of nonflagellated S. Typhimurium inside fibroblasts may account for strategies directed to minimize host cell signaling and to remain hidden in the infected cell. Our study also demonstrates at the protein level the induction in intracellular bacteria of functions encoded by the pSLT virulence plasmid. TlpA, a coiled-coil plasmid protein regulated by PhoP-PhoQ that responds to temperatures found in the host (70, 71), was upregulated in our model by nongrowing intracellular bacteria. TlpA is dispensable for virulence (70), which supports the tempting idea of S. Typhimurium using some functions exclusively to persist asymptomatically in the host (25).
To our knowledge, this study also represents the first comparative transcriptomic analysis of wild-type and phoP mutant bacteria residing in the host cell and in two markedly distinct proliferative states. A relevant aspect of this comparison was the identification of genes regulating PhoP-PhoQ and responding to the nonproliferative intracellular state. This category includes novel PhoP-PhoQ-regulated genes such as ushA, encoding a putative UDP-sugar hydrolase/5′-nucleotidase, which could be induced exclusively by dormant intracellular bacteria. Thus, upregulation of ushA as seen in the fibroblast model was not observed in transcriptomic studies involving bacteria proliferating inside macrophages, epithelial cells, or resting extracellular bacteria in stationary phase (46, 53, and this study). Our transcriptomic study therefore provides a valuable source of data to identify and analyze in detail novel functions that may be used by S. Typhimurium to restrain growth inside host cells.
Transcriptomic analyses were completed with comparative studies to monitor the relative levels of individual PhoP-PhoQ-regulated proteins in intracellular and extracellular S. Typhimurium. To our knowledge, this is another experimental approach that has no precedents in the literature regarding the regulatory function of PhoP-PhoQ in intracellular S. Typhimurium. The data led us to tentatively shape the intracellular PhoP-PhoQ regulon and to dissect other aspects, including the regulation exerted by this system over the stationary-phase sigma factor RpoS, the postinfection time at which the system is activated in intracellular bacteria, and the phagosomal signals sensed by bacteria for such an activation. RpoS, known to play an important role in bacterial adaptation to stationary phase (72), is produced at higher levels by nongrowing intracellular bacteria than by the overgrowing phoP mutant. This observation is consistent with our early genetic screenings that identified RpoS as a factor involved in attenuating S. Typhimurium growth inside fibroblasts (33). The data also fit with a model in which dormant intracellular bacteria could use PhoP-PhoQ to positively regulate the levels of this alternative sigma factor. A regulatory pattern of this kind has been shown for S. Typhimurium and E. coli (57, 73, 74).
The data obtained at the protein level with reporter proteins such as MgtC and PagC allowed us to define the time at which PhoP-PhoQ is induced by dormant intracellular bacteria, an event estimated to occur at 1 to 2 h postinfection of the fibroblast. This timing is somehow delayed compared to the 30 min reported to be required for PhoP-PhoQ induction in S. Typhimurium residing within macrophages (55). A different kinetics of vacuolar acidification, which occurs more rapidly in macrophages than in nonphagocytic cells (6), or processes related to the peculiar growth status adopted by the bacteria inside the fibroblast may explain the extra time required for nongrowing bacteria to activate the PhoP-PhoQ system. Differences in the type of signals sensed by the system cannot be discarded. However, the data obtained with individual PhoP-PhoQ-regulated proteins support that, similarly to what has been shown in macrophages in different studies (35, 55), vacuolar acidification could be an essential signal stimulating the PhoP-PhoQ system in dormant intracellular S. Typhimurium. Several lines of evidence support this conclusion. First, in marked contrast to VirK and PagN, the PhoP-PhoQ-regulated protein PagP involved in lipid A modification (75, 76) was notably induced in a low-Mg2+ environment but not in dormant intracellular bacteria or in extracellular bacteria exposed to acid pH (Fig. 4). Second, dissipation of intravacuolar pH abrogated the induction of the PhoP-PhoQ system in nongrowing intracellular bacteria.
In addition to providing clues on the mode of induction of the system, the estimation of physiological levels of individual proteins unraveled new aspects of the biology of dormant intracellular S. Typhimurium. An example is the massive production of PagN, an intriguing finding considering that this outer membrane protein is used by extracellular S. Typhimurium to adhere and invade host cells (77, 78). PagN in intracellular bacteria therefore must accomplish an additional yet-unknown function. Our data also unveiled that VirK, a protein required for S. Typhimurium resistance to antimicrobial peptides and survival inside macrophages (79), is upregulated by nongrowing intracellular bacteria persisting inside fibroblasts. Taken together, these observations indicate that the establishment of a persistence state within the fibroblast relies on a delicate balance between the response of the infected cell to the intruder bacteria and the counterresponse of the invading bacteria. Most probably, the interconnection between the two responses results in the unique dormant intracellular lifestyle of S. Typhimurium that this study has started to decipher.
Supplementary Material
ACKNOWLEDGMENTS
We thank Javier López-Garrido, Gadea Rico, Javier Mariscotti, and Reini Hurme for sending antibodies and constructing strains that were used in some of the experiments. We also thank Juan Carlos Oliveros (CNB-CSIC) for helping us in the bioinformatics analyses and Diana Barroso and Pablo García for technical assistance.
This work was supported by grants BIO2010-18885 (to F.G-P.), CSD2008-00013-INTERMODS (to F.G.-P. and J.C.), and BIO2010-15023 (to J.C.) from the Spanish Ministry of Economy and Competitiveness and grant P10-CVI-5879 from the Junta of Andalucía (to J.C.). C.N.-H. and A.T. held fellowships from the Consejería de Educación de la Comunidad de Madrid.
Footnotes
Published ahead of print 22 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.01080-12.
REFERENCES
- 1. Haraga A, Ohlson MB, Miller SI. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53–66 [DOI] [PubMed] [Google Scholar]
- 2. Tsolis RM, Xavier MN, Santos RL, Baumler AJ. 2011. How to become a top model: impact of animal experimentation on human Salmonella disease research. Infect. Immun. 79:1806–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Valdez Y, Ferreira RB, Finlay BB. 2009. Molecular mechanisms of Salmonella virulence and host resistance. Curr. Top. Microbiol. Immunol. 337:93–127 [DOI] [PubMed] [Google Scholar]
- 4. Kaiser P, Diard M, Stecher B, Hardt WD. 2012. The streptomycin mouse model for Salmonella diarrhea: functional analysis of the microbiota, the pathogen's virulence factors, and the host's mucosal immune response. Immunol. Rev. 245:56–83 [DOI] [PubMed] [Google Scholar]
- 5. Valdez Y, Grassl GA, Guttman JA, Coburn B, Gros P, Vallance BA, Finlay BB. 2009. Nramp1 drives an accelerated inflammatory response during Salmonella-induced colitis in mice. Cell. Microbiol. 11:351–362 [DOI] [PubMed] [Google Scholar]
- 6. Steele-Mortimer O. 2008. The Salmonella-containing vacuole: moving with the times. Curr. Opin. Microbiol. 11:38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kato A, Groisman EA. 2008. The PhoQ/PhoP regulatory network of Salmonella enterica. Adv. Exp. Med. Biol. 631:7–21 [DOI] [PubMed] [Google Scholar]
- 8. Prost LR, Miller SI. 2008. The Salmonellae PhoQ sensor: mechanisms of detection of phagosome signals. Cell. Microbiol. 10:576–582 [DOI] [PubMed] [Google Scholar]
- 9. Mastroeni P, Grant AJ. 2011. Spread of Salmonella enterica in the body during systemic infection: unravelling host and pathogen determinants. Expert Rev. Mol. Med. 13:e12 doi:10.1017/S1462399411001840 [DOI] [PubMed] [Google Scholar]
- 10. Watson KG, Holden DW. 2010. Dynamics of growth and dissemination of Salmonella in vivo. Cell. Microbiol. 12:1389–1397 [DOI] [PubMed] [Google Scholar]
- 11. Monack DM, Bouley DM, Falkow S. 2004. Salmonella typhimurium persists within macrophages in the mesenteric lymph nodes of chronically infected Nramp1+/+ mice and can be reactivated by IFNgamma neutralization. J. Exp. Med. 199:231–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Richter-Dahlfors A, Buchan AM, Finlay BB. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186:569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Salcedo SP, Noursadeghi M, Cohen J, Holden DW. 2001. Intracellular replication of Salmonella typhimurium strains in specific subsets of splenic macrophages in vivo. Cell. Microbiol. 3:587–597 [DOI] [PubMed] [Google Scholar]
- 14. Sheppard M, Webb C, Heath F, Mallows V, Emilianus R, Maskell D, Mastroeni P. 2003. Dynamics of bacterial growth and distribution within the liver during Salmonella infection. Cell. Microbiol. 5:593–600 [DOI] [PubMed] [Google Scholar]
- 15. Monack DM, Mueller A, Falkow S. 2004. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat. Rev. Microbiol. 2:747–765 [DOI] [PubMed] [Google Scholar]
- 16. Gog JR, Murcia A, Osterman N, Restif O, McKinley TJ, Sheppard M, Achouri S, Wei B, Mastroeni P, Wood JL, Maskell DJ, Cicuta P, Bryant CE. 2012. Dynamics of Salmonella infection of macrophages at the single cell level. J. R. Soc. Interface. 9:2696–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Helaine S, Thompson JA, Watson KG, Liu M, Boyle C, Holden DW. 2010. Dynamics of intracellular bacterial replication at the single cell level. Proc. Natl. Acad. Sci. U. S. A. 107:3746–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Knodler LA, Vallance BA, Celli J, Winfree S, Hansen B, Montero M, Steele-Mortimer O. 2010. Dissemination of invasive Salmonella via bacterial-induced extrusion of mucosal epithelia. Proc. Natl. Acad. Sci. U. S. A. 107:17733–17738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. García-del Portillo F. 2008. Heterogeneity in tissue culture infection models: a source of novel host-pathogen interactions? Microbes Infect. 10:1063–1066 [DOI] [PubMed] [Google Scholar]
- 20. García-del Portillo F, Cossart P. 2012. A new view to intracellular pathogens and host responses in the South of Spain. EMBO Mol. Med. 4:160–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sturm A, Heinemann M, Arnoldini M, Benecke A, Ackermann M, Benz M, Dormann J, Hardt WD. 2011. The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1. PLoS Pathog. 7:e1002143 doi:10.1371/journal.ppat.1002143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kumar D, Rao KV. 2011. Regulation between survival, persistence, and elimination of intracellular mycobacteria: a nested equilibrium of delicate balances. Microbes Infect. 13:121–133 [DOI] [PubMed] [Google Scholar]
- 23. Gopinath S, Carden S, Monack D. 2012. Shedding light on Salmonella carriers. Trends Microbiol. 20:320–327 [DOI] [PubMed] [Google Scholar]
- 24. Monack DM. 2012. Salmonella persistence and transmission strategies. Curr. Opin. Microbiol. 15:100–107 [DOI] [PubMed] [Google Scholar]
- 25. Ruby T, McLaughlin L, Gopinath S, Monack D. 2012. Salmonella's long-term relationship with its host. FEMS Microbiol. Rev. 36:600–615 [DOI] [PubMed] [Google Scholar]
- 26. Kingsley RA, Humphries AD, Weening EH, De Zoete MR, Winter S, Papaconstantinopoulou A, Dougan G, Baumler AJ. 2003. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype typhimurium: identification of intestinal colonization and persistence determinants. Infect. Immun. 71:629–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McLaughlin LM, Govoni GR, Gerke C, Gopinath S, Peng K, Laidlaw G, Chien YH, Jeong HW, Li Z, Brown MD, Sacks DB, Monack D. 2009. The Salmonella SPI2 effector SseI mediates long-term systemic infection by modulating host cell migration. PLoS Pathog. 5:e1000671 doi:10.1371/journal.ppat.1000671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nix RN, Altschuler SE, Henson PM, Detweiler CS. 2007. Hemophagocytic macrophages harbor Salmonella enterica during persistent infection. PLoS Pathog. 3:e193 doi:10.1371/journal.ppat.0030193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Menendez A, Arena ET, Guttman JA, Thorson L, Vallance BA, Vogl W, Finlay BB. 2009. Salmonella infection of gallbladder epithelial cells drives local inflammation and injury in a model of acute typhoid fever. J. Infect. Dis. 200:1703–1713 [DOI] [PubMed] [Google Scholar]
- 30. Lawley TD, Chan K, Thompson LJ, Kim CC, Govoni GR, Monack DM. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11 doi:10.1371/journal.ppat.0020011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gal-Mor O, Gibson DL, Baluta D, Vallance BA, Finlay BB. 2008. A novel secretion pathway of Salmonella enterica acts as an antivirulence modulator during salmonellosis. PLoS Pathog. 4:e1000036 doi:10.1371/journal.ppat.1000036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Parsons DA, Heffron F. 2005. sciS, an icmF homolog in Salmonella enterica serovar Typhimurium, limits intracellular replication and decreases virulence. Infect. Immun. 73:4338–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cano DA, Martinez-Moya M, Pucciarelli MG, Groisman EA, Casadesus J, García-del Portillo F. 2001. Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect. Immun. 69:6463–6474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tierrez A, García-del Portillo F. 2005. New concepts in Salmonella virulence: the importance of reducing the intracellular growth rate in the host. Cell. Microbiol. 7:901–909 [DOI] [PubMed] [Google Scholar]
- 35. Alpuche Aranda CM, Swanson JA, Loomis WP, Miller SI. 1992. Salmonella typhimurium activates virulence gene transcription within acidified macrophage phagosomes. Proc. Natl. Acad. Sci. U. S. A. 89:10079–10083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoiseth SK, Stocker BA. 1981. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291:238–239 [DOI] [PubMed] [Google Scholar]
- 37. Ortega A, Gonzalo-Asensio J, García-del Portillo F. 2012. Dynamics of Salmonella small RNA expression in non-growing bacteria located inside eukaryotic cells. RNA Biol. 9:469–488 [DOI] [PubMed] [Google Scholar]
- 38. Paradela A, Mariscotti JF, Navajas R, Ramos-Fernandez A, Albar JP, García-del Portillo F. 2011. Inverse regulation in the metabolic genes pckA and metE revealed by proteomic analysis of the Salmonella RcsCDB regulon. J. Proteome Res. 10:3386–3398 [DOI] [PubMed] [Google Scholar]
- 39. Snavely MD, Miller CG, Maguire ME. 1991. The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J. Biol. Chem. 266:815–823 [PubMed] [Google Scholar]
- 40. Tierrez A, García-del Portillo F. 2004. The Salmonella membrane protein IgaA modulates the activity of the RcsC-YojN-RcsB and PhoP-PhoQ regulons. J. Bacteriol. 186:7481–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lober S, Jackel D, Kaiser N, Hensel M. 2006. Regulation of Salmonella pathogenicity island 2 genes by independent environmental signals. Int. J. Med. Microbiol. 296:435–447 [DOI] [PubMed] [Google Scholar]
- 42. Uzzau S, Figueroa-Bossi N, Rubino S, Bossi L. 2001. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. U. S. A. 98:15264–15269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Strong SA, Pizarro TT, Klein JS, Cominelli F, Fiocchi C. 1998. Proinflammatory cytokines differentially modulate their own expression in human intestinal mucosal mesenchymal cells. Gastroenterology 114:1244–1256 [DOI] [PubMed] [Google Scholar]
- 44. Aiastui A, Pucciarelli MG, García-del Portillo F. 2010. Salmonella enterica serovar Typhimurium invades fibroblasts by multiple routes differing from the entry into epithelial cells. Infect. Immun. 78:2700–2713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. García-del Portillo F, Pucciarelli MG, Casadesus J. 1999. DNA adenine methylase mutants of Salmonella typhimurium show defects in protein secretion, cell invasion, and M cell cytotoxicity. Proc. Natl. Acad. Sci. U. S. A. 96:11578–11583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Eriksson S, Lucchini S, Thompson A, Rhen M, Hinton JC. 2003. Unravelling the biology of macrophage infection by gene expression profiling of intracellular Salmonella enterica. Mol. Microbiol. 47:103–118 [DOI] [PubMed] [Google Scholar]
- 47. Mariscotti JF, García-del Portillo F. 2009. Genome expression analyses revealing the modulation of the Salmonella Rcs regulon by the attenuator IgaA. J. Bacteriol. 191:1855–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pucciarelli MG, García-del Portillo F. 2003. Protein-peptidoglycan interactions modulate the assembly of the needle complex in the Salmonella invasion-associated type III secretion system. Mol. Microbiol. 48:573–585 [DOI] [PubMed] [Google Scholar]
- 49. Adegboyega PA, Mifflin RC, DiMari JF, Saada JI, Powell DW. 2002. Immunohistochemical study of myofibroblasts in normal colonic mucosa, hyperplastic polyps, and adenomatous colorectal polyps. Arch. Pathol. Lab. Med. 126:829–836 [DOI] [PubMed] [Google Scholar]
- 50. Springer TA. 1990. Adhesion receptors of the immune system. Nature 346:425–434 [DOI] [PubMed] [Google Scholar]
- 51. Huntington ND, Tarlinton DM. 2004. CD45: direct and indirect government of immune regulation. Immunol. Lett. 94:167–174 [DOI] [PubMed] [Google Scholar]
- 52. Powell DW, Mifflin RC, Valentich JD, Crowe SE, Saada JI, West AB. 1999. Myofibroblasts. II. Intestinal subepithelial myofibroblasts. Am. J. Physiol. 277:C183–201 [DOI] [PubMed] [Google Scholar]
- 53. Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, Bongaerts RJ, Ahmad N, Rhen M, Hinton JC. 2008. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell. Microbiol. 10:958–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Innes D, Beacham IR, Beven CA, Douglas M, Laird MW, Joly JC, Burns DM. 2001. The cryptic ushA gene [ushA (c)] in natural isolates of Salmonella enterica (serotype Typhimurium) has been inactivated by a single missense mutation. Microbiology 147:1887–1896 [DOI] [PubMed] [Google Scholar]
- 55. Martin-Orozco N, Touret N, Zaharik ML, Park E, Kopelman R, Miller S, Finlay BB, Gros P, Grinstein S. 2006. Visualization of vacuolar acidification-induced transcription of genes of pathogens inside macrophages. Mol. Biol. Cell 17:498–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu. Rev. Microbiol. 65:189–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tu X, Latifi T, Bougdour A, Gottesman S, Groisman EA. 2006. The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc. Natl. Acad. Sci. U. S. A. 103:13503–13508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Garcia Vescovi E, Soncini FC, Groisman EA. 1996. Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84:165–174 [DOI] [PubMed] [Google Scholar]
- 59. Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092–5099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bader MW, Navarre WW, Shiau W, Nikaido H, Frye JG, McClelland M, Fang FC, Miller SI. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50:219–230 [DOI] [PubMed] [Google Scholar]
- 61. Bader MW, Sanowar S, Daley ME, Schneider AR, Cho U, Xu W, Klevit RE, Le Moual H, Miller SI. 2005. Recognition of antimicrobial peptides by a bacterial sensor kinase. Cell 122:461–472 [DOI] [PubMed] [Google Scholar]
- 62. Prost LR, Daley ME, Le Sage V, Bader MW, Le Moual H, Klevit RE, Miller SI. 2007. Activation of the bacterial sensor kinase PhoQ by acidic pH. Mol. Cell 26:165–174 [DOI] [PubMed] [Google Scholar]
- 63. Groisman EA, Mouslim C. 2006. Sensing by bacterial regulatory systems in host and non-host environments. Nat. Rev. Microbiol. 4:705–709 [DOI] [PubMed] [Google Scholar]
- 64. García-del Portillo F, Zwick MB, Leung KY, Finlay BB. 1993. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 90:10544–10548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rathman M, Sjaastad MD, Falkow S. 1996. Acidification of phagosomes containing Salmonella typhimurium in murine macrophages. Infect. Immun. 64:2765–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Muller AJ, Kaiser P, Dittmar KE, Weber TC, Haueter S, Endt K, Songhet P, Zellweger C, Kremer M, Fehling HJ, Hardt WD. 2012. Salmonella gut invasion involves TTSS-2-dependent epithelial traversal, basolateral exit, and uptake by epithelium-sampling lamina propria phagocytes. Cell Host Microbe 11:19–32 [DOI] [PubMed] [Google Scholar]
- 67. Hebrard M, Kroger C, Sivasankaran SK, Handler K, Hinton JC. 2011. The challenge of relating gene expression to the virulence of Salmonella enterica serovar Typhimurium. Curr. Opin. Biotechnol. 22:200–210 [DOI] [PubMed] [Google Scholar]
- 68. Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, Stumpf MP, Buck M. 2010. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34:797–827 [DOI] [PubMed] [Google Scholar]
- 69. Sun YH, Rolan HG, Tsolis RM. 2007. Injection of flagellin into the host cell cytosol by Salmonella enterica serotype Typhimurium. J. Biol. Chem. 282:33897–33901 [DOI] [PubMed] [Google Scholar]
- 70. Gal-Mor O, Valdez Y, Finlay BB. 2006. The temperature-sensing protein TlpA is repressed by PhoP and dispensable for virulence of Salmonella enterica serovar Typhimurium in mice. Microbes Infect. 8:2154–2162 [DOI] [PubMed] [Google Scholar]
- 71. Hurme R, Berndt KD, Normark SJ, Rhen M. 1997. A proteinaceous gene regulatory thermometer in Salmonella. Cell 90:55–64 [DOI] [PubMed] [Google Scholar]
- 72. Hengge-Aronis R. 2002. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol. Mol. Biol. Rev. 66:373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Eguchi Y, Ishii E, Hata K, Utsumi R. 2011. Regulation of acid resistance by connectors of two-component signal transduction systems in Escherichia coli. J. Bacteriol. 193:1222–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yu JL, Guo L. 2011. Quantitative proteomic analysis of Salmonella enterica serovar Typhimurium under PhoP/PhoQ activation conditions. J. Proteome Res. 10:2992–3002 [DOI] [PubMed] [Google Scholar]
- 75. Gunn JS, Belden WJ, Miller SI. 1998. Identification of PhoP-PhoQ activated genes within a duplicated region of the Salmonella typhimurium chromosome. Microb. Pathog. 25:77–90 [DOI] [PubMed] [Google Scholar]
- 76. Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250–253 [DOI] [PubMed] [Google Scholar]
- 77. Lambert MA, Smith SG. 2009. The PagN protein mediates invasion via interaction with proteoglycan. FEMS Microbiol. Lett. 297:209–216 [DOI] [PubMed] [Google Scholar]
- 78. Lambert MA, Smith SG. 2008. The PagN protein of Salmonella enterica serovar Typhimurium is an adhesin and invasin. BMC Microbiol. 8:142 doi:10.1186/1471-2180-8-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Brodsky IE, Ghori N, Falkow S, Monack D. 2005. Mig-14 is an inner membrane-associated protein that promotes Salmonella typhimurium resistance to CRAMP, survival within activated macrophages and persistent infection. Mol. Microbiol. 55:954–972 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.