Abstract
Ileal lesions of patients with Crohn's disease are colonized by adherent-invasive Escherichia coli (AIEC) bacteria that are able to adhere to and invade intestinal epithelial cells (IEC), to replicate within macrophages, and to form biofilm. Clinical observations showed that bacterial biofilms were associated with the mucosa of inflammatory bowel disease patients. In the present study, we analyzed the relationship between AIEC colonization of the gut and the formation of biofilm, focusing on the involvement of the σE pathway in the AIEC-IEC interaction. We observed that σE pathway inhibition in AIEC reference strain LF82 led to an impaired ability to adhere to and invade IEC but also induced a large decrease in the abilities to colonize the intestinal mucosa and form biofilm. This indicates that targeting of the σE pathway could be a very potent therapeutic strategy by which to interfere with the ability of AIEC to form biofilm on the gut mucosa of Crohn's disease patients.
INTRODUCTION
Crohn's disease (CD) is an inflammatory bowel disease (IBD) that occurs in individuals with a genetic predisposition in whom an environmental or infectious trigger causes an abnormal immune response (1–3). Several lines of evidence suggest that bacteria play a role in the onset and perpetuation of IBD (4), and clinical observations have shown that bacterial biofilms are associated with the mucosa of IBD patients, since the mean density of the mucosal biofilm was 2-fold higher in IBD patients than in patients with irritable bowel syndrome (IBS) or controls, and that the bacteria were mostly adherent (5). Escherichia coli has been assigned a putative role in CD. These bacteria are abnormally predominant in early and chronic ileal lesions of CD, and most E. coli strains isolated from the ileal mucosa of CD patients adhere to intestinal epithelial cells (IEC) (6–8). In addition to their ability to adhere to IEC, E. coli cells are able to invade IEC and belong to the pathogenic group of adherent-invasive E. coli (AIEC) (9). Many independent studies have reported the abnormal presence of AIEC bacteria associated with the ileal mucosa of CD patients (8, 10–14) owing to increased ileal expression of CEACAM6 (carcinoembryonic antigen-related cell adhesion molecule 6), which acts as a receptor for AIEC binding to the intestinal mucosa (15, 16). The adhesion-and-invasion process of AIEC reference strain LF82 involves, in addition to type 1 pili, flagella, outer membrane proteins (OMPs), and outer membrane vesicles (17–21). The LF82 invasion process occurs via the interaction between the endoplasmic-reticulum-localized stress response chaperone Gp96 and the OMP OmpA expressed at the surface of outer membrane vesicles (OMVs), allowing OMVs to fuse with IEC and deliver vesicle components and virulence factors to or into host cells (21).
The σE factor, also called RpoE, is activated by stresses that interfere with the folding of OMPs such as heat shock, overexpression of OMP-encoding genes, and mutations in genes that encode chaperones required for OMP folding (22–24). As expected from its role in the stress response, the σE regulon includes genes that encode periplasmic foldases, proteases, and chaperones that aid in OMP folding. In addition, σE transcribes an array of biosynthetic enzymes that are involved in phospholipid, fatty acid, lipopolysaccharide, and membrane-derived oligosaccharide synthesis and transport and a number of other cell envelope proteins, including lipoproteins, inner membrane proteins, and envelope proteins of unknown function (25–27). In AIEC strains, a model proposed by Rolhion and collaborators indicated that, at high osmolarity similar to that of the gastrointestinal tract, increased expression of OmpC in AIEC LF82 bacteria led to the activation of the σE regulatory pathway, a pathway that could regulate type 1 pilus and flagellum expression (19).
The aim of the present study was to investigate the activation of the σE regulatory pathway in AIEC bacteria and its involvement in AIEC virulence. We report here that the σE-mediated pathway is directly involved in the ability of AIEC strains to adhere to and invade IEC, and we demonstrate that, interestingly, the σE pathway is fully required for biofilm formation and colonization of the intestinal mucosa by AIEC strains. We also demonstrate that the σE-mediated pathway is activated during the adhesion and biofilm formation processes, shedding light on an original mechanism by which the σE-mediated pathway is induced by and required for the adhesion and biofilm formation of AIEC strains.
MATERIALS AND METHODS
Ethics statement.
The animal protocols used in this study were approved by the CEMEA Auvergne committee for ethical issues (permit CEMEAA CE16-0927-2956), and all animals were used in accordance with the European Community guidelines for the care and use of laboratory animals (86/609/CEE).
Reference bacterial strains, plasmids, and cell lines.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Bacteria were grown routinely in LB broth overnight at 37°C without shaking. Antibiotics were added to the medium at the following concentrations: ampicillin, 50 mg · ml−1; kanamycin, 50 mg · ml−1; chloramphenicol, 25 mg · ml−1.
Intestine-407 (I-407) cells (derived from human intestinal embryonic jejunum and ileum tissues) were purchased from Flow Laboratories, Inc., McLean, VA. Cultured cells were maintained in an atmosphere containing 5% CO2 at 37°C in modified Eagle medium (Seromed; Biochrom KG, Berlin, Germany) supplemented with 10% (vol/vol) fetal bovine serum (Lonza); 1% nonessential amino acids (Lonza); 1% l-glutamine (Lonza); 200 U of penicillin, 50 mg of streptomycin, and 0.25 mg of amphotericin B per liter; and 1% minimal essential medium (MEM) vitamin mix X-100 (Lonza).
Bacterial growth.
Each strain was tested for the ability to grow under static conditions in cell culture medium (MEM supplemented with 10% heat-inactivated fetal calf serum [FCS]) at 37°C for at least 10 h. Growth was monitored by measuring the optical density at 620 nm (OD620).
Promoter expression assay.
To generate promoter fusion constructs, the rpoE and rpoH promoters of strains LF82 were amplified by PCR using primers BamHI-rpoE/EcoRI-rpoE and BamHI-rpoH/EcoRI-rpoH, respectively (see Table S2 in the supplemental material). The resulting fragments contained the entire RpoE binding site previously described as present in these promoters (24, 28). These PCR fragments were ligated into plasmid vector pRS550 (29), and the results were designated pRS550-rpoE and pRS550-rpoH (see Table S1 in the supplemental material). β-Galactosidase activity was analyzed with a β-galactosidase assay kit (Qiagen), and strains harboring construct pRS550-rpoE or pRS550-rpoH were grown in LB culture medium containing arabinose (0, 0.02, 0.2, 1.0, or 2.0% l-arabinose; Sigma-Aldrich). The β-galactosidase activity of each sample was determined by measuring the OD420 at 24 h, and the number of bacteria in each sample was calculated by the OD620 measurement for Miller unit determination.
Adhesion and invasion assays.
The bacterial adhesion assay was performed as described previously (9). Briefly, I-407 cells were seeded into 24-well tissue culture plates at 4 × 105 per well. Monolayers were then infected at a multiplicity of infection of 10 bacteria per cell in 1 ml of cell culture medium without antibiotics and with heat-inactivated FCS (PAA Laboratories). When needed, adhesion and invasion assays were performed after centrifugation for 8 min at 1,000 × g. After 3 h of incubation at 37°C, monolayers were washed three times in phosphate-buffered saline (pH 7.2). Epithelial cells were then lysed with 1% Triton X-100 (Euromedex) in deionized water. Samples were diluted and plated onto Mueller-Hinton agar plates to determine the number of CFU corresponding to the total number of cell-associated bacteria (adherent and intracellular bacteria). To determine the number of intracellular bacteria, fresh cell culture medium containing 100 mg · ml−1 gentamicin was added for 1 h of incubation to kill extracellular bacteria. Monolayers were then lysed with 1% Triton X-100, and bacteria were quantified as described above. Invasion inhibition assays were performed after a 30-min pretreatment of cells at 37°C using anti-Gp96 antibody (H-212; Santa Cruz Biotech).
Construction of isogenic mutants and transcomplementation assays.
Isogenic mutants were generated with a PCR product by using the method described by Datsenko et al. (30) and modified by Chaveroche et al. (31). The primers used are listed in Table S2 in the supplemental material. For transcomplementation assays, a PCR product containing the entire 1,614-bp rseAB operon or the entire 575-bp rpoE gene were cloned into the pBAD24 and pBAD30 vectors, respectively (32) (see Tables S1 and S2 in the supplemental material).
RNA manipulations, reverse transcription (RT), and RT-PCR.
Cultures were grown at 37°C in LB with or without NaCl or in Eagle medium supplemented with 10% (vol/vol) of heat-inactivated fetal bovine serum at normal pH (7.0 to 7.5), at pH 6, or with 2% bile salts (sodium cholate; Sigma-Aldrich). At an OD620 of 0.2 and when needed, l-arabinose was added to induce the overexpression of RseAB. Total RNAs were extracted from bacteria cultured overnight and treated with DNase (Roche Diagnostics) to remove any contaminating genomic DNA.
For RNA isolation from bacteria associated with epithelial I-407 cells, at 4 h postinfection, I-407 epithelial cell monolayers were washed twice and lysed by exposure to 0.1% (wt/vol) sodium dodecyl sulfate, 0.1% (vol/vol) acidic phenol, and 19% (vol/vol) ethanol in water for 30 min on ice (33). Bacteria were not lysed by this procedure, and mRNA was stabilized and protected from degradation. After centrifugation (10 min, 6,000 × g), the bacterial pellet was subjected to RNA extraction.
For RNA extraction of biofilm-associated bacteria, strains were grown overnight in Luria-Bertani broth with 5 g · liter−1 glucose (Euromedex) at 35.5°C, after which 1/100 dilutions were made in M63 minimal medium (U.S. Biological) supplemented with 8 g · liter−1 (0.8%) glucose. Aliquots of 15 ml were then placed into wells of non-cell-treated polystyrene petri plates and incubated at 30°C without shaking. At different time points, the plates were washed once, bacteria were harvested using a scraper, and RNA was extracted as previously described.
The RNA was reverse transcribed and amplified using primers specific for rseA, rpoE, or the 16S rRNA gene (see Table S2 in the supplemental material). Amplification of a single expected PCR product was confirmed by electrophoresis on a 2% agarose gel. RT-PCR was performed using an Eppendorf Realplex, and RNA levels were quantified using RNA master SYBR green I (Roche Diagnostic) with 0.25 mg of total RNA.
Motility assay.
Bacterial strains were grown overnight at 37°C without agitation in LB broth, and 2-μl portions of the culture were inoculated into the centers of 0.3% LB agar plates. The plates were incubated at 37°C, and motility was assessed quantitatively by examining the circular swimming motion of the growing motile bacterial cells every other hour.
Biofilm formation assays.
Biofilm formation assays were performed as previously described (34). Strains were grown overnight in Luria-Bertani broth with 5 g · liter−1 glucose (Euromedex) at 35.5°C, after which 1/100 dilutions were made in M63 minimal medium (U.S. Biological) supplemented with 8 g · liter−1 (0.8%) glucose. When required, arabinose was added to the medium. Aliquots of 130 μl were then placed in wells of non-cell-treated polystyrene microtiter plates and incubated overnight at 30°C without shaking. Afterwards, growth OD630s were read. The wells were washed once, adherent bacteria were stained with 1% crystal violet solubilized in ethanol, and OD570s were read. Biofilm measurements were calculated using the formula SBF = (AB − CW)/G, where SBF is specific biofilm formation, AB is the OD570 of the attached and stained bacteria, CW is the OD570 of the stained control wells containing only bacterium-free medium (to eliminate unspecific or abiotic OD values), and G is the OD630 of cell growth in broth (35, 36). Assays were performed in triplicate.
Biofilm formation assays were also performed using paraformaldehyde (PFA)-fixed I-407 IEC monolayers. Briefly, confluent I-407 monolayers were fixed for 15 min in 4% PFA, and after washing, bacterial strains expressing green fluorescent protein (GFP) (37) in M63 minimal medium were applied as previously described and the mixture was incubated overnight at 30°C without shaking. For visualization, IEC monolayers were fixed for 15 min in 4% PFA and phalloidin-tetramethyl rhodamine isocyanate (TRITC) was used to visualize actin and Hoechst stain was used to visualize nuclei. The slides were examined with a Zeiss LSM 510 Meta confocal microscope.
Mouse ileal loop experiments.
Six-week-old FVB wild-type male mice were starved for 24 h before surgery, with water available ad libitum. They were anesthetized, and their intestines were exteriorized through a midline incision (38). Two or three intestinal segments (about 1 cm) without Peyer's patches were ligated and inoculated with mixed inoculums comprising equivalent numbers (5 × 107 CFU) of two bacterial strains (LF82 and LF82, LF82 and LF82/pBAD24, or LF82 and LF82/pBAD24-rseAB) in the presence of 2% arabinose, and the number of each bacterial strain associated with the mucosa of ligated loops was determined to establish the competitive index (CI), which provides a sensitive measurement of the relative degree of attenuation (39). The animal protocols used were approved by the CEMEA Auvergne committee for ethical issues (permit CEMEAA, CE16-0927-2956), and all animals were used in accordance with the European Community guidelines for the care and use of laboratory animals (86/609/CEE). Surgery was performed under ketamine-xylazine anesthesia, and all efforts were made to minimize suffering. Mice were killed by cervical dislocation according to animal care procedures.
Statistical analysis.
Numerical values are expressed as means with standard errors of the means. Statistical comparisons were performed by using the two-tailed Student t test, unless the variables required a two-tailed Fisher exact test. A P value of <0.05 was considered statistically significant.
RESULTS
σE pathway activation during the AIEC strain LF82 adhesion process.
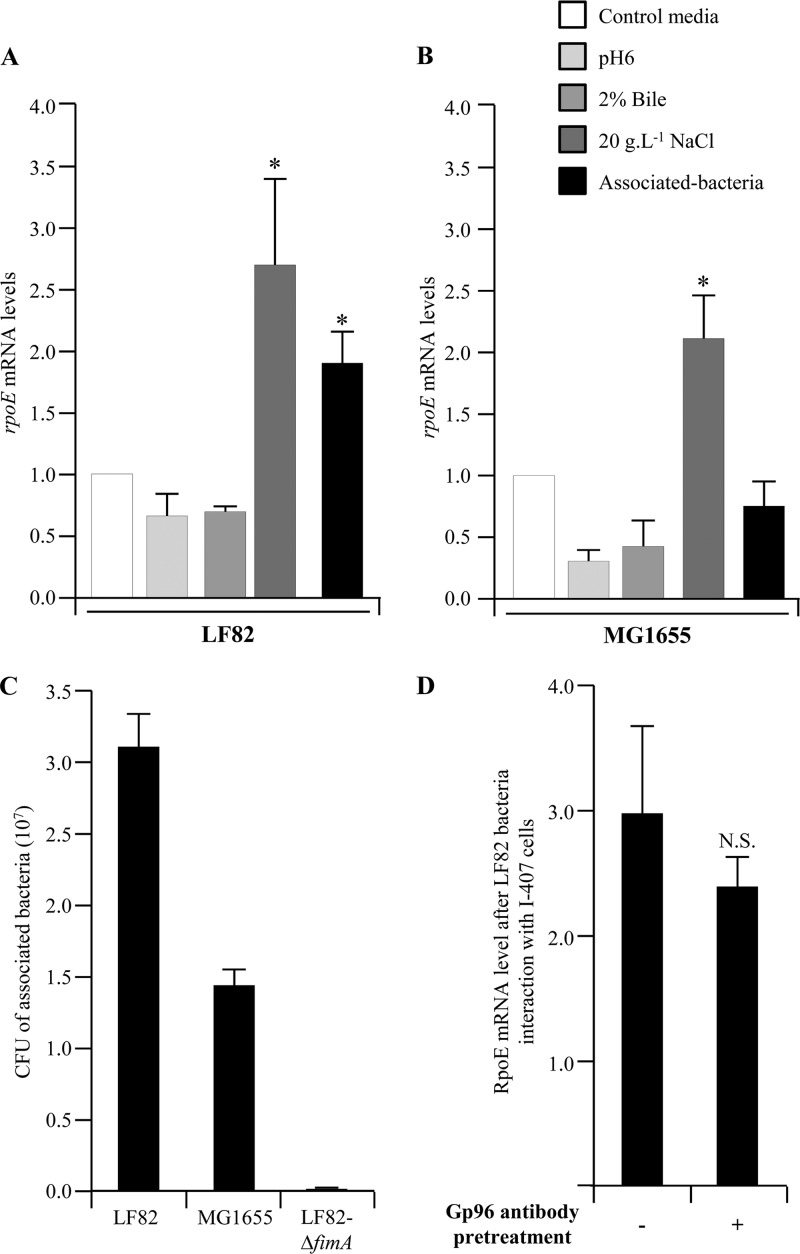
σE pathway activation was analyzed by quantification of rpoE mRNA, since it is well established that σE upregulates its own transcription (25). After the growth of AIEC strain LF82 or an E. coli K-12 strain in acid culture medium or in the presence of 2% bile salts, the rpoE mRNA levels were similar to those observed in bacteria grown in cell culture medium. In contrast, 2.7- and 2.3-fold increases in the rpoE mRNA levels were observed after the growth of LF82 and MG1655 bacteria at high osmolarity, respectively (Fig. 1A and B). Interestingly, when we analyzed σE pathway activation during the adhesion process, we also observed a 1.9-fold increase in the rpoE mRNA level in AIEC LF82 bacteria associated with IEC. No such increase was observed for MG1655 bacteria associated with IEC, although MG1655 is also able to adhere to I-407 cells, as shown in comparison with the nonadherent LF82-ΔfimA isogenic mutant (Fig. 1C). To address whether the σE pathway was activated during the adhesion or invasion step of AIEC LF82 bacteria, we pretreated IEC monolayers with anti-Gp96 antibodies, since it had previously been reported that Gp96 molecule expression at the IEC surface is required for the invasion process of AIEC strain LF82 and that such a pretreatment strongly decreased the invasion ability of strain LF82 (21). After Gp96 blockade, we still observed a high, 2.4-fold, increase in the rpoE mRNA level in AIEC LF82 bacteria associated with IEC (Fig. 1D). Altogether, these findings show for both the pathogenic LF82 and nonpathogenic MG1655 E. coli strains that high osmolarity activates the σE pathway but that the adhesion process activates the σE pathway in AIEC strain LF82 but not in E. coli K-12 strain MG1655.
Fig 1.
(A, B) Activation of the σE pathway in AIEC strain LF82 and nonpathogenic E. coli strain MG1655. Fold variation of rpoE mRNA levels in wild-type strains LF82 (A) and MG1655 (B) grown in medium at pH 6, in medium with 2% bile salts, or in medium with 20 g · liter−1 NaCl or adherent to I-407 epithelial cells, relative to that in wild-type strains grown in classic medium. 16S rRNA levels were measured as controls. Data are the mean ± the SEM of three separate experiments. *, P < 0.05. (C) Adhesion of AIEC strain LF82, nonpathogenic E. coli K-12 strain MG1655, and isogenic mutant LF82-ΔfimA to I-407 cells. Cell-associated bacteria were quantified after a 3-h infection period. Each value is the mean number of CFU ± the SEM of at least four separate experiments. (D) Activation of the σE pathway in AIEC strain LF82 associated with I-407 cells after anti-Gp96 antibody pretreatment. Fold variation of rpoE mRNA levels in I-407 epithelial-cell-adhering bacteria of wild-type strain LF82 with or without a 30-min pretreatment of cell monolayers with anti-Gp96 antibody. N.S., not statistically significant.
Involvement of the σE pathway in the ability of LF82 to interact with host cells.
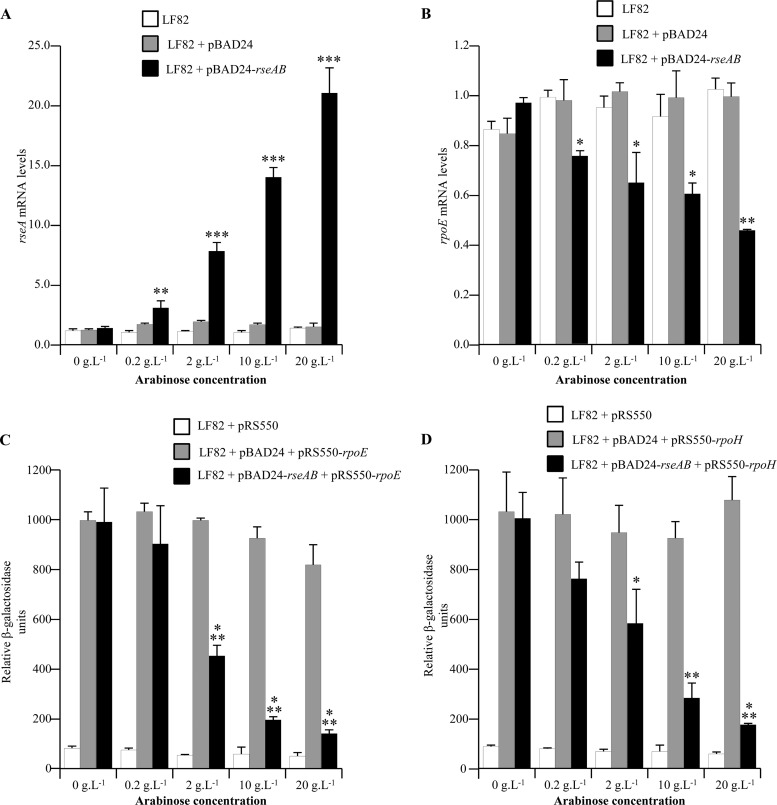
We next addressed the involvement of the σE pathway in the adhesion-and-invasion process of AIEC strain LF82. For this purpose, we decided to create a mutant of AIEC strain LF82 with a deletion of the σE-encoding gene. We obtained no mutant, which suggests that such a mutation is probably lethal in this strain. To counteract this methodology problem, we generated strains LF82 and MG1655 transformed with plasmid pBAD24-rseAB, which allows the expression of anti-sigma factors RseA and RseB, which prevents σE interaction with RNA polymerase (40, 41). As controls, we analyzed the growth curves of these constructs, and this showed that overexpression of RseAB led to no significant difference between the growth curves (see Fig. S1 in the supplemental material). AIEC LF82 bacteria with rseAB cloned into the arabinose-inducible pBAD24 vector were grown in the presence of various arabinose concentrations, and the levels of rseA mRNA were analyzed by quantitative RT-PCR. We observed arabinose dose-dependent expression of rseA (Fig. 2A). In parallel, we observed an arabinose dose-dependent decrease in rpoE mRNA levels (Fig. 2B). In addition, since an RpoE binding site was previously reported to be present in the rpoE and rpoH promoters (24, 28), we analyzed the activities of these promoters by generating constructs by using plasmid pRS550 to obtain β-galactosidase expression driven by the rpoE or rpoH promoter. In the presence of increased arabinose concentrations, we observed decreased activities of both the rpoE and rpoH promoters (Fig. 2C and D). When a 2% dose of arabinose was used, the overexpression of RseAB led to significant 7.81- and 5.38-fold decreases in the rpoE and rpoH promoter activities, respectively. These data confirm that overexpression of RseAB leads to a significant decrease in σE pathway activity, and an arabinose concentration of 2% was used in all experiments requiring overexpression of RseAB to allow σE pathway inhibition.
Fig 2.
(A, B) Fold variation of rseA (A) and rpoE (B) mRNAs levels in strains LF82, LF82/pBAD24, and LF82/pBAD24-rseAB in the presence of various doses of arabinose. Results are expressed as relative expression compared to that of wild-type strain LF82 in the absence of arabinose. 16S rRNA levels were measured as controls. Data are the mean ± the SEM of three separate experiments. (C, D) Activation of the rpoE (C) and rpoH (D) promoters in strains LF82, LF82/pBAD24, and LF82/pBAD24-rseAB in the presence of various doses of arabinose. Shown is the β-galactosidase activity per OD620 unit resulting from the expression of lacZ driven by the DNA sequence upstream of the rpoE or rpoH gene. Data are the mean ± the SEM of four separate experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
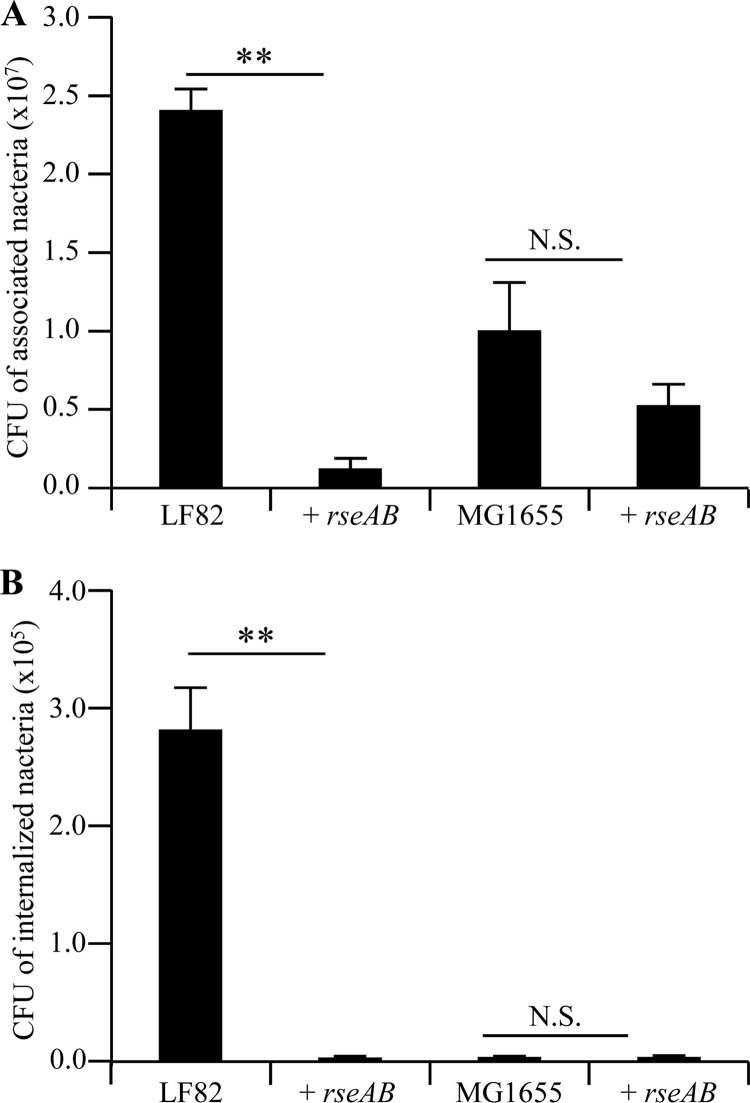
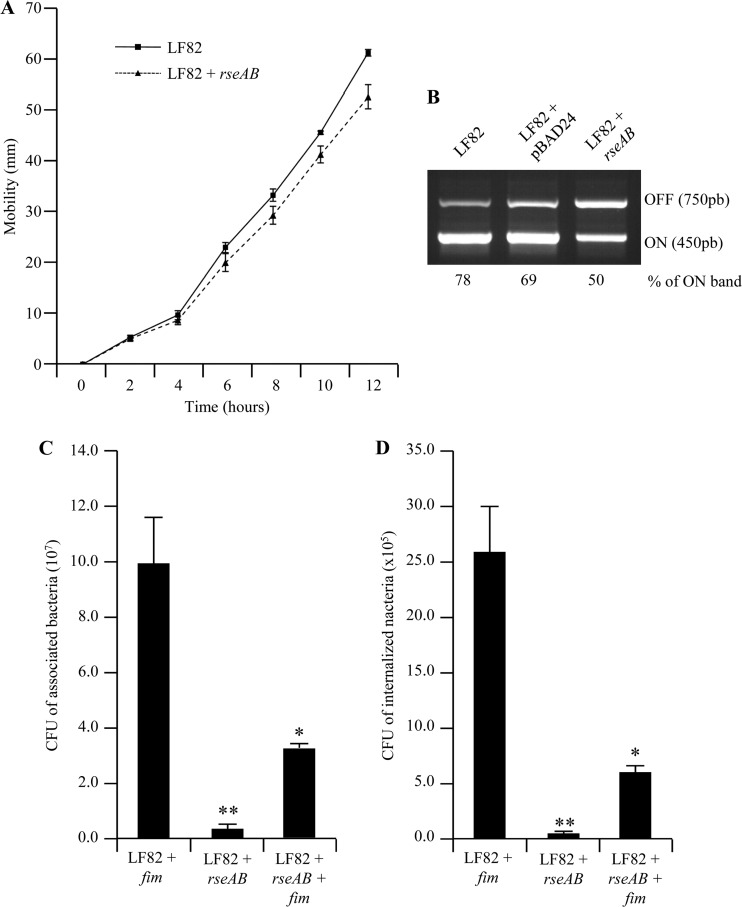
AIEC LF82 bacteria overexpressing the σE inhibitory complex RseAB had a significantly decreased ability to adhere to and invade IEC (Fig. 3A and B), with 20.51- and 66.99-fold decreases in adhesion and invasion, respectively, compared to those of wild-type strain LF82. Interestingly, the overexpression of the σE inhibitory complex RseAB had no effect on the ability of nonpathogenic E. coli strain MG1655 to interact with IEC. These findings demonstrate for the first time that the σE pathway is directly involved in the adhesion-and-invasion process of AIEC strain LF82. To further address the mechanism causing the decreased interaction of AIEC LF82 bacteria overexpressing the σE inhibitory complex RseAB with IECs, we analyzed the expression of flagella and type 1 pili. Flagellum expression was determined by analyzing the motility of bacteria on 0.3% LB agar, and we observed that motility was only slightly decreased in AIEC strain LF82 overexpressing the σE inhibitory complex RseAB, compared to that of the wild-type strain (Fig. 4A). Type 1 pilus expression was determined by analyzing type 1 pilus phase variation. This showed that 50% of the bacteria expressed the ON phase of the fim promoter when RseAB was overexpressed, compared to 78% and 69% in the wild-type strain or the wild-type strain complemented with the empty vector, respectively (Fig. 4B). Following these observations, we performed adhesion-and-invasion experiments with an LF82/pBAD24-rseAB strain transformed with the cloned fim operon in order to force the bacteria to express type 1 pili. A centrifugation step was performed to establish close contact between bacteria and epithelial cells in order to abrogate any defect in bacterial motility. Interestingly, even with constitutive type 1 pilus expression and with a centrifugation step, the adhesion-and-invasion levels of the LF82/pBAD24-rseAB construct were only partially restored compared to those of the LF82 wild-type strain (Fig. 4C and D). These results demonstrate that type 1 pili and flagella are involved in the decreased adhesion and invasion abilities observed during σE pathway inhibition.
Fig 3.
(A, B) Abilities of strains LF82, LF82/pBAD24-rseAB, MG1655, and MG1655/pBAD24-rseAB to adhere to (A) and invade (B) I-407 IEC. Each value is the mean ± the SEM of at least four separate experiments. **, P < 0.01; N.S., not statistically significant.
Fig 4.
(A) Motility assay of wild-type strain LF82 and strain LF82/pBAD24-rseAB on 0.3% agar at 37°C. (B) Regulation of type 1 pili in strain LF82/pBAD24-rseAB. PCR analysis was used to determine the invertible element orientation of the fim operon in strains LF82, LF82/pBAD24, and LF82/pBAD24-rseAB. A 450-bp product revealed the ON orientation of the invertible element, and a 750-bp product revealed its OFF orientation. (C, D) The abilities of strains LF82 + pHSG575-fim, LF82/pBAD24-rseAB, and LF82/pBAD24-rseAB/pHSG575-fim to adhere to (C) and invade (D) I-407 IEC. Centrifugation was performed to force contact between bacteria and I-407 IEC. Each value is the mean ± the SEM of at least four separate experiments. *, P < 0.05; **, P < 0.01.
Involvement of the σE pathway in the ability of LF82 to form biofilm.
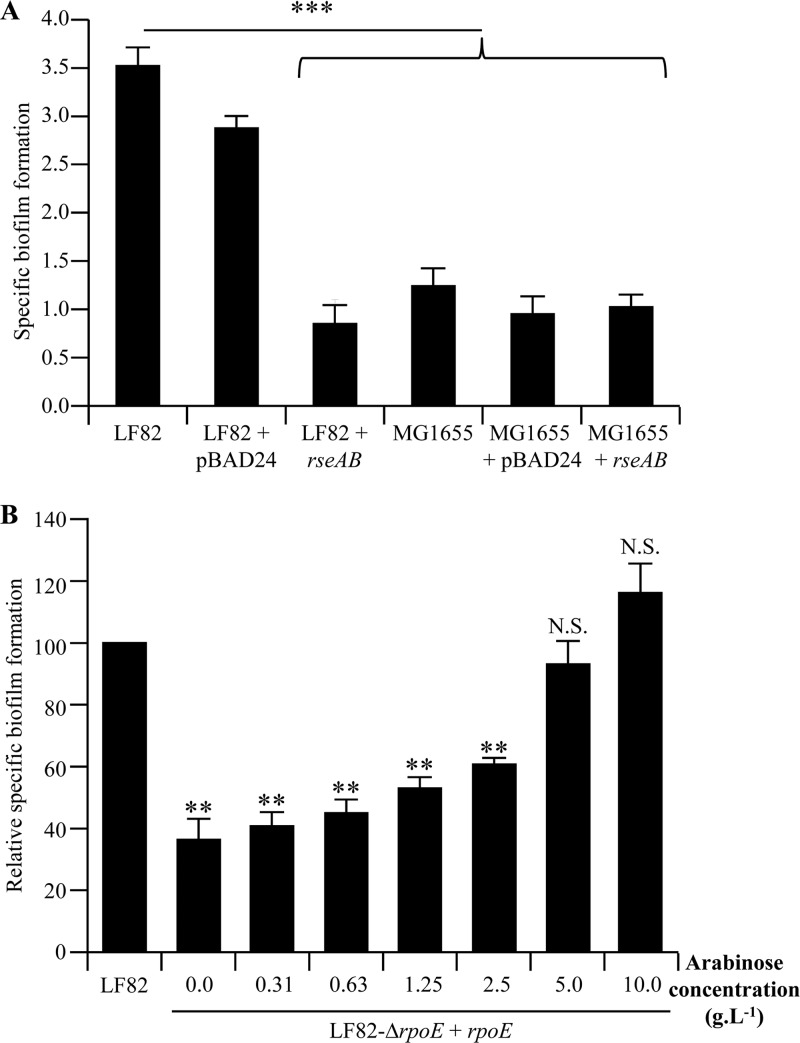
While we demonstrated that the σE-mediated pathway was involved in the interaction of AIEC strain LF82 with host cells (Fig. 3A and B), and since bacterial biofilms were associated with the mucosa of IBD patients (5) and biofilm formation capacity is a novel pathogenic feature of the AIEC pathovar (34), we analyzed whether the σE-mediated pathway is involved in AIEC biofilm formation. For this purpose, we compared the biofilm production of wild-type strain LF82 and that of strain LF82 overexpressing RseA and -B on a plastic surface. Inhibition of the σE pathway led to a significant decrease in the ability of strain LF82 to form biofilm, since the LF82/pBAD24-rseAB strain had a mean SBF index of 0.79 ± 0.14, compared to 3.23 ± 0.14 for wild-type strain LF82 (P = 0.00013) (Fig. 5A). Interestingly, inhibition of the σE pathway in E. coli K-12 strain MG1655 had no effect on its ability to form biofilm even if overexpression of the σE pathway inhibitory complex RseAB led to similar decreased expression of rpoE mRNA in both AIEC strain LF82 and E. coli K-12 strain MG1655. In addition, we observed that strain MG1655 had an SBF index (1.14 ± 0.11) similar to that of strain LF82 overexpressing RseAB (0.79 ± 0.14). To confirm these data and counteract the lethality due to σE deletion, we generated the σE deletion in strain LF82 already transformed with plasmid pBAD30-rpoE. With this construction, in the absence of arabinose, a significant decrease in biofilm formation was observed, with a residual level 35.60% ± 7.39% of that of wild-type strain LF82 (Fig. 5B). Interestingly, when the arabinose concentration increased, biofilm formation increased in a dose-response manner, and in the presence of 5 and 10 g · liter−1 arabinose, no difference between LF82-ΔrpoE/pBAD30-rpoE and the wild-type strain was observed. These data suggest that the σE pathway is involved in the ability of AIEC strain LF82 to form biofilm.
Fig 5.
(A) SBF indexes of AIEC strain LF82 and nonpathogenic E. coli strain MG1655 with or without RseAB overexpression. Data are the mean ± the SEM of three separate experiments. (B) SBF index of isogenic mutant LF82-ΔrpoE transcomplemented with pBAD30-rpoE and grown in the presence of 0.00, 0.31, 0.63, 1.25, 2.50, 5.00, or 10.00 g · liter−1 arabinose. Data are the mean ± the SEM of three separate experiments. **, P < 0.01; ***, P < 0.001; N.S., not statistically significant.
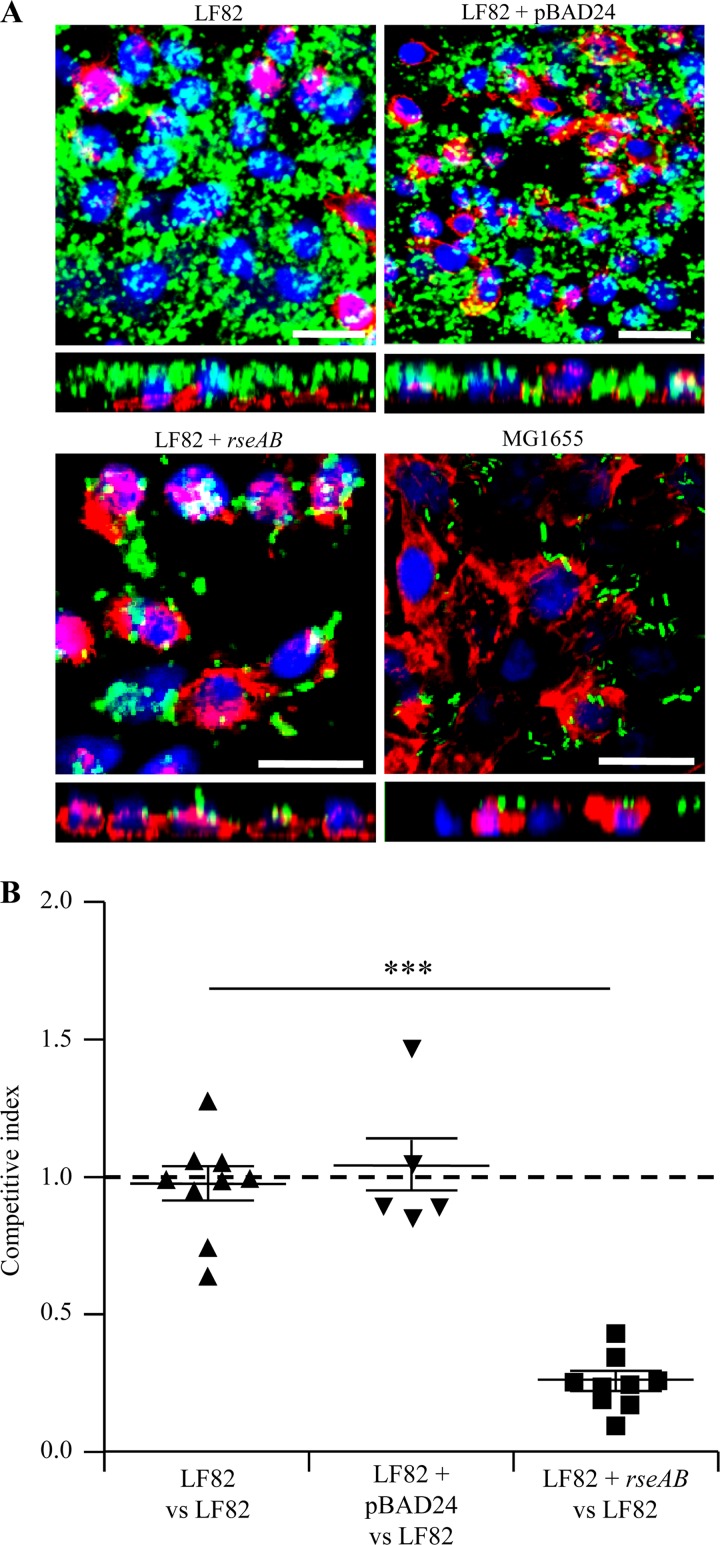
The abilities of strain LF82 and strain LF82 overexpressing RseAB to form biofilm were also analyzed by using an original method that we developed for this study with PFA-fixed IEC on glass coverslips. Results presented in Fig. 6A show that strain LF82 was able to form biofilm at the surface of fixed IEC. In contrast, with strain LF82/pBAD24-rseAB and E. coli K-12 strain MG1655, only a few diffusely adhering bacteria were observed at the IEC surface.
Fig 6.
(A) Confocal analysis of LF82, LF82/pBAD24, LF82/pBAD24-rseAB, and MG1655 biofilm formation at the surface of a PFA-fixed monolayer of I-407 IEC. Bacteria expressing GFP were used, actin is stained red with phalloidin-TRITC, and nuclei are stained blue with Hoechst. Representative z sections were visualized under each confocal slice. Bars, 50 μm. (B) CI of strain LF82/pBAD24-rseAB compared to that of wild-type strain LF82. Intestinal ileal loops were inoculated with mixed inoculums comprising equivalent numbers of the wild-type and LF82 pBAD24-rseAB strains, and their presence was compared by CI analysis, which provides a sensitive measurement of the relative degree of attenuation. **, P < 0.01; ***, P < 0.001.
Finally, the phenotype of strain LF82/pBAD24-rseAB was analyzed by another approach using an intestinal ileal loop assay as an in vivo model to assess the interaction of bacteria with intestinal mucosa. Intestinal ileal loops were inoculated with a mixed inoculum comprising equivalent numbers of wild-type LF82 and LF82/pBAD24-rseAB bacteria, and their presence was compared by CI analysis, which provided a sensitive measurement of the relative degree of attenuation (39). Strain LF82 overexpressing RseAB had a mean CI of 0.27 ± 0.033, indicating that its ability to interact with the surface of an intestinal ileal biopsy specimen was greatly impaired compared to that of the wild-type strain (P < 0.001) (Fig. 6B). The analyses of in vitro cocultures of LF82 wild-type and LF82/pBAD24-rseAB mutant bacteria revealed that both strains remained stable over time (see Fig. S2 in the supplemental material). Altogether, these data indicate that inhibition of the σE pathway led to a decreased ability of AIEC strain LF82 to form biofilm at the IEC surface.
The σE pathway was activated in AIEC strain LF82 during biofilm formation.
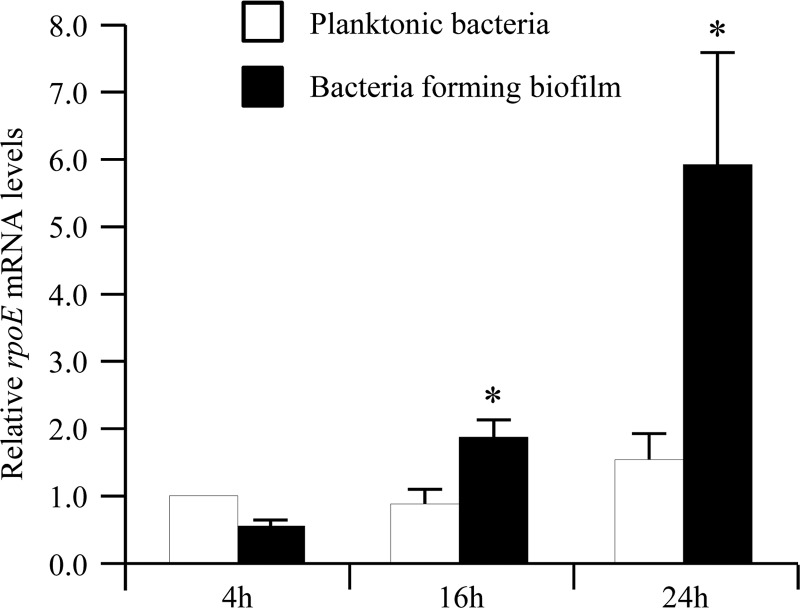
Since we demonstrate in the present study that the σE pathway is involved in the biofilm formation process, we analyzed whether the σE pathway was activated during biofilm formation by AIEC strain LF82 compared to planktonic bacteria. Results presented in Fig. 7 revealed that the σE pathway was highly activated during biofilm formation in AIEC strain LF82, with 1.71- and 5.79-fold increases, compared to planktonic bacteria at 4 h postinoculation, taken as 1. This activation of the σE pathway during the biofilm formation process was similar to the one previously observed during the adhesion process, as shown in Fig. 1.
Fig 7.
Activation of the σE pathway in AIEC strain LF82 during the biofilm formation process. Shown is the n-fold variation of rpoE mRNA levels in wild-type strain LF82 during biofilm formation (4, 16, and 24 h) relative to that of the wild-type strain grown for 4 h in classic medium. 16S rRNA levels were measured as controls. Data are the mean ± the SEM of three separate experiments. *, P < 0.05; **, P < 0.01.
DISCUSSION
Clinical observations showed that bacterial biofilms were associated with the mucosa of IBD patients, since the mean density of the mucosal biofilm was 2-fold higher in IBD patients than in patients with IBS or in controls, and that the bacteria were mostly adherent (5). Among the bacteria highly adherent to the ileal mucosa of CD patients, AIEC has been observed (10), in which we have identified type 1 pili and flagella as important virulence factors mediating the interaction of the bacteria with IEC (17, 18). In AIEC strain LF82, a model elaborated by Rolhion et al. proposed that, at a high osmolarity similar to that of the gastrointestinal tract, activation of the σE regulatory pathway could modulate the expression of genes involved in AIEC interactions with host cells (19). Such involvement of the σE pathway has been reported in various pathogens. For example, it is involved in the virulence of Salmonella enterica serovar Typhimurium, since after σE inactivation, salmonellae were no longer able to survive inside macrophages and had highly attenuated virulence in mice (42). A Vibrio cholerae ΔrpoE mutant had a highly attenuated ability to colonize the intestine and a highly attenuated lethal effect on mice (43).
The role of the σE pathway in the ability of E. coli to interact with IEC was observed for AIEC strain LF82 but not for nonpathogenic E. coli K-12 strain MG1655. In the present study, comparison of σE pathway activation in AIEC strain LF82 and in nonpathogenic E. coli K-12 strain MG1655 showed that when the bacteria interact with IEC, the σE mRNA level increased in AIEC strain LF82 but not in K-12 strain MG1655. Since it is well established that σE upregulates its own transcription (25), this indicates that AIEC adhesion to host cells leads to activation of the σE pathway. This is the first report indicating such an activation of the σE pathway in bacteria interacting with host cells and is in contrast with a previous report concerning Neisseria gonorrhoeae in which the analysis of global gene expression during the interaction of the bacteria with IEC showed that there was no activation of the σE pathway during the adhesion process (44).
Analysis of the relationship between the AIEC phenotype and activation of the σE-mediated pathway indicated that σE plays a crucial and direct role in AIEC strain LF82 since inhibition of this pathway greatly decreased the adhesion-and-invasion process. The analysis of the bacterial factors involved in this phenotype revealed that type 1 pili and flagella, previously reported to play a key role in the adhesion and invasion processes (17, 18), were partially involved in the decreased adhesion and invasion abilities observed during σE pathway inhibition. This indicates that another bacterial factor(s), regulated by the σE pathway, is (are) involved in these phenotypes. In addition, when we analyzed another phenotypic characteristic of AIEC strains, i.e., the ability to form bacterial biofilm, as described by Martinez-Medina et al. (34), we observed that inhibition of the σE pathway led to a greatly decreased ability of AIEC strains LF82 to form biofilm on plastic surfaces and on fixed IEC. In addition, we developed an in vivo intestinal ileal loop assay model to perform a CI analysis of wild-type strain LF82 and strain LF82 overexpressing RseAB. CI determination provides a sensitive measurement of the relative degree of attenuation of mutants compared to wild-type strains, and such an analyses were very helpful in the identification of virulence factors in Salmonella spp. (39) and Listeria monocytogenes (45). Results obtained with AIEC strain LF82 revealed that, after inhibition of the σE pathway, the presence of LF82 bacteria at the surface of the murine intestinal mucosa was greatly decreased. Such a phenotype was not observed with K-12 strain MG1655. These findings indicate that some genes whose transcription is under the control of σE should be involved in biofilm formation by AIEC strains and that such factors or their σE-dependent expression are absent in K-12 strain MG1655.
In conclusion, our findings demonstrate for the first time a specific activation of the σE-mediated pathway during the adhesion process of AIEC strain LF82 that results in the increased abilities of bacteria to adhere to and invade IEC and to form biofilm. These results also shed light on an original mechanism by which the σE-mediated pathway is induced in bacteria during biofilm formation and controls the expression of a gene(s) required for this process. Altogether, these data suggest that both the adhesion and biofilm formation processes (i) lead to activation of the σE pathway and (ii) are σE-dependent mechanisms. This circle could explain the high capacities of AIEC strains to colonize the intestinal mucosa and to form biofilm, as previously described in patients with CD (10, 34). Finally, our study indicates that targeting of the σE pathway could be a very potent therapeutic strategy by which to interfere with ability of AIEC to form biofilm on the gut mucosa of CD patients and to prevent subsequent chronic intestinal inflammation.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by the Ministère de la Recherche et de la Technologie, by Inserm (U1071), by INRA (USC-2018), and by grants from the Association F. Aupetit (AFA) and the European Commission through the FP7 IBDase project.
We thank the ICCF platform from the Université d'Auvergne for confocal microscopy and A. R. Arnold (Georgia State University, Atlanta, GA) for critically reading the manuscript.
B.C. and A.D.-M. designed the research, performed experiments, analyzed the data, and wrote the manuscript. A.D.-M. obtained funding.
We have no financial conflicts of interest.
Footnotes
Published ahead of print 26 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01079-12.
REFERENCES
- 1. Kaser A, Zeissig S, Blumberg RS. 2010. Inflammatory bowel disease. Annu. Rev. Immunol. 28:573–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Strober W, Fuss I, Mannon P. 2007. The fundamental basis of inflammatory bowel disease. J. Clin. Invest. 117:514–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Xavier RJ, Podolsky DK. 2007. Unravelling the pathogenesis of inflammatory bowel disease. Nature 448:427–434 [DOI] [PubMed] [Google Scholar]
- 4. Chassaing B, Darfeuille-Michaud A. 2011. The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology 140:1720–1728 [DOI] [PubMed] [Google Scholar]
- 5. Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. 2005. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43:3380–3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Di Martino P, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel JF. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn's disease. Gastroenterology 115:1405–1413 [DOI] [PubMed] [Google Scholar]
- 7. Swidsinski A, Ladhoff A, Pernthaler A, Swidsinski S, Loening-Baucke V, Ortner M, Weber J, Hoffmann U, Schreiber S, Dietel M, Lochs H. 2002. Mucosal flora in inflammatory bowel disease. Gastroenterology 122:44–54 [DOI] [PubMed] [Google Scholar]
- 8. Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. 2004. Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology 127:80–93 [DOI] [PubMed] [Google Scholar]
- 9. Boudeau J, Glasser AL, Masseret E, Joly B, Darfeuille-Michaud A. 1999. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect. Immun. 67:4499–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Darfeuille-Michaud A, Boudeau J, Bulois P, Neut C, Glasser AL, Barnich N, Bringer MA, Swidsinski A, Beaugerie L, Colombel JF. 2004. High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn's disease. Gastroenterology 127:412–421 [DOI] [PubMed] [Google Scholar]
- 11. Martinez-Medina M, Aldeguer X, Lopez-Siles M, Gonzalez-Huix F, Lopez-Oliu C, Dahbi G, Blanco JE, Blanco J, Garcia-Gil LJ, Darfeuille-Michaud A. 2009. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm. Bowel Dis. 15:872–882 [DOI] [PubMed] [Google Scholar]
- 12. Baumgart M, Dogan B, Rishniw M, Weitzman G, Bosworth B, Yantiss R, Orsi RH, Wiedmann M, McDonough P, Kim SG, Berg D, Schukken Y, Scherl E, Simpson KW. 2007. Culture independent analysis of ileal mucosa reveals a selective increase in invasive Escherichia coli of novel phylogeny relative to depletion of Clostridiales in Crohn's disease involving the ileum. ISME J. 1:403–418 [DOI] [PubMed] [Google Scholar]
- 13. Sasaki M, Sitaraman SV, Babbin BA, Gerner-Smidt P, Ribot EM, Garrett N, Alpern JA, Akyildiz A, Theiss AL, Nusrat A, Klapproth JM. 2007. Invasive Escherichia coli are a feature of Crohn's disease. Lab. Invest. 87:1042–1054 [DOI] [PubMed] [Google Scholar]
- 14. Eaves-Pyles T, Allen CA, Taormina J, Swidsinski A, Tutt CB, Eric Jezek G, Islas-Islas M, Torres AG. 2008. Escherichia coli isolated from a Crohn's disease patient adheres, invades, and induces inflammatory responses in polarized intestinal epithelial cells. Int. J. Med. Microbiol. 298:397–409 [DOI] [PubMed] [Google Scholar]
- 15. Barnich N, Carvalho FA, Glasser AL, Darcha C, Jantscheff P, Allez M, Peeters H, Bommelaer G, Desreumaux P, Colombel JF, Darfeuille-Michaud A. 2007. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J. Clin. Invest. 117:1566–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carvalho FA, Barnich N, Sivignon A, Darcha C, Chan CH, Stanners CP, Darfeuille-Michaud A. 2009. Crohn's disease adherent-invasive Escherichia coli colonize and induce strong gut inflammation in transgenic mice expressing human CEACAM. J. Exp. Med. 206:2179–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Barnich N, Boudeau J, Claret L, Darfeuille-Michaud A. 2003. Regulatory and functional co-operation of flagella and type 1 pili in adhesive and invasive abilities of AIEC strain LF82 isolated from a patient with Crohn's disease. Mol. Microbiol. 48:781–794 [DOI] [PubMed] [Google Scholar]
- 18. Boudeau J, Barnich N, Darfeuille-Michaud A. 2001. Type 1 pili-mediated adherence of Escherichia coli strain LF82 isolated from Crohn's disease is involved in bacterial invasion of intestinal epithelial cells. Mol. Microbiol. 39:1272–1284 [DOI] [PubMed] [Google Scholar]
- 19. Rolhion N, Carvalho FA, Darfeuille-Michaud A. 2007. OmpC and the sigma(E) regulatory pathway are involved in adhesion and invasion of the Crohn's disease-associated Escherichia coli strain LF82. Mol. Microbiol. 63:1684–1700 [DOI] [PubMed] [Google Scholar]
- 20. Rolhion N, Barnich N, Claret L, Darfeuille-Michaud A. 2005. Strong decrease in invasive ability and outer membrane vesicle release in Crohn's disease-associated adherent-invasive Escherichia coli strain LF82 with the yfgL gene deleted. J. Bacteriol. 187:2286–2296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rolhion N, Barnich N, Bringer MA, Glasser AL, Ranc J, Hebuterne X, Hofman P, Darfeuille-Michaud A. 2010. Abnormally expressed ER stress response chaperone Gp96 in CD favours adherent-invasive Escherichia coli invasion. Gut 59:1355–1362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mecsas J, Welch R, Erickson JW, Gross CA. 1995. Identification and characterization of an outer membrane protein, OmpX, in Escherichia coli that is homologous to a family of outer membrane proteins including Ail of Yersinia enterocolitica. J. Bacteriol. 177:799–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Missiakas D, Betton JM, Raina S. 1996. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 21:871–884 [DOI] [PubMed] [Google Scholar]
- 24. Rouvière PE, De Las PEñas A, Mecsas J, Lu CZ, Rudd KE, Gross CA. 1995. rpoE, the gene encoding the second heat-shock sigma factor, sigma E, in Escherichia coli. EMBO J. 14:1032–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. 2006. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 4:e2 doi:10.1371/journal.pbio.0040002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dartigalongue C, Missiakas D, Raina S. 2001. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 276:20866–20875 [DOI] [PubMed] [Google Scholar]
- 27. Rezuchova B, Miticka H, Homerova D, Roberts M, Kormanec J. 2003. New members of the Escherichia coli sigmaE regulon identified by a two-plasmid system. FEMS Microbiol. Lett. 225:1–7 [DOI] [PubMed] [Google Scholar]
- 28. Erickson JW, Vaughn V, Walter WA, Neidhardt FC, Gross CA. 1987. Regulation of the promoters and transcripts of rpoH, the Escherichia coli heat shock regulatory gene. Genes Dev. 1:419–432 [DOI] [PubMed] [Google Scholar]
- 29. Simons RW, Houman F, Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85–96 [DOI] [PubMed] [Google Scholar]
- 30. Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chaveroche MK, Ghigo JM, d'Enfert C. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97 doi:10.1093/nar/28.22.e97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lucchini S, Liu H, Jin Q, Hinton JC, Yu J. 2005. Transcriptional adaptation of Shigella flexneri during infection of macrophages and epithelial cells: insights into the strategies of a cytosolic bacterial pathogen. Infect. Immun. 73:88–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Martinez-Medina M, Naves P, Blanco J, Aldeguer X, Blanco JE, Blanco M, Ponte C, Soriano F, Darfeuille-Michaud A, Garcia-Gil LJ. 2009. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC). BMC Microbiol. 9:202 doi:10.1186/1471-2180-9-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Naves P, del Prado G, Huelves L, Gracia M, Ruiz V, Blanco J, Rodriguez-Cerrato V, Ponte MC, Soriano F. 2008. Measurement of biofilm formation by clinical isolates of Escherichia coli is method-dependent. J. Appl. Microbiol. 105:585–590 [DOI] [PubMed] [Google Scholar]
- 36. Niu C, Gilbert ES. 2004. Colorimetric method for identifying plant essential oil components that affect biofilm formation and structure. Appl. Environ. Microbiol. 70:6951–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Valdivia RH, Hromockyj AE, Monack D, Ramakrishnan L, Falkow S. 1996. Applications for green fluorescent protein (GFP) in the study of host-pathogen interactions. Gene 173:47–52 [DOI] [PubMed] [Google Scholar]
- 38. Hitotsubashi S, Fujii Y, Yamanaka H, Okamoto K. 1992. Some properties of purified Escherichia coli heat-stable enterotoxin II. Infect. Immun. 60:4468–4474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beuzón CR, Holden DW. 2001. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 3:1345–1352 [DOI] [PubMed] [Google Scholar]
- 40. De Las Peñas A, Connolly L, Gross CA. 1997. The sigmaE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of sigmaE. Mol. Microbiol. 24:373–385 [DOI] [PubMed] [Google Scholar]
- 41. Missiakas D, Mayer MP, Lemaire M, Georgopoulos C, Raina S. 1997. Modulation of the Escherichia coli sigmaE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24:355–371 [DOI] [PubMed] [Google Scholar]
- 42. Humphreys S, Stevenson A, Bacon A, Weinhardt AB, Roberts M. 1999. The alternative sigma factor, sigmaE, is critically important for the virulence of Salmonella Typhimurium. Infect. Immun. 67:1560–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kovacikova G, Skorupski K. 2002. The alternative sigma factor sigma(E) plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect. Immun. 70:5355–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Du Y, Lenz J, Arvidson CG. 2005. Global gene expression and the role of sigma factors in Neisseria gonorrhoeae in interactions with epithelial cells. Infect. Immun. 73:4834–4845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Auerbuch V, Lenz LL, Portnoy DA. 2001. Development of a CI assay to evaluate the virulence of Listeria monocytogenes actA mutants during primary and secondary infection of mice. Infect. Immun. 69:5953–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.