Abstract
The anaerobe Bacteroides fragilis is a highly aerotolerant, opportunistic pathogen that is an important component of the human intestinal microbiota. Aerotolerance has been linked to a robust oxidative stress response, which in turn is necessary for maximal virulence in a mouse intra-abdominal abscess model. During oxidative stress, there is a dynamic change in gene expression that encompasses a third of the genome, but there is a paucity of information on factors that control this response. A large number of transcription regulators, including about 14 extracytoplasmic function (ECF) sigma factors, are affected by oxidative stress, and one of these, EcfO, was used as a model of ECF sigma factor activity during stress. Genetic and biochemical experiments showed that EcfO was located in an operon with a structurally unique anti-sigma factor, Reo. Cells deleted for EcfO were impaired during exposure to oxygen or other forms of oxidative stress, whereas reo mutants were more resistant to stress. Protein-protein interaction experiments demonstrated that Reo directly interacts with and regulates the activity of EcfO. Expression microarray and chromatin affinity precipitation assays were used to identify target genes regulated by EcfO, and an EcfO recognition sequence was identified. The results revealed that EcfO controls a regulon of novel lipoproteins whose distribution in nature is restricted to members of the Bacteroidetes phylum.
INTRODUCTION
The obligate anaerobe Bacteroides fragilis is an indigenous inhabitant of the gastrointestinal tract of humans, where it contributes to normal host function, but it is also a significant opportunistic pathogen (1–3). B. fragilis is the anaerobe most frequently isolated from clinical specimens, and it is usually associated with peritonitis, intra-abdominal abscess, and bacteremia. Infection generally occurs when organisms are translocated from the gut as a result of some underlying disease, surgery, or trauma (4, 5). Thus, during the course of infection, B. fragilis must adapt from the anaerobic, highly reducing colon to a new, more oxidizing environment. The significance of this adaptation was shown previously when we demonstrated that resistance to oxidative stress is required for full virulence in a mouse abscess model (6).
B. fragilis is highly aerotolerant, being able to survive about 3 days in the presence of atmospheric levels of oxygen. A number of studies have shown that aerotolerance is linked to a comprehensive oxidative stress response (OSR) that is similar in scope to that seen in many facultative or aerobic organisms. During the initial stages of the OSR, a LysR family regulator, OxyR, controls the induction of genes for detoxification enzymes such as catalase, Dps, thioredoxin (Trx) peroxidase, and alkyl hydroperoxide reductase (7, 8). If the stress is not corrected, there is a truly global transcriptional response that modulates the expression of more than 1,000 genes coding for a wide range of cellular functions, including central metabolism, carbohydrate utilization, biosynthesis, and energy generation (6, 9, 10). This response to more prolonged stress may be important during the transition from the anaerobic colonic environment, but little is known about how it is regulated.
Among the B. fragilis genes affected by aerobic exposure were a large number of transcriptional regulators, including 14 extracytoplasmic function (ECF) sigma factors (see the supplemental material in reference6). ECF sigma factors are known to transmit changes in environmental signals to specific responses in gene expression. Upon induction, they bind to core RNA polymerase and alter promoter selectivity, thus allowing differential gene expression in response to changes in the environment. There are a number of examples in which ECF sigma factors modulate responses to oxidative stress. In Mycobacterium tuberculosis, both σE and σH play a role in protection against oxidative stress induced by a range of compounds such as H2O2, cumene hydroperoxide, plumbagin, and diamide (11–13). In the alphaproteobacterium Caulobacter crescentus, σT plays a role in both H2O2 stress and osmotic stress, and σF controls the stationary-phase expression of a regulon including msrA and sodA, which help protect against oxidative stress (14, 15). In both of these examples, the ECF sigma factors appear to contribute to multiple-stress resistance, indicating broad overlapping functions in the regulons of many of these stress responses.
Several mechanisms by which the activities of ECF sigma factors are regulated have been described. In most cases, a general mode of control involves specific anti-sigma factors, which function as negative regulators by sequestering the sigma factors, preventing both binding to DNA and associations with core RNA polymerase under noninducing conditions (16). Upon the perception of an inducing signal, anti-sigma factor regulation is mitigated by diverse mechanisms, which include but are not limited to intramembrane proteolysis (17), sigma factor mimicry/partner switching by an anti-anti-sigma factor (18), redox-mediated conformational switching (19), and the export of the anti-sigma factor (20). Most ECF sigma factor genes are located in the same operon as their cognate anti-sigma factor.
In this report, one of the aerobically induced sigma factors, EcfO (Ecf sigma factor in oxidative stress), and its anti-sigma factor, Reo (regulator of EcfO), were used as a model to study the role of these regulators in the oxidative stress response of B. fragilis. We show that the deletion of EcfO affects sensitivity to several oxidative stressors and the ability to survive aerobic exposure. We also show with genetic and biochemical experiments that Reo interacts with and regulates the activity of EcfO. Data from expression microarray and real-time PCR experiments using ecfO and reo mutants defined a set of novel target genes which are unique to Bacteroides and are regulated by EcfO. Chromatin affinity precipitation (ChAP) assays of EcfO in combination with real-time PCR (ChAP-RT) were used to distinguish direct promoter targets of EcfO from its secondary downstream targets. We hope to gain a better understanding of how EcfO regulates its targets under different growth conditions and the role of this regulon in resistance to oxidative stress.
MATERIALS AND METHODS
Strains, plasmids, and growth media.
Relevant characteristics of the bacterial strains and plasmids used in this study are described in Table 1. Bacteroides strains were routinely grown in an anaerobic chamber at 37°C in TYG (tryptone, yeast extract, and glucose) medium (27). For some experiments, maltose replaced glucose as the carbon source (TYM medium). Rifampin (20 μg ml−1), gentamicin (50 μg ml−1), tetracycline (5 μg ml−1), and erythromycin (5 μg ml−1) were added to the medium when required. Escherichia coli strains were routinely grown in LB medium containing the following antibiotics when needed: ampicillin (100 μg ml−1), carbenicillin (50 μg ml−1), spectinomycin (50 μg ml−1), and kanamycin (35 μg ml−1).
Table 1.
Plasmids and bacterial strains used in this study
| Strain or plasmid | Descriptiona | Reference(s) or source |
|---|---|---|
| Strains | ||
| 638Rb | B. fragilis clinical isolate; Rfr Gmr | 21 |
| ABD77 | B. fragilis TM4000 (638R) thyA; Rfr Gmr Tpr | 22 |
| IB524 | ABD77 ecfO thyA+; Rfr Gmr | This study |
| IB525 | 638R reo::tetQ; Tcr Rfr Gmr | This study |
| IB526 | IB524 ecfO reo::tetQ; Tcr Rfr | This study |
| Plasmids | ||
| pFD516 | Bacteroides suicide vector, 7.7 kb; (Spr) Emr | 23 |
| pFD340 | Bacteroides constitutive expression vector, 8.8 kb; (Apr) Emr | 24 |
| pFD1146 | Bacteroides IPTG-inducible expression vector, 9.4 kb; (Spr) Emr | 25 |
| pHT1146 | His-tagged vector derived from pFD1146, 9,906 kb; (Spr) Emr | This studyc |
| pFD1045 | Bacteroides maltose-inducible expression vector containing the osuA promoter, 7.4 kb; (Apr) Emr | 26, this studyc |
| pHT1045 | His-tagged vector, derived from pFD1045, 7.9 kb; (Apr) Emr | This studyc |
| pGEX4T-1 | E. coli GST expression vector expressing glutathione S-transferase, 4.9 kb; (Apr) | GE Healthcare |
| pET32a(+) | E. coli His-tagged expression vector containing a thioredoxin tag, 5.9 kb; (Apr) | Novagen |
Antibiotic resistance markers in parentheses are expressed in E. coli strains, and antibiotic resistance markers without parentheses are expressed in Bacteroides strains. Rfr, rifampin resistance; Emr, erythromycin resistance; Tcr, tetracycline resistance; Gmr, gentamicin resistance; Spr, spectinomycin resistance; Apr, ampicillin resistance.
NCBI accession number FQ312004.
See the supplemental material for additional details.
Construction of mutants.
The construction of the ecfO deletion mutant was carried out by allelic exchange using the positive-selection vector pYT102, as described previously (22). Briefly, PCR (see Table S1 in the supplemental material for primers) was used to amplify regions upstream and downstream of the EcfO gene to create a 237-bp in-frame deletion lacking codons 45 to 123, and this was cloned into pYT102. The plasmid was mobilized into B. fragilis ADB77 (isogenic with 638R), and following confirmation of the double-crossover allelic exchange, the mutant strain was reverted back to thymine prototrophy (22). In experiments described below, the DNA sequence that encompassed the ecfO gene fragment with the in-frame deletion was cloned as a negative control, and it is designated the ΔecfO mutant when associated with a plasmid construct.
The reo mutant was created by using a similar strategy, except that the deleted portion of the gene was replaced by a tetracycline resistance cassette (tetQ), and the suicide vector used was pFD516 (23). In this case, double-crossover mutations were identified by screening for tetracycline-resistant, erythromycin-sensitive colonies. Evidence for the mutations was shown by the analysis of PCR products using genomic DNA and cDNA templates from these strains.
Three additional reo mutations were constructed for protein-protein interaction studies: Reo-ΔTM was a deletion of the first 78 amino acids; Reo-N1-TM contained only the first 48 amino acids of Reo; and Reo-N1+TM contained the first 78 amino acids of Reo, including the trans-membrane domain. These proteins were cloned into pGEX4T-1 and pET32a for in-frame expression with N-terminal tags, glutathione S-transferase (GST), and Trx-6×His, respectively.
Oxidative stress sensitivity assays.
For disk diffusion assays overnight, anaerobic cultures (0.1 ml) were spread onto TYG plates without cysteine. A sterile 6-mm filter disk was placed onto the center of each plate, and 10 μl of the stressing agent was added to the disk (28). Following overnight incubation, the diameters of the zones of growth inhibition were measured for triplicate experiments.
Another assay was used to measure recovery after aerobic stress. Cultures grown overnight were removed from the anaerobic chamber and serially diluted in sterile phosphate-buffered saline (PBS), and 5 μl of each dilution was spotted onto multiple TYG plates lacking cysteine. The plates were incubated under aerobic conditions at 37°C and then moved to anaerobic conditions after different exposure times to allow for the recovery of cells.
Protein expression and purification.
The overexpression of proteins in E. coli was conducted by using pGEX4T-1 for the isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible expression of N-terminally GST-tagged proteins or pET32a for the IPTG-inducible expression of N-terminal thioredoxin-His-tagged proteins. Proteins were routinely expressed in BL21(DE3) cells (Promega) by the induction of mid-log-phase cells with 1 mM IPTG for 3 h. Cells were extracted in extraction buffer (5 M NaCl, 50 mM NaH2PO4 [pH 8.0]) containing 1× protease inhibitor cocktail (catalog number P8465; Sigma), 0.5% Triton X-100, and 1 mM phenylmethylsulfonyl fluoride (PMSF). Cells were lysed by a French press, and after centrifugation at 27,000 × g for 20 min at 4°C, the soluble supernatant was used for affinity purification.
Two systems were used for the inducible expression of His-tagged proteins in B. fragilis: plasmid pHT1045, for maltose-inducible expression, or plasmid pHT1146, for IPTG-inducible expression (Table 1; see also Fig. S1 and S2 and procedures in the supplemental material for detailed descriptions of plasmids used for the expression of His-tagged proteins in B. fragilis).
In vitro protein-protein interaction assays.
In vitro protein-protein interaction experiments, or pulldown assays, were performed between GST-bound proteins and His-tagged proteins. All GST-tagged and His-tagged proteins were readily purified from the soluble fraction of the cell lysate and were not found in inclusion bodies. For purification, the GST-tagged proteins were affinity bound to glutathione-Sepharose 4B resin, and the Trx-His-tagged proteins were eluted in imidazole elution buffer after binding on Ni-nitrilotriacetic acid (NTA) resin (Qiagen, Valencia, CA). All of the affinity-tagged proteins were stable upon isolation, as shown by SDS-PAGE analysis. Following the quantification of protein, between 1:1 and 4:1 molar equivalents of GST-tagged protein (bound to glutathione-Sepharose resin) and the His-tagged partner protein, respectively, were incubated in the presence of 10% glycerol and protein extraction buffer at 4°C overnight with gentle shaking. Controls were included to exclude nonspecific interactions with the affinity tags. The unbound fraction was removed, and the resin was washed in extraction buffer containing 0.1% Triton X. The proteins were eluted at 95°C for 5 min with 300 μl elution buffer and 300 μl of 2× SDS loading buffer. Bound and unbound proteins were analyzed by Coomassie staining and Western blotting with mouse monoclonal antibody to the His tag (ABM, Inc., Richmond, Canada).
Subcellular fractionation.
Cytoplasmic and membrane fractions were prepared, as described previously (29), from log-phase cultures grown in 300 ml of TYG medium. Cells were lysed by a French press, and cellular debris was removed by low-speed centrifugation. The membrane fraction in the supernatant was separated from the cytoplasmic fraction by ultracentrifugation in a 70.1 fixed-angle Ti rotor at 55,000 × g for 2 h at 4°C. His-tagged proteins present in the fractions were concentrated on Ni-NTA columns and eluted in comparable volumes. His-tagged proteins in the membrane pellets were suspended in denaturing 6 M guanidine HCl-phosphate buffer containing 0.5 M NaCl, purified under denaturing buffered conditions on Ni-NTA columns, and eluted under native conditions after washing with native phosphate wash buffers containing 20 mM imidazole.
Chromatin affinity purification.
Chromatin immunoprecipitation and affinity purification assays using affinity-tagged proteins were described previously and were optimized for B. fragilis, as follows (30–32). A maltose-inducible plasmid expressing the His-tagged EcfO protein was mobilized into either the ecfO or ecfO reo mutant. For use as a negative control, a plasmid containing the ecfO gene fragment with the 237-bp deletion was mobilized into the ecfO reo mutant. These strains were grown in TYG medium containing 0.25% glucose and 5 μg ml−1 erythromycin. To induce protein expression, a culture grown overnight was diluted 1:100 into 500 ml of prewarmed TYM medium and incubated for about 8 h until the log phase. Chloramphenicol was added to a final concentration of 100 μg ml−1, and an aliquot of 25 ml was collected for RNA preparation. In the remaining culture, protein-DNA complexes were cross-linked by the addition of formaldehyde to a final concentration of 1% and incubated on ice with periodic shaking for 30 min. The reaction was quenched with glycine at a final concentration of 125 mM at room temperature for 5 min. The cells were collected at 8,000 rpm for 5 min and washed twice in PBS buffer (1.36 mM NaCl, 27 μM KCl, 17.6 μM KH2PO4, 100 μM Na2HPO4 [pH 7.4]).
Cells were lysed in 20 ml of native extraction buffer (5 M NaCl, 50 mM NaH2PO4 [pH 8.0]) containing 1× protease inhibitor cocktail (Sigma-Aldrich, Inc., St. Louis, MO), 1 mM PMSF, 1 mg ml−1 lysozyme, and RNase A (EMD-Millipore [Novagen], Billerica, MA). After incubation at room temperature for 20 min, cells were sonicated in a 50-ml tube on ice using the large tip of a Branson 450 sonifier (output 5, 90% duty cycle) 7 times with 30 1-s pulses, with 2 min between each time. Triton was added to 1%, and three more rounds of sonication were performed. The DNA fragment size range was verified by analysis on a 1% agarose gel, and samples containing fragments of between 200 bp and 2,000 bp were used for affinity purification.
The sheared lysate was centrifuged at 27,000 × g at 4°C for 20 min to remove unbroken cells and cell debris. The supernatant was filtered and incubated with 500 μl of preequilibrated Ni-NTA beads overnight at 4°C with gentle shaking. A sample of unbound lysate was collected and treated in exactly the same way as for the downstream eluted protein. The beads were washed three times with extraction buffer, once in extraction buffer containing 1% Triton, and three times in extraction buffer containing 10 mM imidazole. Bound protein was eluted twice with 400 μl of phosphate buffer at pH 8.0 containing 500 mM imidazole, 1% SDS, and 10 mM dithiothreitol (DTT). Fifty microliters of 5 M NaCl and 1 μl of RNase A were added, and the samples were un-cross-linked at 65°C overnight. The samples were centrifuged at 27,000 × g at 4°C, sodium acetate was added to the supernatant to a final concentration of 0.3 M, and the DNA was ethanol precipitated. The DNA pellet was suspended in 50 mM Tris buffer containing 25 mM EDTA at pH 7.5 and treated with 5 μl of 20 mg ml−1 proteinase K at 55°C for 1 h. After 5 μl of 3 M sodium acetate was added, the Qiagen PCR cleanup kit was used to obtain a clean DNA preparation. A 1:10-diluted aliquot of the preparation was used for real-time quantitative PCR (qPCR) to verify the enrichment of control genes.
RNA isolation and qRT-PCR analysis.
RNA isolation was carried by out using the hot-phenol method, as described previously (33). The samples were treated three times with Turbo DNA-free DNase and passed on a silica column (Invitrogen, Carlsbad, CA). Twenty micrograms of RNA was used for first-strand cDNA synthesis in a reaction mixture with 13 ng μl−1 random hexamers, 0.5 mM deoxynucleoside triphosphates (dNTPs), 1× first-strand buffer, and 1 μl Superscript II RNase H-reverse transcriptase I (Invitrogen, Carlsbad, CA).
A cDNA preparation containing 2 ng/μl was used for the quantitative reverse transcriptase PCR (qRT-PCR) reactions. The qPCRs for each primer set were performed on serial dilutions of a template plasmid as a PCR standard. The copy number for each standard was estimated according to Avogadro calculations, based on the length of the DNA fragment and its concentration in the sample: [copy number per/g] = [6.022E+23 (number/mole)]/[fragment length (bp) × 650 (g/mole)].
The reaction mixture contained 12.5 μl of 2× iQ SYBR green Supermix, 1.5 μl of 5 mM forward primer, 1.5 μl of 5 mM reverse primer, 4.5 μl H2O, and 5 μl of cDNA template (diluted to about 2 ng μl−1 RNA) per well. All samples were run in triplicate, and DNase-cleaned RNA (diluted to the same starting quantity) was run as a control for genomic DNA contamination. In cases where a standard was absent, the relative expression level was determined by the Pfaffl method (34), using 16S RNA as a reference.
Expression microarrays.
Second-strand cDNA for expression microarray analysis was prepared as follows: 30 μl of second-strand reaction buffer, 3 μl of 10 mM dNTP, 1 μl of 10 U μl−1 E. coli DNA ligase, 4 μl of 10 U μl−1 E. coli DNA polymerase, 1 μl of 2 U μl−1 E. coli RNase H, and water were added to the product of the first-strand cDNA synthesis reaction mixture described above to a final volume of 150 μl. After 2 h of incubation at 16°C, 2 μl (10 units) of T4 DNA polymerase was added, and the mixture was further incubated at 16°C for 5 min. The reaction was stopped by the addition of 10 μl of 0.5 M EDTA to the mixture. RNA digestion was carried out by the addition of 1 μl of 10 mg ml−1 RNase A. Double-stranded cDNA was cleaned up by extraction with phenol-chloroform-isoamyl alcohol (25:24:1) and ethanol precipitation in the presence of 0.75 M ammonium acetate. One microgram of purified double-stranded cDNA was used for labeling with cy3 and expression array hybridization. A 365,000-molecular-weight (385K) high-density-oligonucleotide whole-genome expression array (Roche/NimbleGen Systems, Inc., Madison, WI) with 5 technical replicates of each probe per slide was designed for B. fragilis 638R using the genome sequence and open reading frame (ORF) predictions. Labeling, hybridization, and processing of the arrays were performed at the Florida State University Roche/NimbleGen Microarray Facility.
In order to elucidate the EcfO regulon, three independent cultures of the ecfO mutant plus the empty vector and four independent cultures of the reo mutant containing pFD1146-ecfO were started from single colonies and grown overnight in the absence of IPTG. These cultures were then diluted in TYG medium containing 1 mM IPTG and grown to the mid-logarithmic phase for RNA isolation and microarray analysis, as described above. The raw microarray expression data were normalized together by using the robust multiarray average (RMA) algorithm, as implemented in Roche DEVA 1.1 software. The normalized data were analyzed by using ArrayStar software (Dnastar, Inc., Madison, WI). During the course of these experiments, several of the cultures exhibited random phase variation of the capsule polysaccharide loci for polysaccharide A (PSA), PSF, and PSG, as described previously (35–37). In order to better visualize the EcfO regulon, the expression values for these capsule operons were removed from the ArrayStar expression analysis. However, the complete set of microarray expression data have been deposited in the NCBI Gene Expression Omnibus, and the top 100 induced genes also are shown in Table S2 in the supplemental material. Preliminary microarray expression data with different strains or plasmid expression systems (see Table S3 in the supplemental material) did not show obvious capsule phase variation.
5′ RACE mapping.
The 5′ ends of mRNA were mapped by 5′ rapid amplification of cDNA ends (RACE), using commercial reagents (Invitrogen, Carlsbad, CA) and primers described in Table S1 in the supplemental material. Primers 1335-GSP1.1 and sigOD-GSP2.2were used to generate the first-strand cDNA. Primers sigOD-RT2 and sigOD-GSP2.2 were used for nested PCR in combination with the RACE abridged anchor primer to amplify RACE products for EcfO. Primers 1335-GSP2 and 1335-GSP2.2 were used in combination with the RACE abridged anchor primer to amplify RACE products for BF638R_1335. The PCR products were cloned into pGEM-T, and at least 4 colonies from each were sequenced to determine the transcription initiation sites.
Microarray data accession numbers.
Microarray expression data described in the text and in the supplemental material have been deposited in the NCBI Gene Expression Omnibus database under accession numbers GSE39231 and GSE35536.
RESULTS
ECF sigma factors in Bacteroides fragilis.
The genome sequence of B. fragilis revealed 43 predicted ECF sigma factors; 1 primary sigma factor, σ70; and 1 RpoN-like sigma factor, σ54 (38). A primary sequence alignment and phylogenetic analyses of the ECF sigma factors showed significant structural diversity (see Fig. S3 in the supplemental material). These proteins were spread over 6 of the >40 major conserved ECF families described previously by Staron et al. (39), and we also noted that 6 of these sigma factors either were previously ungrouped or were assigned to one of the minor ECF families. More than half (25/43) of the B. fragilis ECF sigma factors belonged to the ECF10 group, which has been associated with the uptake and metabolism of complex carbohydrates (39). ECF10 family members in Bacteroides thetaiotaomicron have been shown to participate in trans-envelope signaling cascades, which modulate both polysaccharide metabolism and capsule biosynthesis (40).
In a previous microarray study, the expressions of 14 of the ECF sigma factors genes or their associated anti-sigma factors were modulated during exposure to oxygen (see the supplemental material in reference 6). The affected genes are representative of most of the B. fragilis ECF sigma factor families probably reflecting diversity in sensory perception and target response genes (see Fig. S3 in the supplemental material). Five of these genes coded for ECF sigma factors that were induced 3- to 8-fold by oxygen, and one of these ECF sigma factors, EcfO, is associated with the ECF21 family, which has not been previously investigated for any other organism (39). The ecfO gene was induced 3.4-fold after exposure to air, and interestingly, it was located in close proximity to oxyR, which encodes a key regulator of the OSR. These observations suggested that EcfO might be important for the B. fragilis oxidative stress response.
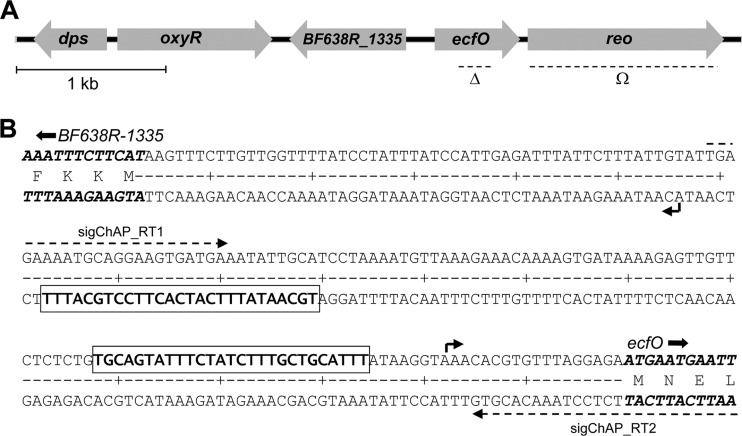
The ecfO gene is cotranscribed with BF638R_1337, located downstream and encoding a putative anti-sigma factor, Reo (Fig. 1; see also Fig. S4 in the supplemental material). The BF638R_1335 gene is divergently transcribed from the ecfO promoter region, and it is adjacent to but transcribed in the opposite direction of oxyR (Fig. 1). BF638R_1335 is a conserved protein in Bacteroides but is of unknown function. The C terminus of the pflA gene is located 237 bp downstream of reo and is transcribed in the opposite orientation.
Fig 1.
Genetic organization and nucleotide sequence of the ecfO locus. (A) The dashed lines below the arrows show the regions either deleted (Δ) or deleted and replaced with the tetQ gene (Ω). The unmarked deletion of ecfO lacks amino acids 45 to 123. The reo mutation was generated by replacing nearly the entire gene (after amino acid 15) with a tetracycline cassette. (B) The transcription initiation sites, shown by the bent arrows, for BF638R_1335 and for the ecfO-reo operon were mapped by using a standard 5′ RACE protocol. The conserved EcfO binding sites (Fig. 7) are boxed and in boldface type. Primer binding sites in the region used for either RACE mapping or ChAP-RT analysis are shown as dashed arrows, and the N-terminal portions of the coding sequences for the genes are indicated.
The EcfO mutant has a defective response to oxidative stress.
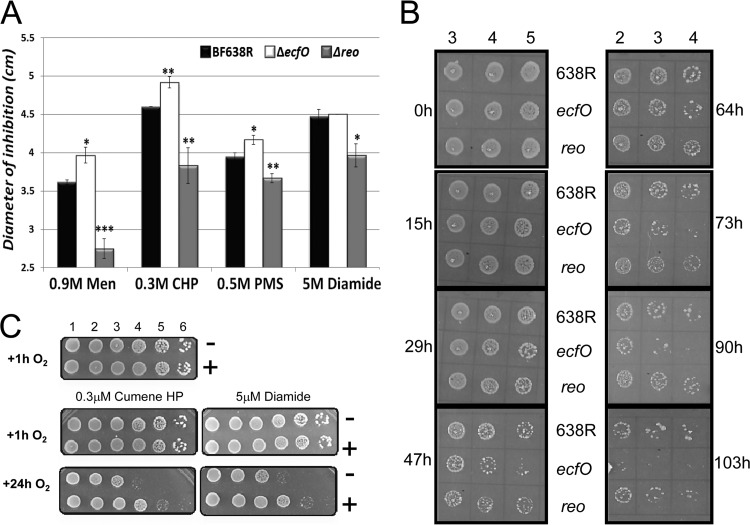
It was suspected that EcfO has a role in the OSR since its transcription level increased during exposure to air (6). To test this, the viability of the ecfO mutant was determined in disk diffusion assays with the oxidative stress agents cumene hydroperoxide, diamide, phenazine methosulfate, and menadione. As shown in Fig. 2A, the absence of EcfO reduced the ability of B. fragilis to defend itself against oxidative stress, as demonstrated by the significantly larger inhibition zones in the disk diffusion assays for three of the stressing agents tested. In contrast, a reo mutant was more resistant to stress than the wild type, with significantly smaller inhibition zones for each of the oxidative stressors, especially menadione, which had an extremely reduced zone of inhibition. Based on these results, we hypothesized that Reo is an anti-sigma factor which inhibits EcfO activity.
Fig 2.
Phenotypic analysis of ecfO mutants reveals sensitivity to oxidative stress. (A) The growth inhibition zones of the wild type (BF638R) and the ecfO and reo mutants were compared in disk diffusion assays with different oxidative stress agents. Compared to the wild type, the two-tailed t test statistics are as follows: P values of <0.01 (*), P values of <0.005 (**), and P values of <0.0001 (***). Men, menadione; CHP, cumene hydroperoxide; PMS, phenazine methosulfate (PMS). (B) Aerobic exposure stress tests comparing wild-type strain BF638R to the ecfO and reo mutants incubated in air for different times. The log dilution is shown at the top, and the incubation times are on the side. (C) Preinduction of plasmid-encoded, His-tagged EcfO in the ecfO background mutant complemented the ecfO mutant phenotype. ecfO(pHT1045-ecfO) cells were grown in glucose-repressing (−) or maltose-inducing (+) medium for 6 h to induce the expression of His-tagged EcfO, and serial dilutions (shown at the top) were then spotted onto TYM agar plates containing cumene hydroperoxide or diamide. The plates were exposed to air at 37°C for 1 or 24 h, and cell survival was then determined after anaerobic incubation to allow for the rescue of cells.
To test aerotolerance, wild-type (638R) and ecfO and reo mutant cultures were grown to the mid-log phase in an anaerobic chamber, serially diluted, and spotted onto TYG plates. After exposure to air at 37°C for different time points, the plates were transferred into the anaerobic chamber for outgrowth and recovery. The results showed that the survival of the different mutants in air reflected the results that we observed with the disk diffusion assays; that is, the EcfO-deficient mutant was more sensitive to extended aerobic exposure than the wild-type cells (Fig. 2B). The Reo mutant had a survival profile similar to that of the parent and was somewhat more resistant to this stress.
Complementation of the ecfO mutation restores resistance to oxidative stress.
An experiment was designed to determine if the loss of EcfO could be complemented by introducing the gene under the control of an inducible promoter (Fig. 2C). For this work, ecfO containing a dual N-terminal/C-terminal His tag was cloned into the maltose-inducible vector pHT1045 and transferred into the ecfO mutant. The ecfO(pHT1045-ecfO) cells were grown overnight in glucose medium and then subcultured in medium containing either maltose (inducing) or glucose. At the mid-log phase, cells were serially diluted and spotted onto maltose-containing stress plates. To ameliorate the oxidative stress conditions, the stress plates contained 0.3 μM cumene or 5 μM diamide and were exposed to air prior to anaerobic recovery. We hypothesized that cells grown in the presence of glucose would have little or no available EcfO at the time of stress but that when induced with maltose, the excess EcfO would have stimulated an effective defense response. Results showed that with a minimal aerobic stress of 1 h of air exposure, there was little difference in survival, but cells preinduced for EcfO and exposed to air for 24 h showed about a 10-fold difference in survival compared to cells from the noninducing glucose medium. It can be concluded that EcfO induced under these conditions is sufficient to complement the phenotype of the ecfO mutation. In addition, the data indicate that the dual-His-tagged EcfO protein was functional in B. fragilis.
The N terminus of Reo interacts with EcfO.
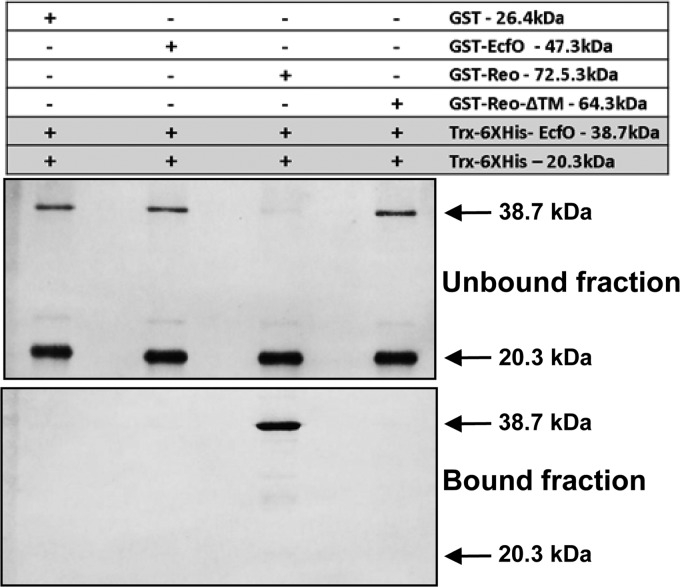
It is common that ECF sigma factors are regulated by an anti-sigma factor located within the same genetic locus and are either cotranscribed or divergently transcribed from the sigma factor gene. The reo gene is in an operon with ecfO, and the phenotype of a reo mutant suggested that it may regulate EcfO activity. Reo was annotated as a hypothetical protein with no conserved anti-sigma factor domains, but it is predicted to have a single transmembrane region (amino acids [aa] 49 to 71). To test if Reo is an anti-sigma factor for EcfO, we constructed EcfO and Reo fusion proteins with affinity tags for thioredoxin (Trx)-His and GST, respectively, and then tested if they were able to physically interact by affinity pulldown assays. Each interaction assay included a purified Trx-His tag in the binding reaction mixture to exclude nonspecific interactions due to the affinity tag. Purified GST-tagged proteins were bound to glutathione-Sepharose resin and incubated with equivalent amounts of Trx-His-tagged proteins. After incubation, washing, and elution, protein profiles of fractions bound or unbound to the glutathione beads were compared by Coomassie staining and Western blotting. As observed by Western blotting (Fig. 3), the GST controls did not bind to EcfO, nor did EcfO form dimers with itself. The full-length Reo protein was able to bind to EcfO, depleting it from the unbound fraction. Interestingly, when the N terminus of Reo was deleted, the mutated protein (Reo-ΔTM, with an N-terminal deletion of aa 1 to 73) lost its EcfO binding activity. This finding suggests that Reo possesses a cytoplasmic sigma factor binding domain linked by a trans-membrane domain to a periplasmic sensor domain.
Fig 3.
Western blot probed with anti-His tag antibody used to test for EcfO-Reo interactions and map the Reo interaction domain. GST-tagged proteins were affinity bound to glutathione-Sepharose resin and incubated with purified Trx-His-tagged EcfO (38.7 kDa). The Trx-His protein (20.3 kDa) was also included in each experiment as a negative control. Following incubation, the unbound fraction was collected, and the resin was washed. The pulled-down proteins (bound fraction) were then eluted from the Sepharose beads. His-tagged proteins collected in the unbound and bound fractions were detected by Western blotting using a mouse monoclonal antibody to the His tag.
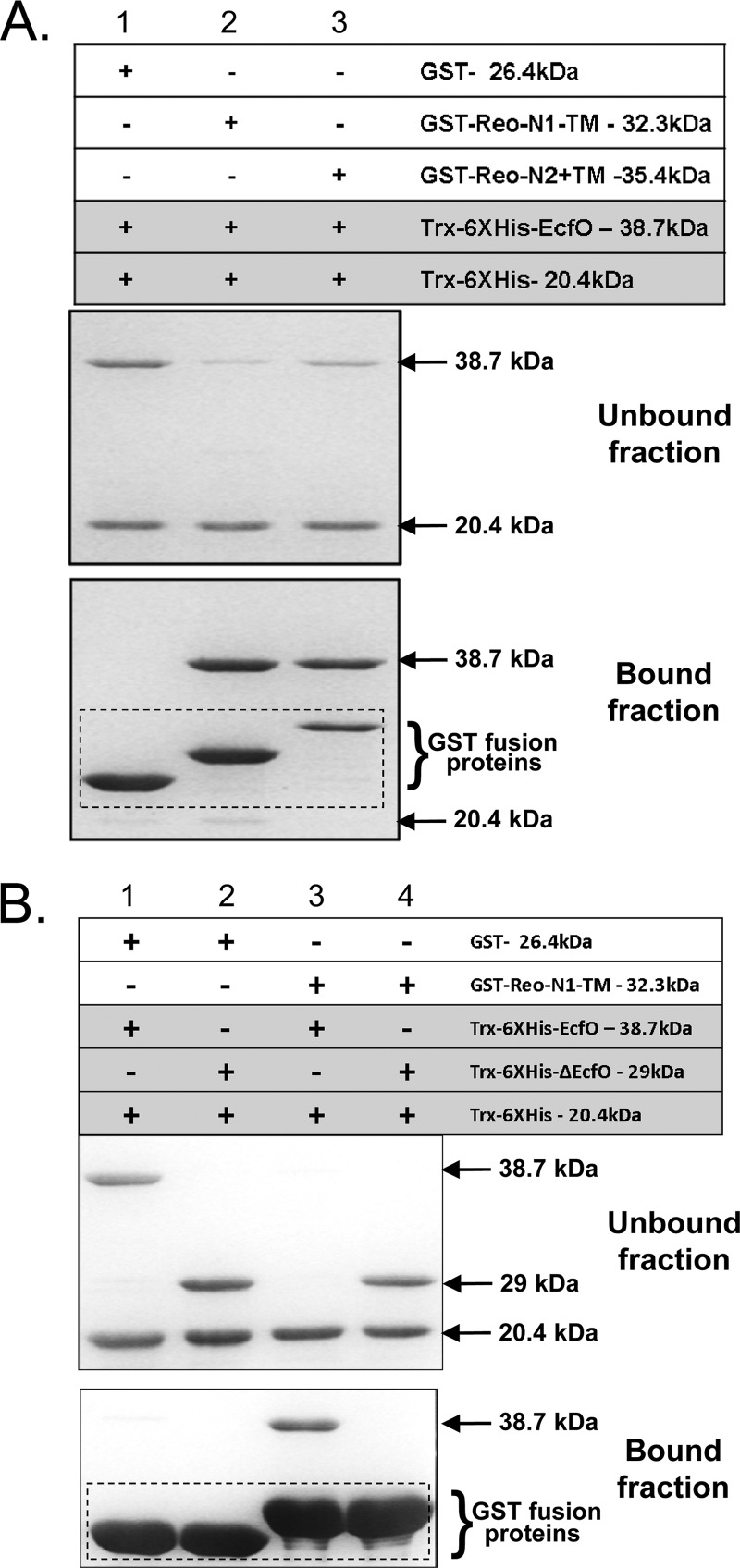
Further evidence that the N-terminal amino acids of Reo are sufficient to interact with EcfO was obtained by pulldown experiments using the Reo N1-TM protein, which contains just the first 48 aa of Reo (Fig. 4A, lane 2). The GST fusion to these amino acids was sufficient to pull down EcfO from the soluble protein suspension, and the presence of the trans-membrane domain did not interfere with this interaction (Fig. 4A, lane 3). A negative control containing only GST did not interact with EcfO. Another experiment was designed to determine if mutated EcfO containing an in-frame deletion of aa 44 to 123 (ΔEcfO) was able to bind to the N terminus of Reo (Fig. 4B). The ΔecfO mutation from the strain IB524 chromosome was used as a template to generate this deleted protein construct. Unlike the wild-type protein, the ΔEcfO protein was unable to bind to the GST-bound N terminus of Reo.
Fig 4.
Identification of Reo and EcfO interaction domains. (A) Coomassie-stained SDS-PAGE gels confirming the EcfO interaction domain of Reo. The following GST-tagged proteins were used: GST-Reo-N1-TM is the N-terminal 48 amino acids of Reo without a transmembrane domain, and GST-Reo-N2+TM is the N-terminal 78 amino acids of Reo with a transmembrane domain. GST alone was used as a negative control. GST-tagged proteins were affinity purified and incubated with affinity-purified Trx-His-tagged EcfO. Unbound proteins (Trx-6×His tagged) in the flowthrough and proteins eluted after pulldowns were visualized by Coomassie staining. (B) Coomassie-stained SDS-PAGE gels mapping the Reo interaction domain in EcfO. GST-tagged proteins (Reo-N1-TM and GST, as a negative control) were affinity purified and incubated with purified Trx-6×His-EcfO or the deletion derivative Trx-6×His-ΔEcfO, derived from strain IB524. Unbound proteins (Trx-6×His tagged) in the flowthrough and proteins in the bound fraction eluted after pulldowns were visualized by Coomassie staining.
Reo and EcfO interact on the cytoplasmic membrane.
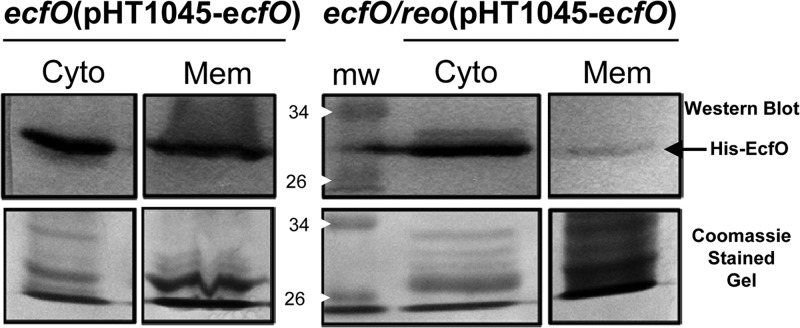
Evidence for in vivo interactions was obtained from an experiment designed to identify the subcellular localization of EcfO and to infer the localization of Reo. A maltose-inducible His-tagged EcfO protein was introduced on a plasmid into two stains, either the ecfO mutant or the ecfO reo double mutant. Cultures were grown anaerobically in maltose-containing medium, and membrane and cytoplasmic fractions were then isolated. We predicted that if Reo was localized to the membrane, the overexpression of EcfO would result in it being found in both the membrane and the cytoplasm fractions. In contrast, we expected to see only a cytoplasmic localization of EcfO in the Reo mutant. As shown in Fig. 5, EcfO was enriched in the expected fractions for both strains. It is notable that the membrane fraction for the ecfO reo(pHT1045-ecfO) mutant was intentionally overloaded relative to the cytoplasmic fraction, but this resulted in only a weak signal for EcfO in the membrane.
Fig 5.
Dynamic localization of EcfO in the membrane and cytoplasm in the presence of a membrane-associated anti-sigma factor. The subcellular localization of EcfO was investigated by using affinity-tagged EcfO expressed in either the ecfO mutant or the ecfO reo double mutant. Cells were grown anaerobically to the mid-logarithmic phase in TYM medium to induce pHT1045-ecfO expression and then fractionated into cytoplasmic and membrane fractions. Following affinity purification of His-tagged EcfO and Western blotting with anti-His tag antibodies (top), EcfO was observed in both the cytoplasmic and membrane fractions. In the absence of its anti-sigma factor Reo, EcfO remained predominantly free in the cytoplasm. Coomassie-stained gels (bottom) show the total protein loaded in the 26- to 34-kDa range that corresponds to His-tagged EcfO. Cyto, cytoplasmic fraction; Mem, membrane fraction; mw, molecular weight marker (in thousands).
Expression arrays reveal that EcfO regulates a discrete set of target genes.
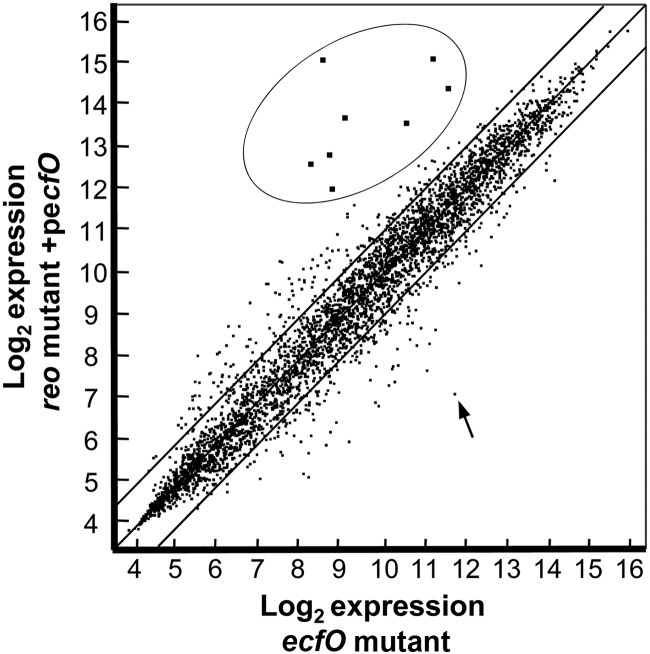
Expression microarrays were used to help identify genes activated downstream of EcfO. The experimental strategy was designed to induce EcfO expression in the absence of its inhibitor Reo. To implement this strategy, the ecfO gene was placed under the control of an IPTG-inducible promoter and then expressed under anaerobic conditions in the reo mutant background. RNA obtained from these strains was compared to that of the ecfO mutant containing the empty vector. Genome expression levels were visualized by scatter plots of the log2-normalized data, and the results showed that there was a tight correlation (R = 0.95) between the two data sets, except for one clearly distinct set of induced genes in the EcfO overexpression strain (Fig. 6). There were eight genes induced by 9-fold or more, and this set included the ecfO gene, which was induced 11-fold (Table 2). As expected, the expression level for the reo gene was considerably repressed, and the low background of reo expression that was observed was due to the presence of the intact N terminus of the gene in the reo mutant (Fig. 1 and Table 2). Six of the induced genes code for novel lipoproteins of unknown function which are related only to other proteins of Bacteroidetes lineage, and the seventh is a radical S-adenosylmethionine (SAM) superfamily member. Three of the encoded lipoproteins (BF638R_0588, BF638R_0743, and BF638R_1335) are similar to NigD, a protein associated with the Prevotella nigrescens bacteriocin nigrescin (41, 42). The results from these microarray experiments were in close agreement with data from preliminary microarray experiments using either a maltose-inducible ecfO construct or a constitutively expressed ecfO construct. These earlier data showed that among the most highly expressed genes was the same set of novel lipoprotein and radical SAM genes as seen with the IPTG-inducible system shown here (NCBI Gene Expression Omnibus accession number GSE35536) (see Table S3 in the supplemental material).
Fig 6.
Scatter plot of genome expression values comparing the ecfO mutant to the reo mutant containing ecfO in an IPTG-inducible plasmid. The RMA-normalized log2 expression values from at least 3 independent experiments comparing IPTG-induced pFD1146-ecfO in the reo mutant background to the ecfO mutant with an empty vector control were averaged and plotted by using ArrayStar software. The EcfO regulon of induced genes is circled, and an arrow indicates the repressed expression of the reo gene.
Table 2.
Genes induced following ectopic expression of ecfO in an reo null mutant
| Gene | Function or domain | Fold inductiona |
|---|---|---|
| BFR638R_0588 | NigD family lipoprotein with secretion signal sequence | 112 |
| BFR638R_1335 | NigD family lipoprotein with secretion signal sequence | 29 |
| BFR638R_4448 | Putative lipoprotein, radical SAM/lipoprotein system | 25 |
| BFR638R_4447 | Radical SAM family lipoprotein | 20 |
| BFR638R_0743 | NigD family lipoprotein with secretion signal sequence | 18 |
| BFR638R_2785 | Lipoprotein with possible secretion signal sequence | 9 |
| BFR638R_2513 | Lipoprotein with secretion signal sequence | 9 |
| ecfO | ECF sigma factor | 11 |
| reo | Anti-sigma factor | 0.04b |
Fold induction was determined from the normalized log2 expression data averaged from all experiments. The normalized microarray expression data are available from the NCBI Gene Expression Omnibus under accession number GSE39231. The values represent the averages from at least 3 independent biological replicates, each with five technical replicates.
There was some low-level expression observed for the reo gene due to mRNA from the 5′ end of the mutated Δreo gene which was detected by microarray probes.
The EcfO regulon is comprised of 7 direct target operons.
The sequences upstream of each putative EcfO regulon gene were examined in search of an EcfO consensus promoter recognition motif. The transcription start sites for ecfO and BF638R_1335 were mapped by using 5′ RACE. The sites were located 18 and 53 bp upstream of the EcfO and BF638R_1335 translation start sites, respectively. A comparison of these two divergent promoters to those of the other regulon genes identified in the expression microarrays revealed conservation, and a consensus promoter recognition sequence emerged. Strongly conserved −10- and −35-like regions were observed in the upstream noncoding regions of BF638R_4448, BF638R_0743, BF638R_2513, and BF638R_2785 and within the BF638R_2478 gene (Fig. 7). All of these except the latter were identified by expression microarray data as being regulated by EcfO.
Fig 7.
Logo of the promoter recognition sequence of EcfO. Based on the data from expression and ChAP experiments defining the EcfO regulon (Tables 2 to 4), the target promoter sequences were aligned, and a consensus promoter was identified by using CLC Main Workbench v6.5 software.
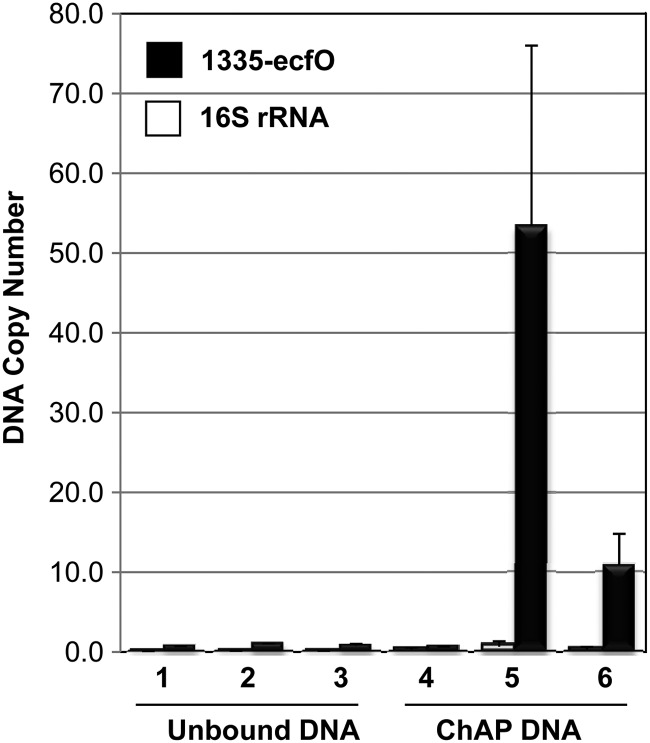
We designed a chromatin affinity purification (ChAP) experiment to address whether EcfO can be enriched in vivo at the promoters of the target genes. This method has not been used previously for B. fragilis and was adapted from methods described previously (30, 43). Because of the absence of specific antibodies to EcfO, we used an affinity-tagged approach with pHT1045-ecfO (dually His tagged), which was expressed in either the ecfO or the ecfO reo mutant (31, 32). Based on results from the complementation experiment (Fig. 2C), it was clear that the dual His tag did not interfere significantly with the function of EcfO. As a negative control, we used the ecfO reo double mutant strain, carrying the same vector but containing the ΔecfO gene with the in-frame deletion. From these same experimental samples, we also prepared RNA in order to correlate the gene expression profiles to the chromatin enrichment profiles. After the amplification of ChAP and unbound DNA, enrichment at the BF638R_1335 ecfO promoter region was examined by using qPCR. The results in Fig. 8 show that samples containing His-tagged EcfO pulled down between 1 × 106 and 6 × 106 copies of the promoter fragment per ng of total DNA, representing a 10- to 20-fold enrichment of the promoter region in the ChAP samples compared to the negative controls. In contrast, there was no change between samples in the unbound fractions. 16S rRNA was not enriched in either the unbound DNA fraction or the ChAP DNA fractions (Fig. 8). These ratios reflected the enrichments observed prior to DNA amplification (data not shown) and in three other independent experiments, including two with IPTG-inducible pHT1146-ecfO.
Fig 8.
Affinity enrichment of EcfO at the divergent target promoters of BF638R_1335 and ecfO. Maltose-inducible His-tagged EcfO (pHT1045-ecfO) was expressed in the background strain of the ecfO deletion mutant (IB524) (lanes 3 and 6) or the ecfO reo double mutant (IB526) (lanes 2 and 5). For a negative control, the ecfO gene fragment containing the 237-bp internal deletion was expressed from pHT1045 in IB526 (lanes 1 and 4). After chromatin affinity precipitation (ChAP) and DNA amplification of unbound and affinity-purified (ChAP) DNA, qPCR was used to determine the fold enrichment of EcfO at the divergent BF638R_1335-ecfO promoter region in the samples. The DNA copy number is ×105 ng−1 of DNA template after EcfO-ChAP. The 16S rRNA locus was used as a negative control to demonstrate the lack of enrichment.
Amplified DNA was used in a ChAP/qPCR assay to quantify EcfO enrichment at the promoter sites of other predicted target genes. The relative fold gene expression and the corresponding enrichment of EcfO at the promoters were confirmed by qRT-PCR estimations of copy number(Tables 3 and 4). There was a close correlation between the PCR data for both the ChAP and gene expression experiments. As a negative control, BF638R_2478 showed neither an enrichment of EcfO nor an increased accumulation of transcript. It was also observed that the presence of the EcfO inhibitor Reo reduced the overall promoter enrichment and gene expression by over 2-fold. The promoter sites of several other genes, including three ECF sigma factors, were also tested and showed no enrichment by EcfO-ChAP (data not shown).
Table 3.
Affinity enrichment of EcfO at promoter binding sites
| Straina | Relative fold enrichment at promoters of target genesb |
|||||||
|---|---|---|---|---|---|---|---|---|
| BF638R_16S | BF638R_2478 | BF638R_4448 | BF638R_0588 | BF638R_2785 | BF638R_0743 | BF638R_1335 | ecfO | |
| ecfO reo(pHT-ΔecfO) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ecfO reo(pHT-ecfO) | 2 | 1 | 6 | 10 | 6 | 8 | 37 | 37 |
| ecfO(pHT-ecfO) | 2 | 1 | 2 | 3 | 2 | 2 | 14 | 14 |
pHT1045-ecfO or pHT1045-ΔecfO was harbored in either the ecfO reo double mutant (IB526) or the ecfO mutant (IB524), as indicated.
qPCR was used to quantify the enrichment of EcfO at target genes after ChAP. The fold enrichment compared to the negative controls is shown based on the copy number per ng of DNA.
Table 4.
Regulation of gene expression
| Straina | Relative gene expression (copies/ng)b |
|||||||
|---|---|---|---|---|---|---|---|---|
| BF638R_16S | BF638R_2478 | BF638R_4448 | BF638R_0588 | BF638R_2785 | BF638R_0743 | BF638R_1335 | ecfO | |
| ecfO reo(pHT-ΔecfO) | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| ecfO reo(pHT-ecfO) | 1 | 1 | 19 | 2,508 | 307 | 603 | 619 | 86,911 |
| ecfO(pHT-ecfO) | 1 | 1 | 6 | 1,302 | 60 | 214 | 212 | 57,068 |
pHT1045-ecfO or pHT1045-ΔecfO was harbored in either the ecfO reo double mutant (IB526) or the ecfO mutant (IB524), as indicated.
qRT-PCR showing EcfO target gene regulation of RNA isolated from the same samples used for ChAP. The gene expression values compared to the negative control are shown based on copy number per ng of RNA.
DISCUSSION
B. fragilis encodes a large number of sensory signaling systems, which include two-component system regulators and ECF sigma factors. These systems are critical for survival in their normal but ever-changing intestinal environment and for adaption to extraintestinal sites as an opportunistic pathogen. In the related organism B. thetaiotaomicron, about half of the ECF sigma factors are associated with polysaccharide utilization loci, and they regulate the use of both host- and dietary-derived polysaccharides (40). Likewise, in B. fragilis, 58% of ECF sigma factors may be involved with polysaccharide utilization based on their similarity to those in B. thetaiotaomicron and their resemblance to the ECF10 sigma factor family (see Fig. S3 in the supplemental material) (39). In contrast, EcfO is a member of an uncharacterized ECF sigma factor family, ECF21. Furthermore, EcfO was of interest because its gene was induced by aerobic exposure, and it is located adjacent to the gene for a key regulator of oxidative stress, OxyR. B. fragilis OxyR has been implicated in the rapid induction of catalase, alkyl-hydroperoxide reductase, Dps, and thioredoxin peroxidase, which are the first responders to oxidative stress (6, 8). The expression of these enzymes provides rapid protection from free radical damage, but little is known about the mechanisms that control other facets of the OSR and which may be necessary for prolonged protection or recovery from stress. A number of ECF sigma factors will likely play a role in the OSR, and in this report, we demonstrate a link between EcfO activity and protection against prolonged oxidative stress.
The ecfO mutant demonstrated greater sensitivity than the wild type to redox cycling agents and organic peroxides (Fig. 2A), and this phenotype was consistent with its proposed role as an oxidative stress response regulator. Extended aerobic exposure, which may induce a wide range of stresses, also caused a greater inhibition of the mutant than the parent strain. The protection afforded by EcfO against such a variety of oxidative stress agents suggests that this regulator likely does not control a specific response to reactive oxygen species (ROS) but may in fact be induced by the downstream toxic effects of this stress. This is supported by the observation that the defect in the aerobic survival of the ecfO mutant did not manifest until about 48 h of exposure (Fig. 2B). It will be interesting to determine if other stresses will induce the EcfO protective response. The maltose-inducible complementation of the ecfO mutant stress phenotype supported the importance of EcfO in the response to oxidative stress conditions.
Reo is proposed to be a negative regulator, anti-sigma factor, for EcfO. The reo null mutation would remove the inhibition of EcfO, which would lead to significantly higher levels of EcfO and the accumulation of the target response gene products prior to induction by any stress. This explains the exceptional ability of the reo mutant to respond to oxidative stress. The response to menadione, cumene hydroperoxide, phenazine methosulfate, and diamide stress was better than that of the wild-type strain, quite comparable to the overexpression of EcfO (Fig. 2A and data not shown). The role of anti-sigma factors as principal regulators of sigma factor activity has been comprehensively reviewed, and a general trend in membrane-bound anti-sigma factors is that the N-terminal cytoplasmic domain interacts with the cognate sigma factor (16, 44, 45). The mechanism that regulates EcfO activity shares some characteristics with these systems. The N-terminal 48 amino acids of Reo which interact with EcfO (Fig. 4) have weakly conserved identity to the anti-sigma K factor RSK of Mycobacterium tuberculosis, which negatively regulates the activity of sigma factor K (46). There is also a trans-membrane domain between amino acid residues 49 and 71, and at the C-terminal end, there is an outer membrane protein β-barrel domain with a partial conservation of porin proteins and OmpA. This arrangement has not been described previously for an anti-sigma factor, and it is likely that Reo is sensing some signal in the outer membrane which it then transmits to the N terminus, resulting in the release of EcfO. The nature of the signal and the role of the β-barrel domain are unknown.
The precise mechanism by which Reo regulates EcfO is still under investigation, but our results confirm that Reo is a negative regulator that can physically interact with it using the N-terminal 48 amino acids (Fig. 3 and 4). Furthermore, in the absence of Reo, EcfO was no longer found to be associated with the membrane fraction, suggesting that it is more available to initiate transcription at its promoter target sites (Fig. 5). Typical mechanisms used to control regulators of oxidative stress responses often rely on some form of thiol-based redox switching, and this can include anti-sigma factors (19, 47). The Streptomyces coelicolor anti-sigma factor RsrA is responsive to redox changes and was previously described (19). A Zn(II) ion coordinates cysteine and histidine residues to form a structural conformation which allows binding to its sigma factor σA (48). The oxidation of two cysteine residues leads to disulfide bond formation and the release of Zn with a concomitant conformational change releasing σA. Interestingly, Reo contains two cysteine residues in the C terminus with potential for disulfide bond formation.
The combined approach of expression microarrays and chromatin affinity purification allowed us to establish the direct targets of EcfO. We first determined a putative regulon using microarray studies with ectopically induced EcfO, and we then used ChAP/qPCR to establish a direct interaction with the promoter. The positive control for chromatin enrichment was the shared ecfO and BF638R_1335 promoter region. As a negative control, we used the 16S rRNA gene, and our controls confirmed that EcfO was enriched neither in unbound DNA fractions nor at the 16S gene locus (Fig. 8). ChAP/qPCR data and qRT-PCR of the same cultures did verify the 7 transcriptional units of the EcfO regulon initially predicted from the expression microarray (Tables 3 and 4).
The function of the EcfO regulon is not known, and all of the direct target genes code for proteins of unknown function. A common feature of these proteins is that all but one are lipoproteins, and three (BF638R_0588, BF638R_0743, and BF638R_1335) are members of the novel NigD superfamily found only in the Bacteroidetes. The original NigD homologue was identified in a genetic locus associated with the production of nigrescin, a bacteriocin secreted by Prevotella nigrescens (42). The nigD gene was located adjacent to the bacteriocin gene, and thus, it was proposed that NigD is a bacteriocin immunity protein. X-ray structures (generated by the Joint Center For Structural Genomics [JCSG]) for this protein homologue in Bacteroides vulgatus and Parabacteroides distasonis are available (Protein Data Bank [PDB] accession numbers 3K6O and 3K0Y, respectively), but they have not yet been associated with any particular function. It seems unlikely that the NigD homologues described here are related directly to bacteriocin immunity, but given the protective function of BF638R_1335 (data not shown), perhaps they play a common role in responding to specific types of extracytoplasmic stresses caused by the perturbation of the outer membrane structure.
BF638R_4447 is a member of the radical SAM oxidoreductase superfamily, and BF638R_4448 is a member of the radical SAM lipoproteins, which are Bacteroides sp.-specific proteins found to be associated only with radical SAM enzymes. The radical SAM proteins are a large superfamily containing iron sulfur centers and with diverse functions, including protein modification and general metabolism, such as menaquinone biosynthesis and pyruvate metabolism. It is possible that these proteins work in concert with the NigD homologues in a specialized function associated with protein modifications in response to stress conditions.
EcfO is the first characterized member of the ECF21 family, and its activity is controlled by an anti-sigma factor with a unique protein architecture, but overall, it demonstrates classical ECF sigma factor function. EcfO and Reo have been highly conserved in Bacteroides species. Interestingly, this conservation includes an association with a divergently transcribed nigD gene and a genome location near oxyR (in those Bacteroides species that have an oxyR). EcfO is required for optimal resistance to oxidative stress, and it mediates resistance through the regulation of novel target genes, which likely are associated with the cell envelope. The EcfO regulon appears to be required as part of a concerted response to effectively manage stress and assist cells in coping with their changing environment. It is interesting to note that the EcfO regulon was not previously found to be induced by exposure to 5% O2 (30 min), 100 μM H2O2 (30 min), or air (60 min) (6). We currently speculate that this regulon may have a greater role during more extended exposures to oxidative stress (or other stresses), and a preliminary expression microarray using cultures exposed to air for 3 h showed a strong induction of the regulon (data not shown). Future experiments will focus on the identification of the signal or signals that modulate the activity of Reo and further elucidate the protective role of this regulon.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant AI40588 to C.J.S.
We thank E. R. Rocha and A. C. Parker for helpful discussions and advice.
Footnotes
Published ahead of print 26 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01491-12.
REFERENCES
- 1.Finegold SM, George WL. 1989. Anaerobic infections in humans. Academic Press, New York, NY [Google Scholar]
- 2.Finegold SM. 1983. Normal indigenous microflora, p 3–31. In Finegold S. (ed), Human intestinal microflora in health and disease. Academic Press, New York, NY [Google Scholar]
- 3.Smith CJ, Rocha ER, Paster BJ. 2006. The medically important Bacteroides spp. in health and disease, p 381–427. In Dworkin M. (ed), The prokaryotes: an evolving electronic resource for the microbiological community, vol 7. Springer Verlag, New York, NY [Google Scholar]
- 4.Lassmann B, Gustafson DR, Wood CM, Rosenblatt JE. 2007. Reemergence of anaerobic bacteremia. Clin. Infect. Dis. 44:895–900 [DOI] [PubMed] [Google Scholar]
- 5.McClean KL, Sheehan GJ, Harding GK. 1994. Intra-abdominal infection: a review. Clin. Infect. Dis. 19:100–116 [DOI] [PubMed] [Google Scholar]
- 6.Sund CJ, Rocha ER, Tzianabos AO, Wells WG, Gee JM, Reott MA, O'Rourke DP, Smith CJ. 2008. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol. Microbiol. 67:129–142 [DOI] [PubMed] [Google Scholar]
- 7.Rocha ER, Herren CD, Smalley DJ, Smith CJ. 2003. The complex oxidative stress response of Bacteroides fragilis: the role of OxyR in control of gene expression. Anaerobe 9:165–173 [DOI] [PubMed] [Google Scholar]
- 8.Rocha ER, Owens G, Jr, Smith CJ. 2000. The redox-sensitive transcriptional activator OxyR regulates the peroxide response regulon in the obligate anaerobe Bacteroides fragilis. J. Bacteriol. 182:5059–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smalley D, Rocha ER, Smith CJ. 2002. Aerobic-type ribonucleotide reductase in the anaerobe Bacteroides fragilis. J. Bacteriol. 184:895–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spence C, Wells WG, Smith CJ. 2006. Characterization of the primary starch utilization operon in the obligate anaerobe Bacteroides fragilis: regulation by carbon source and oxygen. J. Bacteriol. 188:4663–4672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manganelli R, Voskuil MI, Schoolnik GK, Dubnau E, Gomez M, Smith I. 2002. Role of the extracytoplasmic-function sigma factor sigma(H) in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365–374 [DOI] [PubMed] [Google Scholar]
- 12.Manganelli R, Voskuil MI, Schoolnik GK, Smith I. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423–437 [DOI] [PubMed] [Google Scholar]
- 13.Raman S, Song T, Puyang X, Bardarov S, Jacobs WR, Jr, Husson RN. 2001. The alternative sigma factor SigH regulates major components of oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119–6125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alvarez-Martinez CE, Baldini RL, Gomes SL. 2006. A Caulobacter crescentus extracytoplasmic function sigma factor mediating the response to oxidative stress in stationary phase. J. Bacteriol. 188:1835–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alvarez-Martinez CE, Lourenco RF, Baldini RL, Laub MT, Gomes SL. 2007. The ECF sigma factor sigma(T) is involved in osmotic and oxidative stress responses in Caulobacter crescentus. Mol. Microbiol. 66:1240–1255 [DOI] [PubMed] [Google Scholar]
- 16.Osterberg S, del Peso-Santos T, Shingler V. 2011. Regulation of alternative sigma factor use. Annu. Rev. Microbiol. 65:37–55 [DOI] [PubMed] [Google Scholar]
- 17.Heinrich J, Wiegert T. 2009. Regulated intramembrane proteolysis in the control of extracytoplasmic function sigma factors. Res. Microbiol. 160:696–703 [DOI] [PubMed] [Google Scholar]
- 18.Francez-Charlot A, Frunzke J, Reichen C, Ebneter JZ, Gourion B, Vorholt JA. 2009. Sigma factor mimicry involved in regulation of general stress response. Proc. Natl. Acad. Sci. U. S. A. 106:3467–3472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang JG, Paget MS, Seok YJ, Hahn MY, Bae JB, Hahn JS, Kleanthous C, Buttner MJ, Roe JH. 1999. RsrA, an anti-sigma factor regulated by redox change. EMBO J. 18:4292–4298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. 1993. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science 262:1277–1280 [DOI] [PubMed] [Google Scholar]
- 21.Privitera G, Dublanchet A, Sebald M. 1979. Transfer of multiple antibiotic resistance between subspecies of Bacteroides fragilis. J. Infect. Dis. 139:97–101 [DOI] [PubMed] [Google Scholar]
- 22.Baughn AD, Malamy MH. 2002. A mitochondrial-like aconitase in the bacterium Bacteroides fragilis: implications for the evolution of the mitochondrial Krebs cycle. Proc. Natl. Acad. Sci. U. S. A. 99:4662–4667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith CJ, Rollins LA, Parker AC. 1995. Nucleotide sequence determination and genetic analysis of the Bacteroides plasmid, pBI143. Plasmid 34:211–222 [DOI] [PubMed] [Google Scholar]
- 24.Smith CJ, Rogers MB, McKee ML. 1992. Heterologous gene expression in Bacteroides fragilis. Plasmid 27:141–154 [DOI] [PubMed] [Google Scholar]
- 25.Parker AC, Smith CJ. 2012. Development of an IPTG inducible expression vector adapted for Bacteroides fragilis. Plasmid 68:86–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobo LA, Smith CJ, Rocha ER. 2011. Flavin mononucleotide (FMN)-based fluorescent protein (FbFP) as reporter for gene expression in the anaerobe Bacteroides fragilis. FEMS Microbiol. Lett. 317:67–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bacic MK, Smith CJ. 1 May 2008, posting date Laboratory maintenance and cultivation of Bacteroides species. Curr. Protoc. Microbiol. Chapter 13: Unit 13C.1. doi:10.1002/9780471729259.mc13c01s9. [DOI] [PMC free article] [PubMed]
- 28.Sund CJ, Wells WG, Smith CJ. 2006. The Bacteroides fragilis P20 scavengase homolog is important in the oxidative stress response but is not controlled by OxyR. FEMS Microbiol. Lett. 261:211–217 [DOI] [PubMed] [Google Scholar]
- 29.Parker AC, Smith CJ. 1993. Genetic and biochemical analysis of a novel ambler class A β-lactamase responsible for cefoxitin resistance in Bacteroides species. Antimicrob. Agents Chemother. 37:1028–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Efromovich S, Grainger D, Bodenmiller D, Spiro S. 2008. Genome-wide identification of binding sites for the nitric oxide-sensitive transcriptional regulator NsrR. Methods Enzymol. 437:211–233 [DOI] [PubMed] [Google Scholar]
- 31.Ishikawa S, Ogura Y, Yoshimura M, Okumura H, Cho E, Kawai Y, Kurokawa K, Oshima T, Ogasawara N. 2007. Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res. 14:155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamimi Y, Lines M, Coca-Prados M, Walter MA. 2004. Identification of target genes regulated by FOXC1 using nickel agarose-based chromatin enrichment. Invest. Ophthalmol. Vis. Sci. 45:3904–3913 [DOI] [PubMed] [Google Scholar]
- 33.Rocha ER, Smith CJ. 1997. Regulation of Bacteroides fragilis katB mRNA by oxidative stress and carbon limitation. J. Bacteriol. 179:7033–7039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 doi:10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coyne MJ, Weinacht KG, Krinos CM, Comstock LE. 2003. Mpi recombinase globally modulates the surface architecture of a human commensal bacterium. Proc. Natl. Acad. Sci. U. S. A. 100:10446–10451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krinos CM, Coyne MJ, Weinacht KG, Tzianabos AO, Kasper DL, Comstock LE. 2001. Extensive surface diversity of a commensal microorganism by multiple DNA inversions. Nature 414:555–558 [DOI] [PubMed] [Google Scholar]
- 37.Weinacht KG, Roche H, Krinos CM, Coyne MJ, Parkhill J, Comstock LE. 2004. Tyrosine site-specific recombinases mediate DNA inversions affecting the expression of outer surface proteins of Bacteroides fragilis. Mol. Microbiol. 53:1319–1330 [DOI] [PubMed] [Google Scholar]
- 38.Cerdeno-Tarraga AM, Patrick S, Crossman LC, Blakely G, Abratt V, Lennard N, Poxton I, Duerden B, Harris B, Quail MA, Barron A, Clark L, Corton C, Doggett J, Holden MT, Larke N, Line A, Lord A, Norbertczak H, Ormond D, Price C, Rabbinowitsch E, Woodward J, Barrell B, Parkhill J. 2005. Extensive DNA inversions in the B. fragilis genome control variable gene expression. Science 307:1463–1465 [DOI] [PubMed] [Google Scholar]
- 39.Staron A, Sofia HJ, Dietrich S, Ulrich LE, Liesegang H, Mascher T. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 74:557–581 [DOI] [PubMed] [Google Scholar]
- 40.Martens EC, Roth R, Heuser JE, Gordon JI. 2009. Coordinate regulation of glycan degradation and polysaccharide capsule biosynthesis by a prominent human gut symbiont. J. Biol. Chem. 284:18445–18457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaewsrichan J, Douglas CW, Teanpaisan R. 2005. Characterization of minimal bacteriocin operon from Prevotella nigrescens ATCC 25261. Lett. Appl. Microbiol. 40:138–145 [DOI] [PubMed] [Google Scholar]
- 42.Teanpaisan R, Narawatthana S, Utarabhand P. 2009. The gene coding for nigrescin produced by Prevotella nigrescens ATCC 25261. Lett. Appl. Microbiol. 49:293–298 [DOI] [PubMed] [Google Scholar]
- 43.Cho BK, Barrett CL, Knight EM, Park YS, Palsson BO. 2008. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:19462–19467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hughes KT, Mathee K. 1998. The anti-sigma factors. Annu. Rev. Microbiol. 52:231–286 [DOI] [PubMed] [Google Scholar]
- 45.Yoshimura M, Asai K, Sadaie Y, Yoshikawa H. 2004. Interaction of Bacillus subtilis extracytoplasmic function (ECF) sigma factors with the N-terminal regions of their potential anti-sigma factors. Microbiology 150:591–599 [DOI] [PubMed] [Google Scholar]
- 46.Said-Salim B, Mostowy S, Kristof AS, Behr MA. 2006. Mutations in Mycobacterium tuberculosis Rv0444c, the gene encoding anti-SigK, explain high level expression of MPB70 and MPB83 in Mycobacterium bovis. Mol. Microbiol. 62:1251–1263 [DOI] [PubMed] [Google Scholar]
- 47.Antelmann H, Helmann JD. 2011. Thiol-based redox switches and gene regulation. Antioxid. Redox Signal. 14:1049–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li W, Bottrill AR, Bibb MJ, Buttner MJ, Paget MS, Kleanthous C. 2003. The role of zinc in the disulphide stress-regulated anti-sigma factor RsrA from Streptomyces coelicolor. J. Mol. Biol. 333:461–472 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.