Abstract
Streptococcus mutans, a member of the human indigenous oral microbiome, produces a quorum-sensing peptide called the competence-stimulating peptide (CSP) pheromone. We previously demonstrated that S. mutans expresses its CSP pheromone under specific stresses and responds to high levels of CSP by inducing cell death in a fraction of the bacterial population. Streptococci lack the classical SOS response, and the induction of the SigX regulon has been proposed to act as a general stress response in Gram-positive bacteria. We show here that inactivation of SigX abolished the CSP-induced cell death phenotype. Among SigX-regulated genes, SMU.836 (now named lytFSm), encoding a conserved streptococcal protein, is a functional peptidoglycan hydrolase involved in CSP-induced cell lysis. We also demonstrated that LytFSm is most likely a self-acting autolysin, since LytFSm produced by attacker cells cannot trigger CSP-induced lysis of LytFSm-deficient target cells present in the same environment. Electron microscopy revealed important morphological changes accompanying autolysis of CSP-induced wild-type cultures that were absent in the LytFSm-deficient mutant. The LytFSm promoter was activated in the physiological context of elevated concentrations of the CSP pheromone under stress conditions, such as exposure to heat, hydrogen peroxide, and acid. In a long-term survival assay, the viability of a mutant deficient in LytFSm autolysin was significantly lower than that observed for the wild-type strain. The results of this study suggest that cell death of S. mutans induced by its quorum-sensing CSP pheromone may represent a kind of altruistic act that provides a way for the species to survive environmental stresses at the expense of some of its cells.
INTRODUCTION
Quorum sensing is a system used by bacteria for communication between cells using small diffusible molecules secreted by individual cells. These signaling molecules increase in concentration as a function of the bacterial cell density, allowing a coordinated population response. Many collective behaviors performed by bacteria, such as biofilm formation, bioluminescence, swarming motility, competence for DNA uptake, and virulence factor secretion, are often regulated by quorum sensing (1, 2). It is now well known that bacterial quorum-sensing mechanisms can go well beyond the counting of cell numbers. Indeed, there is growing evidence that quorum-sensing constitutes a global regulatory system in many different bacterial species (3, 4).
Gram-positive bacteria primarily use short peptides that can be posttranslationally modified as signaling molecules (5). In streptococci, there are two general activation pathways for intraspecies communication. Streptococcal cells utilize nonmodified small oligopeptides that are reimported into the cells. Once internalized, the signal peptide interacts with a transcriptional regulator, thereby modifying the expression of target genes directly or indirectly (6, 7). The other pathway involves the competence-stimulating peptide (CSP), also called peptide pheromone. These peptide pheromones are usually the products of propeptides containing a double glycine leader at the N terminus, which is processed concomitantly with export by specific transporters. At a threshold concentration, the secreted peptides are detected by membrane-bound two-component signaling proteins, and signal transduction occurs by a phosphorylation cascade (8).
Streptococcus mutans, a member of the human indigenous oral microbiota, is considered a major etiological agent of dental caries (9, 10). Recent studies have shown that certain strains of S. mutans can be highly invasive and could play a significant role in the development of certain systemic diseases, such as cardiovascular diseases (11). In S. mutans, the intraspecies quorum-sensing system mediated by the CSP-ComDE pathway consists of the CSP pheromone, a membrane-bound histidine kinase sensor (ComD), and its cognate cytoplasmic response regulator (ComE) (12). When CSP molecules accumulate above a threshold level required for detection, extracellular CSP pheromones bind to the ComD sensor, initiating the phosphorelay cascade that leads to transcriptional activation of the alternative sigma factor SigX (also known as ComX), of the sigma-70 family. SigX assembles with core polymerase to recognize a set of noncanonical promoter sites possessing a CIN box consensus sequence (13, 14). SigX is responsible for induction of the CSP-dependent com regulon and is also considered a key factor in general stress responses in Gram-positive bacteria (15). In Streptococcus pneumoniae, a subset of CSP-responsive genes includes a few that encode proteins involved in a competence-programmed mechanism of predation of noncompetent cells. This mechanism is called microbial fratricide, or allolysis, and is thought to be used by competent cells to acquire DNA from their noncompetent siblings (16, 17). In S. mutans, the CSP-ComDE quorum-sensing system regulates several phenotypes, including genetic competence (18, 19), biofilm formation (12), acid tolerance (20), bacteriocin production (21–23), and formation of stress-induced multidrug-tolerant persister cells (24).
In the oral cavity, bacteria are constantly exposed to a wide range of environmental stresses (e.g., constant cycles of “famine and feast,” fluctuations in pH, and oxidative stress resulting from hydrogen peroxide present in certain oral care products used for oral hygiene and tooth whitening). S. mutans has evolved several mechanisms allowing survival and persistence of the species in its natural habitat (25, 26). Work done by our group has shown that particular stressful conditions, such as exposure to acid, activate CSP production. We demonstrated that the stress-inducible CSP pheromone (or alarmone) triggered death of a subpopulation in the physiological context of an elevated concentration of CSP (23, 27). Using DNA microarrays, we identified a small antimicrobial peptide (CipB, mutacin V) as a major regulatory factor involved in CSP-induced cell death. We showed that death occurs in a subpopulation of S. mutans culture through the intracellular accumulation of CipB and is prevented by the action of the immunity protein CipI, which resides in a separate unlinked genetic locus to allow its differential regulation (23, 28). Recently, we demonstrated that under CSP-induced conditions, the CipB peptide directly and/or indirectly regulated ∼130 genes, among which are the SigX-regulated genes (29). The aim of the present study was to continue our investigation of the factor(s) responsible for the death of S. mutans cells through its stress-inducible CSP pheromone and CSP-regulated CipB peptide. Our results suggest that in S. mutans, the intraspecies quorum-sensing CSP-ComDE pathway overlaps the bacterial stress response to induce autolysis via a conserved streptococcal autolysin. We also provide evidence that the altruistic behavior of lytic cells can be beneficial to the population as a whole.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. S. mutans strains were grown in Todd-Hewitt medium supplemented with 0.3% yeast extract (THYE) in the absence or presence of 2 μM synthetic CSP (sCSP; Advanced Protein Technology Centre, Hospital for Sick Children, Toronto, Ontario, Canada) and incubated statically at 37°C in air with 5% CO2. Deletion mutants were constructed in the wild-type (WT) S. mutans strain UA159 by PCR ligation mutagenesis. Escherichia coli strains were cultivated aerobically in Luria-Bertani (LB) medium at 37°C. When needed, antibiotics were added as follows: ampicillin (100 μg/ml) or chloramphenicol (20 μg/ml) for E. coli and erythromycin (10 μg/ml) or spectinomycin (1,000 μg/ml) for S. mutans. Growth was monitored using a microbiology workstation (Bioscreen C; Growth Curves USA) unless otherwise specified.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| S. mutans | ||
| UA159 | Wild-type S. mutans reference strain | Lab stock |
| ΔsigX mutant | In-frame SMU.1997 deletion mutant derived from UA159; Emr or Spr | 23 |
| ΔlytFSm mutant | In-frame SMU.836 deletion mutant derived from UA159; Emr or Spr | This study |
| Δ574 mutant | In-frame SMU.574 deletion mutant derived from UA159; Emr | This study |
| Δ609 mutant | In-frame SMU.609 deletion mutant derived from UA159; Emr | This study |
| Δ689 mutant | In-frame SMU.689 deletion mutant derived from UA159; Emr | This study |
| Δ704 mutant | In-frame SMU.704 deletion mutant derived from UA159; Emr | This study |
| Δ772 mutant | In-frame SMU.772 deletion mutant derived from UA159; Emr | 29 |
| Δ1700 mutant | In-frame SMU.1700 deletion mutant derived from UA159; Emr | This study |
| UA159(pIB187) | UA159 harboring pIB187; Emr | 31 |
| ΔsigX(pIB187) mutant | ΔsigX mutant harboring pIB187; Spr Emr | This study |
| ΔlytFSm(pIB187) mutant | ΔlytFSm mutant harboring pIB187; Spr Emr | This study |
| UA159(pDDK5) | UA159 harboring pDDK5; Emr | This study |
| UA159(pDDK6) | UA159 harboring pDDK6; Emr | This study |
| UA159(pDDK7) | UA159 harboring pDDK7; Emr | This study |
| ΔsigX(pDDK5) mutant | ΔsigX mutant harboring pDDK5; Spr Emr | This study |
| ΔsigX(pDDK6) mutant | ΔsigX mutant harboring pDDK6; Spr Emr | This study |
| ΔlytFSm(pDDK8) mutant | ΔlytFSm mutant harboring pDDK8; Spr Emr | This study |
| E. coli | ||
| DH10B | Host strain for cloning and plasmid production | Lab stock |
| BL21(DE3) | Host strain for pET-15b LIC TEV expression | Novagen |
| BL21(DE3)(pLysS) | BL21(DE3) harboring pLysS; Cmr | Novagen |
| BL21(DE3)(pLysS)(pDDK1) | BL21(DE3)(pLysS) harboring pDDK1; Apr Cmr | This study |
| BL21(DE3)(pDDK2) | BL21(DE3) harboring pDDK2; Apr | This study |
| BL21(DE3)(pDDK3) | BL21(DE3) harboring pDDK3; Apr | This study |
| BL21(DE3)(pDDK4) | BL21(DE3) harboring pDDK4; Apr | This study |
| Plasmids | ||
| pET-15b LIC TEV | Expression vector derived from pET-15b; Apr | A. Savchenkob |
| pIB187 | Shuttle plasmid containing the gusA gene encoding the ß-glucuronidase enzyme under the control of P23; Emr | 30 |
| pDDK1 | lytFSm cloned into pET-15b LIC TEV; Apr | This study |
| pDDK2 | GBS-Bsp4 and CHAP domain of LytFSm cloned into pET-15b LIC TEV; Apr | This study |
| pDDK3 | CHAP domain of LytFSm cloned into pET-15b LIC TEV; Apr | This study |
| pDDK4 | LytFSm without its CHAP domain cloned into pET-15b LIC TEV; Apr | This study |
| pDDK5 | Promoterless gusA pIB187 vector; Emr | This study |
| pDDK6 | Promoter of lytFSm transcriptionally fused to gusA into pDDK5; Emr | This study |
| pDDK7 | Promoter of sigX transcriptionally fused to gusA into pDDK5; Emr | This study |
| pDDK8 | lytFSm cloned under the control of its own promoter into pDDK5; Emr | This study |
Apr, ampicillin resistance; Emr, erythromycin resistance; Cmr, chloramphenicol resistance; Spr, spectinomycin resistance.
A. Savchenko (Structural Proteomics, Toronto, Canada).
Expression of recombinant proteins.
Recombinant plasmids expressing LytFSm (SMU.836) and its truncated derivatives (Fig. 1) were constructed by PCR cloning of the relevant DNA fragments into the ligation-independent vector pET-15b LIC TEV under the control of the IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter. The full-length (FL) coding region of lytFSm was amplified using UA159 genomic DNA as the template and the primers CMT-567 (5′-GGAGATATACATATGAGAAAACTTAAAGTGGCAC-3′) and CMT-568 (5′-GAAGTACAGGTTCTCGTCAGGATAAATATAAG-3′). To construct the truncated His6-tagged LytFSm derivatives representing group B streptococcus (GBS) Bsp4 with the CHAP (cysteine- and histidine-dependent aminohydrolase/peptidase) domain (B4C), the CHAP domain only (C), and the protein with the CHAP domain deleted (ØC), primers CMT-569 (5′-GGAGATATACATATGATTAACTATGAAGATCCTCAG-3′) and CMT-568, primers CMT-570 (5′-GGAGATATACATATGGTCCATTATCAACCTTCTG-3′) and CMT-568, and primers CMT-567 and CMT-571 (5′-GAAGTACAGGTTCTCGACAATTTGATTGAGCTGC-3′) were used, respectively. The plasmids designated pDDK1 (FL), pDDK2 (B4C), pDDK3 (C), and pDDK4 (ØC) were sequenced on both strands for confirmation. The recombinant plasmids were then transferred into electrocompetent BL21(DE3)(pLysS) (for the pDDK1 construct) or BL21(DE3) (for pDDK2, pDDK3, and pDDK4 constructs) cells to express the His6-tagged recombinant proteins. E. coli cells were incubated aerobically at 37°C in LB medium containing the appropriate antibiotic(s). When the optical density at 600 nm (OD600) reached ∼0.3, the cultures were transferred to 30°C and incubated until an OD600 of ∼0.5 was reached. IPTG (1 mM) was then added to induce expression of the recombinant proteins. Protein expression was verified by SDS-PAGE using the Laemmli buffer system. The protein bands were visualized by staining with Coomassie brilliant blue. In addition, the expression of recombinant proteins was confirmed with anti-His antibodies.
Fig 1.
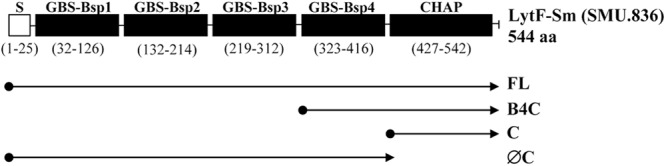
Schematic representation of LytFSm protein and its truncated derivatives. The numbers indicate the positions of amino acids. S, signal peptide; GBS Bsp, group B streptococcus Bsp-like domain; CHAP, cysteine- and histidine-dependent aminohydrolase/peptidase domain. Four constructs were expressed in E. coli: FL (full-length gene), B4C (GBS Bsp4 and CHAP domain), C (CHAP domain only), and ØC (CHAP domain deleted). Protein domains were identified with the Pfam database.
Promoter fusion analysis.
Reporter strains were constructed by cloning the putative promoter region of lytFSm and sigX genes upstream of a promoterless gusA gene into pDDK5 plasmid generated by deleting the P23 promoter from the pIB187 vector. Overnight cultures of the reporter strains were diluted (1:100) into fresh THYE-erythromycin broth in the absence and presence of 2 μM sCSP and incubated statically at 37°C. Cells were harvested in early log, mid-log, early stationary, and late stationary phases. Cells were then lysed by sonication, and intracellular β-glucuronidase (GUS) activity was determined. Briefly, intracellular fractions were combined in equal parts with GUS buffer (100 mM Na2HPO4 [pH 7.0], 20 mM β-mercaptoethanol, 2 mM EDTA, 0.2% Triton X-100, 1 mM para-nitrophenyl-β-d-glucuronide [PNPG] substrate; Sigma). The reaction mixture was incubated at 37°C for 120 min, and the reaction was stopped by the addition of 3.5 mM Na2CO3. The absorbance at 420 nm (A420) was measured after 120 min of color development. The GUS activity was expressed as (1,000 × A420)/(time [min] × OD600) in Miller units (MU). All assays were performed in triplicate from three independent cultures.
For the environmental stress assays, overnight cultures of reporter strain UA159(pDDK6) were diluted (1:100) into fresh THYE-erythromycin medium and exposed to the following stresses: acid shock (THYE broth acidified to pH 5.5 by addition of HCl), amino acid starvation (100 μg/ml of serine hydroxamate), oxidative stress (0.5 mM H2O2), protein synthesis-inhibiting antibiotic (50 μg/ml of spectinomycin), and heat shock (50°C). Cells were then lysed by sonication, and intracellular GUS activity was determined as described above. All assays were performed in triplicate from at least three independent cultures.
Quantification of β-glucuronidase released.
Cell lysis of S. mutans strains was assessed by harvesting the supernatants of cultures expressing the gusA reporter gene cloned into pIB187 vector as described previously (31). Briefly, supernatants were mixed in equal parts with GUS buffer, and the reaction mixture was incubated at 37°C for 120 min. The reaction was stopped by the addition of 3.5 mM Na2CO3, and the GUS activity was calculated as described above. All assays were performed in triplicate from three independent cultures.
Triton X-100-induced autolysis assay.
Overnight cultures of S. mutans strains were diluted (1:100) into fresh THYE broth in the absence and presence of 2 μM sCSP and incubated at 37°C until the mid-log phase of growth was reached. E. coli cells harboring the different pET-15b constructs were cultivated in LB medium containing the appropriate antibiotic(s) and induced with IPTG (1 mM) for 2 h for expression of the recombinant proteins. S. mutans and E. coli cells were then harvested by centrifugation, washed once in ice-cold sterile water, and suspended in 50 mM glycine buffer (pH 8.0) containing 0.1% Triton X-100. Cells were incubated at 37°C, and lysis was assessed by measuring the OD600 of the bacterial suspension. Results were normalized to the OD600 at time zero (OD600 of 0.5). All assays were performed in triplicate from three independent cultures.
Zymogram analysis.
Zymography was used to detect the peptidoglycan hydrolase activity of LytFSm. Cell fractions from S. mutans WT and its LytFSm-deficient mutant were extracted from the cell pellets of cultures grown at 37°C for 5 h in THYE broth in the absence and presence of 2 μM sCSP. Whole-cell extracts were prepared by dissolving the cell pellets in SDS sample buffer (0.25 M Tris [pH 6.8], 2% SDS, and 10% glycerol) and heated at 95°C for 10 min prior to electrophoresis. To prepare the substrates incorporated into the gel, S. mutans cells were cultivated in the absence and presence of 2 μM sCSP at 37°C in THYE broth until an OD600 of 0.2 was reached. Cells were immediately harvested by centrifugation and washed in ice-cold Tris-NaCl buffer (20 mM Tris-HCl [pH 7.4], 100 mM NaCl). Finally, the cell pellet was resuspended in 1.5 M Tris-HCl (pH 8.8). The zymogram was performed according to Berg et al. (32) using the Laemmli buffer system, with gels containing 10% polyacrylamide in the separating gel and 4% polyacrylamide in the stacking gel.
Scanning electron microscopy.
Scanning electron microscopy (SEM) analysis was performed using cells of S. mutans WT and its LytFSm-deficient mutant cultivated statically in THYE broth in the absence and presence of 2 μM sCSP. Overnight cultures were applied to poly-l-lysine-coated small glass discs and incubated at room temperature for 30 min. Cells were then fixed in 3.7% formaldehyde, dehydrated through ethanol rinses, critical-point dried with liquid CO2, mounted, and sputter coated with gold. Samples were examined using a scanning electron microscope (model S-2500; Hitachi Instruments, San Jose, CA).
Long-term survival.
The ability of S. mutans WT strain and its LytFSm-deficient mutant to survive for several days was tested using a long-term survival assay. Overnight cultures were diluted (1:100) with fresh THYE medium in the absence and presence of 2 μM sCSP and incubated statically at 37°C in air with 5% CO2. Samples were first withdrawn when stationary phase was reached (day 0), serially diluted in sterile phosphate-buffered saline (PBS), and plated on THYE agar plates. Aliquots were then withdrawn daily, serially diluted, and spot plated. Colonies were counted after 48 h of incubation. Results were normalized to the CFU value measured on day 0. All assays were performed in triplicate from three independent cultures.
RESULTS
The sigma factor SigX governs the CSP-induced cell death pathway in S. mutans.
We previously demonstrated that the stress-inducible CSP pheromone was capable of causing cell death of a fraction of the S. mutans population at high concentrations (23). Microarray-based expression profiling showed that the CSP pheromone alters the expression (± ≥2-fold) of 277 genes, among which approximately 90% are directly or indirectly controlled by the sigma factor SigX (23). These results prompted us to hypothesize that the CSP-induced growth defects observed were governed by the sigma factor SigX. We thus tested a sigX deletion mutant for its ability to survive under high concentrations of synthetic CSP (sCSP). Our results showed that although there was a SigX-dependent decrease in the growth rate during early log phase, a high concentration of sCSP does not appear to dramatically affect the growth yield of the ΔsigX mutant (Fig. 2). These results strongly support a role for SigX in the regulation of the CSP-induced cell death pathway.
Fig 2.
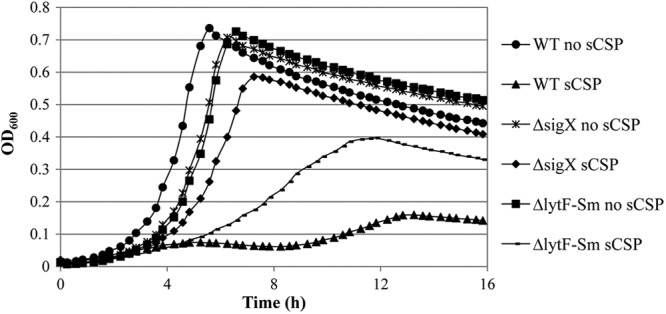
Growth of S. mutans in the absence and presence of the CSP pheromone. Overnight cultures of wild-type (WT) S. mutans UA159, ΔsigX mutant, and ΔlytFSm mutant (SMU.836 deletion mutant) were diluted (1:100) in fresh THYE broth in the absence (no sCSP) and presence (sCSP) of 2 μM sCSP. Growth was monitored using a microbiology workstation. Data are representative of three independent experiments.
In order to investigate whether the growth-inhibitory effect of CSP pheromone leads to lysis of S. mutans cells, we used β-glucuronidase (GUS) as the marker enzyme; the release of cytoplasmic GUS into the extracellular medium was quantified as a measure of cell lysis. S. mutans strains were thus transformed with pIB187 containing a GUS reporter gene under the control of the P23 lactococcal promoter for constitutive expression. Our results showed that mid-log-phase and stationary-phase cultures of WT S. mutans cultivated in the presence of a high concentration of sCSP released GUS (Fig. 3). As expected, GUS activity was not significantly altered in the supernatants of ΔsigX mutant cultures exposed to the same level of sCSP, confirming the role of the sigma factor SigX in regulation of the CSP-induced cell death pathway. Based on these results, we conclude that high concentrations of the CSP pheromone are bactericidal and cause the lysis of S. mutans through a SigX-dependent mechanism.
Fig 3.
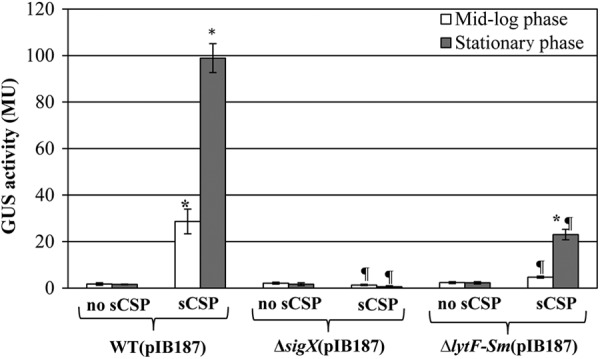
Measure of cytoplasmic GUS enzyme released into the extracellular medium by wild-type (WT) S. mutans UA159, ΔsigX mutant, and ΔlytFSm mutant transformed with plasmid pIB187 containing a GUS reporter gene under the control of a constitutive promoter. The strains were grown in the absence (no sCSP) and presence (sCSP) of 2 μM sCSP. Enzyme activity was measured after 5 h (mid-log phase) and 24 h (stationary phase) of growth in the supernatant of the same number of cells. GUS activity is expressed in Miller units (MU). All assays were performed in triplicate in three independent experiments. Statistical significance was determined by analysis of variance (ANOVA) (*, sCSP versus no sCSP; ¶, mutant plus sCSP versus WT plus sCSP).
The conserved streptococcal protein LytF is a major actor involved in CSP-induced cell death in S. mutans.
Since the process of CSP-induced death leads to cell lysis, we searched the genome of S. mutans UA159 reference strain for lysis effectors. We found two functional murein hydrolases, SMU.609 (33) and AtlA (34, 35), two putative membrane proteins similar to bacteriophages holin proteins, LrgB and CidB (36), and six additional putative autolysins (SMU.22, SMU.76, SMU.704, SMU.707, SMU.772, and SMU.836) (Table 2). In order to determine if one or more of these proteins were responsible for the CSP-induced cell lysis phenotype, S. mutans isogenic mutants defective in SMU.574 (LrgB), SMU.609, SMU.689 (AtlA), SMU.704, SMU.772, SMU.836, and SMU.1700 (CidB) were generated, and their growth kinetics were analyzed in the presence of sCSP. Importantly, inactivation of SMU.836 drastically attenuated the response to sCSP in S. mutans (Fig. 2). SMU.836 encodes a conserved (≥50% amino acid similarity) LytF-like murein hydrolase that is found in the genomes of several streptococcal species, such as S. sanguinis, S. gordonii, S. anginosus, and S. parasanguinis (32, 37, 38). To confirm the role of SMU.836 (referred to here as LytFSm) in CSP-induced cell lysis, we quantified the release of cytoplasmic GUS into the supernatant. Our results showed that GUS activity was decreased in the LytFSm-deficient mutant cultures incubated in the presence of sCSP, the reduction being approximately 5-fold at mid-log and stationary growth phases, corresponding to less than ∼16% and ∼23% of the wild-type values at 5 and 24 h, respectively (Fig. 3).
Table 2.
S. mutans genes encoding putative and known cell wall hydrolases
| NCBI locus | Description | Fold change in the presence of sCSPa | Cin boxb | Role in CSP-induced cell lysisc |
|---|---|---|---|---|
| SMU.22 | Glucan-binding protein; CHAP domain | No change | No | NT |
| SMU.76 | Putative N-acetyl-muramidase | No change | No | NT |
| SMU.574 | Effector of murein hydrolase; LrgB | No change | No | − |
| SMU.609 | N-Acetyl-muramidase | No change | No | − |
| SMU.689 | Autolysin AtlA | No change | No | − |
| SMU.704 | Autolysin | No change | No | − |
| SMU.707 | Putative endolysin | No change | No | NT |
| SMU.772 | Putative glucan binding protein | +10.00 | Yes | − |
| SMU.836 | LytF-like murein hydrolase | +18.34 | Yes | + |
| SMU.1700 | Putative murein hydrolase; CidB | No change | No | − |
We used scanning electron microscopy to investigate whether there were noticeable differences between WT and ΔlytFSm mutant cells grown in the absence and presence of sCSP. When cells were cultivated in the absence of sCSP, no differences between the WT and ΔlytFSm mutant samples were observed (Fig. 4A). For WT cells grown in the presence of sCSP, however, an amorphous mass of cell debris arising from lysis of cells was detected, as well as some ruptured cells. In contrast, almost no lysis of the cells was observed in samples from ΔlytFSm mutant cells incubated with sCSP, and the majority of the cells looked healthy (Fig. 4A). Interestingly, we also observed that some sCSP-induced WT cells, but not LytFSm-deficient cells, became enlarged, or swollen, and some were very much elongated (Fig. 4B), suggesting a possible role for LytFSm protein in the disruption of cellular integrity in the presence of CSP. Altogether, these results demonstrated that lytFSm, encoding a LytF-like murein hydrolase, is a major effector during CSP-induced cell lysis in S. mutans.
Fig 4.
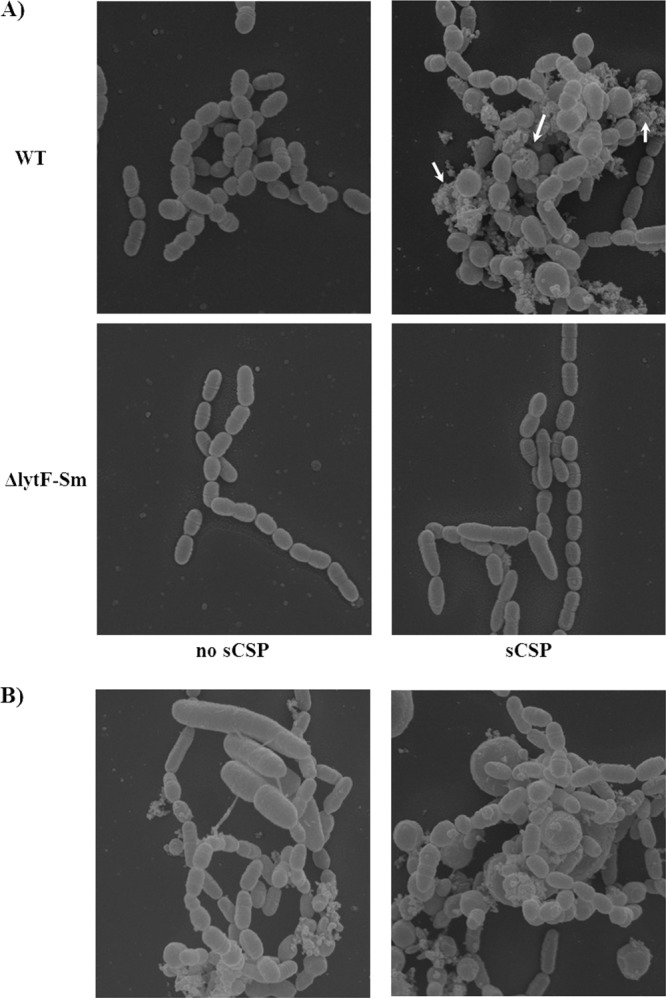
Effect of the CSP pheromone on S. mutans cultures. (A) S. mutans UA159 (WT) and ΔlytFSm mutant were cultivated in the absence (no sCSP) and presence (sCSP) of 2 μM sCSP. There was formation of an amorphous mass of cell debris (arrows) in WT cultures following incubation in the presence of sCSP arising from lysis of cells. (B) Some WT cells grown in the presence of sCSP became enlarged and very elongated. Magnification, ×15,000. SEM was performed three times with duplicate samples on separate days. Similar results were obtained in all experiments.
LytFSm is a functional cell wall hydrolase.
LytFSm protein possesses four GBS (group B streptococcus) Bsp-like peptidoglycan-binding domains containing conserved aromatic and charged residues for interaction with the cell wall and a C-terminal CHAP (cysteine- and histidine-dependent aminohydrolase/peptidase) domain that functions in peptidoglycan hydrolysis (32, 37) (Fig. 1). In order to determine the potential role of LytFSm in peptidoglycan hydrolysis, we first performed a Triton X-100-induced autolysis assay using bacterial cells harvested at mid-log phase. Following the addition of Triton X-100, WT and ΔlytFSm cultures grown in the absence of sCSP were found to be relatively resistant to Triton X-100-mediated lysis (Fig. 5A). As expected, Triton X-100 stimulated lysis activity of WT cells grown in the presence of sCSP, while this lytic response was completely abolished in the LytFSm-deficient strain. Based on these results, we conclude that LytFSm is a functional cell wall hydrolase involved in the CSP-induced cell lysis process.
Fig 5.
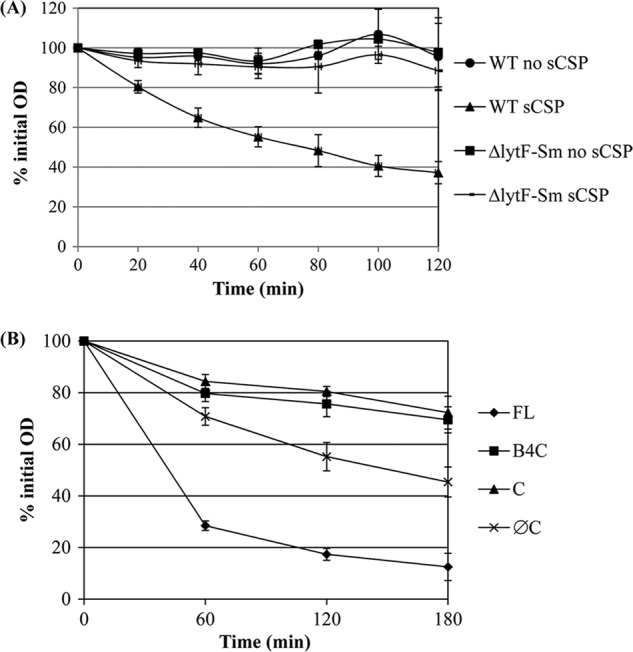
Triton X-100-induced autolysis assay. (A) Autolytic activity of S. mutans UA159 wild-type (WT) strain and ΔlytFSm mutant pregrown in the absence (no sCSP) and presence of 2 μM sCSP (sCSP). (B) Autolytic activity of E. coli cells expressing full-length LytFSm (FL), GBS Bsp4 plus the CHAP domain (B4C), the CHAP domain only (C), and LytFSm without its CHAP domain (ØC). All assays were performed in triplicate in three independent experiments.
In order to identify the domains required for the lytic activity, a series of truncated derivatives of LytFSm were constructed (Fig. 1). The effects of full-length LytFSm and its derivatives on cell lysis were then analyzed by transferring each of the constructs into E. coli, followed by induction with IPTG. As shown in Fig. 6, only the full-length LytFSm protein caused lysis of E. coli cells. No lysis was observed in cells expressing LytFSm with its CHAP domain deleted, confirming the importance of this domain for lytic activity. Interestingly, the C-terminal CHAP domain alone was not sufficient to induce cell damage. Moreover, addition of the fourth GBS Bsp domain did not restore the activity, suggesting that the CHAP domain requires more than one GBS Bsp domain for specific recognition of the peptidoglycan. We also evaluated the effect of autolysis by Triton X-100 on E. coli cells harvested 2 h after IPTG treatment to induce expression of LytFSm recombinant proteins. As shown in Fig. 5B, cultures expressing the LytFSm full-length protein induced rapid autolytic activity, confirming the role of LytFSm in peptidoglycan hydrolysis.
Fig 6.
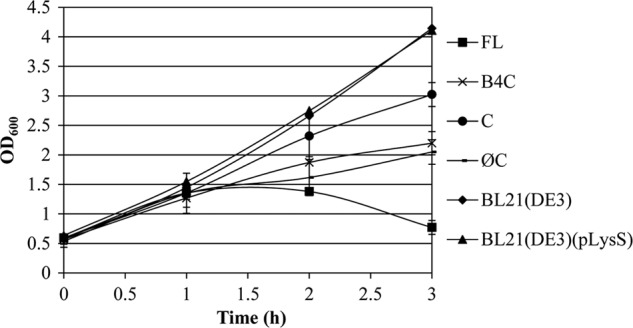
Induction of recombinant plasmids expressing LytFSm protein and its derivatives. The growth characteristics of E. coli cells expressing full-length (FL) LytFSm, GBS Bsp4 plus the CHAP domain (B4C), the CHAP domain only (C), and LytFSm without its CHAP domain (ØC) were analyzed by measuring the absorbance (OD600) after every 30 min following IPTG induction. Control experiments include growth of BL21(DE3) and BL21(DE3)(pLysS) in the presence of IPTG. All assays were performed in triplicate from three independent experiments.
LytFSm is active against S. mutans cells able to undergo CSP-induced lysis.
The murein hydrolase activity of LytFSm was also determined by zymographic analyses using different strains of S. mutans as substrates. We first performed a zymogram analysis of total protein extracts prepared from S. mutans WT strain cultivated in the presence of sCSP. Proteins in the extracts were separated by SDS-PAGE on a gel containing heat-killed cells from WT S. mutans cultivated in THYE broth in the absence of sCSP. Although two bands corresponding to the processed and unprocessed forms of the major autolysin AtlA (34) were clearly visible, no clear zones associated with LytFSm (molecular mass, ∼60 kDa) were detected (Fig. 7A). This finding raised the question of whether LytFSm targets only a CSP-responsive cell population. To answer this, we first compared LytFSm activity using sCSP-treated WT cells incorporated into the SDS-PAGE gel (Fig. 7B) and activity using nontreated WT cells incorporated into the SDS-PAGE gel (Fig. 7A). In this case, an ∼60-kDa lytic band corresponding to LytFSm was detected (Fig. 7B). We also tested the ΔlytFSm mutant strain cultivated in the presence of sCSP to ascertain that the hydrolysis band was due to the action of the LytFSm enzyme. As expected, no band at ∼60 kDa was detected (Fig. 7C). In addition, we performed zymography analysis using isogenic ΔsigX and ΔlytFSm mutant strains treated with sCSP. Interestingly, the lytic zone was absent when these CSP-responsive cells were used as substrates (data not shown). Table 3 summarizes the results obtained by zymographic analysis. Together, our results suggest that LytFSm is a functional murein hydrolase directed against S. mutans cells able to undergo CSP-induced lysis.
Fig 7.
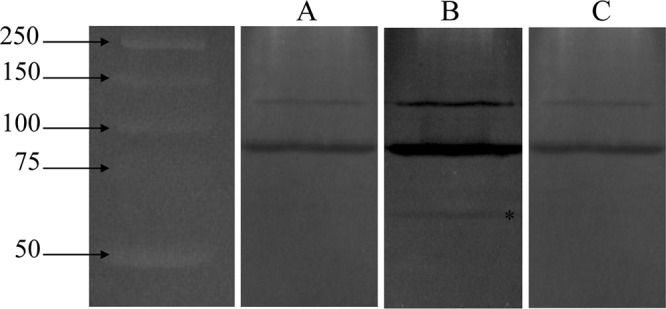
Zymogram activity gel. Heat-killed cells of WT S. mutans cultivated in the absence (A) and presence (B and C) of 2 μM sCSP were used as the substrate and were incorporated into a 10% SDS-PAGE gel (see Materials and Methods for details). Whole-cell extracts of UA159 cultivated in the absence of sCSP (A), UA159 cultivated in the presence of sCSP (B), and the ΔlytFSm mutant cultivated in the presence of sCSP (C) were used. The asterisk indicates the band associated with LytFSm. The two major autolysin bands observed with each substrate correspond mostly to the unprocessed and processed forms of AtlA. Sizes (in kilodaltons) of Precision Plus protein standards (Bio-Rad) are on the left.
Table 3.
Identification of LytFSm murein hydrolase by zymography
| Substrateb | Presence of clearing zone in whole-cell extracts ofa: |
||
|---|---|---|---|
| UA159 WT, no sCSP | UA159 WT + sCSP | ΔlytFSm mutant + sCSP | |
| UA159 WT, no sCSP | − | − | − |
| UA159 WT + sCSP | − | + | − |
| ΔlytFSm, no sCSP | − | − | − |
| ΔlytFSm + sCSP | − | − | − |
| ΔsigX, no sCSP | − | − | − |
| ΔsigX + sCSP | − | − | − |
− and +, absence and presence of a clearing zone caused by the action of LytFSm.
Strains listed were sources of cells used in SDS-PAGE gels as substrates.
LytFSm autolysin participates in self-inflicted lysis.
In S. pneumoniae, the secreted peptidoglycan hydrolase CbpD produced by competent cells lyses noncompetent siblings in order to release DNA that can be taken up by CbpD-producing cells. This competence-induced cell lysis phenotype is termed fratricide (16, 17). Recently, Berg et al. (32) demonstrated that the genomes of nearly all streptococcal species lacking the CbpD fratricin encode the unrelated competence-regulated murein hydrolase LytF, which could have the same function. In order to test this hypothesis, we performed cocultivation experiments to compare the ability of LytFSm-producing cells (attacker cells) to mediate lysis of LytFSm-deficient cells (target cells). We constructed a ΔlytFSm mutant containing the pIB187 plasmid constitutively expressing cytoplasmic GUS (release of GUS was quantified as a measure of cell lysis) and mixed the ΔlytFSm(pIB187) mutant with WT strain containing the promoterless pDDK5 plasmid at a one-to-one ratio in THYE broth. The cocultures were cultivated in the presence of sCSP to induce the production of LytFSm by WT cells before the release of cytoplasmic GUS by ΔlytFSm(pIB187) mutant cells into the supernatant was quantified at mid-log and stationary growth phases. Our results showed that GUS activity was not increased in cocultures of the ΔlytFSm(pIB187) mutant and WT(pDDK5) compared with the monocultures of ΔlytFSm(pIB187) (Fig. 8). We then speculated that cell-cell contact might be required for the delivery of LytFSm enzyme to the target cells. We cocultured sCSP-induced WT(pDDK5) cells with ΔlytFSm(pIB187) mutant cells to provide cell contact but did not observed any functional complementation of the mutant (data not shown). The fact that cocultivation experiments failed to elicit the lysis of S. mutans target cells suggested that CSP-induced lysis could not be triggered by LytFSm enzyme produced by neighboring attacker cells. Altogether, these results strongly indicate that LytFSm is most probably not secreted into the extracellular environment or tethered to the cell wall after export to kill neighboring cells. LytFSm autolysin most likely causes cell lysis via intracellular (self) action.
Fig 8.
Measurement of GUS released into the extracellular medium by S. mutans strains transformed with plasmid pIB187 containing a GUS reporter gene under the control of a constitutive promoter. For the coculture experiments, WT cells were transformed with a promoterless GUS construct (pDDK5). The strains were grown in the presence of 2 μM sCSP, and the enzyme activity (expressed in Miller units [MU]) was measured after 5 h (mid-log phase) and 24 h (stationary phase) of growth from the supernatant of the same number of cells. The units of GUS activity were normalized to CFU/ml. All assays were performed in triplicate in three independent experiments.
lytFSm is SigX dependent and stress inducible.
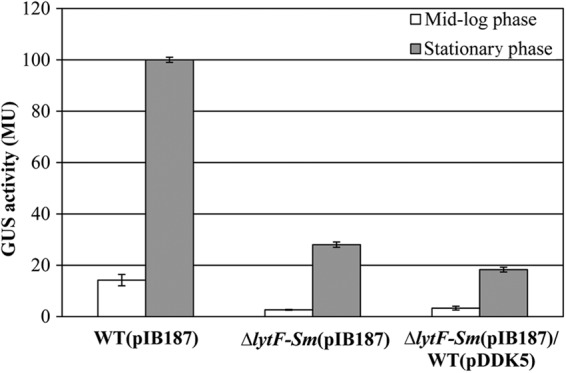
We previously demonstrated by transcriptome analysis that the SMU.836 gene, encoding a LytF-like murein hydrolase, was strongly upregulated under sCSP-induced conditions (Table 2), and this regulation was dependent on the presence of a functional sigX gene (23). Recently, Lemme et al. (18) confirmed these results using a combination of flow cytometry sorting of cells guided by a green fluorescent protein-labeled SigX reporter and DNA microarray analysis. Bioinformatic analysis of the promoter region of lytFSm revealed an obvious CIN box, suggesting a SigX-dependent regulation. Consequently, we were first interested in determining whether lytFSm was regulated similarly to sigX in the presence of sCSP. As shown in Table 4, PlytF and PsigX were active and inducible in the WT strain throughout growth. GUS levels in sCSP-induced WT cultures containing the PlytF and PsigX fusions increased rapidly during exponential growth to reach a maximum during the transition from exponential to stationary phase. In contrast, both promoters displayed minimum activity under uninduced (no sCSP) conditions. PlytF activity was also measured in the sigX mutant background in the absence and presence of sCSP. No PlytF activity was detected in the ΔsigX mutant under any of the tested conditions. These results demonstrated that lytFSm had a strict requirement for the sigma factor SigX.
Table 4.
Activity of PlytF and PsigX in the absence and presence of sCSPa
| Growth phase | GUS activity (MU) of: |
|||||
|---|---|---|---|---|---|---|
| PlytF |
PsigX |
|||||
| WT, no sCSP | WT + sCSP | ΔsigX strain, no sCSP | ΔsigX strain + sCSP | WT, no sCSP | WT + sCSP | |
| Early log | 0.82 ± 0.34 | 51.55 ± 7.77 | NDb | ND | 0.43 ± 0.08 | 14.16 ± 2.42 |
| Mid-log | 0.43 ± 0.04 | 63.35 ± 6.98 | ND | ND | 0.27 ± 0.02 | 15.28 ± 1.48 |
| Early stationary | 0.42 ± 0.16 | 61.36 ± 2.38 | ND | ND | 0.19 ± 0.02 | 13.94 ± 1.97 |
| Late stationary | 0.29 ± 0.05 | 9.50 ± 0.43 | ND | ND | 0.29 ± 0.09 | 5.02 ± 0.77 |
The gusA gene was used as a reporter gene to analyze promoter activity.
ND, not detected.
Previous work done by our group demonstrated that S. mutans integrates its response to specific environmental stresses with its intraspecies CSP-ComDE quorum-sensing system (23, 24). Therefore, we hypothesized that certain stresses could activate the expression of the CSP-inducible lytFSm gene. S. mutans WT cultures were exposed to heat, acidic pH, oxidative stress, the protein synthesis inhibitor spectinomycin, and serine hydroxylate (to mimic amino acid starvation). The PlytF activity in WT strain was significantly increased by heat at 50°C, acidic conditions at pH 5.5, and the presence of hydrogen peroxide (Table 5). PlytF activity was, however, unchanged in the presence of the antimetabolite serine hydroxylate and the protein synthesis inhibitor spectinomycin. These data suggest that S. mutans turns on the expression of its LytFSm autolysin gene in response to specific environmental stresses via its CSP alarmone. We also tested the promoter activity of lytFSm gene using ΔsigX mutant cells exposed to the same stresses (Table 5). No detectable activity could be found in a ΔsigX mutant confirming the regulatory link between lytFSm expression and SigX factor.
Table 5.
Effects of environmental stressors on PlytF activity
| Stress | Change in straina |
|
|---|---|---|
| WT | ΔsigX | |
| Heat shock at 50°C | 2.99 ± 0.08 | ND |
| Acid stress at pH 5.5 | 1.98 ± 0.35 | ND |
| H2O2-induced oxidative stress | 2.61 ± 0.97 | ND |
The gusA gene was used as a reporter gene to analyze the activity of PlytF. Promoter activity is presented as the average fold change ± SD compared with the activity in the absence of the stress. ND, not detected.
Gene inactivation of LytFSm affects long-term survival.
Taking into account that LytFSm is a stress-inducible self-acting murein hydrolase, we hypothesized that CSP-induced autolysis could constitute an altruistic strategy enabling a fraction of the S. mutans population to survive by feeding on the nutrients released from their dead siblings. In order to test if the action of LytFSm autolysin could be advantageous for the bacterial population as a whole, we conducted a long-term survival assay using monocultures of WT strain and its LytFSm-deficient mutant cultivated in the presence of sCSP. After 2 days, there was no difference in survival between WT and mutant strains (Fig. 9). However, after several days (≥5 days) of incubation in stationary phase, the viability of the LytFSm-deficient mutant was significantly lower than that observed for the WT strain, an approximately 5-fold reduction after 8 days. To confirm the role of CSP-induced LytFSm in enhanced survival of S. mutans, we also conducted the experiment using WT and ΔlytFSm cells cultivated in the absence of sCSP. The results obtained showed no significant difference in cell survival between WT and ΔlytFSm mutant (data not shown).
Fig 9.
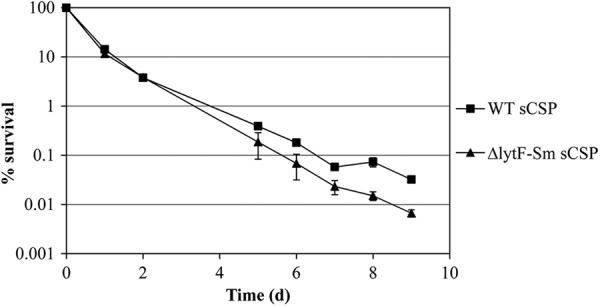
Long-term survival of S. mutans WT and its ΔlytFSm mutant in THYE broth containing 2 μM sCSP. Samples were first taken when stationary phase was reached (day 0), serially diluted in sterile PBS, and plated on THYE agar plates. Aliquots were then taken daily, serially diluted, and spot plated. Colonies were counted after 48 h of incubation. Results were normalized to the CFU value measured on day 0. The data are the averages and standard deviations of results from three independent cultures.
DISCUSSION
Bacterial peptidoglycan hydrolases have a variety of physiological functions in the autolysin-producing cell and in the cell population. They are essential for maintaining cellular integrity during processes which are assumed to require localized and/or transient autolytic activity, such as cell wall turnover, separation of daughter cells following cell division, sporulation and germination, assembly of secretion systems, resuscitation of dormant cells, and microbial fratricide (39). The selective killing of siblings by a subpopulation of bacteria committing fratricide is a highly evolved and complex mechanism. During fratricide, pneumococci that are competent for natural genetic transformation kill and lyse noncompetent siblings that are present in the same environment (17). This mechanism enables cells in the competent state to take up exogenous DNA, which might then be integrated into the bacterial chromosome. The amidase CbpD activates two specific autolysins, LytA and LytC, which in the presence of CbpD and the two-peptide bacteriocin CibAB efficiently degrade the cell walls of noncompetent siblings (40). The lytic enzymes CbpD and LytA and the CibAB bacteriocin are encoded by late competence genes and are consequently an integral part of the CSP-ComDE quorum-sensing system of the bacteria (41, 42).
In S. mutans, the intraspecies CSP-ComDE quorum-sensing system controls important virulence factors (43). We have recently shown that the CSP pheromone is also stress inducible and triggers death in a fraction of the S. mutans population at high concentrations (23). In our previous studies, we clearly showed that CipB (mutacin V) does participate in the CSP-induced cell death pathway (23) but at the transcriptional level, by indirectly regulating SigX-dependent genes (29). In this study, we continued our investigation of the factor(s) responsible for the death of S. mutans through its stress-inducible CSP pheromone. We focused our investigation on SMU.836, a SigX-regulated gene that was found to be negatively regulated in a ΔcipB mutant array analyzed under conditions of sCSP induction (29). We first confirmed that CSP-induced cell death leads to lysis of the cells. We also showed that the CSP pheromone acts via the X-state regulatory cascade (SigX regulon) to trigger a bacterial autolytic pathway through the activation of at least one peptidoglycan hydrolase, LytFSm, encoded by SMU.836. LytFSm, a conserved streptococcal lytic enzyme, is a major factor responsible for approximately 80% of the CSP-induced cell lysis observed. Electron microscopy also revealed important morphological changes (increase in size, loss of regular morphology) accompanying the lysis of WT cells treated with the stress-inducible CSP pheromone. Qi et al. also observed similar morphological changes using S. mutans UA140 treated with sCSP (27). Interestingly, these morphological changes are reminiscent of those associated with the lytic response to treatment with β-lactam antibiotics in E. coli (44). The fact that a mutant deficient in LytFSm autolysin is still slightly susceptible to CSP-induced lysis strongly suggested the presence of an additional minor actor(s). Our search for other effectors among the CSP-induced genes identified by our microarray analysis yielded no other SigX-regulated lytic enzymes other than those tested in this study. We can, however, hypothesize that the CSP-induced cell lysis mechanism involves a bacteriocin system, as is the case for allolysis in pneumococci (40). Bacteriocins generally act by creating channels or pores in the cell membrane that destroy the membrane potential (45, 46). In S. mutans, the biosynthesis of its class II bacteriocins is regulated by the CSP-ComDE quorum-sensing system (47, 48). Although our previous data demonstrated that the bacteriolytic activity of two nonlantibiotics (mutacin IV and VI) did not play a major role in the CSP-induced death process (23), we cannot rule out the possibility that an undescribed bacteriocin system could be involved. In fact, bioinformatic analysis of S. mutans UA159 genome has revealed several additional genes potentially coding for bacteriocin-like peptides under the control of the CSP-ComDE quorum-sensing pathway (29, 47). We are investigating whether these putative antimicrobial peptides contribute to the CSP-induced cell lysis phenomenon. Alternatively, it is possible that yet-uncharacterized CSP-inducible genes could play a role in CSP-induced lysis. Interestingly, the intergenic region IGR662 immediately located downstream of LytFSm gene was also found to be differentially regulated in the presence of CSP by Lemme et al. (18). This intergenic region may represent a regulatory RNA molecule that could play a role in the regulation of CSP-induced lysis. Further experiments will be required to explore this possibility.
A fundamental property of bacteria is their ability to regulate growth when faced with a fluctuating environment. S. mutans must face numerous environmental stresses to survive in the oral cavity. Under specific stresses, S. mutans expresses its CSP pheromone and responds to increased concentrations of CSP by inducing cell lysis in a fraction of the population following activation of LytFSm murein hydrolase. Like apoptosis in plants, animals, and mammalian cells, quorum-sensing-induced cell death may represent a form of programmed cell death mediated by an intracellular program. In this form of bacterial suicide, the bacterial cell death machinery usually remains latent, awaiting activation by a trigger factor. For example, a bacterial cell may commit suicide before damage becomes incompatible with vitality. In this instance, CSP-induced cell death functions to eliminate damaged cells from the population after exposure to detrimental environmental conditions. Autolysis, which appears undesirable to a single-celled organism, may, however, be advantageous to a bacterial population at the multicellular level. In this case, quorum-sensing-induced cell death could represent a kind of altruistic act that provides a way for a bacterial population to survive environmental stresses at the expense of some if its cells. Interestingly, the viability of a LytFSm-deficient mutant in a long-term survival assay was significantly lower than that observed for the wild-type strain, strongly suggesting that surviving cells may benefit from the nutrients released during CSP-induced cell lysis. The lysis of a portion of the bacterial population under unfavorable conditions can thus represent an ecological advantage, where some cells die to benefit the whole population. If this is the case, the activity of autolysins must be highly regulated in order to prevent the eradication of the bacterial population. A number of regulatory mechanisms could be proposed, including physical and chemical alterations to the peptidoglycan substrate. Work done with E. coli has shown that growth at low pH inhibits bacteriolysis, suggesting that changes in the wall's ionic environment may affect autolysis activity (49). Interestingly, preliminary work done in our lab demonstrated that acidic conditions affected the ability of S. mutans cells to trigger a CSP-induced cell death response. Indeed, growth at a pH of ≤5.0 drastically attenuated the response to CSP pheromone, since no significant growth defect relative to the wild-type strain was observed at high levels of sCSP (our unpublished data). It can be speculated that the energized state of the membrane is also important in regulation of CSP-induced autolysis.
The fact that LytFSm murein hydrolase was active against CSP-responsive S. mutans cells able to undergo CSP-induced lysis is also intriguing and strongly supports the notion that this autolytic enzyme is a component of a complex regulatory circuit. It also suggests that LytFSm most likely participates in self-inflicted lysis (cis-acting LytFSm). Interestingly, we monitored the growth kinetics of S. mutans in the presence of increasing concentrations of the recombinant LytFSm proteins. Our results showed that the addition of exogenous autolysin (full-length and truncated forms) did not affect the growth of S. mutans, suggesting that the autolysin could require a specific substrate not accessible from outside the cell to initiate lysis. These results also support a cis-acting mechanism. Alternatively, we can speculate that LytFSm would need to be present in the cell walls of cells attacked by LytFSm-producing cells and could be activated by the action of the enzyme itself (trans-acting LytFSm). These questions will be the subject of future investigations. How a subpopulation of CSP-responsive cells protect themselves from the LytFSm killing factor also remains an open question that warrants further investigation.
Cells monitor their population density by releasing signaling molecules to which they respond. It is now becoming increasingly evident that bacterial quorum-sensing mechanisms can go well beyond the counting of cell numbers. With regard to bacteria that utilize quorum-sensing as part of their pathogenic lifestyle, the results of this study reinforce the idea that the bacterial quorum-sensing system represents an attractive target for new drugs that could disrupt and/or enhance quorum-sensing related phenomena.
ACKNOWLEDGMENTS
We thank Marie-Claude Jobin for technical assistance with the construction of recombinant autolysins and Jiang Wang for the SEM observations. We thank Indranil Biswas and Alexei Savchenko for their generous gifts of plasmid pIB187 and pET-15b LIC TEV, respectively.
This work was supported by a Canadian Institutes of Health Research (CIHR) grant MOP-93555 to C.M.L. C.M.L. is a recipient of a Canada Research Chair.
Footnotes
Published ahead of print 26 October 2012
REFERENCES
- 1. Diggle SP, Crusz S, Cámara M. 2007. Quorum sensing. Curr. Biol. 17: R907– R910 [DOI] [PubMed] [Google Scholar]
- 2. Ng WL, Bassler BL. 2009. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 43: 197– 222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Diggle SP. 2010. Microbial communication and virulence: lessons from evolutionary theory. Microbiology 156: 3503– 3512 [DOI] [PubMed] [Google Scholar]
- 4. Platt TG, Fuqua C. 2010. What's in a name? The semantics of quorum sensing. Trends Microbiol. 18: 383– 387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dunny GM, Leonard BAB. 1997. Cell-cell communication in Gram-positive bacteria. Annu. Rev. Microbiol. 51: 527– 564 [DOI] [PubMed] [Google Scholar]
- 6. Fontaine L, Boutry C, de Frahan MH, Delplace B, Fremaux C, Horvath P, Hols P. 2010. A novel pheromone quorum-sensing system controls the development of natural competence in Streptococcus thermophilus and Streptococcus salivarius. J. Bacteriol. 192: 1444– 1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mashburn-Warren L, Morrison DA, Federle MJ. 2010. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol. Microbiol. 78: 589– 606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horinouchi S, Ueda K, Nakayama J, Ikeda T. 2010. Cell-to-cell communications among microorganisms, p 283–337 In Mander L, Liu HW. (ed), Comprehensive natural products II: chemistry and biology. Elsevier Ltd., New York, NY [Google Scholar]
- 9. Loesche WJ. 1986. Role of Streptococcus mutans in human dental decay. Microbiol. Rev. 50: 353– 380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Takahashi N, Nyvad B. 2008. Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 42: 409– 418 [DOI] [PubMed] [Google Scholar]
- 11. Nakano K, Ooshima T. 2009. Serotype classification of Streptococcus mutans and its detection outside the oral cavity. Future Microbiol. 4: 891– 902 [DOI] [PubMed] [Google Scholar]
- 12. Li YH, Lau PCY, Lee JH, Ellen RP, Cvitkovitch DG. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183: 897– 908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee MS, Morrison DA. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181: 5004– 5016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Oggioni MR, Morrison DA. 2008. Cooperative regulation of competence development in Streptococcus pneumoniae: cell-to-cell signaling via a peptide pheromone and an alternative sigma factor, p 345–362 In Winans SC, Bassler BL. (ed), Chemical communication among bacteria. ASM Press, Washington, DC [Google Scholar]
- 15. Claverys JP, Prudhomme M, Martin B. 2006. Induction of competence regulons as general stress response to stress in Gram-positive bacteria. Annu. Rev. Microbiol. 60: 451– 475 [DOI] [PubMed] [Google Scholar]
- 16. Claverys JP, Havarstein LS. 2007. Cannibalism and fratricide: mechanisms and raisons d'être. Nat. Rev. Microbiol. 5: 219– 229 [DOI] [PubMed] [Google Scholar]
- 17. Guiral S, Mitchell TJ, Martin B, Claverys JP. 2005. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc. Natl. Acad. Sci. U. S. A. 102: 8710– 8715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lemme A, Grobe L, Reck M, Tomasch J, Wagner-Dobler I. 2011. Subpopulation-specific transcriptome analysis of competence-stimulating-peptide-induced Streptococcus mutans. J. Bacteriol. 193: 1863– 1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li YH, Tang N, Aspiras MB, Lau Lee PCJH, Ellen RP, Cvitkovitch DG. 2002. A quorum-sensing signaling system essential for genetic competence in Streptococcus mutans is involved in biofilm formation. J. Bacteriol. 184: 2699– 2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li YH, Lau PCY, Tang N, Svensater G, Ellen RP, Cvitkovitch DG. 2002. Novel two-component regulatory system involved in biofilm formation and acid resistance in Streptococcus mutans. J. Bacteriol. 184: 6333– 6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kreth J, Merritt J, Shi W, Qi F. 2005. Co-ordinated bacteriocin production and competence development: a possible mechanism for taking up DNA from neighbouring species. Mol. Microbiol. 57: 392– 404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kreth J, Merritt J, Zhu L, Shi W, Qi F. 2006. Cell density- and ComE-dependent expression of a group of mutacin and mutacin-like genes in Streptococcus mutans. FEMS Microbiol. Lett. 265: 11– 17 [DOI] [PubMed] [Google Scholar]
- 23. Perry JA, Jones MB, Peterson SN, Cvitkovitch DG, Lévesque CM. 2009. Peptide alarmone signaling triggers an auto-active bacteriocin necessary for genetic competence. Mol. Microbiol. 72: 905– 917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leung V, Lévesque CM. 2012. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J. Bacteriol. 194: 2265– 2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lemos JA, Burne RA. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154: 3247– 3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith EG, Spatafora GA. 2012. Gene regulation in S. mutans: complex control in a complex environment. J. Dent. Res. 91: 133– 141 [DOI] [PubMed] [Google Scholar]
- 27. Qi F, Kreth J, Lévesque CM, Kay O, Mair RW, Shi W, Cvitkovitch DG, Goodman SD. 2005. Peptide pheromone induced cell death of Streptococcus mutans. FEMS Microbiol. Lett. 251: 321– 326 [DOI] [PubMed] [Google Scholar]
- 28. Perry JA, Cvitkovitch DG, Lévesque CM. 2009. Cell death in Streptococcus mutans biofilms: a link between CSP and extracellular DNA. FEMS Microbiol. Lett. 299: 261– 266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dufour D, Cordova M, Cvitkovitch DG, Lévesque CM. 2011. Regulation of the competence pathway as a novel role associated with a streptococcal bacteriocin. J. Bacteriol. 193: 6552– 6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Biswas I, Jha JK, Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154: 2275– 2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Syed MA, Koyanagi S, Sharma E, Jobin MC, Yakunin AF, Lévesque CM. 2011. The chromosomal mazEF locus of Streptococcus mutans encodes a functional type II toxin-antitoxin addiction system. J. Bacteriol. 193: 1122– 1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berg KH, Ohnstad HS, Havarstein LS. 2012. LytF, a novel competence-regulated murein hydrolase in the genus Streptococcus. J. Bacteriol. 194: 627– 635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Catt DM, Gregory RL. 2005. Streptococcus mutans murein hydrolase. J. Bacteriol. 187: 7863– 7865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ahn SJ, Burne RA. 2006. The atlA operon of Streptococcus mutans: role in autolysin maturation and cell surface biogenesis. J. Bacteriol. 188: 6877– 6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shibata Y, Kawada M, Nakano Y, Toyoshima K, Yamashita Y. 2005. Identification and characterization of an autolysin-encoding gene of Streptococcus mutans. Infect. Immun. 73: 3512– 3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ahn SJ, Rice KC, Oleas J, Bayles KW, Burne RA. 2010. The Streptococcus mutans Cid and Lrg systems modulate virulence traits in response to multiple environmental signals. Microbiology 156: 3136– 3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eaton RE, Jacques NA. 2010. Deletion of competence-induced genes overexpressed in biofilms caused transformation deficiencies in Streptococcus mutans. Mol. Oral Microbiol. 25: 406– 417 [DOI] [PubMed] [Google Scholar]
- 38. Okinaga T, Xie Z, Niu G, Qi F, Merritt J. 2010. Examination of the hdrRM regulon yields insight into the competence system of Streptococcus mutans. Mol. Oral Microbiol. 25: 165– 177 [DOI] [PubMed] [Google Scholar]
- 39. Vollmer W, Joris B, Charlier P, Foster S. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32: 259– 286 [DOI] [PubMed] [Google Scholar]
- 40. Eldholm V, Johnsborg O, Haugen K, Ohnstad HS, Havarstein LS. 2009. Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology 155: 2223– 2234 [DOI] [PubMed] [Google Scholar]
- 41. Eldholm V, Johnsborg O, Straume D, Ohnstad HS, Berg KH, Hermoso JA, Havarstein LS. 2010. Pneumococcal CbpD is a murein hydrolase that requires a dual envelope binding specificity to kill target cells during fratricide. Mol. Microbiol. 76: 905– 917 [DOI] [PubMed] [Google Scholar]
- 42. Kausmally L, Johnsborg O, Lunde M, Knutsen E, Havarstein LS. 2005. Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J. Bacteriol. 187: 4338– 4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cvitkovitch DG, Li YH, Ellen RP. 2003. Quorum-sensing and biofilm formation in streptococcal infections. J. Clin. Invest. 112: 1626– 1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. van Heijenoort J. 2011. Peptidoglycan hydrolases of Escherichia coli. Microbiol. Mol. Biol. Rev. 75: 636– 663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Abee T. 1995. Pore-forming bacteriocins of gram-positive bacteria and self-protection mechanisms of producer organisms. FEMS Microbiol. Lett. 129: 1– 10 [DOI] [PubMed] [Google Scholar]
- 46. Hechard Y, Sahl HG. 2002. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 84: 545– 557 [DOI] [PubMed] [Google Scholar]
- 47. Merritt J, Qi F. 2012. The mutacins of Streptococcus mutans: regulation and ecology. Mol. Oral Microbiol. 27: 57– 69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. van der Ploeg JR. 2005. Regulation of bacteriocin production in Streptococcus mutans by the quorum-sensing system required for development of genetic competence. J. Bacteriol. 187: 3980– 3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Goodell EW, Lopez R, Tomasz A. 1976. Suppression of lytic effect of beta lactams on Escherichia coli and other bacteria. Proc. Natl. Acad. Sci. U. S. A. 73: 3293– 3297 [DOI] [PMC free article] [PubMed] [Google Scholar]


