Abstract
Escherichia coli has been used as a platform host for studying the production of free fatty acids (FFA) and other energy-dense compounds useful in biofuel applications. Most of the FFA produced by E. coli are found extracellularly. This finding suggests that a mechanism for transport across the cell envelope exists, yet knowledge of proteins that may be responsible for export remains incomplete. Production of FFA has been shown to cause cell lysis, induce stress responses, and impair basic physiological processes. These phenotypes could potentially be diminished if efflux rates were increased. Here, a total of 15 genes and operons were deleted and screened for their impact on cell viability and titer in FFA-producing E. coli. Deletions of acrAB and rob and, to a lower degree of statistical confidence, emrAB, mdtEF, and mdtABCD reduced multiple measures of viability, while deletion of tolC nearly abolished FFA production. An acrAB emrAB deletion strain exhibited greatly reduced FFA titers approaching the tolC deletion phenotype. Expression of efflux pumps on multicopy plasmids did not improve endogenous FFA production in an acrAB+ strain, but plasmid-based expression of acrAB, mdtEF, and an mdtEF-tolC artificial operon improved the MIC of exogenously added decanoate for an acrAB mutant strain. The findings suggest that AcrAB-TolC is responsible for most of the FFA efflux in E. coli, with residual activity provided by other resistance-nodulation-cell division superfamily-type efflux pumps, including EmrAB-TolC and MdtEF-TolC. While the expression of these proteins on multicopy plasmids did not improve production over the basal level, their identification enables future engineering efforts.
INTRODUCTION
Production of fuels and chemicals in metabolically engineered microbes can induce toxicity and stresses that reduce yields, titers, and productivities (1, 2). Overcoming toxicity is a challenge to developing stable and economically viable production processes. Hydrophobic and lipophilic compounds, including energy-dense biofuels such as n-butanol and hydrocarbons, intercalate in the cytoplasmic membrane (3). Intercalation can alter membrane fluidity, membrane protein function, and aerobic respiration (3–6). To avoid physiological problems caused by these compounds and others, bacteria express transporters that actively cause the efflux of toxic chemicals (2, 7–9). This idea has motivated efforts to engineer cells with enhanced export capabilities (10).
Escherichia coli has been used as a platform host for studying the production of free fatty acids (FFA) (11–20), which can be catalytically converted to alkanes (12, 21), and fatty-acid-derived products such as fatty acid ethyl esters (FAEE) and fatty alcohols (13, 22–24). FFA and FAEE are found predominantly extracellularly, yet no export mechanism has been conclusively demonstrated to date. Despite the apparent ability to excrete FFA, engineered E. coli strains are subject to physiological perturbations resulting in reduced cell viability, reduced membrane integrity, large increases in membrane unsaturated fatty acid content, induction of membrane stress responses, and a loss of proton motive force coupled with increased expression of genes involved in aerobic respiration (25). One strategy to avoid these negative physiological effects is to increase the export of FFA from the cell, but this requires the identification of FFA exporters. In past studies, the percentage of intact and viable cells was shown to greatly decrease in FFA-producing cells as they enter stationary phase, whereas control cells remained fully intact (25). These observations led to the hypothesis that any genes that were essential to FFA export would exacerbate the negative phenotypes when deleted.
In this work, the phenotypes of a set of E. coli deletion mutants were used to identify native E. coli genes involved in FFA export. Selection of candidate genes was based on four criteria (Table 1). First, genes previously observed to be involved in FFA import were targeted, as a dual role in export could also be possible. This encompassed three genes, fadL, tolC, and prc, for which a transposon mutagenesis screening of membrane-bound proteins identified defective or absent growth on oleate (26). Second, genes that encode resistance-nodulation-cell division superfamily (RND)-type multidrug efflux pumps, all of which associate with the TolC outer membrane channel, were targeted on the basis of the ability to confer resistance to sodium dodecyl sulfate (SDS), decanoate, and bile salts (27–29). These included emrAB, acrAB, mdtABCD, acrD, mdtEF, and acrEF. Third, several genes that encode annotated multidrug efflux pumps were identified as having increased expression in fatty-acid-producing strains (25). These include cmr, mdtD, mdtG, mdtK, emrAB, and mdtEF. Fourth, the transcriptional-activator-encoding marA, rob, and soxS genes, many of the members of whose regulons were upregulated in fatty-acid-producing strains, were targeted because of their role in regulating genes that encode drug efflux pumps such as AcrAB (30) and MdtG (31), the TolC outer membrane channel (32), and the outer membrane porin OmpF (33). In our prior study, the deletion of rob had a negative impact on viable cell counts (25). Rob is known to be activated at a high fatty acid concentration (5 mM) and to induce acrAB expression (29, 30). As many genes in the MarA, Rob, and SoxS regulons have unknown functions, it may be possible to identify a new protein potentially involved in fatty acid export by preventing the activation of an entire regulon.
Table 1.
Targeted genes with possible FFA transport roles
| Gene(s) | Rationale for selection | Reference(s) |
|---|---|---|
| tolC | Prior role in alleviating FFA toxicity; outer membrane component for most MFA-type efflux pumps in E. coli; member of Rob regulon | 26, 32, 49–51 |
| acrAB | Member of Rob regulon; confers SDS and decanoate resistance | 28–30 |
| fadL | Necessary role in outer membrane import of long-chain FFA; increased expression in microarray data sets | 26 |
| prc | Identified role in fatty acid import | 26 |
| acrD | Confers SDS resistance | 28 |
| acrEF | Confers SDS resistance | 28 |
| mdtABCD | Confers SDS resistance; increased expression of mdtD in microarray data sets | 25, 28 |
| cmr | Efflux pump; increased expression in both microarray data sets | 25 |
| mdtG | Efflux pump; member of Rob regulon; increased expression in both microarray data sets | 25, 31 |
| mdtK | Efflux pump; decreased expression in one microarray data set and increased expression in another | 25 |
| emrAB | Confers SDS resistance; increased expression in one microarray data sets | 25, 28 |
| mdtEF | Confers SDS resistance; increased expression in one microarray data set | 25, 28 |
| marA | Overlapping regulon with Rob; increased expression in both microarray data sets | 25 |
| soxS | Overlapping regulon with Rob; increased expression in both microarray data sets | 25 |
| rob | Activation by FFA; increased expression in one microarray data set | 25, 30 |
| ompF | Outer membrane protein; indirectly in Rob regulon; strongly decreased expression in both microarray data sets | 25, 33 |
Targeted genes were deleted in a plasmid-free FFA-producing strain (TY05), a strain containing three copies of a codon-optimized acyl-acyl carrier protein thioesterase from Umbellularia californica (BTE) under the control of an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter integrated into chromosomal loci of β-oxidation genes (fadD, fadE, and fadAB) (34). The same gene deletions were also made in a negative-control strain (TY06) containing three copies of BTE with an active-site mutation (BTE-H204A) that renders the protein nonfunctional (35). The deletion strains were screened for decreased viability and increased cell lysis relative to TY05. This screening strategy was necessitated by previous findings that exogenous addition of lauric acid, the major species produced by TY05, elicits lower toxicity than endogenous production (25). Deletions that both decreased viability and increased cell lysis included tolC, acrAB, rob, emrAB, and mdtABCD. These genes were cloned into expression vectors in an effort to increase FFA efflux rates. Cells harboring these plasmids were analyzed for the ability to improve FFA production and cell viability. While all of the expressed drug efflux pump components improved tolerance to SDS in an acrAB deletion strain (i.e., were functionally expressed), none of the drug efflux pumps, when expressed from a medium-copy-number plasmid in a strain with intact chromosomal acrAB, increased the FFA titer, viability, or the MIC of exogenous octanoate or decanoate. However, when chromosomal acrAB was disrupted, the MIC of decanoate was increased by the plasmid-based expression of all of the selected efflux pump genes except mdtABCD. Our findings suggest that FFA export in E. coli is mediated primarily by AcrAB-TolC with additional, but reduced, activity conferred by EmrAB-TolC and possibly the MdtEF-TolC and MdtABC-TolC multidrug efflux systems.
MATERIALS AND METHODS
Chemicals, reagents, enzymes, and oligonucleotide primers.
Chemicals were purchased from Fisher Scientific (Pittsburgh, PA) unless otherwise specified. Cloning reagents were purchased from New England BioLabs (Ipswich, MA), Fermentas (Glen Burnie, MD), Promega (Madison, WI), and Qiagen (Valencia, CA). Oligonucleotides (see Table S1 in the supplemental material) were purchased from Integrated DNA Technologies (Coralville, IA).
Strain construction.
The bacterial strains and plasmids used in this study are listed in Table 2; see also Table S2 in the supplemental material. The primary strains used in this work are E. coli TY05 and TY06 (34). Additional gene deletions were added by P1 phage transduction of Kanr-encoding cassettes from the Keio collection of gene knockout mutants (36) as previously described (12, 37). Deletion of operons (acrAB, mdtEF, emrAB, acrEF, and mdtABCD) was performed by λ Red-mediated recombination of Kanr-encoding cassettes amplified from template plasmid pKD13 (38) by using primers 13 to 22. Recombination of linear cassettes in strain DY330 was performed as described by Thomason et al. (39). Phage P1 lysates were prepared by using a modified liquid procedure (40), with cells grown and infected at 30°C. These lysates were then used to transduce the Kanr-encoding cassettes into strains TY05 and TY06. The Kanr-encoding cassette was removed by FLP recombinase expressed on pCP20 (41). The presence of the desired deletions and BTE or BTE-H204A integrations was confirmed by colony PCR (primers 1 and 2, 9 to 12, and 23 to 50).
Table 2.
Bacterial strains and plasmids used in this studya
| Strain or plasmid | Relevant genotype/propertyb | Source or reference |
|---|---|---|
| Strains | ||
| TY05 | K-12 MG1655 fadD::Ptrc-BTE fadE::Ptrc-BTE fadAB::Ptrc-BTE | 34 |
| TY06 | K-12 MG1655 fadD::Ptrc-BTE-H204A fadE::Ptrc-BTE-H204A fadAB::Ptrc-BTE-H204A | 34 |
| TY05ara | K-12 MG1655 fadD::Ptrc-BTE fadE::Ptrc-BTE fadAB::Ptrc-BTE ΔaraFGH Φ(ΔaraEp PCP18-araE) ΔaraBAD | This work |
| BW25113 | lacIq rrnB3 F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 | 36 |
| JW5249-1 | BW25113 ΔmarA752::kan | 36 |
| JW4359-1 | BW25113 Δrob721::kan | 36 |
| JW4023-5 | BW25113 ΔsoxS756::kan | 36 |
| JW5503-1 | BW25113 ΔtolC732::kan | 36 |
| JW2341-1 | BW25113 ΔfadL752::kan | 36 |
| JW1819-1 | BW25113 Δprc755::kan | 36 |
| JW2454-1 | BW25113 ΔacrD790::kan | 36 |
| JW1040-1 | BW25113 ΔmdtG723::kan | 36 |
| JW1655-1 | BW25113 ΔmdtK740::kan | 36 |
| JW0826-1 | BW25113 Δcmr742::kan | 36 |
| JW0912-1 | BW25113 ΔompF746::kan | 36 |
| DY330 | K-12 W3110 ΔlacU169 gal490 pglΔ8 λ cI857 Δ(cro-bioA) (Tetr) | 39 |
| Plasmids | ||
| pBAD33 | PBAD promoter, pACYC origin, Cmr | 44 |
| pBAD33* | pBAD33 with araC-C280* mutation | 45 |
| pBAD33*-acrAB | pBAD33* carrying acrAB under PBAD control | This work |
| pBAD33*-mdtEF | pBAD33* carrying mdtEF under PBAD control | This work |
| pBAD33*-mdtEF-tolC | pBAD33* carrying mdtEF-tolC artificial operon under PBAD control | This work |
| pBAD33*-emrAB | pBAD33* carrying emrAB under PBAD control | This work |
| pBAD33*-mdtABCD | pBAD33* carrying mdtABCD under PBAD control | This work |
| pKD13 | Template plasmid, R6K gamma origin, Ampr Kanr | 38 |
| pCP20 | Carries yeast FLP recombinase under constitutive promoter control, pSC101 origin, λ cI857+, λ pR Rep(Ts), Ampr Cmr | 41 |
For the full strain and plasmid list, see Table S2 in the supplemental material.
Abbreviations: Tetr, tetracycline resistance; Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Kanr, kanamycin resistance; Ts, temperature sensitive.
Strain TY05ara was constructed by sequential P1 phage transductions using lysates harboring Φ(ΔaraEp kan Pcp8-araE), araFGH::kan, and araBAD::cat loci from strains BW27271, BW27269, and NRD204, respectively (42, 43). These chromosomal modifications allow homogeneous induction with l-arabinose. Antibiotic resistance genes were removed after each transduction using pCP20, and the presence of all of the desired loci was confirmed by colony PCR using the primers listed. TY05ara acrAB::kan was also constructed by P1 phage transduction using a lysate derived from TY05 acrAB::kan. All loci were reconfirmed by colony PCR of the transductant (additional primers 3 to 8).
Plasmid construction.
Plasmid pBAD33* is a modified version of pBAD33 (44) harboring a mutant form of araC with cysteine 280 converted to a premature stop codon (AraC-C280*). This mutation has been previously observed to reduce inhibition of gene expression from the PBAD promoter in the presence of IPTG (45), allowing the use of both inducing agents simultaneously. The plasmid was generated by PCR using primers 51 and 52 with template pBAD33, which introduced the C280* mutation and an XhoI restriction site at the 5′ and 3′ ends. The PCR product was then digested with XhoI and ligated to form plasmid pBAD33*. Selected genes and operons were amplified by PCR from MG1655 genomic DNA with their putative upstream ribosome binding sites and added 5′ and 3′ restriction sites using primers 53 to 62. PCR products were subsequently inserted into pBAD33* between the added restriction sites. Selected operons were also inserted into pBAD33*-tolC to generate artificial operons with tolC.
Cell cultivation.
All strains tested for viability and fatty acid production were grown in an incubator shaker at 37°C and 250 rpm in 250-ml shake flasks with 4× headspace in LB medium supplemented with 0.4% (vol/vol) glycerol. Cultures were induced with 1 mM IPTG at an optical density at 600 nm (OD600) of 0.2 to induce the expression of BTE or BTE-H204A. Strains were grown in biological triplicate from overnight cultures inoculated with independent colonies. For strains harboring transporter expression plasmids, chloramphenicol was added to a concentration of 34 μg/ml and cultures were induced at an OD of 0.2 with 0.2% (wt/vol) l-arabinose in addition to IPTG as described above. These experiments were conducted with culture medium supplemented with 0.5 mM Ca2+ and 0.5 mM Mg2+. It has previously been noted that LB medium is likely divalent cation limited (46) and these species are necessary to stabilize outer membrane lipopolysaccharides (47).
Cell viability measurements from plate counts.
Serial dilutions of cell culture were spread onto LB agar plates as previously described (25) at the times indicated. Individual colonies were counted after overnight incubation at 37°C and one additional night of incubation at room temperature.
SYTOX flow cytometry assays.
Cell permeability was assessed by using SYTOX Green, and green fluorescence histograms were generated as previously described (25). Two distinct populations were evident from the green fluorescence histograms, allowing a logarithmic-scale green fluorescence intensity of 420 to serve as the cutoff between cells counted as intact (≤420) and nonintact (>420) (see Fig. S1 in the supplemental material).
Fatty acid extraction and analysis.
Fatty acids were extracted and methylated from cell cultures (with foam collapsed to obtain accurate titers) as previously described (12, 25). To collapse foam, 200 μl of 1:10-diluted antifoam 204 (Sigma, St. Louis, MO) in ethanol was added to each culture and the culture was heated with gentle swirling in an 85°C water bath for 5 to 10 min. Next, 2.5 ml of cell culture was collected and pentadecanoic and heptadecanoic acid (Fluka, Buchs, Switzerland) internal standards dissolved in ethanol were added, followed by the addition of 0.1 ml of glacial acetic acid and 5 ml of 1:1 (vol/vol) chloroform-methanol. After vigorous vortexing and centrifugation, the upper aqueous layer and cell debris were removed and the chloroform layer was evaporated under a nitrogen stream. Residual water was removed by lyophilization, the dried residue was methylated by the addition of 0.5 ml of 1.25 M HCl in methanol (Fluka), and the reaction was allowed to proceed with overnight incubation at 50°C. The reaction mixtures were quenched by the addition of 5 ml of 100-mg/ml aqueous NaHCO3, and fatty acid methyl esters (FAME) were extracted twice into 0.5 ml hexane. The hexane layers were collected for analysis by gas chromatography-mass spectrometry on a model 7890 Agilent gas chromatograph with an HP-5ms capillary column (30 m by 0.25 mm) and a model 5975 mass spectrometer (Agilent Technologies, Santa Clara, CA) with helium carrier gas. One microliter of sample was injected with a 1:10 split ratio, followed by a second injection with a 1:100 split ratio for BTE-expressing cultures. The oven temperature program was 100°C for 2 min, 150°C for 4 min, and a ramp to 250°C at a rate of 4°C/min. Peak identification was achieved by normalizing peak areas to internal standard concentrations and comparison with calibration curves of an FAME standard (Supelco catalog no. 18918) with added methyl heptadecanoate and methyl pentadecanoate (Fluka). Unsaturated FAME not present in the standard were identified by comparison to the National Institute of Standards and Technology Mass Spectral Database and quantified by assuming an equivalent sensitivity ratio of the unsaturated to saturated species for C10, C12, and C14 FAME as for C16 species, of which both saturated and monounsaturated species were present in the standard. Each fatty acid titer reported is the mean of three biological replicates.
MIC assays.
Agar plates containing various concentrations of SDS were prepared by mixing equal volumes of autoclaved 2× YT agar (16 g/liter Bacto tryptone, 10 g/liter Bacto yeast extract, 5 g/liter sodium chloride, 30 g/liter agar, adjusted to pH 7.0 with NaOH) and various concentrations of SDS (0, 0.1, 0.5, 1, 5, 10, 15, 25, and 50 mg/ml) in sterile water plus final concentrations of 34 μg/ml chloramphenicol and 0.2% (wt/vol) l-arabinose. Overnight 5-ml cultures of TY05 ΔacrAB harboring pBAD33* or transporter genes cloned into pBAD33* were grown in LB medium containing 34 μg/ml chloramphenicol. These cultures were diluted 1:100 in 5 ml LB medium containing 34 μg/ml chloramphenicol and 0.2% (wt/vol) arabinose and grown in a shaker for 4 h at 37°C and 250 rpm. Serial 10-fold dilutions were prepared in phosphate-buffered saline, and 3.0-μl volumes of 10−4, 10−5, and 10−6 dilutions were spotted onto plates containing SDS.
Agar plates containing various concentrations of FFA were prepared by mixing equal volumes of 2× LB agar with sterile water containing 10% (wt/vol) stocks of FFA in ethanol with equivalent volumes of ethanol in all plates and equimolar concentrations of NaOH to adjust the pH to 7. Pure octanoic acid was added directly. The FFA stock was premixed with 5 ml of 10% Brij 35 per 50 ml of the final LB agar mixture prior to neutralization and addition of extra water or 2× LB agar to assist in dispersion. Where applicable, final concentrations of 34 μg/ml chloramphenicol and 0.2% (wt/vol) l-arabinose or only 0.2% (wt/vol) l-arabinose were also added to the plates. Cultures were grown as described above, except that the TY05 and gene deletion strains were grown in unsupplemented LB medium.
RESULTS
Viability analysis of single gene/operon deletion strains.
The targeted genes and operons listed in Table 1 were deleted from E. coli TY05, a plasmid-free, FFA-producing strain, and TY06, a control strain expressing nonfunctional thioesterases. These strains do not require antibiotics for maintenance of thioesterase expression. Therefore, the probability of nonspecific effects resulting from altered antibiotic efflux or altered membrane permeability to antibiotics caused by FFA production was reduced. The viability of these strains was measured at 8 h postinoculation by two methods. First, serial dilutions of culture were plated and numbers of CFU per milliliter were calculated. Second, a flow cytometry assay was performed by staining cells with SYTOX Green nucleic acid dye, which is unable to permeate intact inner membranes (48). While the number of CFU per milliliter is dependent on both the absolute number of cells and the percentage of viable and culturable cells, the flow cytometry assay also provides the total number of cells per milliliter. Therefore, the number of CFU per milliliter was normalized to the number of cells per milliliter obtained by flow cytometry to provide a percentage of viable and culturable cells comparable to the SYTOX Green assay (CFU/event ratio).
The data collected were used to identify gene deletions that decreased viability by one or ideally both of the measures in FFA-producing TY05 but had no impact on viability in non-FFA-producing TY06 (Fig. 1a and b). As expected, TY06 was nearly 100% viable, as determined by the CFU/event ratio. In contrast, TY05 exhibited a ratio of 0.43 ± 0.16, with a few deletions exhibiting statistically significantly (P < 0.05) reduced ratios. These were TY05 ΔacrAB, TY05 Δrob, TY05 ΔmdtABCD, and TY05 ΔemrAB. Notably, the corresponding deletions in TY06 exhibited no statistically significant deviation from the CFU/event ratio of the base strain, TY06, indicating that the observed reductions in viability were specific to the condition of FFA production. Some additional gene deletions in TY05 produced reduced average viabilities compared to TY05 but with 0.15 > P > 0.10. These were TY05 ΔmdtEF and TY05 Δprc. The latter strain had plate counts approximately equivalent to those of TY05, but both TY06 Δprc and TY05 Δprc had a smaller cell size and a higher number of cells per milliliter. Furthermore, it should be noted that TY05 ΔtolC exhibited counts of CFU per milliliter much lower than those of TY05, but also greatly reduced flow cytometry counts and OD600 values at 8 h. No reduction in counts of CFU per milliliter, flow cytometry counts, or OD600 values were observed for TY06 ΔtolC compared to TY06.
Fig 1.
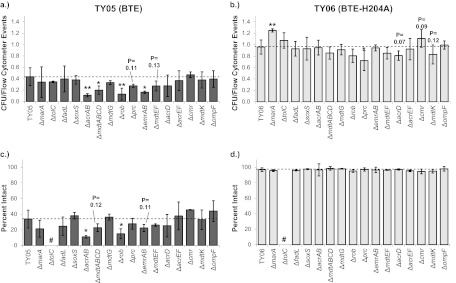
Normalized CFU counts and percentages of intact cells for single gene deletions in strains TY06 and TY05. Panels: a and b, values for strain TY06 (BTE-H204A expressing); c and d, values for strain TY05 (BTE-expressing); a and c, normalized CFU counts (number of CFU per milliliter from plate counts divided by the number of cells per milliliter determined by flow cytometry); b and d, percentages of intact cells measured by SYTOX Green flow cytometry assay. Error bars are standard errors propagated by using cell counts (the number of intact cells with green fluorescence greater than 50, and the number of nonintact cells with green fluorescence greater than 440). *, P < 0.05 (compared to TY06 or TY05); **, P < 0.01 (compared to TY06 or TY05); #, value not calculated (see text).
The results of the SYTOX Green flow cytometry screening were qualitatively similar to the normalized CFU counts (Fig. 1c and d). All TY06 background strains were nearly 100% intact. Conversely, TY05 background strains were less than 50% intact and showed variability that mirrored the data collected in the CFU/event ratio screening. The correlation between normalized CFU counts and percentages of intact cells by SYTOX Green is further supported by scatter plots (see Fig. S2 in the supplemental material), where a linear fit to the points produces R2 values of 0.0016 for TY06 strains and 0.6984 for TY05 strains (excluding TY05 ΔtolC for reasons described below). Compared to TY05, TY05 Δrob and TY05 ΔacrAB exhibited statistically significant (P < 0.05) reductions in the percentage of intact cells. A few additional gene deletions in TY05 produced reduced intact-cell percentages compared to those of TY05 but with 0.15 > P > 0.10, including TY05 ΔmdtABCD and TY05 ΔemrAB. Intact-cell percentages were not calculated for tolC deletion strains because of shifted green fluorescence histograms compared to those of all other strains. Analysis of forward scatter values by flow cytometry indicated a larger average cell size for TY05 ΔtolC than for other TY05 strains, with TY06 ΔtolC exhibiting forward scatter histograms similar to those of other TY06 strains (data not shown). As the outer membrane is permeable to SYTOX Green, a combination of changes in cell size and/or defective efflux of SYTOX Green from the periplasmic space may be responsible for the shifted green fluorescence histograms.
A total of four gene deletions in TY05 resulted in both reduced CFU/event ratios and reduced percentages of intact cells compared to those of TY05. These were TY05 Δrob, TY05 ΔacrAB, TY05 ΔemrAB, and TY05 ΔmdtABCD. Rob is a transcription factor that is known to activate its regulon upon activation by FFA (29, 30), and deletion was previously observed to cause a lower population density (number of CFU per milliliter) of a strain of E. coli (K-12 strain MG1655 ΔfadD ΔaraBAD Δrob) expressing BTE on a plasmid than of the same strain without a rob deletion. Deletion of marA and soxS, which encode transcription factors with regulons overlapping that of Rob but which are activated by different mechanisms, previously produced no significant reduction in the number of CFU per milliliter (25), and the same result was obtained here. Members of the Rob regulon include acrAB and tolC. AcrAB, EmrAB, and MdtABC are all inner membrane and periplasmic linker protein components of drug efflux pumps that interact with an outer membrane component, TolC (49–51). MdtD is an uncharacterized putative inner membrane protein that may function as a separate drug efflux system (51).
To further confirm the role of the identified genes in conferring resistance to FFA, TY05 with deletions in acrAB, rob, tolC, emrAB, mdtEF, and mdtABCD was spotted onto plates containing various concentrations of octanoic and decanoic acids (described in the supplemental material). Reductions of the MIC of octanoate were observed for TY05 ΔacrAB, Δrob, ΔtolC, and ΔmdtABCD, while reductions of the MIC of decanoate were observed for TY05 ΔacrAB, Δrob, and ΔtolC compared to those for baseline strain TY05 (Table 3; see Fig. S3 in the supplemental material). This result is consistent with prior work demonstrating hypersensitivity of an acrAB deletion strain to the exogenous addition of 5 mM decanoate in liquid culture (29).
Table 3.
Maximum tested concentrations of exogenously added FFA at which growth of strain TY05 with or without gene deletions was observed on solid medium with pH adjusted to 7.0a
| Strain | Maximum tested concn (g/liter) at which growth was observed |
|
|---|---|---|
| Octanoate | Decanoate | |
| TY05 | 5 | 5 |
| TY05 ΔacrAB | 3 | 1 |
| TY05 ΔemrAB | 5 | 5 |
| TY05 ΔmdtEF | 5 | 5 |
| TY05 ΔmdtABCD | 4 | 5 |
| TY05 ΔtolC | 0 | None |
| TY05 Δrob | 4 | 3 |
See Fig. S7 in the supplemental material for images.
Fatty acid titers of single gene/operon deletion strains.
Fatty acid titers were analyzed 8 and 24 h postinoculation in separate sets of biological triplicate cultures because of the destructive conditions required to obtain accurate volumetric titers. While it would have been anticipated that an increased percentage of nonintact cells would result in lower fatty acid titers, statistically equivalent titers were observed in strain TY05 with or without all of the gene deletions for which reduced viability was observed (see Fig. S4 in the supplemental material).
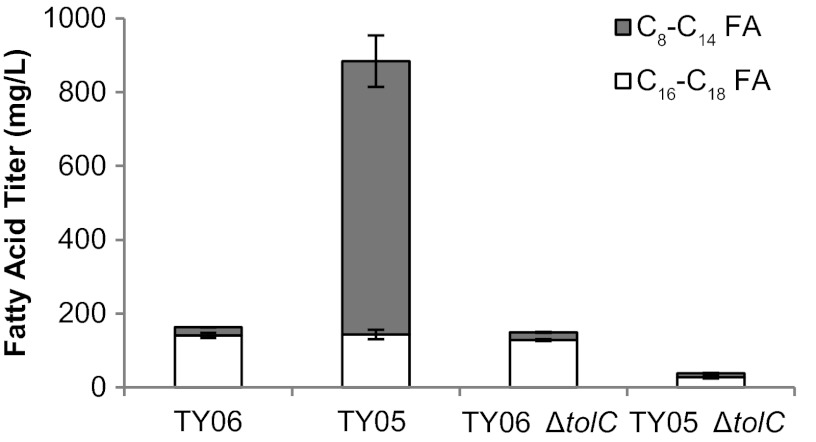
Interestingly, FFA production at both 8 and 24 h in TY05 ΔtolC was nearly abolished (Fig. 2). After 8 h, TY05 produced 428 ± 29 mg/liter C8 to C14 fatty acids, while TY05 ΔtolC produced 12 ± 1 mg/liter. At 24 h, TY05 produced a total of 740 ± 70 mg/liter C8 to C14 fatty acids while TY05 ΔtolC produced 11 ± 2 mg/liter. At 24 h, the composition of fatty acids in TY06 ΔtolC was very similar to that in TY06, with total titers of predominantly C16 to C18 fatty acids of 163 ± 7 and 149 ± 4 mg/liter in TY06 and TY06 ΔtolC, respectively. It is evident that the drastic change in phenotype upon tolC deletion is specific to conditions of FFA production, and tolC appears to be required for FFA production.
Fig 2.
Total fatty acid titers in TY05 and TY05 ΔtolC after 24 h. Fatty acids were extracted from cultures grown for 24 h in LB plus 0.4% glycerol at 37°C. While deletion of tolC from TY06 (non-FFA producing) had virtually no effect, deletion of tolC from TY05 (FFA producing) nearly abolished fatty acid production, particularly of C8 to C14 species (gray).
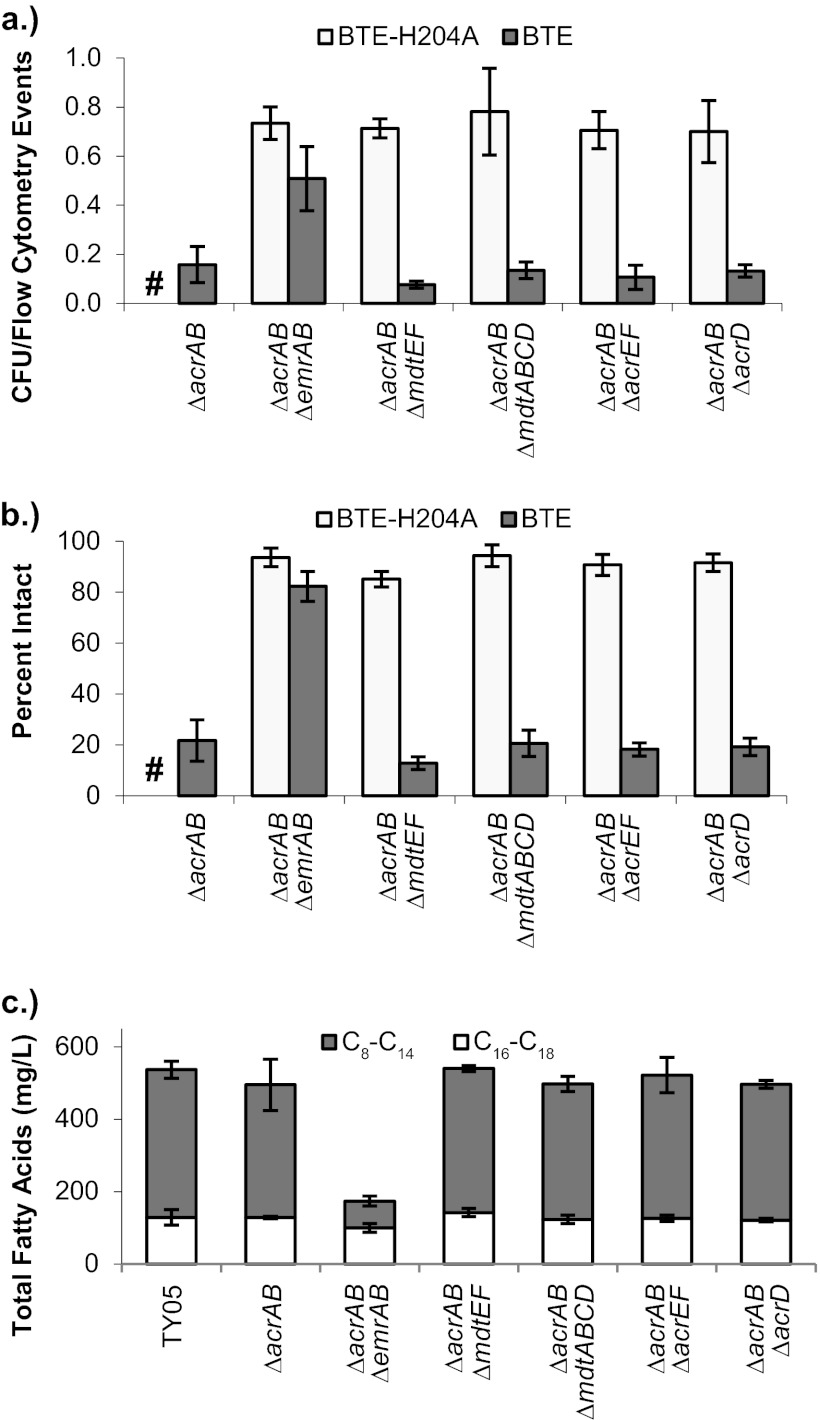
Viability analysis of double transporter gene/operon deletions.
The activities of many efflux pumps are masked by the basal activity of the AcrAB-TolC complex, which confers resistance to the widest characterized range of compounds (27, 28). AcrAB is also responsible for conferring resistance to SDS at concentrations exceeding 50 mg/ml in strain TY05, while the MIC for TY05 ΔacrAB is <0.1 mg/ml (see Fig. S7 in the supplemental material). The structural similarity of SDS and lauric acid and the loss of viability observed under conditions of FFA production in TY05 ΔacrAB implicate AcrAB as a transporter of FFA while also suggesting that AcrAB may be masking the activity of other identified efflux pumps, all of which also confer various degrees of resistance to SDS (27, 28) and possibly FFA. To determine if additional efflux pumps are involved, additional genes were deleted from TY05 ΔacrAB. CFU/event ratios and percentages of intact cells were determined for these strains as described above (Fig. 3). Biological triplicates of TY05 ΔacrAB were independently run as negative controls. All double deletions in TY06 were statistically equivalent and equal in value to TY06 and TY06 ΔacrAB for each of the measures. None of the double deletion TY05 strains exhibited CFU/event ratios or percentages of intact cells lower than those of TY05 ΔacrAB with a P value of <0.05, but TY05 ΔacrAB ΔmdtEF was lower in both measures, with a P value of 0.14 to 0.16. Interestingly, TY05 ΔacrAB ΔemrAB exhibited a phenotype very different from that of either single deletion strain. It exhibited a higher number of CFU per milliliter, a higher CFU/event ratio, and a higher percentage of intact cells than TY05 ΔacrAB. Growth of TY05 ΔacrAB ΔemrAB stalled at a lower final OD600 both in shake flasks at the 8-h sampling point (7.7 ± 0.4 for TY05 ΔacrAB, 5.6 ± 0.5 for TY05 ΔacrAB ΔemrAB) and in a plate reader growth curve compared to other negative-control strains (see Fig. S5 in the supplemental material). This reduced level of growth may prevent the onset of the stationary-phase cell lysis event observed in other FFA-overproducing strains at the 8-h sampling point.
Fig 3.
Normalized CFU counts, percentages of intact cells, and fatty acid titers after 8 h for double efflux pump deletions in strain TY06 (where shown; BTE-H204A) and TY05 (BTE). Cultures were sampled at 8 h postinoculation. (a) Normalized CFU counts (number of CFU per milliliter from plate counts divided by the number of cells per milliliter from flow cytometry counts). (b) Percentages of intact cells determined by SYTOX Green flow cytometry assay. TY05 and TY05 ΔacrAB were independently run as negative controls. (c) Total fatty acid titers for double efflux pump deletions in TY05 at 8 h postinoculation. TY05 ΔacrAB ΔemrAB exhibits much lower fatty acid production (primarily reduced C8 to C14) than other TY05 strains. Titers lower than those of other TY05 strains persist in TY05 ΔacrAB ΔemrAB after 24 h (see Fig. S6 in the supplemental material). #, data not measured.
Fatty acid titers of double transporter gene/operon deletions.
Fatty acid titers were analyzed at 8 h (in TY05 strains only, Fig. 3) and 24 h postinoculation for all double deletions (see Fig. S6 in the supplemental material). Double deletions in strain TY06 all produced fatty acid titers similar to those of TY06 and single deletions in TY06, ranging between 145 and 170 mg/liter at 24 h. At 8 h, similar titers were observed for TY05, TY05 ΔacrAB, and all double deletions (∼500 mg/liter) except TY05 ΔacrAB ΔemrAB, where the titer was significantly reduced (177 ± 26 mg/liter). Most of the fatty acid titer reduction in this strain was in C8 to C14 fatty acids. After 24 h, the fatty acid titer of TY05 ΔacrAB ΔemrAB increased significantly (490 ± 16 mg/liter) but remained lower than those of TY05 and TY05 ΔacrAB (∼700 mg/liter). TY05 ΔmdtEF exhibited a slightly decreased fatty acid titer at 24 h of 638 ± 21 mg/liter.
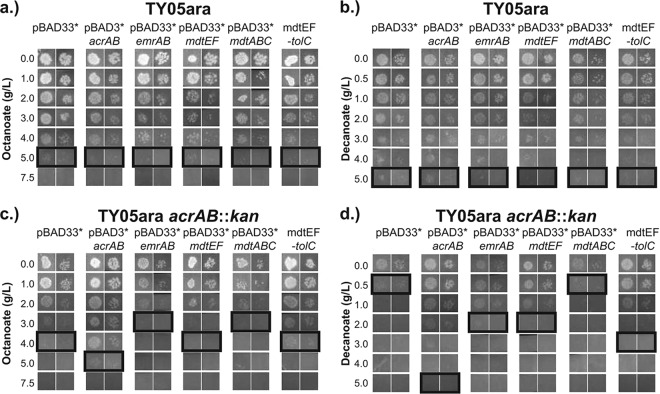
Effects of supplemental expression of drug efflux pumps in FFA-producing strains.
TY05ara strains harboring pBAD33*, pBAD33*-acrAB, pBAD33*-emrAB, pBAD33*-mdtEF, pBAD33*-mdtABCD, or pBAD33*-mdtEF-tolC (plasmids selected because they confer increased resistance to SDS when expressed in TY05 ΔacrAB [see Fig. S7 in the supplemental material]) were grown in LB supplemented with 0.4% glycerol, 34 μg/ml chloramphenicol, 0.5 mM CaCl2, and 0.5 mM MgSO4. While the supplementation of divalent cations succeeded in increasing the percentage of intact cells as measured by SYTOX Green after 8 h (see Fig. S8 in the supplemental material), plasmid-based expression of the drug efflux pumps failed to increase the percentage of intact cells above that in the empty-vector control. Furthermore, no improvement in the total or C8 to C14 fatty acid titer was observed after 8 or 24 h in TY05ara harboring plasmids that encode drug efflux pumps relative to that in TY05ara harboring the empty vector (see Fig. S8 in the supplemental material).
MICs of exogenous FFA for strains expressing drug efflux pumps from multicopy plasmids.
To determine if plasmid-based expression of identified drug efflux pumps confers resistance to exogenously added medium-chain-length FFA, we determined the MICs of octanoate and decanoate for TY05ara and TY05ara acrAB::kan harboring pBAD33*, pBAD33*-acrAB, pBAD33*-emrAB, pBAD33*-mdtEF, pBAD33*-mdtABCD, and pBAD33*-mdtEF-tolC in the absence of BTE induction (Fig. 4). Strains harboring efflux pump expression plasmids were not able to increase the MIC of octanoate or decanoate above that for TY05ara/pBAD33* (which grew on up to 5 g/liter octanoate and 5 g/liter decanoate), consistent with prior endogenous FFA production results. In contrast, TY05ara acrAB::kan/pBAD33* exhibited growth at up to only 4 g/liter octanoate and 0.5 g/liter decanoate. Two efflux pumps (AcrAB and MdtEF-TolC) complemented the acrAB deletion and increased the MIC of octanoate. Upon a challenge with decanoate, no efflux pumps restored the baseline MIC for TY05ara/pBAD33* but most of the efflux pumps expressed from multicopy plasmids enabled growth above the MIC for TY05ara acrAB::kan/pBAD33* (AcrAB, EmrAB, MdtEF, and MdtEF-TolC).
Fig 4.
Assays of octanoate and decanoate MICs for TY05ara and TY05ara acrAB::kan expressing selected efflux pumps from a medium-copy-number plasmid (pBAD33*). TY05ara (a) and TY05ara acrAB::kan (c) harboring drug efflux pumps were plated on various concentrations of octanoate. No efflux pump increased the MIC of octanoate for TY05ara. In TY05ara acrAB::kan, expression of AcrAB and MdtEF-TolC increased the MIC after 24 h and only AcrAB increased the MIC after 1 week of incubation (see the text). (b, d) The same strains plated on various concentrations of decanoate. No efflux pump increased the MIC of decanoate for TY05ara. In TY05ara acrAB::kan, plasmid-based expression of AcrAB, EmrAB, MdtEF, and MdtEF-TolC increased the MIC (see the text). Boxes denote the maximum concentrations at which growth was observed after incubation for 1 night at 37°C and 6 days at room temperature.
In summary, the native level of AcrAB activity is sufficient to confer resistance to exogenous medium-chain-length FFA. Expression of AcrAB from a medium-copy-number plasmid in acrAB deletion strains only restores the wild-type tolerance but does not improve it further. When expressed from plasmids, three drug efflux pumps (EmrAB, MdtEF, and MdtEF-TolC) partially complemented the reduced decanoate tolerance resulting from the deletion of acrAB but had no impact in cells harboring acrAB.
DISCUSSION
Many renewable biochemicals and biofuels are toxic to the host organism at the high concentrations required by economically feasible bioprocesses (1, 2, 52). In particular, endogenous production of biofuels and chemicals may result in toxic effects at concentrations lower than those obtained by exogenous addition of the same compound. When produced inside E. coli, molecules must traverse the inner and outer membranes. Therefore, the cell cannot use strategies that are effective at combating exposure to exogenous agents, such as reduced permeability of the outer membrane through decreased porin expression, lipopolysaccharide modifications, or other modifications of the lipid bilayer. This phenomenon has been observed for lauric acid, as endogenous production via BTE overexpression results in greatly reduced measures of viability and increased cell lysis relative to both exogenous addition of lauric acid to the growth medium (25) and addition of lauric acid to plates at concentrations well above the titers present in strains producing it (data not shown).
It has been suggested that microbial efflux pumps, which confer resistance to a wide range of antibiotics, solvents, and cationic or lipophilic compounds (9, 28, 29, 53, 54), could improve the secretion of endogenously produced compounds and confer resistance to high levels of the compound accumulated in fermentation broth (1, 2, 9). Furthermore, it has been suggested that product secretion would keep intracellular concentrations low, thereby reducing product inhibition and improving flux through reversible reactions (2). Many efflux pumps have been isolated from E. coli and pseudomonads that confer increased resistance to toxic solvents such as toluene and hexane (53, 55, 56). Competition assays have also identified drug efflux pumps from libraries that are enriched following a challenge from a toxic level of an exogenously added hydrophobic compound (10). These strategies are difficult to employ when exogenous addition of the target compound does not elicit significant growth inhibition, as is the case for saturated C12 and longer-chain-length FFA. Additionally, the aforementioned competition assay and other studies performed with E. coli have necessitated deletion of acrAB to render the cells sensitive to most of the compounds of interest (10, 28). In one of these studies, a plasmid that encodes AcrB was found to be highly enriched in competition assays against five advanced biofuels (10).
We surmised that E. coli already possesses the ability to secrete FFA via native drug efflux pumps or other transport machinery and sought to identify the genes responsible in this study such that they could be overexpressed to allow improved production of FFA. The lines of evidence in support of this assumption were that (i) the native enteric environment of E. coli is rich in fatty acids (30); (ii) a number of drug efflux pumps, when overexpressed in an acrAB deletion strain, confer an increased MIC of SDS, which is structurally very similar to FFA (28); (iii) the transcription factor Rob is activated by 5 mM decanoate, which leads to increased transcription of acrAB (29, 30); (iv) the overlapping MarA/Rob/SoxS regulons are upregulated in BTE-expressing cultures, yet only when rob was deleted were decreased measures of viability observed (25); and (v) both acrAB and tolC deletion strains were previously shown to exhibit hypersensitivity to exogenously added decanoate (26, 29). To date, three drug efflux pump systems have been implicated in FFA export in bacteria: AcrAB in E. coli (29), EmhABC in Pseudomonas fluorescens cLP6a (57), and FarAB in Neisseria gonorrhoeae (58). RND-type efflux pumps, as well as other classes of transporters, are also implicated in the secretion of neutral lipid species in Alcanivorax borkumensis SK2 (59). Interestingly, the FarAB system was first identified on the basis of homology to E. coli EmrAB (58). Multidrug efflux systems have previously been proposed to have alternative physiological roles such as membrane lipid turnover (60). This may be another rationale for the observed activity in E. coli and could be related to the induction of many efflux systems under cell envelope stress conditions, as these conditions may necessitate active membrane lipid remodeling.
Since screening could not be performed with exogenously added lauric acid, a screening method was based on the observation that FFA production causes a significant increase in nonintact cells as indicated by SYTOX Green staining during early stationary phase (25). Three out of 15 single gene or operon deletions either resulted in statistically significant reductions of both normalized CFU counts and percentages of intact cells or nearly abolished FFA production in the BTE-expressing cultures. Conversely, no deletion significantly altered viability or fatty acid production in control (non-FFA-producing) cultures. The three hits were rob, acrAB, and tolC. We had previously identified Rob as being important to maintaining viability in another fatty-acid-producing strain (25), likely because of its role in activating its regulon, which includes acrAB but also a number of genes that encode proteins with ill-defined functions. AcrAB appears to be the most important drug efflux pump characterized in E. coli for conferring SDS resistance (28) and resistance to exogenous FFA (29); therefore, it is perhaps unsurprising that it was also the only single efflux pump deletion to render cells more sensitive to endogenous FFA production. As further validation of their roles, rob and acrAB deletion strains also grew at maximum concentrations of exogenously added octanoate and decanoate lower than those at which strains where these genes were intact did.
TolC, which serves as the outer membrane component of the AcrAB drug efflux pump and many other inner membrane drug efflux pumps, had previously been identified in a transposon mutagenesis screening for membrane-associated proteins that are essential for growth on decanoate or oleate as a sole carbon source (26). However, its role was associated not with import of FFA but rather with the remediation of an undefined toxic effect of FFA on growth. One possibility is that TolC was necessary for the efflux of excess toxic quantities of FFA beyond the levels that could be processed by acyl coenzyme A synthetase (FadD). These excess FFA either could have been imported into the periplasm by FadL, the dominant importer of FFA across the outer membrane (61), or could have entered the periplasm and cytosol by diffusive processes. TolC could also play a number of secondary roles that are unrelated to direct FFA efflux, as it has been implicated in the extrusion of intracellular metabolites, including signaling molecules such as cyclic AMP (62), siderophores for iron acquisition (63), and excess cysteine (64). Furthermore, disruption of tolC triggers the induction of phage shock proteins and reduced NADH oxidase activity, suggesting a role for TolC in the maintenance of inner membrane integrity or direct interaction with enzymes of aerobic respiration (65). Thus, while tolC is critical for achieving production of FFA, the precise role of TolC in efflux of FFA remains unresolved.
A number of additional efflux pumps were identified in the single deletion screenings as reducing normalized CFU counts and percentages of intact cells but with P values between 0.05 and 0.15 for one or both measures. These included deletions in emrAB, mdtEF, and mdtABCD. These three efflux pumps are also known to confer a small increase in SDS resistance when overexpressed in an acrAB deletion strain, in addition to acrD and acrEF (28), which did not have any impact on viability in this study as single deletions. To determine if the effects of deleting these genes and operons were masked by the presence of chromosomal acrAB, these deletions were tested in strain TY05 ΔacrAB for their effects on viability and FFA production. No differential effects versus the deletion in acrAB alone were observed, except for in TY05 ΔacrAB ΔemrAB, which exhibited a phenotype approaching that observed for TY05 ΔtolC, with reduced levels of growth and delayed onset of stationary phase, coupled with a dramatic reduction in FFA production after 8 h (but largely recovered at 24 h). Thus, expression of both acrAB and emrAB appears to be important for FFA efflux in BTE-expressing cultures, although deletion of only one of these operons has an impact only on viability and none on FFA titers. We hypothesize that multiple RND-type efflux pumps may play compensatory roles, albeit with various activities, to achieve the same net level of excretion of FFAs until multiple inner membrane components are deleted. Complex transcriptional regulation of genes that encode these drug efflux pumps may be involved in compensation of activities. For example, MprA is inferred to transcriptionally repress both the acrAB and mprA-emrAB operons in the absence of small-molecule ligands (66, 67). If FFAs can serve as ligands for which binding to MprA mediates derepression, transiently increased intracellular levels of FFAs that result from deletion of acrAB could feasibly result in an increased level of expression of emrAB. Further experiments are warranted to probe comparative expression levels of genes that encode other efflux pumps in TY05, TY05 ΔacrAB, and TY05 ΔacrAB ΔemrAB.
In conclusion, AcrAB appears to be providing a native level of FFA efflux that cannot be exceeded by the plasmid-based expression of either acrAB or other targeted efflux pumps. This may be due to an already high basal level of expression or saturation of membrane protein insertion machinery. Other efflux pumps, particularly MdtEF and EmrAB, can complement the activity of AcrAB, albeit with lower degrees of efficacy. New insights were gained, including expansion of the range of substrates for these efflux pumps to include C8 to C12 FFA, providing a newly recognized mechanism for which FFA are actively excreted from cells. Since the expression of AcrAB and other efflux pumps from plasmids failed to increase FFA titers, additional engineering efforts may be required. Targets include overcoming saturation of membrane protein insertion pathways, proton motive force dissipation, and negative consequences of FFA production that remained uncorrected in this study (e.g., elevated unsaturated membrane lipid content [25]).
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by the DOE Great Lakes Bioenergy Research Center (DOE BER Office of Sciences DE-FC02-07ER64494). R.M.L. was supported as a trainee in the Chemistry-Biology Interface Training Program (NIH) and by the Department of Chemical and Biological Engineering Dahlke-Hougen Fellowship. M.A.K. was the recipient of a Holstrom Environmental Research Scholarship (University of Wisconsin-Madison). M.G.P. was supported as a trainee in the Biotechnology Training Program (NIH).
Footnotes
Published ahead of print 26 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01477-12.
REFERENCES
- 1.Nicolaou SA, Gaida SM, Papoutsakis ET. 2010. A comparative view of metabolite and substrate stress and tolerance in microbial bioprocessing: from biofuels and chemicals, to biocatalysis and bioremediation. Metab. Eng. 12:307–331 [DOI] [PubMed] [Google Scholar]
- 2.Dunlop MJ. 2011. Engineering microbes for tolerance to next-generation biofuels. Biotechnol. Biofuels 4:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sikkema J, de Bont JA, Poolman B. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59:201–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ingram LO. 1976. Adaptation of membrane lipids to alcohols. J. Bacteriol. 125:670–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ingram LO, Vreeland NS. 1980. Differential effects of ethanol and hexanol on the Escherichia coli cell envelope. J. Bacteriol. 144:481–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sardessai Y, Bhosle S. 2002. Tolerance of bacteria to organic solvents. Res. Microbiol. 153:263–268 [DOI] [PubMed] [Google Scholar]
- 7.Isken S, de Bont JA. 1998. Bacteria tolerant to organic solvents. Extremophiles 2:229–238 [DOI] [PubMed] [Google Scholar]
- 8.Grkovic S, Brown MH, Skurray RA. 2002. Regulation of bacterial drug export systems. Microbiol. Mol. Biol. Rev. 66:671–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramos JL, Duque E, Gallegos MT, Godoy P, Ramos-Gonzalez MI, Rojas A, Teran W, Segura A. 2002. Mechanisms of solvent tolerance in gram-negative bacteria. Annu. Rev. Microbiol. 56:743–768 [DOI] [PubMed] [Google Scholar]
- 10.Dunlop MJ, Dossani ZY, Szmidt HL, Chu HC, Lee TS, Keasling JD, Hadi MZ, Mukhopadhyay A. 2011. Engineering microbial biofuel tolerance and export using efflux pumps. Mol. Syst. Biol. 7:487 doi:10.1038/msb.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu X, Vora H, Khosla C. 2008. Overproduction of free fatty acids in E. coli: implications for biodiesel production. Metab. Eng. 10:333–339 [DOI] [PubMed] [Google Scholar]
- 12.Lennen RM, Braden DJ, West RM, Dumesic JA, Pfleger BF. 2010. A process for microbial hydrocarbon synthesis: overproduction of fatty acids in Escherichia coli and catalytic conversion to alkanes. Biotechnol. Bioeng. 106:193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardayre SB, Keasling JD. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559–562 [DOI] [PubMed] [Google Scholar]
- 14.Liu T, Vora H, Khosla C. 2010. Quantitative analysis and engineering of fatty acid biosynthesis in E. coli. Metab. Eng. 12:378–386 [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Li M, Agrawal A, San KY. 2011. Efficient free fatty acid production in Escherichia coli using plant acyl-ACP thioesterases. Metab. Eng. 13:713–722 [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Agrawal A, San KY. 2012. Improving fatty acid production in Escherichia coli through the overexpression of malonyl coA-acyl carrier protein transacylase. Biotechnol. Prog. 28:60–65 [DOI] [PubMed] [Google Scholar]
- 17.Yu X, Liu T, Zhu F, Khosla C. 2011. In vitro reconstitution and steady-state analysis of the fatty acid synthase from Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 108:18643–18648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Zhang X, Agrawal A, San KY. 2012. Effect of acetate formation pathway and long chain fatty acid CoA-ligase on the free fatty acid production in E. coli expressing acy-ACP thioesterase from Ricinus communis. Metab. Eng. 14:380–387 [DOI] [PubMed] [Google Scholar]
- 19.Liu H, Yu C, Feng D, Cheng T, Meng X, Liu W, Zou H, Xian M. 2012. Production of extracellular fatty acid using engineered Escherichia coli. Microb. Cell Fact. 11:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. 2011. Engineered reversal of the beta-oxidation cycle for the synthesis of fuels and chemicals. Nature 476:355–359 [DOI] [PubMed] [Google Scholar]
- 21.Maki-Arvela P, Kubickova I, Snare M, Eranen K, Murzin DY. 2007. Catalytic deoxygenation of fatty acids and their derivatives. Energy Fuels 21:30–41 [Google Scholar]
- 22.Kalscheuer R, Stolting T, Steinbuchel A. 2006. Microdiesel: Escherichia coli engineered for fuel production. Microbiology 152:2529–2536 [DOI] [PubMed] [Google Scholar]
- 23.Doan TT, Carlsson AS, Hamberg M, Bulow L, Stymne S, Olsson P. 2009. Functional expression of five Arabidopsis fatty acyl-CoA reductase genes in Escherichia coli. J. Plant Physiol. 166:787–796 [DOI] [PubMed] [Google Scholar]
- 24.Teerawanichpan P, Qiu X. 2010. Fatty acyl-CoA reductase and wax synthase from Euglena gracilis in the biosynthesis of medium-chain wax esters. Lipids 45:263–273 [DOI] [PubMed] [Google Scholar]
- 25.Lennen RM, Kruziki MA, Kumar K, Zinkel RA, Burnum KE, Lipton MS, Hoover SW, Ranatunga DR, Wittkopp TM, Marner WD, II, Pfleger BF. 2011. Membrane stresses induced by endogenous free fatty acid overproduction in Escherichia coli. Appl. Environ. Microbiol. 77:8114–8128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Azizan A, Black PN. 1994. Use of transposon TnphoA to identify genes for cell envelope proteins of Escherichia coli required for long-chain fatty acid transport: the periplasmic protein Tsp potentiates long-chain fatty acid transport. J. Bacteriol. 176:6653–6662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sulavik MC, Houseweart C, Cramer C, Jiwani N, Murgolo N, Greene J, DiDomenico B, Shaw KJ, Miller GH, Hare R, Shimer G. 2001. Antibiotic susceptibility profiles of Escherichia coli strains lacking multidrug efflux pump genes. Antimicrob. Agents Chemother. 45:1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishino K, Yamaguchi A. 2001. Analysis of a complete library of putative drug transporter genes in Escherichia coli. J. Bacteriol. 183:5803–5812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma D, Cook DN, Alberti M, Pon NG, Nikaido H, Hearst JE. 1995. Genes acrA and acrB encode a stress-induced efflux system of Escherichia coli. Mol. Microbiol. 16:45–55 [DOI] [PubMed] [Google Scholar]
- 30.Rosenberg EY, Bertenthal D, Nilles ML, Bertrand KP, Nikaido H. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609–1619 [DOI] [PubMed] [Google Scholar]
- 31.Fábrega A, Martin RG, Rosner JL, Tavio MM, Vila J. 2010. Constitutive SoxS expression in a fluoroquinolone-resistant strain with a truncated SoxR protein and identification of a new member of the marA-soxS-rob regulon, mdtG. Antimicrob. Agents Chemother. 54:1218–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aono R, Tsukagoshi N, Yamamoto M. 1998. Involvement of outer membrane protein TolC, a possible member of the mar-sox regulon, in maintenance and improvement of organic solvent tolerance of Escherichia coli K-12. J. Bacteriol. 180:938–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andersen J, Forst SA, Zhao K, Inouye M, Delihas N. 1989. The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem. 264:17961–17970 [PubMed] [Google Scholar]
- 34.Youngquist JT, Lennen RM, Ranatunga DR, Bothfeld WH, Marner WD, II, Pfleger BF. 2012. Kinetic modeling of free fatty acid production in Escherichia coli based on continuous cultivation of a plasmid free strain. Biotechnol. Bioeng 109:1518–1527 [DOI] [PubMed] [Google Scholar]
- 35.Yuan L, Nelson BA, Caryl G. 1996. The catalytic cysteine and histidine in the plant acyl-acyl carrier protein thioesterases. J. Biol. Chem. 271:3417–3419 [DOI] [PubMed] [Google Scholar]
- 36.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008 doi:10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomason LC, Costantino N, Court DL. 2007. E. coli genome manipulation by P1 transduction. Curr. Protoc. Mol. Biol. Chapter 1: Unit 1.17 [DOI] [PubMed] [Google Scholar]
- 38.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomason L, Court DL, Bubunenko M, Costantino N, Wilson H, Datta S, Oppenheim A. 2007. Recombineering: genetic engineering in bacteria using homologous recombination. Curr. Protoc. Mol. Biol. Chapter 1 :Unit 1.16 [DOI] [PubMed] [Google Scholar]
- 40.Donath MJ, II, Dominguez MA, Withers ST., III 2011. Development of an automated platform for high-throughput P1-phage transduction of Escherichia coli. J. Lab. Autom. 16:141–147 [DOI] [PubMed] [Google Scholar]
- 41.Cherepanov PP, Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 42.Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, Keasling JD. 2001. Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology 147:3241–3247 [DOI] [PubMed] [Google Scholar]
- 43.De Lay NR, Cronan JE. 2007. In vivo functional analyses of the type II acyl carrier proteins of fatty acid biosynthesis. J. Biol. Chem. 282:20319–20328 [DOI] [PubMed] [Google Scholar]
- 44.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SK, Chou HH, Pfleger BF, Newman JD, Yoshikuni Y, Keasling JD. 2007. Directed evolution of AraC for improved compatibility of arabinose- and lactose-inducible promoters. Appl. Environ. Microbiol. 73:5711–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nikaido H, Takatsuka Y. 2009. Mechanisms of RND multidrug efflux pumps. Biochim. Biophys. Acta 1794:769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roth BL, Poot M, Yue ST, Millard PJ. 1997. Bacterial viability and antibiotic susceptibility testing with SYTOX Green nucleic acid stain. Appl. Environ. Microbiol. 63:2421–2431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fralick JA. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803–5805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Borges-Walmsley MI, Beauchamp J, Kelly SM, Jumel K, Candlish D, Harding SE, Price NC, Walmsley AR. 2003. Identification of oligomerization and drug-binding domains of the membrane fusion protein EmrA. J. Biol. Chem. 278:12903–12912 [DOI] [PubMed] [Google Scholar]
- 51.Nagakubo S, Nishino K, Hirata T, Yamaguchi A. 2002. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J. Bacteriol. 184:4161–4167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen CR, Lan EI, Dekishima Y, Baez A, Cho KM, Liao JC. 2011. Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl. Environ. Microbiol. 77:2905–2915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Aono R. 1998. Improvement of organic solvent tolerance level of Escherichia coli by overexpression of stress-responsive genes. Extremophiles 2:239–248 [DOI] [PubMed] [Google Scholar]
- 54.Brown MH, Skurray RA. 2001. Staphylococcal multidrug efflux protein QacA. J. Mol. Microbiol. Biotechnol. 3:163–170 [PubMed] [Google Scholar]
- 55.White DG, Goldman JD, Demple B, Levy SB. 1997. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J. Bacteriol. 179:6122–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramos JL, Duque E, Godoy P, Segura A. 1998. Efflux pumps involved in toluene tolerance in Pseudomonas putida DOT-T1E. J. Bacteriol. 180:3323–3329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adebusuyi AA, Foght JM. 2011. An alternative physiological role for the EmhABC efflux pump in Pseudomonas fluorescens cLP6a. BMC Microbiol. 11:252 doi:10.1186/1471-2180-11-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee EH, Shafer WM. 1999. The farAB-encoded efflux pump mediates resistance of gonococci to long-chained antibacterial fatty acids. Mol. Microbiol. 33:839–845 [DOI] [PubMed] [Google Scholar]
- 59.Manilla-Pérez E, Reers C, Baumgart M, Hetzler S, Reichelt R, Malkus U, Kalscheuer R, Waltermann M, Steinbuchel A. 2010. Analysis of lipid export in hydrocarbonoclastic bacteria of the genus Alcanivorax: identification of lipid export-negative mutants of Alcanivorax borkumensis SK2 and Alcanivorax jadensis T9. J. Bacteriol. 192:643–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poole K. 2008. Bacterial multidrug efflux pumps serve other functions. Microbe 3:179–185 [Google Scholar]
- 61.Black PN. 1990. Characterization of FadL-specific fatty acid binding in Escherichia coli. Biochim. Biophys. Acta 1046:97–105 [DOI] [PubMed] [Google Scholar]
- 62.Hantke K, Winkler K, Schultz JE. 2011. Escherichia coli exports cyclic AMP via TolC. J. Bacteriol. 193:1086–1089 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Bleuel C, Grosse C, Taudte N, Scherer J, Wesenberg D, Krauss GJ, Nies DH, Grass G. 2005. TolC is involved in enterobactin efflux across the outer membrane of Escherichia coli. J. Bacteriol. 187:6701–6707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiriyathanawudhiwong N, Ohtsu I, Li ZD, Mori H, Takagi H. 2009. The outer membrane TolC is involved in cysteine tolerance and overproduction in Escherichia coli. Appl. Microbiol. Biotechnol. 81:903–913 [DOI] [PubMed] [Google Scholar]
- 65.Dhamdhere G, Zgurskaya HI. 2010. Metabolic shutdown in Escherichia coli cells lacking the outer membrane channel TolC. Mol. Microbiol. 77:743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lomovskaya O, Lewis K, Matin A. 1995. EmrR is a negative regulator of the Escherichia coli multidrug resistance pump EmrAB. J. Bacteriol. 177:2328–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rodionov DA, Gelfand MS, Mironov AA, Rakhmaninova AB. 2001. Comparative approach to analysis of regulation in complete genomes: multidrug resistance systems in gamma-proteobacteria. J. Mol. Microbiol. Biotechnol. 3:319–324 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.