Abstract
Enterobacter cloacae GS1 is a plant growth-promoting bacterium which colonizes rice roots. In the rhizosphere environment, N-acyl homoserine lactone (NAHL)-like quorum-sensing signals are known to be produced by host plants and other microbial inhabitants. E. cloacae GS1 was unable to synthesize NAHL quorum-sensing signals but had the NAHL-dependent transcriptional regulator-encoding gene sdiA. This study was aimed at understanding the effects of SdiA and NAHL-dependent cross talk in rice root colonization by E. cloacae GS1. Pleiotropic effects of sdiA inactivation included substantial increases in root colonization and biofilm formation, suggesting a negative role for SdiA in bacterial adhesion. We provide evidence that sdiA inactivation leads to elevated levels of biosynthesis of curli, which is involved in cellular adhesion. Extraneous addition of NAHLs had a negative effect on root colonization and biofilm formation. However, the sdiA mutant of E. cloacae GS1 was insensitive to NAHLs, suggesting that this NAHL-induced inhibition of root colonization and biofilm formation is SdiA dependent. Therefore, it is proposed that NAHLs produced by both plant and microbes in the rice rhizosphere act as cross-kingdom and interspecies signals to negatively impact cellular adhesion and, thereby, root colonization in E. cloacae GS1.
INTRODUCTION
Population density-dependent gene expression in bacteria is controlled by quorum-sensing (QS) regulatory networks (1). Quorum sensing plays a major role in the regulation of factors critical for ecological success, such as bacterial adhesion, biofilm formation, host colonization, and virulence factor production (2, 3). The luxI-luxR QS system was first characterized in the marine bacterium Vibrio harveyi and has since been well studied in several other bacteria. Enteric bacteria such as Escherichia, Klebsiella, and Salmonella are unable to synthesize N-acyl homoserine lactone (NAHL) QS signals, as they lack a luxI homolog, while evolutionarily preserving sdiA, which encodes an NAHL-dependent transcriptional regulator. sdiA-like orphaned luxR homologs are less likely to be remnants of gene acquisition or loss, as no bacterial genome contains an orphaned luxI homolog (4). Orphaned LuxR homologs in bacteria may require NAHL-like inducers to regulate gene expression. When QS signal mimics that are synthesized by higher organisms are available to bacteria in an environmental niche, these signals can act as cross-kingdom signals and regulate QS-dependent functions. The response of Escherichia coli and Salmonella SdiA to NAHLs has been well characterized (see references5 to 7 and references therein). E. coli SdiA, for example, is known to regulate biofilm formation in response to the interspecies signals NAHLs (8).
Many reports suggest that NAHL-like mimics are produced by higher organisms and influence gene expression in bacteria and vice versa. For example, plant-derived flavonoids that are known to induce nodulation were recently reported to increase expression of NAHL biosynthesis genes in symbiotic rhizobia (9). Conversely, unicellular algae (Chlamydomonas reinhardtii) and plants (Pisum sativum) are known to produce NAHL-like substances which affect QS behavior in bacteria (10, 11). Certain unidentified substances from gnotobiotically grown rice plants were able to elicit positive responses in three different NAHL biosensors. These substances were characterized and were found to be sensitive to the lactone ring-specific AiiA lactonase, further confirming their structural similarity to NAHLs (12). The rice rhizosphere is rich in plant-derived nutrients which facilitate plant-associated and free-dwelling soil bacterial colonization of the rhizospheric niche (13). NAHL synthesis is more common in plant-associated bacteria than soilborne bacteria (14). Thus, the rhizosphere environment has several possible sources of NAHLs. Any NAHL-based cross talk between SdiA-containing bacteria and NAHL-producing microbial populations remains to be explored. Earlier, we have shown that Enterobacter cloacae GS1, a plant growth-promoting bacterium, colonizes rice roots as microcolonies and forms biofilm-like structures on the root surface (15). Here, we report that E. cloacae GS1 lacks a functional NAHL-based QS system but harbors the luxR homolog sdiA. We demonstrate for the first time the role of sdiA in root colonization, perception and response to environmental NAHLs, and biofilm formation in E. cloacae GS1, a plant symbiont.
MATERIALS AND METHODS
General growth conditions.
Enterobacter cloacae GS1 was grown in Luria-Bertani (LB) medium or M9 minimal medium (16) at 37°C without or with agitation (200 rpm). Chromobacterium violaceum CV026, E. coli(pSB401), E. coli(pSB406), and E. coli(pSB1075) were grown in LB medium supplemented with kanamycin (30 μg ml−1), tetracycline (12 μg ml−1), or ampicillin (100 μg ml−1). Agrobacterium tumefaciens NT1(pZLR4) was grown in AB minimal medium (17) with gentamicin (15 μg ml−1). Wherever required, N-(3-oxohexanoyl)-l-homoserine lactone (OHHL), N-hexanoyl-dl-homoserine lactone (HHL), and N-octanoyl-dl-homoserine lactone (OHL) (Fluka, Sigma-Aldrich) were dissolved in acetonitrile and used at the required concentrations. To study the effect of NAHLs on root colonization and biofilm formation, media were supplemented with OHHL at a final concentration of 1 μM. Sterile filter paper discs loaded with 200 pmol of each NAHL were used in disc diffusion assays.
Extraction and detection of NAHLs and NAHL-like mimics.
NAHLs from spent media and NAHL-like mimics from sterile rice root tissue and exudate were extracted with an equal volume of acidified ethyl acetate (0.1%, vol/vol) as described earlier (12). Concentrated (100-fold) extracts of spent media or root exudates were separated by thin-layer chromatography using methanol (60%, vol/vol) as the mobile phase. Developed chromatograms were overlaid with soft-agar suspensions of biosensor strains such as A. tumefaciens NT1(pZLR4) (18), C. violaceum CV026 (19), and E. coli(pSB401), E. coli(pSB406), and E. coli(pSB1075) (20) and incubated at 30°C for 12 h. Chromatograms overlaid with A. tumefaciens NT1(pZLR4) and C. violaceum CV026 were monitored for the appearance of blue and purple spots, respectively. Bioluminescence around NAHL spots was monitored using a Biospectrum AC imaging system (UVP, Upland, CA) for chromatograms overlaid with E. coli(pSB401), E. coli(pSB406), and E. coli(pSB1075). Sterile medium extracts concentrated to the same levels served as negative controls, and standard OHHL served as a positive control, while acetonitrile served as a solvent control.
Detection of SdiA in E. cloacae GS1 and construction of an sdiA mutant.
SdiA function in E. cloacae GS1 was detected by an NAHL disc diffusion assay, using the plasmid biosensor pBA405, which contains an SdiA-NAHL complex-activated promoter upstream of the luxCDABE operon (7). To test whether NAHL mimics from rice roots activate SdiA and induce bioluminescence in E. cloacae GS1(pBA405), a soft-agar suspension of this strain was overlaid on surface-sterilized and germinated rice seeds. Bioluminescence was monitored using a Biospectrum AC imaging system (UVP, Upland, CA) after incubation for 6 h. Synthetic OHHL, HHL, and OHL (Fluka, Sigma-Aldrich) at 200 pmol were used as positive controls, while acetonitrile served as the negative control.
A 951-bp region containing sdiA was amplified from the E. cloacae GS1 genome using primers SAF1 and SAF2 (Table 1) on an Eppendorf Mastercycler with the following cycling conditions: 94°C for 10 min; 30 cycles of 95°C for 30 s, 55°C for 30 s, 72°C for 1 min; and 72°C for 5 min. The amplified fragment was cloned into pTZ57R/T (Fermentas, Opelstrasse, Germany) to obtain pTZS, the insert in which was verified by sequencing of both strands (Macrogen, Seoul, South Korea). A kanamycin resistance cassette flanked with SmaI sites was introduced into a unique EcoRV site at position 655 of sdiA in pTZS to yield pTZSK. This inactivated sdiA was amplified with Pfu DNA polymerase and inserted into the SpeI site of suicide vector pIVETP (21) blunted with T4 DNA polymerase, to yield pIVETS, which was then mobilized into E. cloacae GS1 from E. coli S-17. Transconjugants which arose due to a double homologous recombination event were selected on kanamycin (30 μg ml−1) and screened for tetracycline (12 μg ml−1) sensitivity. These mutants were further screened by PCR to verify the insertional inactivation of sdiA on the E. cloacae GS1 genome. The loss of SdiA activity was also confirmed by a pBA405-based NAHL disc diffusion assay with the wild type as the positive control. To eliminate the possibility of secondary mutations, the sdiA mutant was complemented in trans with wild-type sdiA. Wild-type sdiA, along with its native promoter, was amplified from E. cloacae GS1 genomic DNA using primers SABam1 and SABam2 (Table 1) and cloned into the unique BamHI site of the low-copy-number vector pBR322. This construct, pBsdiA, was introduced into E. cloacae GS1 sdiA::Kmr to generate the complemented strain E. cloacae GS1 sdiA::Kmr(pBsdiA).
Table 1.
Primers used in this study
| Primer namea | Targeted gene/region | Sequence (5′–3′)b |
|---|---|---|
| SAF1 | sdiA | TTAGCGTTCAATTTGCTCCAGATG |
| SAF2 | GATATCAGTCAGATAAGCCCCGTC | |
| SABam1 | TTTGGATCCTTTAGCGTTCAATTTGCTCC | |
| SABam2 | AAAGGATCCGATATCAGTCAGATAAGCCC | |
| QcsgBF | csgB | CTGGATAATCATGGCGGTGTT |
| QcsgBR | AACCGCGCGAATGTTGA | |
| QcsgAF | csgA | AAGCCAACCTGGTGCACAGT |
| QcsgAR | CATCGACCAATGGAATAGCAAA | |
| QcsgDF | csgD | AACTGCGTATTGGTGCTTCAAA |
| QcsgDR | CCTGCGTGCGATTTTTAACA | |
| QbcsAF | bcsA | TGCCAATGCCCATATTCTGA |
| QbcsAR | TCGTCACGATGGATCACCAT | |
| QrpoBF | rpoB | GCAACTTGTTGTCGCGGATT |
| QrpoBR | TCGACCGTCGTCGTAAGCT |
Primer names prefixed with Q were used for quantitative RT-PCR.
Relevant restriction sites are underscored.
Rice root colonization assay.
The colonization abilities of E. cloacae GS1 and its sdiA mutant were studied as described earlier (15). In brief, gnotobiotic rice plants grown in nutrient solution at 25 ± 1°C were inoculated with ∼108 CFU of the wild type or mutant. At 7 days postinoculation, the counts of root-colonizing bacteria were determined by vortexing the aseptically excised root in the presence of saline and glass beads and standard plate counting. Wherever necessary, the nutrient solution was supplemented with OHHL (1 μM). To test for competitive root-colonizing ability, ∼108 CFU each of the wild type and mutant was coinoculated. The total E. cloacae population on inoculated roots was recovered on LB medium and screened on LB medium containing kanamycin (30 μg ml−1) to discriminate the mutant from the wild type. Counts of E. cloacae GS1 and its sdiA mutant were expressed as a percentage of the total viable number of CFU recovered from inoculated plants. Colonization experiments, individual or competitive, were performed thrice independently with a minimum of five replicates in each experiment.
Biofilm formation, atomic force microscopy, and Congo red binding assay.
For quantitation of biofilm formation on polystyrene, the assay described earlier (22) was adapted, with modifications. Wild-type, mutant, and complemented sdiA mutant cultures at an optical density at 600 nm (OD600) of 0.8 were diluted 1:100 (vol/vol) into fresh LB medium without NaCl. Wherever necessary, OHHL was supplemented to a final concentration of 1 μM. This diluted culture was then split into aliquots of 2.5 ml and placed into each well of a 24-well microtiter plate. Appropriate sterile medium served as a negative control. After 48 h growth at 25°C, mature biofilms were washed thrice with sterile saline, air dried, stained with crystal violet (0.5%, wt/vol), washed to remove unbound crystal violet, and then air dried. The crystal violet bound on 24-well polystyrene plates was then solubilized with absolute ethanol, and the absorbance at 590 nm was measured. The mean of five replicates from one of the three independent experiments is presented, with error bars indicating standard deviations (SDs). Interface biofilms were cultivated on glass as described earlier (23) in LB medium without NaCl at 25°C. Mature biofilms on glass slides were washed thrice, stained with crystal violet, air dried, observed, and photographed under a light microscope (Eclipse Ti; Nikon, Melville, NY), and the architecture was analyzed. Images of each layer of the biofilm were sewn together using the multiple-images sewing feature available in the NIS elements-D program.
Interface biofilms on glass slides were also visualized using atomic force microscopy to visualize finer details (24). Air-medium interface biofilms were imaged with an A100SGS instrument (APE Research, Trieste, Italy). Imaging was carried out by noncontact mode using a V-shaped silicon nitride cantilever resonating at a frequency of 289 kHz. The images were processed using the Gwyddion program and visually evaluated for aggregation, extracellular matrix, and the robustness of the biofilm. A Congo red binding assay was adapted, with modifications (25). E. cloacae GS1, the sdiA mutant, and the complemented sdiA mutant were streaked onto LB medium without NaCl supplemented with Congo red (50 μg ml−1) and Coomassie brilliant blue (6.25 μg ml−1) and incubated at 25°C for 48 h before being scanned on a high-resolution gel scanner (UTA 1100; Amersham Biosciences, Uppsala, Sweden).
Quantitative reverse transcription-PCR (RT-PCR).
Primer pairs producing ∼120-bp products specific to curli biosynthetic genes csgB, csgA, and csgD, a cellulose biosynthetic gene bcsA, and an endogenous control (rpoB) were designed using Primer Express (version 3.0) software (Table 1). E. cloacae GS1 and its sdiA mutant were grown for 8 h statically at 25°C in LB medium without NaCl. Total RNA was prepared from these cultures using the Qiagen RNeasy protocol. Following DNase treatment, cDNA was synthesized by random priming using a RevertAid first-strand cDNA synthesis kit according to the manufacturer's instructions. Synthesized cDNA was quantified, and 100 ng was supplied as the template for quantitative PCR performed in a total reaction volume of 25 μl, as instructed by the Qiagen QuantiFast SYBR green PCR kit manual, on an Applied Biosystems 7500 real-time PCR system. Two independent experiments were performed, where expression of each gene was measured in triplicate. The wild-type gene expression was used as a calibrator to determine the relative change in gene expression.
RESULTS AND DISCUSSION
Rice root exudate contains NAHL-like substances that activate SdiA in E. cloacae GS1.
Extracts of the spent nutrient solution after rice seedling growth under gnotobiotic conditions and extracts of sterile root tissue were found to activate the traI-traR-based NAHL biosensor A. tumefaciens NT1(pZLR4), as shown by the appearance of a spot slightly below the standard OHHL when analyzed by thin-layer chromatography (Fig. 1a). This confirmed that NAHL-like compounds were produced by rice roots and were found in the root exudates. Production of NAHL mimics by plant tissues has been shown before (10, 12). In the natural environment, the rhizosphere contains a mixture of beneficial and pathogenic bacteria producing their cognate NAHLs (26, 27). Thus, the rice rhizosphere environment is likely to contain NAHLs and NAHL mimics from microbial and plant sources, respectively, which could affect population density-dependent functions in bacteria.
Fig 1.

Thin-layer chromatography of rice root exudate extracts and assay for NAHL mimics. (a) Thin-layer chromatography of standard OHHL (lane 1), extract of sterile nutrient solution (lane 2), extract of the spent nutrient solution in which rice seedlings were grown for 10 days (lane 3), extract of the gnotobiotically grown rice root tissue (lane 4), and acetonitrile (lane 5). Chromatograms were developed by overlaying sheets with a soft-agar suspension of the NAHL biosensor A. tumefaciens NT1(pZLR4). (b) Bioluminescence of E. cloacae GS1(pBA405) (wild type [W]) and the absence of bioluminescence in E. cloacae GS1 sdiA::Kmr(pBA405) (mutant [M]) 6 h after overlaying on 3-day-old, surface-sterilized, and germinated rice seeds. Sterile filter paper discs containing 200 pmol of N-(3-oxohexanoyl)-l-homoserine lactone overlaid with E. cloacae GS1(pBA405) served as a positive control (P).
To test whether E. cloacae GS1 produces NAHLs, we examined the concentrated extracts of E. cloacae GS1 culture supernatants in different media for the presence of NAHLs with biosensors such as A. tumefaciens NT1(pZLR4), C. violaceum CV026, E. coli(pSB401), E. coli(pSB406), and E. coli(pSB1075). None of the NAHL biosensors used responded to the E. cloacae GS1 culture supernatant extracts. Analysis of the recently sequenced genome (GenBank accession number AJXP00000000.1) of E. cloacae GS1 indicated the absence of any luxI homolog which is essential for synthesis of NAHLs but showed the presence of SdiA, one of the well-studied LuxR-family transcriptional regulators. PCR with primers SAF1 and SAF2 produced a single 951-bp amplified product containing sdiA as a 723-bp open reading frame. The deduced protein sequence was 240 amino acids long and contained conserved domains belonging to autoinducer binding (Autoind_bind) and helix-turn-helix (HTH) superfamilies, which are characteristic of LuxR-family transcriptional regulators. Functional SdiA in E. cloacae GS1 was detected by transforming this strain with the biosensor plasmid pBA405 containing an SdiA-NAHL complex-activated promoter upstream of the luxCDABE operon. Observation of bioluminescent zones around discs impregnated with NAHLs overlaid with E. cloacae GS1(pBA405) and the absence of bioluminescent zones when NAHL-impregnated discs were overlaid with E. cloacae GS1 sdiA::Kmr(pBA405) confirmed the SdiA function in the parent strain and the absence of the same in the sdiA mutant (Fig. 2). We wanted to understand why E. cloacae GS1 had preserved a response regulator for a QS signal which it was unable to synthesize. SdiA of E. coli and Salmonella has been reported to regulate gene expression in response to environmental NAHLs. When E. cloacae GS1(pBA405) was overlaid on surface-sterilized and germinated rice seeds, a diffused zone of bioluminescence appeared around the seeds. However, such bioluminescent zones were absent when the sdiA mutant containing pBA405 was overlaid on germinated rice seeds (Fig. 1b). These results confirmed that NAHL mimics were produced by the germinated rice seeds and induced SdiA-dependent bioluminescence in E. cloacae GS1(pBA405), while they were unable to do so in the sdiA mutant background. It is known that cell-cell communications at the cross-kingdom level between plant hosts and their microbial symbionts play a regulatory role in host colonization (see references 28 and 29 and references therein). Thus, it was proposed that E. cloacae GS1 sdiA could be conserved in evolution to sense and regulate gene expression in response to environmental NAHLs in the rice rhizosphere. Thus, to gain a deeper insight into the pleiotropic effects that SdiA may have on root colonization in E. cloacae GS1, sdiA was disrupted and the resulting mutant was characterized in vitro and in planta.
Fig 2.
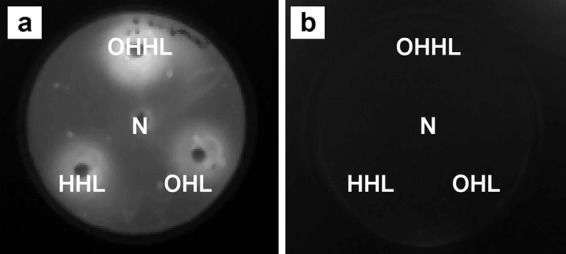
Disc diffusion assay for detection of SdiA activity in E. cloacae GS1. Sterile filter paper discs impregnated with acetonitrile (negative control [N]) or 200 pmol of OHHL, HHL, and OHL were placed on an LB agar plate and overlaid with a soft-agar suspension of E. cloacae GS1(pBA405) (a) or E. cloacae GS1 sdiA::Kmr(pBA405) (b). After incubation at 37°C for 6 h, the plates were imaged with a charge-coupled-device camera to detect SdiA-dependent, NAHL-induced bioluminescence.
The sdiA mutant of E. cloacae GS1 shows improved rice root colonization and biofilm formation.
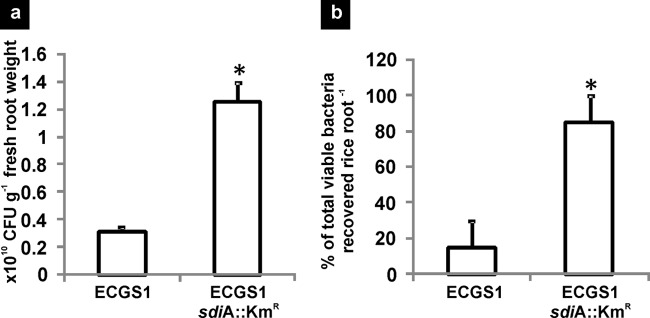
Disruption of sdiA in the mutant strain was confirmed by PCR and an NAHL disc diffusion assay. The absence of bioluminescent zones around discs impregnated with different NAHLs and overlaid with the sdiA mutant containing pBA405 confirmed the loss of SdiA function in E. cloacae GS1 (Fig. 2b). When inoculated on rice roots, the sdiA mutant efficiently colonized rice roots and displayed a 4-fold significant increase in population (P < 0.01) over that of the wild type in 7 days (Fig. 3a). Similarly, when the wild type and sdiA mutant were allowed to compete for the same rhizospheric niche, the sdiA mutant clearly outcompeted the wild type, forming 85% of the total population recovered from the roots, while the wild type formed only 15% of the same (Fig. 3b). These observations imply that inactivation of sdiA improves rice root colonization and also provides a competitive advantage to the sdiA mutant over the wild type during root colonization. The in trans complemented sdiA mutant achieved population sizes similar to those of the wild type on rice roots over the 7-day period, eliminating the possibility of secondary mutations (Table 2). These findings are similar to those in an earlier report (25) on E. coli O157:H7 where inactivation of sdiA led to a significant increase in cellular adhesion and biofilm formation.
Fig 3.
Rice root colonization by wild-type E. cloacae GS1 (ECGS1) and its isogenic sdiA::Kmr mutant (ECGS1 sdiA::Kmr) individually (a) and competitively (b). All colonization experiments were performed thrice independently with at least 4 replicates per experiment. Data shown are the means of replicates from one representative experiment, and error bars indicate SDs. In competitive colonization, counts of the wild type and mutant are expressed as a percentage of the total viable cells recovered per root. *, significant differences in relation to the wild type (P < 0.01).
Table 2.
Sensitivity of E. cloacae GS1 and the complemented sdiA mutant to 1 μM OHHL during root colonization and biofilm formationa
| Strain | Root colonization (no. of CFU g−1 fresh root wt [1010]) with: |
Biofilm formation (OD590) with: |
||
|---|---|---|---|---|
| Acetonitrile | 1 μM OHHL | Acetonitrile | 1 μM OHHL | |
| E. cloacae GS1 | 0.31 ± 0.03 | 0.14 ± 0.02 | 0.87 ± 0.05 | 0.70 ± 0.07 |
| E. cloacae GS1 sdiA::Kmr | 1.25 ± 0.14* | 1.19 ± 0.30* | 1.47 ± 0.13* | 1.50 ± 0.09* |
| E. cloacae GS1 sdiA::Kmr(pBsdiA) | 0.19 ± 0.13 | 0.097 ± 0.08 | 0.72 ± 0.08 | 0.89 ± 0.04 |
The sdiA mutant was insensitive to the presence of OHHL. Biofilm assays and colonization experiments were performed thrice independently with at least 4 replicates each. Data shown are the means from one such experiment. *, significant differences in relation to the wild type (P < 0.01).
Earlier reports have shown that mutations in sdiA significantly alter biofilm formation by E. coli in response to NAHLs (see reference 30 and references therein). Similarly, significant differences in the architecture, robustness, quality, and quantity of E. cloacae GS1 and its sdiA mutant biofilms were visualized by light microscopic observation of the air-medium interface biofilm. A thin layer of cells formed on the air side of the wild-type biofilm, while it was a thick layer of cells cemented together by a matrix that formed on the air side in the case of the sdiA mutant biofilm (Fig. 4a). Quantification of biofilm-forming ability, as described in Materials and Methods, indicated a 1.6-fold, significant increase (P < 0.001) in the biofilm-forming ability of the sdiA mutant over that of the parent strain (Fig. 4b). These findings further substantiated our hypothesis of an overall increase in cellular adhesion to biotic and abiotic surfaces upon sdiA inactivation. Atomic force microscopy analysis of the wild-type and sdiA mutant biofilms revealed that the thick layer observed in the mutant biofilms was a matrix that cemented the cells together. Clear images of cells forming cellular aggregates from the E. cloacae GS1 biofilms are shown in Fig. 4c. It was difficult to image individual cells forming the sdiA mutant aggregates due to this thick extracellular cementing matrix. Enterobacterial adhesion, invasion, and resistance to antibacterials are known to be mediated by formation of an extracellular matrix comprised of cellulose and curli fimbriae (31). In E. coli O157:H7, the expression of genes encoding fimbrial structures that are essential for bacterial adhesion was significantly higher in an sdiA mutant strain than the parent. It was proposed that this increase in curli fimbria production increased attachment of E. coli to HEp-2 cells 2-fold, which may contribute toward better colonization and pathogenesis (25). To verify whether sdiA inactivation in E. cloacae GS1 causes curli fimbria/cellulose overproduction, the wild type and sdiA mutant were plated on medium containing Congo red, which stains both curli and cellulose. E. cloacae GS1 produced pale red colonies, while its isogenic sdiA mutant produced pale red colonies with dark red centers (a fish eye-like appearance), as shown in Fig. 5a, which is characteristic of curli/cellulose overproduction. When complemented with functional sdiA, E. cloacae GS1 sdiA::Kmr exhibited a wild-type-like phenotype. These findings suggest that SdiA has a negative effect on cellular adhesion by its control over cellulose/curli production. SdiA is shown to be a strong transcriptional repressor of the curli biosynthetic operon (csg) in enterohemorrhagic E. coli O157:H7 (25). To check if similar regulatory mechanisms are operational in E. cloacae GS1, we performed quantitative RT-PCR to analyze curli and cellulose biosynthetic gene expression in the wild type and mutant strain (Fig. 5b). We observed an ∼53-fold higher expression level of csgB, the minor subunit of curli fimbriae, and an ∼12-fold elevated expression level of csgA, the major subunit of curli, in the sdiA mutant, which substantiated the curli overproduction phenotype observed for this strain on Congo red agar. We also observed ∼14-fold higher levels of csgD expression in the sdiA mutant. CsgD is a known positive regulator of the curli biosynthetic genes, and its expression is not self-regulated, as in the case of many transcriptional regulators (32). No significant change in the expression of bcsA, coding for the cellulose synthase catalytic subunit, was observed. This finding correlated with our results in a cellulose production assay where no variation in fluorescence was visible when E. cloacae GS1 and its isogenic sdiA mutant were grown on calcofluor white-containing plates (data not shown).
Fig 4.
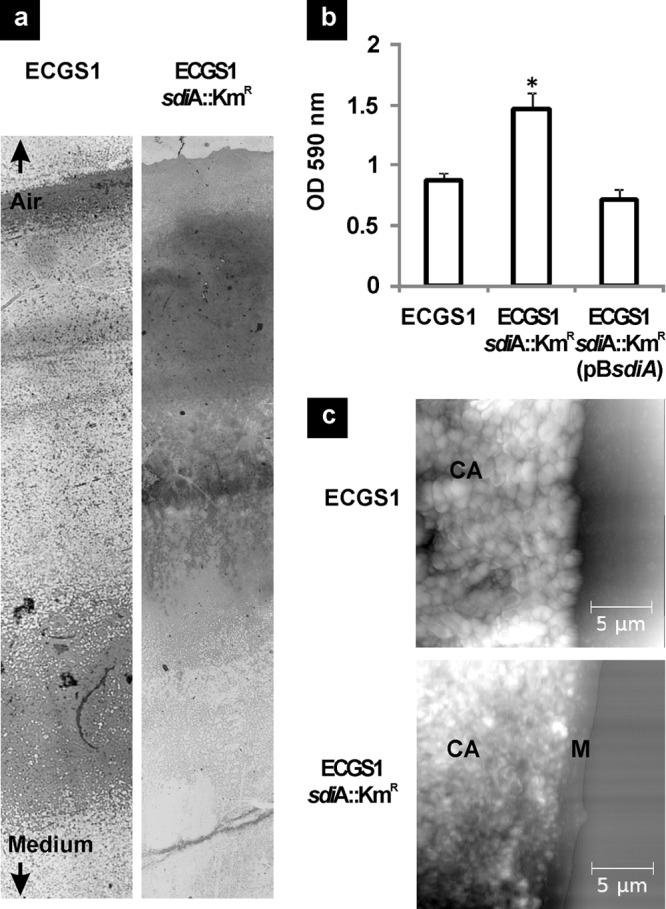
Biofilm formation by wild type and sdiA mutant of E. cloacae GS1. (a) Interface biofilms of E. cloacae GS1 (ECGS1) and its sdiA mutant (ECGS1 sdiA::Kmr) grown on glass slides observed by light microscopy after crystal violet staining showing significant differences in biofilm architecture. (b) Biofilm formation by E. cloacae GS1, its sdiA mutant, and the sdiA mutant complemented in trans on polystyrene quantified by crystal violet staining and measurement of the absorbance at 590 nm. The assay was performed thrice independently with five replicates each. Data shown are the means from one such experiment, and error bars indicate SDs. *, significant differences in relation to the wild type (P < 0.001). (c) Atomic force micrographs of E. cloacae GS1 and its sdiA mutant showing cellular aggregates (CA). A matrix-like structure (M) encapsulating the cellular aggregates of the sdiA mutant is noted.
Fig 5.
Curli overproduction by E. cloacae GS1 sdiA::Kmr compared to the level of production by the parent strain. (a) Curli production phenotype of E. cloacae GS1, its sdiA mutant, and the sdiA mutant complemented in trans on Congo red medium. All three strains were grown on LB agar without NaCl containing Congo red at 25°C for 48 h, and the plate was scanned on a high-resolution gel scanner. E. cloacae GS1 produced pale red colonies (w), while the sdiA mutant produced red colonies with dark red centers showing a typical fish eye-like appearance characteristic of curli overproduction (m). The sdiA mutant complemented in trans appeared similar to the wild type (p). (b) Effect of sdiA inactivation on the expression of curli (csgB, csgA, csgD) and cellulose biosynthesis (bcsA) genes in E. cloacae GS1. Gene expression values of the wild type were set equal to 1 to calculate the relative change in gene expression in the sdiA mutant. Quantitative RT-PCR was performed twice independently, including triplicate measurements of gene expression per gene, and yielded similar results. Data shown are the mean relative change in expression ± standard deviation from one such experiment. *, significant differences in relation to the wild type (P < 0.001).
NAHLs negatively regulate rice root colonization and biofilm formation.
The classical model of NAHL-based quorum sensing involves the LuxI family of proteins, responsible for the synthesis of NAHLs, and the LuxR family of proteins, responsible for regulation of gene expression in an NAHL-dependent manner (1). Recently, many bacterial species were found to contain orphaned LuxR regulators or LuxR-family solos, which are evolutionarily conserved in many bacterial species and may play key roles in sensing and responding to environmental NAHL/NAHL-like signals (see references 33 and 34 and references therein). The effect of NAHLs on root colonization and biofilm formation by E. cloacae GS1 and its sdiA mutant was studied with and without OHHL supplementation. In the presence of OHHL, root colonization by E. cloacae GS1 decreased by 2.2-fold (P < 0.01), while biofilm formation on polystyrene was reduced by 1.2-fold (P < 0.01). This observation is consistent with the findings of an earlier study demonstrating that NAHLs impose a negative effect on biofilm formation in E. coli K-12 BW25113 (8). The E. cloacae GS1 sdiA mutant, however, displayed no significant difference in root colonization and biofilm formation in the presence or absence of OHHL (Table 2). The sdiA mutant's insensitivity to the inhibitory effect of NAHLs on root colonization and biofilm formation suggested that the observed effect was strictly SdiA dependent. Further supporting our inference, the sdiA mutant complemented with wild-type sdiA in trans displayed a sensitivity to the presence of NAHLs during root colonization and biofilm formation. In summary, SdiA of E. cloacae GS1 imposes negative regulatory effects on cellular adhesion in both the presence and absence of NAHL signals. However, the inhibitory effect is intensified in the presence of extraneous NAHLs. Since we find that rice plants synthesize NAHL-like substances, we believe that these NAHL mimics could be cross-kingdom signals which modulate NAHL-inducible gene expression in rhizosphere colonizers. Symbiotic plant-microbe interactions have a certain level of host specificity. Similarly, plants do impose restrictions on the type and number of bacteria that colonize their rhizosphere. This phenomenon, termed the “rhizosphere effect,” heavily influences the bacterial communities associated with the plant, as it allows preferential selection of a few bacterial species, inhibiting the growth of some and not affecting some (35). It is known that root colonization by beneficial bacteria involves an attraction phase, a settlement phase, and a residence phase. The residence phase is when the root surface attains a maximal bacterial population, beyond which the bacterial count becomes dependent on root growth (36). Thus, any plant allows only permissible populations to inhabit its rhizosphere, irrespective of the total inoculated population. In the case of beneficial bacteria, this permissible population is usually sufficient to promote plant growth and improve nutrition. Our findings presented in this article suggest that inactivation of sdiA leads to a 4-fold increase in the permissible population allowed on the rice root surface. We propose that this increase could be a combinatorial effect of derepression of the curli biosynthetic genes and insensitivity to rice-derived NAHL mimics as a result of sdiA inactivation.
ACKNOWLEDGMENTS
This work was financially supported by the Indian Council for Agricultural Research [NBAIM/AMAAS/2007-2012/MG (5)/PG/BG/3]. Central facilities at the Center for Excellence in Genomic Sciences, UGC Networking Resource Center in Biological Sciences, and DBT-IPLS are acknowledged.
M.S. gratefully acknowledges constructive discussions with Hussain Munavar, Manoharan Ramasamy, Chitralekha Manoharan, Madhankumar Anandhakrishnan, and Shanmughapriya Vinod. We thank Shanmugasundaram Sambandam and Balachandar Kalaiarasan for their technical support in atomic force microscopy.
A. tumefaciens NT1(pZLR4) and pBA405 were generous gifts from Stephen Farrand, University of Illinois, and Brian Ahmer, Ohio State University, respectively. pSB401, pSB406, and pSB1075 were obtained from Paul Williams, University of Nottingham. pIVETP was a gift from Gail Preston, University of Oxford.
Footnotes
Published ahead of print 19 October 2012
REFERENCES
- 1. Fuqua WC, Winans SC, Greenberg EP. 1994. Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176: 269–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Antunes LC, Ferreira RB, Buckner MM, Finlay BB. 2010. Quorum sensing in bacterial virulence. Microbiology 156: 2271–2282 [DOI] [PubMed] [Google Scholar]
- 3. Koutsoudis MD, Tsaltas D, Minogue TD, von Bodman SB. 2006. Quorum-sensing regulation governs bacterial adhesion, biofilm development, and host colonization in Pantoea stewartii subspecies stewartii. Proc. Natl. Acad. Sci. U. S. A. 103: 5983–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Case RJ, Labbate M, Kjelleberg S. 2008. AHL-driven quorum-sensing circuits: their frequency and function among the proteobacteria. ISME J. 2: 345–349 [DOI] [PubMed] [Google Scholar]
- 5. Dyszel JL, Soares JA, Swearingen MC, Lindsay A, Smith JN, Ahmer BM. 2010. E. coli K-12 and EHEC genes regulated by SdiA. PLoS One 5: e8946 doi:10.1371/journal.pone.0008946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kanamaru K, Tatsuno I, Tobe T, Sasakawa C. 2000. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohaemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38: 805–816 [DOI] [PubMed] [Google Scholar]
- 7. Michael B, Smith JN, Swift S, Heffron F, Ahmer BM. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183: 5733–5742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lee J, Jayaraman A, Wood TK. 2007. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 7: 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perez-Montano F, Guasch-Vidal B, Gonzalez-Barroso S, Lopez-Baena FJ, Cubo T, Ollero FJ, Gil-Serrano AM, Rodriguez-Carvajal MA, Bellogin RA, Espuny MR. 2011. Nodulation-gene-inducing flavonoids increase overall production of autoinducers and expression of N-acyl homoserine lactone synthesis genes in rhizobia. Res. Microbiol. 162: 715–723 [DOI] [PubMed] [Google Scholar]
- 10. Teplitski M, Robinson JB, Bauer WD. 2000. Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol. Plant Microbe Interact. 13: 637–648 [DOI] [PubMed] [Google Scholar]
- 11. Teplitski M, Chen H, Rajamani S, Gao M, Merighi M, Sayre RT, Robinson JB, Rolfe BG, Bauer WD. 2004. Chlamydomonas reinhardtii secretes compounds that mimic bacterial signals and interfere with quorum sensing regulation in bacteria. Plant Physiol. 134: 137–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Degrassi G, Devescovi G, Solis R, Steindler L, Venturi V. 2007. Oryza sativa rice plants contain molecules that activate different quorum-sensing N-acyl homoserine lactone biosensors and are sensitive to the specific AiiA lactonase. FEMS Microbiol. Lett. 269: 213–220 [DOI] [PubMed] [Google Scholar]
- 13. Bacilio-Jiménez M, Aguilar-Flores S, Ventura-Zapata E, Pérez-Campos E, Bouquelet S, Zenteno E. 2003. Chemical characterization of root exudates from rice (Oryza sativa) and their effects on the chemotactic response of endophytic bacteria. Plant Soil 249: 271–277 [Google Scholar]
- 14. Elasri M, Delorme S, Lemanceau P, Stewart G, Laue B, Glickmann E, Oger PM, Dessaux Y. 2001. Acyl-homoserine lactone production is more common among plant-associated Pseudomonas spp. than among soilborne Pseudomonas spp. Appl. Environ. Microbiol. 67: 1198–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shankar M, Ponraj P, Ilakkiam D, Gunasekaran P. 2011. Root colonization of a rice growth promoting strain of Enterobacter cloacae. J. Basic Microbiol. 51: 523–530 [DOI] [PubMed] [Google Scholar]
- 16. Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 17. Farrand SK, Qin Y, Oger P. 2002. Quorum-sensing system of Agrobacterium plasmids: analysis and utility. Methods Enzymol. 358: 452–484 [DOI] [PubMed] [Google Scholar]
- 18. Shaw PD, Ping G, Daly SL, Cha C, Cronan JE, Jr, Rinehart KL, Farrand SK. 1997. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc. Natl. Acad. Sci. U. S. A. 94: 6036–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, Daykin M, Lamb JH, Swift S, Bycroft BW, Stewart GS, Williams P. 1997. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology 143(Pt 12): 3703–3711 [DOI] [PubMed] [Google Scholar]
- 20. Winson MK, Swift S, Fish L, Throup JP, Jorgensen F, Chhabra SR, Bycroft BW, Williams P, Stewart GS. 1998. Construction and analysis of luxCDABE-based plasmid sensors for investigating N-acyl homoserine lactone-mediated quorum sensing. FEMS Microbiol. Lett. 163: 185–192 [DOI] [PubMed] [Google Scholar]
- 21. Rainey PB. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1: 243–257 [DOI] [PubMed] [Google Scholar]
- 22. Pratt LA, Kolter R. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol. Microbiol. 30: 285–293 [DOI] [PubMed] [Google Scholar]
- 23. Kawarai T, Furukawa S, Narisawa N, Hagiwara C, Ogihara H, Yamasaki M. 2009. Biofilm formation by Escherichia coli in hypertonic sucrose media. J. Biosci. Bioeng. 107: 630–635 [DOI] [PubMed] [Google Scholar]
- 24. Jonas K, Tomenius H, Kader A, Normark S, Romling U, Belova LM, Melefors O. 2007. Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol. 7: 70 doi:10.1186/1471-2180-7-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharma VK, Bearson SM, Bearson BL. 2010. Evaluation of the effects of sdiA, a luxR homologue, on adherence and motility of Escherichia coli O157:H7. Microbiology 156: 1303–1312 [DOI] [PubMed] [Google Scholar]
- 26. Pierson LS, III, Wood DW, Pierson EA. 1998. Homoserine lactone-mediated gene regulation in plant-associated bacteria. Annu. Rev. Phytopathol. 36: 207–225 [DOI] [PubMed] [Google Scholar]
- 27. Steidle A, Sigl K, Schuhegger R, Ihring A, Schmid M, Gantner S, Stoffels M, Riedel K, Givskov M, Hartmann A, Langebartels C, Eberl L. 2001. Visualization of N-acylhomoserine lactone-mediated cell-cell communication between bacteria colonizing the tomato rhizosphere. Appl. Environ. Microbiol. 67: 5761–5770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Quinones B, Dulla G, Lindow SE. 2005. Quorum sensing regulates exopolysaccharide production, motility, and virulence in Pseudomonas syringae. Mol. Plant Microbe Interact. 18: 682–693 [DOI] [PubMed] [Google Scholar]
- 29. Wei HL, Zhang LQ. 2006. Quorum-sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Antonie Van Leeuwenhoek 89: 267–280 [DOI] [PubMed] [Google Scholar]
- 30. Lee J, Maeda T, Hong SH, Wood TK. 2009. Reconfiguring the quorum-sensing regulator SdiA of Escherichia coli to control biofilm formation via indole and N-acylhomoserine lactones. Appl. Environ. Microbiol. 75: 1703–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zogaj X, Bokranz W, Nimtz M, Romling U. 2003. Production of cellulose and curli fimbriae by members of the family Enterobacteriaceae isolated from the human gastrointestinal tract. Infect. Immun. 71: 4151–4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hammar M, Arngvist A, Bian Z, Oslen A, Normark S. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18: 661–670 [DOI] [PubMed] [Google Scholar]
- 33. Patankar AV, Gonzalez JE. 2009. An orphan LuxR homolog of Sinorhizobium meliloti affects stress adaptation and competition for nodulation. Appl. Environ. Microbiol. 75: 946–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Subramoni S, Venturi V. 2009. LuxR-family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology 155: 1377–1385 [DOI] [PubMed] [Google Scholar]
- 35. Green SJ, Michel FC, Hadar Y, Minz D. 2007. Contrasting patterns of seed and root colonization by bacteria from the genus Chryseobacterium and from the family Oxalobacteriaceae. ISME J. 1: 291–299 [DOI] [PubMed] [Google Scholar]
- 36. Urgel ME, Kolter R, Ramos JL. 2002. Root colonization by Pseudomonas putida: love at first sight. Microbiology 148: 341–343 [DOI] [PubMed] [Google Scholar]