Abstract
The DedA protein family is a highly conserved and ancient family of membrane proteins with representatives in most sequenced genomes, including those of bacteria, archaea, and eukarya. The functions of the DedA family proteins remain obscure. However, recent genetic approaches have revealed important roles for certain bacterial DedA family members in membrane homeostasis. Bacterial DedA family mutants display such intriguing phenotypes as cell division defects, temperature sensitivity, altered membrane lipid composition, elevated envelope-related stress responses, and loss of proton motive force. The DedA family is also essential in at least two species of bacteria: Borrelia burgdorferi and Escherichia coli. Here, we describe the phylogenetic distribution of the family and summarize recent progress toward understanding the functions of the DedA membrane protein family.
INTRODUCTION
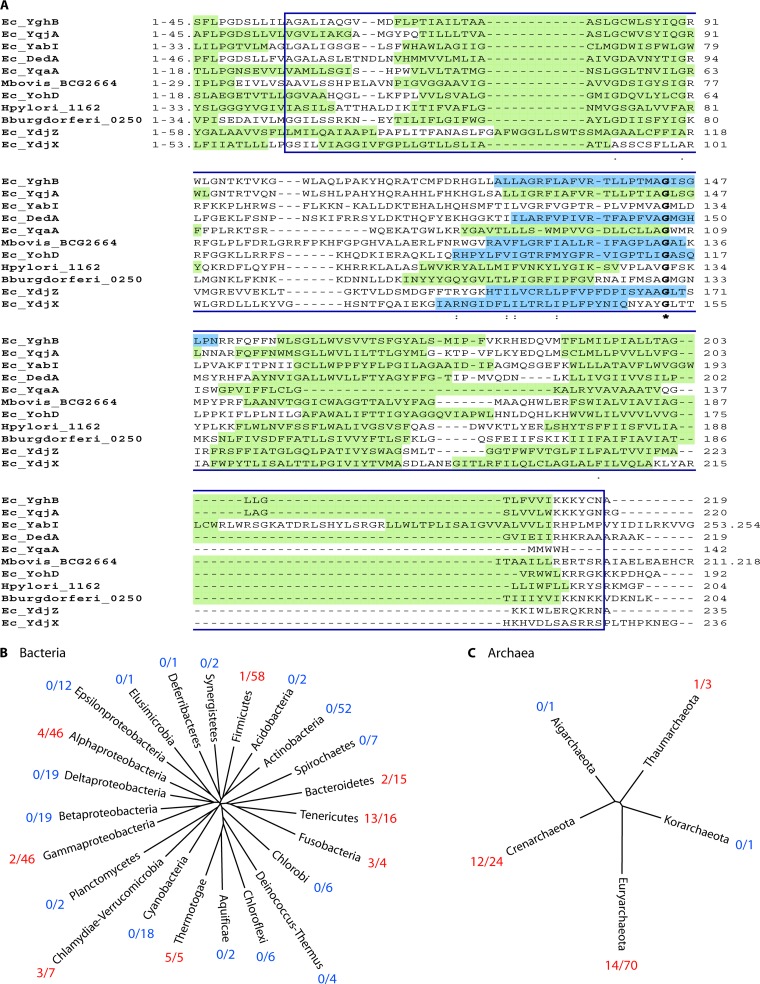
It is predicted that roughly 20 to 25% of polypeptides are integral membrane proteins (1). In contrast, fewer than 1% of all known protein structures in the Protein Data Bank are membrane proteins, and the functions of many are only poorly understood (2, 3). The Escherichia coli dedA gene (EcdedA) was given its name in a 1987 publication for its presence in the DNA sequence of a 9.7-kb fragment between hisT and purF (downstream [of hisT] E. coli DNA gene A) (4). For clarity, we will refer to the DedA family to describe the protein family and EcDedA or EcdedA to describe specifically the E. coli DedA protein or gene. Despite these rather mundane origins, it is now appreciated that the DedA family is a highly conserved protein family represented in most sequenced genomes encoding membrane proteins of unknown function (5). There are virtually thousands of prokaryotic homologs of bacterial DedA proteins currently found in the NCBI protein database, and many sequenced bacterial genomes encode multiple family members (Table 1) (8). However, they remain difficult to classify, as the polypeptides do not resemble known enzymes, transporters, channels, or signaling proteins. While there are examples of multidomain secondary transporters and enzymes in the database containing DedA or “SNARE-associated” domains (9), DedA family proteins are, in most cases, unique polypeptides with no other commonly associated domains. The polypeptides belonging to the DedA family typically contain 4 to 6 predicted transmembrane domains, between 200 and 250 amino acids, and a conserved domain. This DedA domain contains both transmembrane and cytoplasmic domains, as well as a likely amphipathic helix. A strictly conserved amino acid sequence is not present across the entire domain; however, there is at least one universally conserved glycine residue which occurs in or near the potential amphipathic helix (Fig. 1A) and is found in all defined DedA proteins (NCBI Clusters of Orthologous Groups, COG0586; the DedA domain). Here, we summarize recent progress toward understanding the functions of DedA family membrane proteins.
Table 1.
Numbers of DedA family proteins (amino acid BLAST E value, <0.02) found in sequenced genomes of representative bacterial and archaeal speciesa
| Bacterial strain | No. of DedA family homologs |
|---|---|
| Escherichia coli K-12 | 8c |
| Salmonella enterica SL480 | 6 |
| Pseudomonas aeruginosa PAO1 | 5 |
| Helicobacter pylori J99 | 2 |
| Vibrio cholerae El Tor N16961 | 3 |
| Caulobacter crescentus CB15 | 3 |
| Neisseria meningitidis Z2491 | 3 |
| Borrelia burgdorferi B31 | 1b |
| Bacillus subtilis strain 168 | 6 |
| Bacillus anthracis strain Ames | 8 |
| Mycobacterium tuberculosis H37Rv | 4 |
| Chlamydia trachomatis D/UW-3/CX | 0 |
| Synechocystis sp. strain PCC6803 | 3 |
| Halobacterium salinarum NRC-1 | 1 |
Significant homologs (Protein BLAST E value, <0.02) of E. coli YqjA, YghB, DedA, YohD, YabI, YdjZ, YdjX, and YqaA were included in the numbers of proteins displayed in the second column.
The B. burgdorferi B31DedA family gene BB0250 has been demonstrated to be essential (7).
Fig 1.
Phylogenetic analysis of the DedA protein family. (A) Alignment of E. coli DedA proteins and homologs found in Borrellia burgdorferi, Mycobacterium bovis, and Helicobacter pylori from the NCBI database to illustrate the DedA domain, COG0586 (boxed in region). Predicted transmembrane (TM) domains are highlighted in green, and partial TM regions, possibly amphipathic helixes, are highlighted in blue. The singularly conserved amino acid residue of the DedA domain (glycine) is in bold. Of interest, the only conserved glycine residue is in or near the amphipathic helix for all aligned members. The TM prediction software used was TMHMM (84). (B and C) Bacterial (B) and archaeal (C) domain representative trees are shown. Numbers in red demonstrate the proportion of species lacking a significant DedA homolog (Protein BLAST E value, ≤10−4) in each bacterial (B) or archaeal (C) phylum. Otherwise, all species of each phylum contain at least one significant DedA homolog (numbers in blue). Phylogenetic trees were constructed with MEGA (85), using a single 16S rRNA sequence from a representative species of each phylum. Previously published phylogenetic trees for both Bacteria (350 species) (10) and Archaea (100 species) (11) were used as a basis for phylogenetic analyses, though additional species were investigated; presented values are solely from published trees. Significant DedA homologs are found within the Thermotogae phylum although not in the five completed genomes analyzed.
PHYLOGENETIC DISTRIBUTION OF THE DedA FAMILY
To investigate the distribution of the DedA protein family, we scanned a total of 350 sequenced bacterial genomes (10) and 100 sequenced archaeal genomes (11) found in the NCBI database with Protein BLAST using amino acid sequences of all eight E. coli DedA proteins. Using a very conservative Protein BLAST score (E value, ≤10−4) as significant, we found that 33 (9.2%) bacterial species and 27 (27%) archaeal species lack a significant DedA homolog (Fig. 1B and C). The largest proportion of bacterial species that lack a significant DedA homolog can be found in the phylum Tenericutes (with 13/16 species lacking a clear DedA homolog), followed by the Thermotogae (5/5) and Alphaproteobacteria (4/46). As for the Archaea domain, the largest proportion of species lacking a significant DedA homolog is in the Euryarchaeota (14/70) and Crenarchaeota (12/24) phyla. However, it is important to note that the majority of sequenced archaeal species fall within the Euryarchaeota phylum (11).
The presence of a significant DedA homolog is not consistent among organisms of similar habitats; for example, a significant DedA homolog is present within Neisseria spp., Mycoplasma synoviae/M. fermentans, Anaplasma phagocytophilum, and Ehrlichia chaffeensis but absent in Chlamydia, Ureaplasma, and Neorickettsia sennetsu (12–14). In fact, a DedA member is not present among any of the sequenced Chlamydia/Chlamydophilia spp. or Ureaplasma spp.; however, Neisseria spp. have multiple, and some very significant (E value, <10−100) DedA homologs. Interestingly, the majority of Mycoplasma spp. and Rickettsia spp. do not have any significant DedA members, but DedA homologs are present within the genera, e.g., Mycoplasma synoviae/M. fermentans and Rickettsia felis/R. bellii (15, 16). The majority of Clostridium spp. (including C. botulinum) do have a DedA homolog, whereas Clostridium thermocellum does not (17–20). There are many additional genera in which the presence of a DedA homolog is variable, for example, Psychrobacter, Bartonella, and Mycobacterium (21, 22). The significance of the observed distribution of the DedA protein family is as yet unclear.
Another inconsistency in the distribution of DedA homologs is among the reduced genome symbionts and obligate symbionts of various organisms. The DedA family is found in the genomes of several symbionts, including Wigglesworthia glossinidia (23) and Buchnera spp. (24). Some symbionts that lack a DedA homolog are “Candidatus Sulcia muelleri,” “Candidatus Amoebobhilus asiaticus,” “Candidatus Phytoplasma mali,” “Candidatus Zinderia insecticola,” “Candidatus Carsonella ruddii,” “Candidatus Hodgkinia cicadicola,” and “Candidatus Tremblaya princeps” (reviewed in reference 25). The possibility exists that this variability of the DedA distribution is related to the genetic makeup and/or physiology of the symbionts' host species (26–28).
As for the archaeal domain, the distribution of DedA members is quite unpredictable; one note of interest is that no sequenced genomes within the Halobacteriales or Sulfolobales order lack a significant DedA homolog. Also, the reduced genomes of archaeal species “Candidatus Parvarchaeum spp.” and “Candidatus Micrarchaeum acidiphilum” all contain a significant DedA homolog (29). The presence of DedA homologs within several reduced genomes, both bacterial and archaeal, further supports the essentiality of the DedA protein family for species viability. Published phylogenetic trees of the bacterial and archaeal domains were used as a guide for investigating the distribution of DedA members (10, 11).
In regard to the distribution of the DedA protein family, there may be subtle differences between organisms that lack a DedA homolog, which has allowed for a select few to counteract the necessity of DedA proteins. For example, it is possible that other proteins have taken over their role or that symbiotic relationships enable this selective genotypic evolution. Regardless, these species have found a way to exist without the DedA protein family. It is important to note, however, that the identification of significant DedA homologs using a strict BLAST score cutoff (E value, ≤10−4) may have overlooked DedA family members with lower sequence identity to the E. coli DedA proteins.
MUTATIONS IN E. COLI yghB AND yqjA ARE SYNTHETICALLY LETHAL AT ELEVATED TEMPERATURES AND LEAD TO DEFECTS IN CELL DIVISION, ELEVATED STRESS, AND MEMBRANE DEFECTS
The E. coli genome encodes eight predicted members of the DedA family (yqjA, yghB, yabI, yohD, EcdedA, ydjX, ydjZ, and yqaA). Our interest in the DedA family was initiated by the observation that simultaneous deletion of yqjA and yghB from E. coli results in a strain (named BC202) that is temperature sensitive for growth and displays striking defects in cell division (Fig. 2A and B) (5, 30). YghB and YqjA are proteins of 219 and 220 amino acids, respectively, displaying 61% amino acid identity to each other and possessing likely four membrane-spanning domains with cytoplasmic N and C termini (Fig. 3). The other six E. coli homologs display roughly 25 to 30% amino acid identity with each other and YghB/YqjA.
Fig 2.
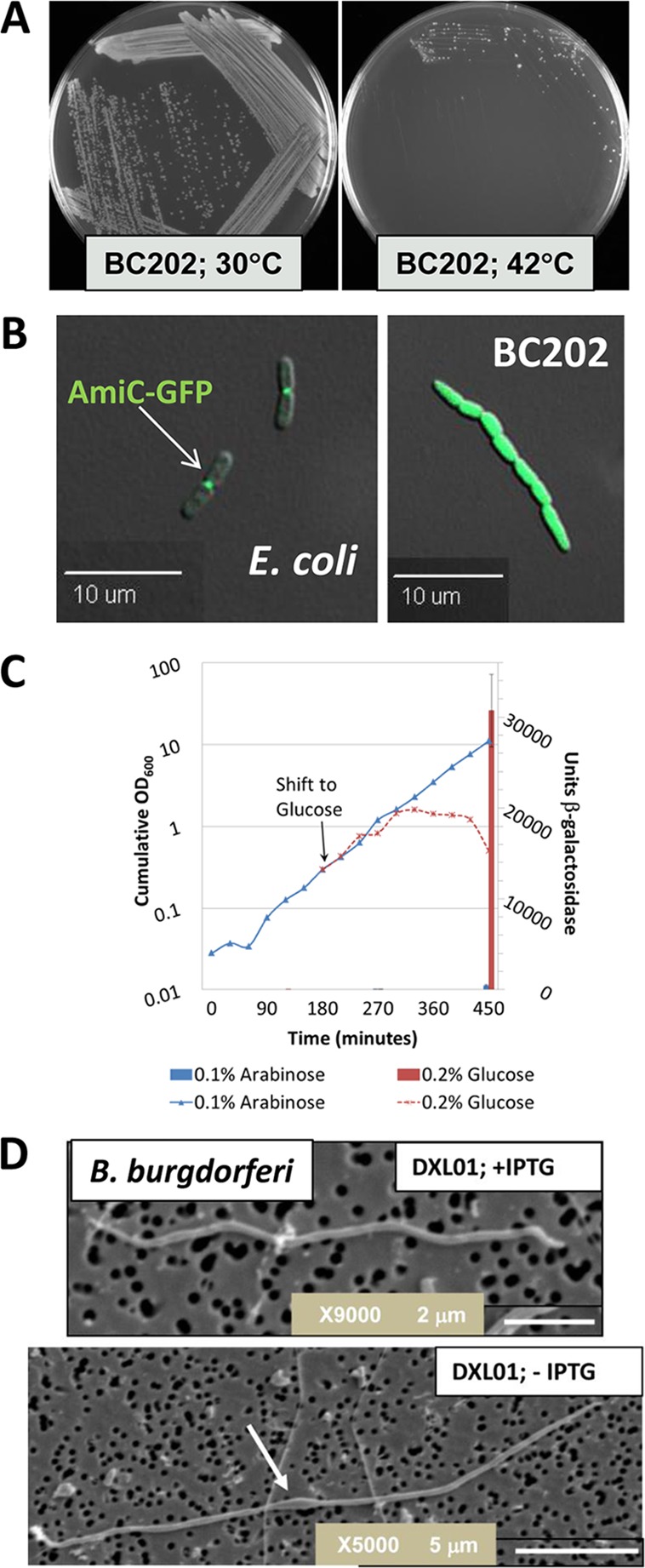
Characteristics of DedA family mutants. (A) BC202 (ΔyghB::Kanr ΔyqjA::Tetr) grows at 30°C but not at 42°C (5). The ΔyghB and ΔyqjA single mutants grow at all temperatures (not shown) (5, 62). (B) Cell division defects of BC202 are caused by failure to export periplasmic amidases via the twin arginine transport pathway. W3110 (left) and BC202 (right) transformed with plasmid pTB28 expressing AmiC-green fluorescent protein (GFP) fusion protein (31) were grown at 30°C and visualized with differential interference contrast (DIC) and fluorescence microscopy. The bright fluorescence observed in the right panel is due to cytoplasmic accumulation of AmiC-GFP (30). Expression of wild-type yghB from a plasmid restores wild-type appearance and AmiC export to BC202 (not shown) (30). (C) The E. coli DedA family is collectively essential at all temperatures. BAL801 (W3110, ΔydjXYZ::cam ΔyabI772 ΔEcdedA726 ΔyohD762 ΔyqjA785 ΔyqaA770 ΔyghB781::kan pBAD-EcdedA) fails to grow when EcDedA expression from a plasmid is repressed by growth in glucose (6). (D) The sole DedA family member of Borrelia burgdorferi is essential (not shown), and depletion of the protein causes cell division defects (7). B. burgdorferi DXL-01 (Δbb0250, pWTD0250 expressing bb0250 behind a borrelia-optimized lac promoter [86]) was cultured with either 1 mM (top) or 0 mM (bottom) IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 to 5 days and visualized by scanning electron microscopy. Depletion of BB0250 causes membrane bulging (arrow) and cell division defects. Images A to D are reproduced with permission (5–7, 30).
Fig 3.
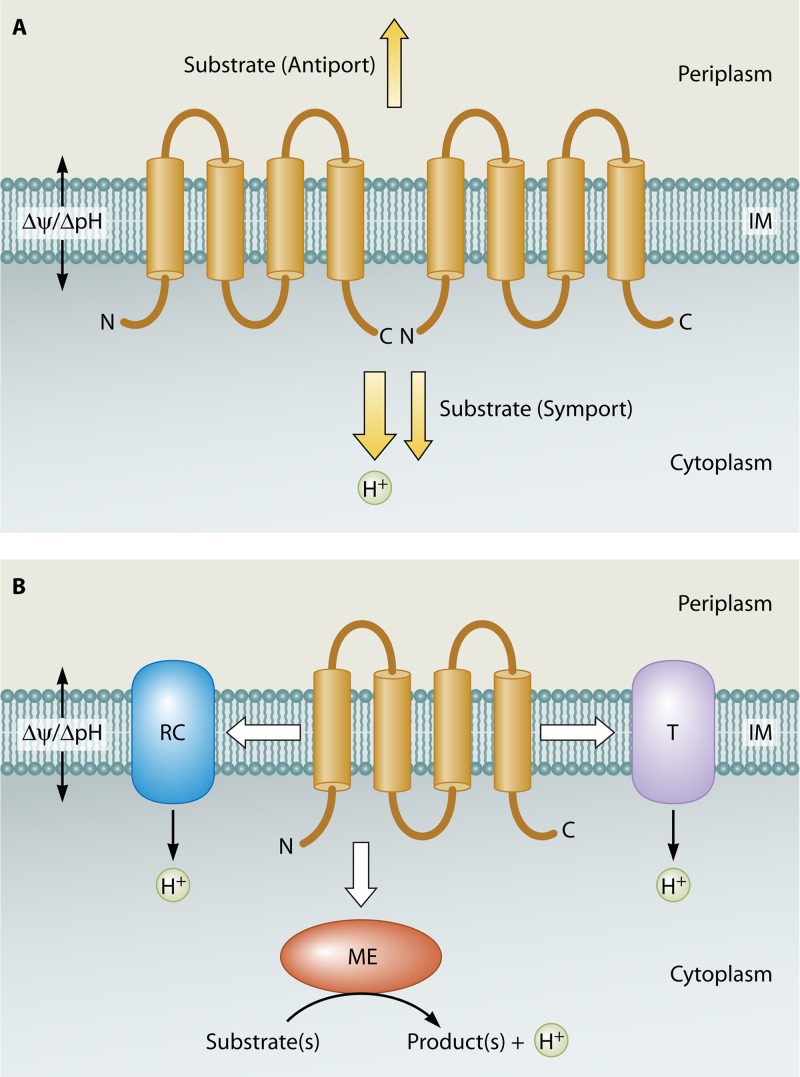
Potential physiological roles of E. coli YqjA and YghB. (A) Formation of YqjA homodimer as proposed for LeuT family members (8) transports protons into the cell coupled with symport or antiport of an uncharacterized substrate. The proposed function of YqjA here is similar to that of the Na+/K+-H+ antiporter MdfA (61, 74) with a significant role in pH/PMF homeostasis. Heterodimerization with YghB or an unknown partner is also a possibility here. (B) An alternative model demonstrating an indirect role for YqjA in pH/PMF homeostasis. Regulation of activity or functional modulation of RC (respiratory complexes), ME (metabolic enzymes), or certain classes of transporters (T) such as MdfA, all of which participate in the pH/PMF homeostasis mechanism in E. coli. The topological model of YqjA, comprising 4-transmembrane helices with cytoplasmic N and C termini, is derived from previously published data (87), SOSUI topological prediction software (88), and unpublished observations. IM, inner membrane.
The E. coli mutant BC202 (ΔyqjA ΔyghB) displays several intriguing phenotypes that reflect important functions for the DedA family. As mentioned, BC202 grows at 30°C but not 42°C (Fig. 2A) and does not complete cell division, forming chains of cells (5). We have demonstrated that the periplasmic amidases AmiA and AmiC are not exported to the periplasm in BC202 and that this is responsible for the cell division defect (Fig. 2B) (30). These amidases are normally exported across the inner membrane via the twin arginine transport (Tat) pathway in E. coli (31), a Sec-independent protein export pathway found in many bacteria and also present in archaea and plants (32–35). AmiA and AmiC are required for normal cell division and envelope integrity (36), and Δtat mutants display cell division defects due to loss of amidase export (31, 37). However, E. coli Δtat (T. Palmer, personal communication) and Δami mutants are not temperature sensitive for growth, unlike BC202. Therefore, the temperature sensitivity of BC202 is independent of the cell division phenotype. BC202 is also not hypersensitive to antibiotics or detergents (5), likely signifying the presence of an intact outer membrane, unlike the situation with Δtat mutants (37, 38) and mutants lacking periplasmic amidases (36). Interestingly, while the Borrelia Δbb0250 strain (described below) also exhibits cell division defects (Fig. 2D), the B. burgdorferi genome does not encode a functional twin arginine pathway or any predicted Tat substrates (39), indicating that the DedA family is involved in functions independent of the Tat pathway.
A number of extracytoplasmic stress response pathways are activated in BC202 under permissive growth conditions (see the accompanying paper by Sikdar et al [40]). E. coli responds to extracytoplasmic stress by activating one or more of several well-known stress response pathways such as the σE, Cpx, Psp, Bae, and Rcs pathways, which help the cells detect and combat alterations in their cell envelope when challenged with conditions that compromise envelope integrity (41). The Cpx and σE pathways are activated in response to disruptions in the folding of envelope proteins and loss of outer membrane integrity, and they have partially overlapping regulons (41–44). The Psp (phage shock protein) response is activated by perturbations in the integrity of the inner membrane by conditions that result in dissipation of the proton motive force (PMF) and/or change the physiological redox state of the cell (extreme heat shock, exposure to ethanol, ionophores, and pIV secretin stress) (45–47). The Bae stress response is induced by toxic compounds such as indole and ethanol (41, 48, 49). Finally, the Rcs pathway is activated by stresses that affect envelope composition (50–53) or the integrity of the peptidoglycan layer (54).
We observed that the Cpx, Psp, Rcs, and Bae stress responses are induced in BC202 under permissive growth conditions while σ32 (controlling the cytoplasmic heat shock response) and σE are not significantly induced (40). This nonspecific induction of multiple envelope stress responses is reminiscent of a general loss of envelope integrity when challenged with certain stresses such as growth in 5% ethanol (41). These findings demonstrate the critical importance of certain DedA family members in proper envelope function of E. coli.
It has been previously demonstrated that dissipation of the PMF results in induction of the Psp stress response (45, 55). Efficient Tat-mediated export of substrates also relies on optimal PMF (56, 57). The PMF in E. coli is comprised of two components—the transmembrane electrical membrane potential difference, Δψ (= ψin − ψout), with ψin (electrical potential inside) being more negative than ψout (electrical potential outside), and the transmembrane pH difference, ΔpH (= pHin − pHout), with pHin (intracellular pH) being more alkaline than pHout (extracellular pH) under normal growth conditions. We found that BC202 exhibits a significant loss of membrane potential compared to parent strain W3110 using the membrane-permeating dye JC-1, consistent with the observed activation of the Psp pathway in BC202 (40). BC202 (as well as the single ΔyqjA mutant [58]) cannot survive at elevated pH, but temperature sensitivity of BC202 is rescued when it is grown at a lower pH (pH 6.0) or when the mdfA (cmr) gene is overexpressed (40). MdfA is a member of the major facilitator superfamily involved in drug efflux and is an Na+/K+-H+ antiporter (59). It is likely that BC202 is unable to maintain the PMF and requires protons outside the cell to be exchanged with sodium/potassium ions. The single ΔmdfA mutant is also sensitive to mild alkaline pH and grows poorly at neutral pH (60). MdfA is unique in that it is capable of transporting diverse substrates across the membrane using both components of the electrochemical gradient (ΔΨ and ΔpH) for electrogenic transport of neutral compounds, while using only ΔpH for electroneutral transport of cationic compounds (61). These observations collectively suggest that YqjA/YghB may play a more general role in membrane protein function or quality control and may be necessary for the homeostasis of the PMF. Whether YqjA/YghB are true transporters or are required for sensing and/or maintaining the PMF is not yet clear (see below and Fig. 3).
DedA FAMILY GENES ARE COLLECTIVELY OR INDIVIDUALLY ESSENTIAL IN BACTERIA
As stated above, the E. coli genome contains eight members of the DedA family. Each of these DedA homologs (yqjA, yghB, yabI, yohD, EcdedA, ydjX, ydjZ, and yqaA) is individually nonessential, as the single gene knockouts have been made and are available from the Keio collection (62). The study of the essentiality of the DedA family in E. coli was stymied due to this high level of genetic redundancy. However, we have recently succeeded in creating a number of strains with deletions of all members of this family (BAL800 series) (6). Each BAL800 strain requires expression of a DedA family member in trans from a hybrid plasmid. Growth of these strains is dependent upon the presence of an inducing agent (in this case, arabinose) in the growth media. Growth in the presence of glucose results in cell lysis (Fig. 2C). Further analysis of the BAL800 series mutants promises to provide a greater understanding of the essential functions of the DedA membrane protein family.
The genome of Borrelia burgdorferi, the cause of Lyme disease, possesses only a single DedA family gene, annotated bb0250 (Table 1) (63). In order to investigate the essentiality of the DedA family and to expand our knowledge of DedA family function, we created a B. burgdorferi Δbb0250 knockout. Strikingly, bb0250 is essential in its host organism and depletion of BB0250 protein, expressed behind an inducible promoter, results in cell death preceded by defects in cell division (Fig. 2D) (7). In other words, the Borrelia mutant phenotypes resemble those of BC202, with the exception being that bb0250 is essential at all temperatures. In addition, cloned bb0250 can fully complement the growth and cell division phenotypes of BC202 even though BB0250 displays only ∼19% amino acid identity to E. coli YqjA (and less to YghB) (7). These results demonstrate conservation of function of DedA family proteins found in two distantly related species of Gram-negative bacteria.
Four of the eight E. coli DedA family genes can restore normal growth and cell division to BC202 when expressed from a plasmid (yqjA, yghB, yabI, and yohD), while four cannot (ydjX, ydjZ, EcdedA, and yqaA). We have categorized the eight E. coli DedA family genes as C (complements BC202) or NC (does not complement BC202). Interestingly, the plasmid required for isolation of BAL800 mutants can harbor a gene from either the C group (i.e., yqjA) or the NC group (i.e., EcdedA). In regard to cell division and temperature sensitivity, the phenotype of the BAL800 family mutants depends upon whether the cloned DedA family gene belongs to the C or NC group. When yqjA (C group) is expressed from the inducible promoter in such a strain, the cell division defects and temperature sensitivity are corrected. When EcdedA (NC group) is expressed from the inducible promoter, the mutant still exhibits cell division defects and is temperature sensitive for growth but is viable at 30°C as long as an inducing agent is supplied. Thus, we can create a BAL800 series mutant just as easily if a gene is expressed belonging to the C group as from the NC group (though not all possible mutants have been isolated to date). These results suggest that all E. coli DedA family genes share a function that is required for survival and is independent of the cell division defects and temperature sensitivity of BC202 (6). Why are there so many DedA proteins in E. coli and other organisms (Table 1)? We cannot answer this question at present, but we may speculate that we are witnessing protein evolution in progress, with certain members duplicated and acquiring new functions (i.e., C group) while still retaining older ancestral functions.
DedA PROTEINS AS POTENTIAL DRUG TARGETS AND ROLES IN VIRULENCE
Recent reports suggest that members of the DedA family may represent potential drug targets and may play roles in virulence in some species. The genomes of most Mycobacterium species encode multiple DedA proteins (Table 1), and one DedA homolog (BCG2664) from M. bovis is possibly the target for the antibiotic halicyclamine A, as bcg2664 confers resistance to this drug when overexpressed in M. smegmatis (64). Halicyclamine A was first isolated from the marine sponge Haliclona sp. and was originally thought to inhibit IMP dehydrogenase (IMPDH), but this turned out not to be the case (65, 66). There exists a possibility that halicyclamine A and/or derivatives of this drug may act as general inhibitors of the DedA membrane protein family. Alternatively, DedA proteins may promote the efflux of antibiotics when overexpressed in these species.
Cationic peptides are important components of the host innate immune system. DedA family proteins appear to be required for resistance to cationic peptides in both Salmonella enterica and Neisseria meningitidis. In Salmonella, YqjA is regulated by PhoP and required for resistance to protamine and the alpha helical cationic peptide magainin 2 (but not polymyxin) (67). Often, resistance to cationic peptides requires covalent modifications to lipid A (68), but the Salmonella ΔyqjA strain exhibits a wild-type lipid A profile (67). Therefore, it is not yet clear what role YqjA plays in cationic peptide resistance in Salmonella. Similarly, a Neisseria NMB1052 (dedA) mutant was found to be hypersensitive to polymyxin (69). Neisseria uses a combination of lipopolysaccharide (LPS) modification, efflux pumps, and type IV pilin secretion to resist effects of cationic peptides (69, 70). Again, the role of DedA genes in promoting resistance to cationic peptides remains unclear, but the similarity of these two mutants in their cationic peptide sensitivity is intriguing.
Type III secretion is used by a number of Gram-negative pathogens to deliver effector proteins to host cells (71, 72). A screen for Yersinia pestis insertion mutants defective in type III secretion identified the DedA family gene ctgA (formerly y0447, encoding a polypeptide most closely related to E. coli YabI). This mutant, termed CHI 1345, was found to have impaired secretion of Yops and attenuated virulence in a mouse infection model (73). While likely not playing a direct role in this protein secretion pathway, CtgA may be required to maintain specific membrane properties that are required for the efficient assembly and/or operation of the type III secretion system (see next section).
PUTATIVE FUNCTIONS FOR DedA FAMILY MEMBERS
Based upon the mutant phenotypes described above, we can hypothesize potential functions of DedA family members. In E. coli, YqjA and YghB are together required for several cellular functions, all involving inner membrane proteins or protein complexes. For example, BC202 is defective in cell division due to inefficient function of the Tat pathway (30) and has an altered membrane phospholipid composition possibly due to an inefficiency in certain lipid synthesis pathways (5). The altered membrane composition is a property shared with the B. burgdorferi DedA family mutant (7). In addition, BC202 cannot survive at elevated pH (pH 8.8), but temperature sensitivity and cell division are rescued when it is grown at a lower pH (pH 6.0) or when the mdfA gene encoding an Na+/K+-H+ antiporter is overexpressed (40). These observations collectively suggest that these DedA family proteins may play a more general role in membrane protein function or quality control and may play a role in maintenance of the PMF.
The membrane potential, Δψ, of BC202 is significantly lower than that of the wild type, resulting in induction of the Psp stress response under permissive growth conditions (40). It is possible that under permissive growth conditions, the PMF homeostasis mechanism is inefficient in BC202 while under nonpermissive conditions the PMF falls below the minimum necessary threshold, leading to cell death. The Na+/K+-H+ antiporter MdfA participates in PMF homeostasis by catalyzing the import of protons into the cytoplasm coupled with the export of monovalent cations (60). The resulting influx of protons lowers the cytoplasmic pH and necessitates bacterial adaptation and survival during exposure to alkaline conditions. Similarly, growth of E. coli in media of low pH reinforces the PMF by providing an enhanced ΔpH component and promotes the influx of protons into the cytoplasm (74). As the pH homeostasis mechanism is physiologically linked to the PMF homeostasis mechanism (74), it is probable that the intracellular pH of BC202 is altered (likely more alkaline than normal). This also explains why conditions promoting proton influx in BC202 alleviate temperature sensitivity and cell division defects.
An analysis of protein evolutionary relationships using a novel software program called AlignMe revealed that bacterial DedA family proteins may share structural motifs with the LeuT protein superfamily (8). LeuT is a bacterial homolog of the neurotransmitter sodium symporter (75) and vSGLT of the solute:sodium symporter family. These protein families share certain structural similarities, including two sets of five transmembrane helices that share a pseudo 2-fold axis of symmetry along the plane of the membrane (76). It is important to note that the evolutionary relationship of the DedA and LeuT families is derived not from amino acid content but from hydrophobicity profiles and therefore would not turn up in a simple BLAST search. The data from the study by Khafizov et al. also suggest that DedA family proteins may adopt dual topologies in the membrane (8).
Since DedA family proteins may share structural motifs with LeuT-type transporters (8), it is possible that E. coli YqjA and YghB form homo- and/or heterodimers and participate in PMF homeostasis in a manner similar to that of MdfA. This would require these functional complexes to display a proton symporter or antiporter activity (Fig. 3A). This is also consistent with a proposed role for a DedA family protein in the uptake of selenite in Ralstonia metallidurans (77) and the occurrence of DedA domains in secondary transporters of the tripartite ATP-independent periplasmic transporter (TRAP-T) family (9). A second possibility is that these proteins regulate the function/activity of a crucial component(s) of the PMF homeostasis mechanism—such as respiratory complexes, metabolic enzymes, or distinct Na+/K+-H+ transporters like MdfA (Fig. 3B) (74). This model also derives support from the regulation of yqjA by the CpxAR two-component system. YqjA is an important member of the Cpx regulon in E. coli (58, 78), and deletion of either cpxR or yqjA renders E. coli sensitive to alkaline pH, demonstrating the necessity of YqjA for E. coli to adapt and survive under alkaline conditions (58). yqjA is also in an operon with mzrA (previously known as yqjB or ecfM), encoding a protein that links the Cpx pathway to the EnvZ/OmpR two-component system (79). (Remarkably, while yghB is not part of the Cpx regulon, its transcription is strongly induced by the quorum-sensing molecule autoinducer-2 [AI-2] [80].)
EUKARYOTIC DedA GENES
While this review focuses upon the functions of the bacterial DedA family proteins, some information on the roles DedA proteins play in multicellular organisms is available. A BLAST search reveals the presence of hundreds of DedA family homologs in eukaryotes (most closely related to E. coli YdjX or YdjZ). Many DedA genes, even in bacteria, are annotated “SNARE-associated Golgi protein.” This is in reference to the reported physical association of protein Tvp38 (an E. coli YdjX homolog) from Saccharomyces cerevisiae with T-SNARE found in purified Tgl2-containing (late) Golgi compartments (81). It was hypothesized that Tvp38, and other unknown proteins isolated in this proteomic approach, may be involved in maintenance of late Golgi/endosomal compartments. The functional significance of this interaction remains to be elucidated, but it clearly suggests that DedA family members play some role in the eukaryotic secretory pathway.
An interesting screen for Caenorhabditis elegans mutants (generated by MosI mutagenesis) resistant to the bacterial pathogen Microbacterium nematophilum revealed the involvement of a DedA family member named Bus-19 (bacterially unswollen-19; with similarity to E. coli YdjX) (82). M. nematophilum is able to colonize the rectum of susceptible worms and induce an inflammatory response that requires the extracellular signal-regulated kinase/mitogen-activated protein (ERK/MAP) kinase pathway (83). The Δbus-19 worms were resistant to infection because the bacteria were unable to adhere to the surface of the rectum, perhaps due to loss of a nematode cell surface receptor. Bus-19 also contains a putative endoplasmic reticulum (ER) localization signal. Again, this study supports the notion that DedA family proteins may play a role in proper functioning of the eukaryotic secretory pathway. Vertebrates, including mice and humans, also harbor DedA family homologs. The Mus musculus and Homo sapiens gene products are annotated Tmem41A, and they share about 25% amino acid identity with E. coli YdjX. Whether the functions of eukaryotic DedA family members are similar to their functions in prokaryotes is one of the more interesting questions in regard to this ancient family and, sadly, cannot be answered at this time.
CONCLUSIONS
Genetics and biochemistry, coupled with insight provided from proteomics and genome sequencing projects, have provided a glimpse into the wide distribution of and important functions carried out by members of the highly conserved DedA membrane protein family. In E. coli, some DedA family members appear to be required to maintain the membrane proton motive force. Others may play different roles in bacterial physiology. These functions have important implications, not just for elucidation of a potential drug target but for the insight that may be provided into the process of protein evolution.
ACKNOWLEDGMENTS
We thank Patrick Lane for valuable assistance in preparation of figures and anonymous reviewers for helpful comments. We thank Aaron Smith for critical reading of the manuscript. We thank Holger Kneuper and Tracy Palmer for sharing unpublished observations regarding growth of Δtat mutants.
Financial support has been provided by the National Science Foundation (MCB-0841853, to W.T.D.).
Biographies

William T. Doerrler is an Associate Professor in the Department of Biological Sciences at LSU. He received his B.S. from Stevens Institute of Technology in 1987 and spent time as a lab technician in the labs of Kenneth O. Lloyd (Sloan-Kettering) and Carl Grunfeld (UCSF) before working on his Ph.D. thesis in the area of glycobiology in the lab of Mark A. Lehrman (University of Texas Southwestern Medical Center). He carried out postdoctoral work in the lab of the late Christian R. H. Raetz at Duke University before arriving at LSU in 2004. In addition to being a father and husband, he has been interested in the DedA family and LSU Athletics in recent years.

Rakesh Sikdar recently was awarded his Ph.D. degree and carried out his graduate work with William Doerrler at LSU. He received his B.Tech (Hons.) in Biotechnology and Biochemical Engineering at the Indian Institute of Technology, Kharagpur, in West Bengal, India, before arriving at LSU in 2007. His research in Escherichia coli seeks to discover the roles of the proteins YqjA and YghB belonging to the DedA protein family. When not actively pursuing his prominent academic and research interests, he is a hobbyist photographer and indulges in extensive travelling and adventure hiking across the country.

Sujeet Kumar received a B.Tech degree in Biotechnology at Vellore Institute of Technology University, Vellore, India, in 2011. Thereafter, he joined the Biological Sciences Department at LSU. Currently, he is a doctoral candidate working with William Doerrler and investigating the roles of essential DedA protein family members in Escherichia coli.

Lisa A. Boughner is a doctoral candidate in William Doerrler's lab at Louisiana State University. Lisa obtained a B.Sc. in Honours Biological Sciences with Thesis from the University of Windsor, Windsor, Ontario, followed by working as a field and laboratory assistant at the Greenhouse and Processing Crops Research Center in Harrow, Ontario, Canada. Lisa joined LSU in 2007, and her doctoral research consists of investigating the essential role of the E. coli DedA protein family. In her “spare” time, Lisa enjoys playing both the classical and bass guitar, as well as traveling.
Footnotes
Published ahead of print 19 October 2012
REFERENCES
- 1. Elofsson A, von Heijne G. 2007. Membrane protein structure: prediction vs reality. Annu. Rev. Biochem. 76: 125–140 [DOI] [PubMed] [Google Scholar]
- 2. Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, Young J, Yukich B, Zardecki C, Berman HM, Bourne PE. 2011. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 39: D392–D401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. White SH. 2009. Biophysical dissection of membrane proteins. Nature 459: 344–346 [DOI] [PubMed] [Google Scholar]
- 4. Nonet ML, Marvel CC, Tolan DR. 1987. The hisT-purF region of the Escherichia coli K-12 chromosome. Identification of additional genes of the hisT and purF operons. J. Biol. Chem. 262: 12209–12217 [PubMed] [Google Scholar]
- 5. Thompkins K, Chattopadhyay B, Xiao Y, Henk MC, Doerrler WT. 2008. Temperature sensitivity and cell division defects in an Escherichia coli strain with mutations in yghB and yqjA, encoding related and conserved inner membrane proteins. J. Bacteriol. 190: 4489–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boughner LA, Doerrler WT. 2012. Multiple deletions reveal the essentiality of the DedA membrane protein family in Escherichia coli. Microbiology 158: 1162–1171 [DOI] [PubMed] [Google Scholar]
- 7. Liang FT, Xu Q, Sikdar R, Xiao Y, Cox JS, Doerrler WT. 2010. BB0250 of Borrelia burgdorferi is a conserved and essential inner membrane protein required for cell division. J. Bacteriol. 192: 6105–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khafizov K, Staritzbichler R, Stamm M, Forrest LR. 2010. A study of the evolution of inverted-topology repeats from LeuT-fold transporters using AlignMe. Biochemistry 49: 10702–10713 [DOI] [PubMed] [Google Scholar]
- 9. Barabote RD, Tamang DG, Abeywardena SN, Fallah NS, Fu JY, Lio JK, Mirhosseini P, Pezeshk R, Podell S, Salampessy ML, Thever MD, Saier MH., Jr 2006. Extra domains in secondary transport carriers and channel proteins. Biochim. Biophys. Acta 1758: 1557–1579 [DOI] [PubMed] [Google Scholar]
- 10. Wu DY, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, Hooper SD, Pati A, Lykidis A, Spring S, Anderson IJ, D'Haeseleer P, Zemla A, Singer M, Lapidus A, Nolan M, Copeland A, Han C, Chen F, Cheng JF, Lucas S, Kerfeld C, Lang E, Gronow S, Chain P, Bruce D, Rubin EM, Kyrpides NC, Klenk HP, Eisen JA. 2009. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462: 1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brochier-Armanet C, Forterre P, Gribaldo S. 2011. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr. Opin. Microbiol. 14: 274–281 [DOI] [PubMed] [Google Scholar]
- 12. Detels R, Green AM, Klausner JD, Katzenstein D, Gaydos C, Handsfield HH, Pequegnat W, Mayer K, Hartwell TD, Quinn TC. 2011. The incidence and correlates of symptomatic and asymptomatic Chlamydia trachomatis and Neisseria gonorrhoeae infections in selected populations in five countries. Sex. Transm. Dis. 38: 503–509 [PMC free article] [PubMed] [Google Scholar]
- 13. Hotopp JCD, Lin MQ, Madupu R, Crabtree J, Angiuoli SV, Eisen J, Seshadri R, Ren QH, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang CB, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou LW, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2(2): e21 doi:10.1371/journal.pgen.0020021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Larsen B, Hwang J. 2010. Mycoplasma, ureaplasma, and adverse pregnancy outcomes: a fresh look. Infect. Dis. Obstet. Gynecol. 2010: 521921 doi:10.1155/2010/521921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kowalczewska M, Vellaiswamy M, Nappez C, Vincentelli R, Scola BL, Raoult D. 2012. Protein candidates for the serodiagnosis of rickettsioses. FEMS Immunol. Med. Microbiol. 64: 130–133 [DOI] [PubMed] [Google Scholar]
- 16. Sprong H, Wielinga PR, Fonville M, Reusken C, Brandenburg AH, Borgsteede F, Gaasenbeek C, van der Giessen JWB. 2009. Ixodes ricinus ticks are reservoir hosts for Rickettsia helvetica and potentially carry flea-borne Rickettsia species. Parasit. Vectors 2: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nerandzic MM, Cadnum JL, Eckart KE, Donskey CJ. 2012. Evaluation of a hand-held far-ultraviolet radiation device for decontamination of Clostridium difficile and other healthcare-associated pathogens. BMC Infect. Dis. 12: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shao XJ, Raman B, Zhu MJ, Mielenz JR, Brown SD, Guss AM, Lynd LR. 2011. Mutant selection and phenotypic and genetic characterization of ethanol-tolerant strains of Clostridium thermocellum. Appl. Microbiol. Biotechnol. 92: 641–652 [DOI] [PubMed] [Google Scholar]
- 19. Siddiqui AR, Bernstein JM. 2010. Chronic wound infection: facts and controversies. Clin. Dermatol. 28: 519–526 [DOI] [PubMed] [Google Scholar]
- 20. Zhang W, Ma J, Zang C, Song Y, Liu P. 2011. The fur transcription regulator and fur-regulated genes in Clostridium botulinum A ATCC 3502. J. Biomed. Biotechnol. 2011: 934756 doi:10.1155/2011/934756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu QY, Eremeeva ME, Li DM. 2012. Bartonella and Bartonella infections in China: from the clinic to the laboratory. Comp. Immunol. Microbiol. Infect. Dis. 35: 93–102 [DOI] [PubMed] [Google Scholar]
- 22. Wirth SE, Ayala-del-Rio HL, Cole JA, Kohlerschmidt DJ, Musser KA, Sepulveda-Torres LD, Thompson LM, Wolfgang WJ. 2012. Psychrobacter sanguinis sp nov., recovered from four clinical specimens over a 4-year period. Int. J. Syst. Evol. Microbiol. 62: 49–54 [DOI] [PubMed] [Google Scholar]
- 23. Akman L, Yamashita A, Watanabe H, Oshima K, Shiba T, Hattori M, Aksoy S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32: 402–407 [DOI] [PubMed] [Google Scholar]
- 24. Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407: 81–86 [DOI] [PubMed] [Google Scholar]
- 25. McCutcheon JP, Moran NA. 2012. Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10: 13–26 [DOI] [PubMed] [Google Scholar]
- 26. McCutcheon JP, Moran NA. 2010. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol. Evol. 2: 708–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Moran NA, Tran P, Gerardo NM. 2005. Symbiosis and insect diversification: an ancient symbiont of sap-feeding insects from the bacterial phylum Bacteroidetes. Appl. Environ. Microbiol. 71: 8802–8810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Penz T, Horn M, Schmitz-Esser S. 2010. The genome of the amoeba symbiont “Candidatus Amoebophilus asiaticus” reveals common mechanisms for host cell interaction among amoeba-associated bacteria. Virulence 1: 541–545 [DOI] [PubMed] [Google Scholar]
- 29. Baker BJ, Comolli LR, Dick GJ, Hauser LJ, Hyatt D, Dill BD, Land ML, VerBerkmoes NC, Hettich RL, Banfield JF. 2010. Enigmatic, ultrasmall, uncultivated Archaea. Proc. Natl. Acad. Sci. U. S. A. 107: 8806–8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sikdar R, Doerrler WT. 2010. Inefficient Tat-dependent export of periplasmic amidases in an Escherichia coli strain with mutations in two DedA family genes. J. Bacteriol. 192: 807–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bernhardt TG, de Boer PA. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol. Microbiol. 48: 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Berks BC, Palmer T, Sargent F. 2005. Protein targeting by the bacterial twin-arginine translocation (Tat) pathway. Curr. Opin. Microbiol. 8: 174–181 [DOI] [PubMed] [Google Scholar]
- 33. Berks BC, Sargent F, Palmer T. 2000. The Tat protein export pathway. Mol. Microbiol. 35: 260–274 [DOI] [PubMed] [Google Scholar]
- 34. De Buck E, Lammertyn E, Anne J. 2008. The importance of the twin-arginine translocation pathway for bacterial virulence. Trends Microbiol. 16: 442–453 [DOI] [PubMed] [Google Scholar]
- 35. Lee PA, Tullman-Ercek D, Georgiou G. 2006. The bacterial twin-arginine translocation pathway. Annu. Rev. Microbiol. 60: 373–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heidrich C, Ursinus A, Berger J, Schwarz H, Holtje JV. 2002. Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. J. Bacteriol. 184: 6093–6099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stanley NR, Findlay K, Berks BC, Palmer T. 2001. Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotropic defects in the cell envelope. J. Bacteriol. 183: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ize B, Stanley NR, Buchanan G, Palmer T. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 48: 1183–1193 [DOI] [PubMed] [Google Scholar]
- 39. Dilks K, Rose RW, Hartmann E, Pohlschroder M. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185: 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sikdar R, Simmons AR, Doerrler WT. 2013. Multiple envelope stress response pathways are activated in an Escherichia coli strain with mutations in two members of the DedA membrane protein family. J. Bacteriol. 195: 12–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bury-Mone S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, Jacq A, Bouloc P. 2009. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet. 5: e1000651 doi:10.1371/journal.pgen.1000651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Connolly L, De Las Penas A, Alba BM, Gross CA. 1997. The response to extracytoplasmic stress in Escherichia coli is controlled by partially overlapping pathways. Genes Dev. 11: 2012–2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Raivio TL, Silhavy TJ. 1999. The sigmaE and Cpx regulatory pathways: overlapping but distinct envelope stress responses. Curr. Opin. Microbiol. 2: 159–165 [DOI] [PubMed] [Google Scholar]
- 44. Vogt SL, Raivio TL. 2012. Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol. Lett. 326: 2–11 [DOI] [PubMed] [Google Scholar]
- 45. Engl C, Beek AT, Bekker M, de Mattos JT, Jovanovic G, Buck M. 2011. Dissipation of proton motive force is not sufficient to induce the phage shock protein response in Escherichia coli. Curr. Microbiol. 62: 1374–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Joly N, Engl C, Jovanovic G, Huvet M, Toni T, Sheng X, Stumpf MP, Buck M. 2010. Managing membrane stress: the phage shock protein (Psp) response, from molecular mechanisms to physiology. FEMS Microbiol. Rev. 34: 797–827 [DOI] [PubMed] [Google Scholar]
- 47. Jovanovic G, Lloyd LJ, Stumpf MP, Mayhew AJ, Buck M. 2006. Induction and function of the phage shock protein extracytoplasmic stress response in Escherichia coli. J. Biol. Chem. 281: 21147–21161 [DOI] [PubMed] [Google Scholar]
- 48. Hirakawa H, Inazumi Y, Masaki T, Hirata T, Yamaguchi A. 2005. Indole induces the expression of multidrug exporter genes in Escherichia coli. Mol. Microbiol. 55: 1113–1126 [DOI] [PubMed] [Google Scholar]
- 49. Nishino K, Honda T, Yamaguchi A. 2005. Genome-wide analyses of Escherichia coli gene expression responsive to the BaeSR two-component regulatory system. J. Bacteriol. 187: 1763–1772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ebel W, Vaughn GJ, Peters HK, III, Trempy JE. 1997. Inactivation of mdoH leads to increased expression of colanic acid capsular polysaccharide in Escherichia coli. J. Bacteriol. 179: 6858–6861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu. Rev. Microbiol. 59: 379–405 [DOI] [PubMed] [Google Scholar]
- 52. Parker CT, Kloser AW, Schnaitman CA, Stein MA, Gottesman S, Gibson BW. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174: 2525–2538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shiba Y, Yokoyama Y, Aono Y, Kiuchi T, Kusaka J, Matsumoto K, Hara H. 2004. Activation of the Rcs signal transduction system is responsible for the thermosensitive growth defect of an Escherichia coli mutant lacking phosphatidylglycerol and cardiolipin. J. Bacteriol. 186: 6526–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J. Bacteriol. 190: 2065–2074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jovanovic G, Engl C, Mayhew AJ, Burrows PC, Buck M. 2010. Properties of the phage-shock-protein (Psp) regulatory complex that govern signal transduction and induction of the Psp response in Escherichia coli. Microbiology 156: 2920–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Alder NN, Theg SM. 2003. Energetics of protein transport across biological membranes. a study of the thylakoid DeltapH-dependent/cpTat pathway. Cell 112: 231–242 [DOI] [PubMed] [Google Scholar]
- 57. Bageshwar UK, Musser SM. 2007. Two electrical potential-dependent steps are required for transport by the Escherichia coli Tat machinery. J. Cell Biol. 179: 87–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Price NL, Raivio TL. 2009. Characterization of the Cpx regulon in Escherichia coli strain MC4100. J. Bacteriol. 191: 1798–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Krulwich TA, Lewinson O, Padan E, Bibi E. 2005. Do physiological roles foster persistence of drug/multidrug-efflux transporters? A case study. Nat. Rev. Microbiol. 3: 566–572 [DOI] [PubMed] [Google Scholar]
- 60. Lewinson O, Padan E, Bibi E. 2004. Alkalitolerance: a biological function for a multidrug transporter in pH homeostasis. Proc. Natl. Acad. Sci. U. S. A. 101: 14073–14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lewinson O, Adler J, Poelarends GJ, Mazurkiewicz P, Driessen AJ, Bibi E. 2003. The Escherichia coli multidrug transporter MdfA catalyzes both electrogenic and electroneutral transport reactions. Proc. Natl. Acad. Sci. U. S. A. 100: 1667–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2: 2006 0008 doi:10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, Gwinn M, Dougherty B, Tomb JF, Fleischmann RD, Richardson D, Peterson J, Kerlavage AR, Quackenbush J, Salzberg S, Hanson M, van Vugt R, Palmer N, Adams MD, Gocayne J, Weidman J, Utterback T, Watthey L, McDonald L, Artiach P, Bowman C, Garland S, Fuji C, Cotton MD, Horst K, Roberts K, Hatch B, Smith HO, Venter JC. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390: 580–586 [DOI] [PubMed] [Google Scholar]
- 64. Arai M, Liu L, Fujimoto T, Setiawan A, Kobayashi M. 2011. DedA protein relates to action-mechanism of halicyclamine A, a marine spongean macrocyclic alkaloid, as an anti-dormant mycobacterial substance. Mar. Drugs 9: 984–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Arai M, Sobou M, Vilcheze C, Baughn A, Hashizume H, Pruksakorn P, Ishida S, Matsumoto M, Jacobs WR, Jr., Kobayashi M. 2008. Halicyclamine A, a marine spongean alkaloid as a lead for anti-tuberculosis agent. Bioorg. Med. Chem. 16: 6732–6736 [DOI] [PubMed] [Google Scholar]
- 66. Jaspars M, Pasupathy V, Crews P. 1994. A tetracyclic diamine alkaloid, halicyclamine A, from the marine sponge Haliclona sp. J. Org. Chem. 59: 3253–3255 [Google Scholar]
- 67. Shi Y, Cromie MJ, Hsu FF, Turk J, Groisman EA. 2004. PhoP-regulated Salmonella resistance to the antimicrobial peptides magainin 2 and polymyxin B. Mol. Microbiol. 53: 229–241 [DOI] [PubMed] [Google Scholar]
- 68. Raetz CR, Reynolds CM, Trent MS, Bishop RE. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76: 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tzeng YL, Ambrose KD, Zughaier S, Zhou X, Miller YK, Shafer WM, Stephens DS. 2005. Cationic antimicrobial peptide resistance in Neisseria meningitidis. J. Bacteriol. 187: 5387–5396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lewis LA, Choudhury B, Balthazar JT, Martin LE, Ram S, Rice PA, Stephens DS, Carlson R, Shafer WM. 2009. Phosphoethanolamine substitution of lipid A and resistance of Neisseria gonorrhoeae to cationic antimicrobial peptides and complement-mediated killing by normal human serum. Infect. Immun. 77: 1112–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cornelis GR. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4: 811–825 [DOI] [PubMed] [Google Scholar]
- 72. Galan JE, Wolf-Watz H. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature 444: 567–573 [DOI] [PubMed] [Google Scholar]
- 73. Houppert AS, Kwiatkowski E, Glass EM, DeBord KL, Merritt PM, Schneewind O, Marketon MM. 2012. Identification of chromosomal genes in Yersinia pestis that influence type III secretion and delivery of Yops into target cells. PLoS One 7: e34039 doi:10.1371/journal.pone.0034039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Krulwich TA, Sachs G, Padan E. 2011. Molecular aspects of bacterial pH sensing and homeostasis. Nat. Rev. Microbiol. 9: 330–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. 2005. Crystal structure of a bacterial homologue of Na+/Cl–dependent neurotransmitter transporters. Nature 437: 215–223 [DOI] [PubMed] [Google Scholar]
- 76. Faham S, Watanabe A, Besserer GM, Cascio D, Specht A, Hirayama BA, Wright EM, Abramson J. 2008. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science 321: 810–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Ledgham F, Quest B, Vallaeys T, Mergeay M, Coves J. 2005. A probable link between the DedA protein and resistance to selenite. Res. Microbiol. 156: 367–374 [DOI] [PubMed] [Google Scholar]
- 78. Yamamoto K, Ishihama A. 2006. Characterization of copper-inducible promoters regulated by CpxA/CpxR in Escherichia coli. Biosci. Biotechnol. Biochem. 70: 1688–1695 [DOI] [PubMed] [Google Scholar]
- 79. Gerken H, Charlson ES, Cicirelli EM, Kenney LJ, Misra R. 2009. MzrA: a novel modulator of the EnvZ/OmpR two-component regulon. Mol. Microbiol. 72: 1408–1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. DeLisa MP, Wu CF, Wang L, Valdes JJ, Bentley WE. 2001. DNA microarray-based identification of genes controlled by autoinducer 2-stimulated quorum sensing in Escherichia coli. J. Bacteriol. 183: 5239–5247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Inadome H, Noda Y, Kamimura Y, Adachi H, Yoda K. 2007. Tvp38, Tvp23, Tvp18 and Tvp15: novel membrane proteins in the Tlg2-containing Golgi/endosome compartments of Saccharomyces cerevisiae. Exp. Cell Res. 313: 688–697 [DOI] [PubMed] [Google Scholar]
- 82. Yook K, Hodgkin J. 2007. MosI mutagenesis reveals a diversity of mechanisms affecting response of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics 175: 681–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Nicholas HR, Hodgkin J. 2004. The ERK MAP kinase cascade mediates tail swelling and a protective response to rectal infection in C. elegans. Curr. Biol. 14: 1256–1261 [DOI] [PubMed] [Google Scholar]
- 84. Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580 [DOI] [PubMed] [Google Scholar]
- 85. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28: 2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Blevins JS, Revel AT, Smith AH, Bachlani GN, Norgard MV. 2007. Adaptation of a luciferase gene reporter and lac expression system to Borrelia burgdorferi. Appl. Environ. Microbiol. 73: 1501–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Daley DO, Rapp M, Granseth E, Melen K, Drew D, von Heijne G. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308: 1321–1323 [DOI] [PubMed] [Google Scholar]
- 88. Hirokawa T, Boon-Chieng S, Mitaku S. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–379 [DOI] [PubMed] [Google Scholar]