Abstract
In order to identify Trichosporon species in formalin-fixed and paraffin-embedded sections from which visual discrimination of non-glabrata Candida species is mostly ineffective but critical for the choice of antifungals, we tested the usefulness of a newly designed peptide nucleic acid probe (PNA) for in situ hybridization (ISH). Results confirmed the usefulness of ISH with our PNA probe in identifying Trichosporon species from Candida albicans.
TEXT
Trichosporon species have been reported as the second or third most common agents of yeast fungemia (1–3), and the prevalence has increased, particularly in patients with hematologic malignancies (4–6). Since Trichosporon spp. exhibit low susceptibility to candins (7), histopathological examination is important as one of the useful diagnostic procedures, even though it is regarded as poor for the identification of Trichosporon species from other dimorphic yeasts, namely, non-glabrata Candida, species, because of their overall similarities (8–10). Therefore, the establishment of an auxiliary diagnostic method for use in routine pathological laboratories is required for diagnosis of disseminated trichosporonosis. In the present study, we report attempts to develop a specific peptide nucleic acid (PNA) probe to Trichosporon spp. and evaluate this method for identification of the fungus in formalin-fixed and paraffin-embedded (FFPE) tissue sections by using in situ hybridization (ISH).
We employed FFPE tissues both from experimentally infected mice and autopsies with a proven diagnosis. Specific-pathogen-free male, 8-week-old Institute of Cancer research mice were injected intravenously with 3 × 107 yeast cells of Trichosporon asahii (strain 015), T. asahii (strain 336), or Candida albicans (J2-15), and their kidneys were obtained 3 days after infection and processed by a conventional method. Lungs from autopsies with disseminated candidiasis and trichosporonosis were also used. Trichosporonosis was diagnosed by DNA sequence analysis. Candidiasis was culture proven (EC Toho approved; 20047).
The antisense PNA probe targeting the 26S rRNA of Trichosporon spp. (N terminus-CGG ACA ATC GAA GAC) was hypothetically designed based on a comparison of the sequences of 26S rRNA genes of Trichosporon spp. and other pathogenic fungi available in the GenBank database. To identify C. albicans, we also used an antisense PNA probe targeting the 26S rRNA of C. albicans (N terminus-TAC TTG TGC GCT ATC GGT) (11). Furthermore, to estimate retention and hybridizability of the target RNA in samples, we used a panfungal antisense PNA probe (N terminus-TAC TTG TGC GCT ATC GGT) (12). The oligonucleotide probes used in this study were made by Fasmac Co., Ltd. (Kanagawa, Japan), and the N terminus of the PNA probes was conjugated to fluorescein isothiocyanate (FITC). The process of obtaining FFPE tissues and the ISH procedure were performed as described previously (12, 13).
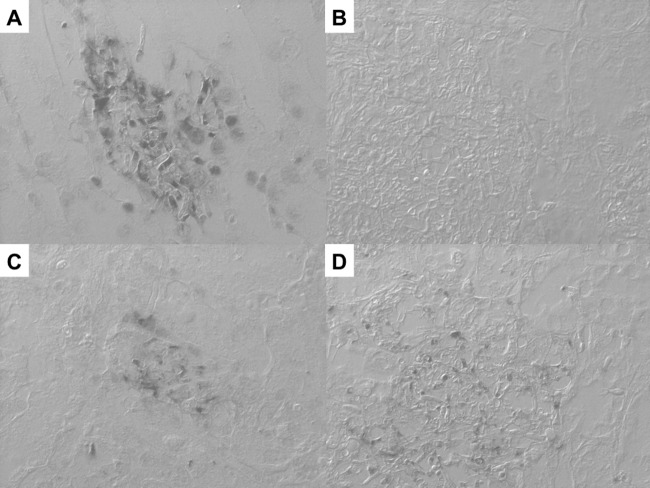
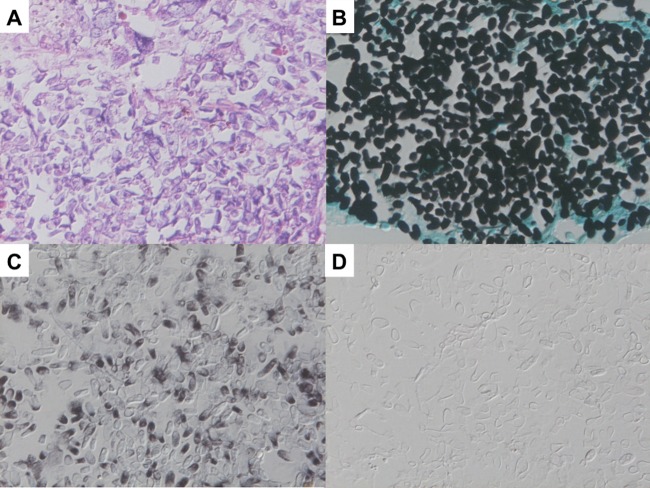
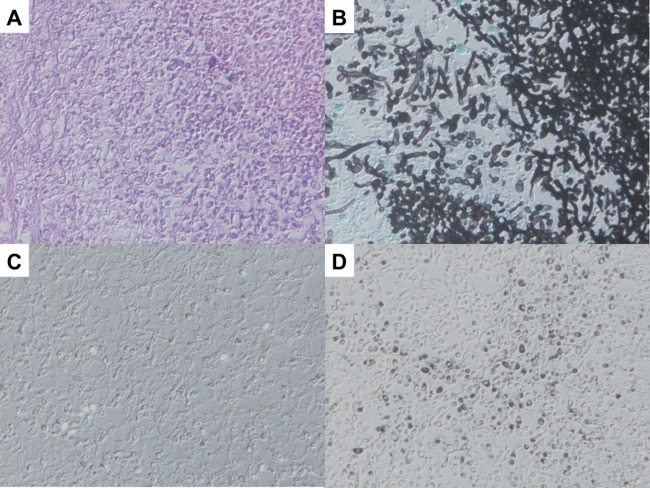
ISH showed strong positive signals against Trichosporon spp. 26S rRNA within yeast-like elements present in renal tissues from mice infected with T. asahii (Fig. 1A), whereas these signals were not observed in specimens derived from mice infected with C. albicans (Fig. 1B). On the other hand, the panfungal PNA probe reacted with T. asahii and C. albicans (Fig. 1C and D) and confirmed the retention and hybridizability of rRNA. In an additional evaluation using autopsy samples, ISH preparation showed that the PNA probe against Trichosporon spp. was strongly reactive with yeast-like elements of Trichosporon spp. (Fig. 2C), whereas the PNA probe against C. albicans was not reactive with any Trichosporon spp. (Fig. 2D). Conversely, the PNA probe against Trichosporon spp. was not reactive with organisms from subjects with candidiasis (Fig. 3C), but its organisms showed strong positive signals when the PNA probe targeting C. albicans was applied (Fig. 3D). Whereas the Trichosporon species-specific probe we designed in the study showed acceptably strong signals for T. asahii in tissue sections from both experimental infections and autopsy samples, it should be confirmed whether the probe actually reacted for non-asahii Trichosporon species in FFPE tissues.
Fig 1.
Specificity verification of the Trichosporon spp. PNA probe and assessments of rRNA retention and its hybridizability in experimentally infected mice. (A) ISH using the Trichosporon spp. PNA probe in renal tissue from mice infected with T. asahii. Strong positive signals against 28S rRNA of Trichosporon spp. were observed in the specimen. (B) ISH using the Trichosporon spp. PNA probe in renal tissue from mice infected with C. albicans. Positive signals were not observed in the specimen. (C) ISH result with the panfungal PNA probe in renal tissue from mice infected with T. asahii. Strong positive signals were observed in the specimen. (D) ISH result with the panfungal PNA probe in renal tissue from mice infected with C. albicans. Strong positive signals were observed in the specimen. Magnification, ×400.
Fig 2.
Results of ISH with a pulmonary lesion of disseminated trichosporonosis confirmed by DNA sequence analysis. (A) Pathological findings with hematoxylin and eosin stain. Histological examination revealed foci consisting of yeast formations of organisms. (B) Findings with Grocott's stain. Grocott's stain showed oval or square yeast-like elements within foci of infection. (C) Result of ISH with the Trichosporon spp. PNA probe. The PNA probe against Trichosporon spp. was strongly reactive with the yeast-like elements of Trichosporon spp. (D) ISH result with the C. albicans PNA probe. The PNA probe against C. albicans was not reactive with any Trichosporon spp. organisms.
Fig 3.
Results of ISH for pulmonary lesions in the case of culture-proven C. albicans. (A) Pathological findings with hematoxylin and eosin stain. Histological examination revealed foci consisting of pseudohyphal and yeast formations of organisms. (B) Results with Grocott's stain. Grocott's stain showed oval yeast-like and pseudohyphal elements within the foci of infection. (C) ISH result with the Trichosporon spp. PNA probe. The PNA probe against Trichosporon spp. was not reactive with any organisms of C. albicans. (D) ISH result with the C. albicans PNA probe. The PNA probe against C. albicans was strongly reactive with pseudohyphal and yeast-like elements of C. albicans.
Trichosporon spp. present certain morphological features in pathological specimens (14). However, their morphological similarities to other fungi, especially non-glabrata Candida species, lead to difficulties in the identification of trichosporonosis. Hence, the establishment of an auxiliary diagnostic method for use in routine pathological laboratories would be useful for a diagnosis of disseminated trichosporonosis with histopathological differentiation from candidiasis. Although a few studies have attempted to identify other fungi in histopathological specimens by using ISH (15–17), no investigations have utilized ISH for the diagnosis of trichosporonosis.
The diagnosis of trichosporonosis by immunohistochemistry using a self-made antibody to Trichosporon has been reported (18, 19); however, these antibodies are not available for commercial use, there are limitations for their use, and their specificity could not be confirmed. Therefore, we developed the auxiliary utility of ISH for the pathological diagnosis of trichosporonosis from FFPE tissues. Although PCR has been regarded as a sensitive and useful assay for the detection of Trichosporon species (20, 21), the application of this technique to pathological specimens has the disadvantage of being highly susceptible to contamination and formalin fixation, potentially leading to diagnostic mistakes. In addition, PCR-based molecular techniques have the difficulty of DNA release in DNA extraction due to the rigid fungal cell wall (22). Accordingly, we now have to regard that FFPE tissue also limits the use of PCR because of DNA degradation and low yield on extraction protocols. On the other hand, ISH has little contamination risk and does not require nucleic acid extraction. From the viewpoint of the above-mentioned properties, ISH may overcome the disadvantage of PCR-based molecular techniques that use FFPE sections. Recently, ISH techniques employing PNA probes for rRNA have been developed as useful techniques for the differentiation of medically important Candida spp. (11, 23). These novel properties enabled PNA probes to hybridize to complementary nucleic acid targets with high specificity and rapid binding kinetics (24, 25).
In conclusion, we wish to emphasize that ISH with our probe can be valuable in distinguishing Trichosporon spp. from non-glabrata Candida species in FFPE tissues, since we demonstrated that our newly designed PNA probes targeting the 26S rRNA showed a specific signal intensity for Trichosporon spp. in various kinds of tissue sections.
ACKNOWLEDGMENTS
This work was supported by Health Science Research Grants for Research on Emerging and Re-emerging Infectious Diseases (H22-Shinko-Ippan-8 and H23-Shinko-Ippan-18) from the Ministry of Health, Labor and Welfare of Japan, grants from the Strategic Basis on Research Grounds for Non-Governmental Schools at Heisei 20th and 23rd and KAKENHI (24790364) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and grants from the Yokohama Foundation for Advancement of Medical Science.
M.S. and Y.O. wrote the manuscript as major and equal contributors. D.S., H.N., and T.I. sampled publications. S.Y.M. advised the first author on ISH. N.T., M.W., and T.N. carried out the histopathological evaluation. K.S. integrated the data and gave final approval to the manuscript as a corresponding author. All authors contributed towards the conceptualization, writing, reading, and approval of the final manuscript.
K. Shibuya reports receiving a research grant from Pfizer Inc. All authors declare that they have no competing interests.
Footnotes
Published ahead of print 24 October 2012
REFERENCES
- 1.Nahass GT, Rosenberg SP, Leonardi CL, Penneys NS. 1993. Disseminated infection with Trichosporon beigelii. Report of a case and review of the cutaneous and histologic manifestations. Arch. Dermatol. 129:1020–1023 [DOI] [PubMed] [Google Scholar]
- 2.Walsh TJ. 1989. Trichosporonosis. Infect. Dis. Clin. North Am. 3:43–52 [PubMed] [Google Scholar]
- 3.Walsh TJ, Groll A, Hiemenz J, Fleming R, Roilides E, Anaissie E. 2004. Infections due to emerging and uncommon medically important fungal pathogens. Clin. Microbiol. Infect. 10(Suppl. 1):48–66 [DOI] [PubMed] [Google Scholar]
- 4.Kume H, Yamazaki T, Togano T, Abe M, Tanuma H, Kawana S, Okudaira M. 2011. Epidemiology of visceral mycoses in autopsy cases in Japan: comparison of the data from 1989, 1993, 1997, 2001, 2005 and 2007 in the Annual of Pathological Autopsy Cases in Japan. Med. Mycol. J. 52:117–127 [DOI] [PubMed] [Google Scholar]
- 5.Shimodaira K, Okubo Y, Nakayama H, Wakayama M, Shinozaki M, Ishiwatari T, Sasai D, Nemoto T, Takahashi K, Ishii T, Saji T, Shibuya K. 2012. Trends in the prevalence of invasive fungal infections from an analysis of annual records of autopsy cases of Toho University. Mycoses 55:435–443 [DOI] [PubMed] [Google Scholar]
- 6.Suzuki K, Nakase K, Kyo T, Kohara T, Sugawara Y, Shibazaki T, Oka K, Tsukada T, Katayama N. 2010. Fatal Trichosporon fungemia in patients with hematologic malignancies. Eur. J. Haematol. 84:441–447 [DOI] [PubMed] [Google Scholar]
- 7.Erer B, Galimberti M, Lucarelli G, Giardini C, Polchi P, Baronciani D, Gaziev D, Angelucci E, Izzi G. 2000. Trichosporon beigelii: a life-threatening pathogen in immunocompromised hosts. Bone Marrow Transplant. 25:745–749 [DOI] [PubMed] [Google Scholar]
- 8.Shibuya K, Ando T, Hasegawa C, Wakayama M, Hamatani S, Hatori T, Nagayama T, Nonaka H. 2004. Pathophysiology of pulmonary aspergillosis. J. Infect. Chemother. 10:138–145 [DOI] [PubMed] [Google Scholar]
- 9.Shibuya K, Coulson WF, Wollman JS, Wakayama M, Ando T, Oharaseki T, Takahashi K, Naoe S. 2001. Histopathology of cryptococcosis and other fungal infections in patients with acquired immunodeficiency syndrome. Int. J. Infect. Dis. 5:78–85 [DOI] [PubMed] [Google Scholar]
- 10.Shibuya K, Hirata A, Omuta J, Sugamata M, Katori S, Saito N, Murata N, Morita A, Takahashi K, Hasegawa C, Mitsuda A, Hatori T, Nonaka H. 2005. Granuloma and cryptococcosis. J. Infect. Chemother. 11:115–122 [DOI] [PubMed] [Google Scholar]
- 11.Oliveira K, Haase G, Kurtzman C, Hyldig-Nielsen JJ, Stender H. 2001. Differentiation of Candida albicans and Candida dubliniensis by fluorescent in situ hybridization with peptide nucleic acid probes. J. Clin. Microbiol. 39:4138–4141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinozaki M, Okubo Y, Nakayama H, Mitsuda A, Ide T, Yamagata Murayama S, Shibuya K. 2009. Application of in situ hybridization to tissue sections for identification of molds causing invasive fungal infection. Nihon Ishinkin Gakkai. Zasshi. 50:75–83 [DOI] [PubMed] [Google Scholar]
- 13.Shinozaki M, Okubo Y, Sasai D, Nakayama H, Murayama SY, Ide T, Wakayama M, Hiruta N, Shibuya K. 2011. Identification of Fusarium species in formalin-fixed and paraffin-embedded sections by in situ hybridization using peptide nucleic acid probes. J. Clin. Microbiol. 49:808–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sano M, Sugitani M, Ishige T, Homma T, Kikuchi K, Sunagawa K, Obana Y, Uehara Y, Kumasaka K, Uenogawa K, Kobayashi S, Hatta Y, Takeuchi J, Nemoto N. 2007. Supplemental utility of nested PCR for the pathological diagnosis of disseminated trichosporonosis. Virchows Arch. 451:929–935 [DOI] [PubMed] [Google Scholar]
- 15.Hanazawa R, Murayama SY, Yamaguchi H. 2000. In-situ detection of Aspergillus fumigatus. J. Med. Microbiol. 49:285–290 [DOI] [PubMed] [Google Scholar]
- 16.Hayden RT, Isotalo PA, Parrett T, Wolk DM, Qian X, Roberts GD, Lloyd RV. 2003. In situ hybridization for the differentiation of Aspergillus, Fusarium, and Pseudallescheria species in tissue section. Diagn. Mol. Pathol. 12:21–26 [DOI] [PubMed] [Google Scholar]
- 17.Montone KT. 2009. Differentiation of Fusarium from Aspergillus species by colorimetric in situ hybridization in formalin-fixed, paraffin-embedded tissue sections using dual fluorogenic-labeled LNA probes. Am. J. Clin. Pathol. 132:866–870 [DOI] [PubMed] [Google Scholar]
- 18.Fukuzawa M, Inaba H, Hayama M, Sakaguchi N, Sano K, Ito M, Hotchi M. 1995. Improved detection of medically important fungi by immunoperoxidase staining with polyclonal antibodies. Virchows Arch. 427:407–414 [DOI] [PubMed] [Google Scholar]
- 19.Tashiro T, Nagai H, Kamberi P, Goto Y, Kikuchi H, Nasu M, Akizuki S. 1994. Disseminated Trichosporon beigelii infection in patients with malignant diseases: immunohistochemical study and review. Eur. J. Clin. Microbiol. Infect. Dis. 13:218–224 [DOI] [PubMed] [Google Scholar]
- 20.Nagai H, Yamakami Y, Hashimoto A, Tokimatsu I, Nasu M. 1999. PCR detection of DNA specific for Trichosporon species in serum of patients with disseminated trichosporonosis. J. Clin. Microbiol. 37:694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugita T, Nakajima M, Ikeda R, Niki Y, Matsushima T, Shinoda T. 2001. A nested PCR assay to detect DNA in sera for the diagnosis of deep-seated trichosporonosis. Microbiol. Immunol. 45:143–148 [DOI] [PubMed] [Google Scholar]
- 22.Karakousis A, Tan L, Ellis D, Alexiou H, Wormald PJ. 2006. An assessment of the efficiency of fungal DNA extraction methods for maximizing the detection of medically important fungi using PCR. J. Microbiol. Methods 65:38–48 [DOI] [PubMed] [Google Scholar]
- 23.Shepard JR, Addison RM, Alexander BD, Della-Latta P, Gherna M, Haase G, Hall G, Johnson JK, Merz WG, Peltroche-Llacsahuanga H, Stender H, Venezia RA, Wilson D, Procop GW, Wu F, Fiandaca MJ. 2008. Multicenter evaluation of the Candida albicans/Candida glabrata peptide nucleic acid fluorescent in situ hybridization method for simultaneous dual-color identification of C. albicans and C. glabrata directly from blood culture bottles. J. Clin. Microbiol. 46:50–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demidov VV, Yavnilovich MV, Belotserkovskii BP, Frank-Kamenetskii MD, Nielsen PE. 1995. Kinetics and mechanism of polyamide (“peptide”) nucleic acid binding to duplex DNA. Proc. Natl. Acad. Sci. U. S. A. 92:2637–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nielsen PE, Egholm M, Berg RH, Buchardt O. 1991. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 254:1497–1500 [DOI] [PubMed] [Google Scholar]