Abstract
Knowledge concerning stability is important in the development and assessment of microbial molecular typing systems and is critical for the interpretation of their results. Typing system stability is usually measured as the fraction of isolates that change type after several in vivo passages, but this does not necessarily reflect in vivo stability. The aim of this study was to utilize survival analysis to provide an informative quantitative measure of in vivo stability and to compare the stabilities of various techniques employed in typing methicillin-resistant Staphylococcus aureus (MRSA). We identified 100 MRSA pairs (isolated from the same patient ≥1 month apart) and typed them using multilocus sequence typing (MLST), phage-derived open reading frame (PDORF) typing, toxin gene profiling (TGP), staphylococcal cassette chromosome mec (SCCmec) subtyping, pulsed-field gel electrophoresis (PFGE), and spa sequence typing. Discordant isolate pairs, belonging to different MLST clonal complexes, were excluded, leaving 81 pairs for analysis. The stabilities of these methods were examined using Kaplan-Meier survival analysis, and discriminatory power was measured by Simpson's index of diversity. The probability percentages that the type remained unchanged at 6 months for spa sequence typing, TGP, multilocus variable number of tandem repeats analysis (MLVA), SCCmec subtyping, PDORF typing, and PFGE were 95, 95, 88, 82, 71, and 58, respectively, while the Simpson's indices of diversity were 0.48, 0.47, 0.70, 0.72, 0.89, and 0.88, respectively. Survival analysis using sequential clinical isolates adds an important quantitative dimension to the measurement of stability of a microbial typing system. Of the methods compared here, PDORF typing provides high discriminatory power, comparable with that of PFGE, and a level of stability suitable for MRSA surveillance and outbreak investigations.
INTRODUCTION
The stability of a new typing method is one of the key parameters defining its utility, along with discriminatory power, reproducibility, ease of use and interpretation, cost, throughput, and concordance with both epidemiologic data and existing typing systems (1). Discriminatory power is usually estimated by Simpson's index of diversity (2), concordance is usually measured by Wallace or Rand coefficients (3), and reproducibility is usually measured by Cohen's kappa.
Stability relates to the likelihood that the measured characteristic of an organism will change with time or in subsequent generations of the organism. If a typing system has low stability, it may incorrectly identify related isolates as being unrelated. It is important to define the stability of a typing system so that guidelines for the interpretation of results can be established. If isolates typed by a highly stable system differ at one locus, it may indicate that they are unrelated strains, whereas if they are typed by an unstable system, they may need to differ at multiple loci before nonrelatedness can be inferred.
Stability has been most commonly measured by observing the fraction of isolates in which the genotype remains unchanged after a fixed number of in vitro passages (1). However, this approach is less applicable to epidemiologic studies and does not provide much relevant information about the natural rate of change over time. Here, we describe a method of measuring stability using Kaplan-Meier survival analysis and its application to the estimation of in vivo stability of typing methods used to study the short-term epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) in hospital and community settings.
MATERIALS AND METHODS
Isolate collection.
A collection of MRSA isolates from clinical and screening samples obtained between July 2005 and March 2009, routinely stored at the Centre for Infectious Diseases and Microbiology, Westmead Hospital, Sydney, Australia, was used in the study. This collection was surveyed to find multiple isolates from the same patient; patients were enrolled if they had two isolates collected ≥1 month apart, irrespective of the anatomical site or type of specimen from which they were isolated. The multilocus sequence type (MLST) was predicted using kinetic PCR for informative single-nucleotide polymorphisms (SNPs) (4), and patients were excluded if their isolates belonged to different MLST clonal complexes (as it was assumed that this represented acquisition of a new strain rather than evolution of the initial strain).
DNA preparation.
For PCR-based methods, one or two colonies from a pure subculture were suspended in 400 μl of molecular-grade water, which was boiled for 10 min and frozen. After thawing, the suspension was centrifuged at 10,000 rpm for 5 min, and the supernatant was used for a DNA template. The same lysate was used for all methods. A sweep of the pure subculture was used to make suspensions for pulsed-field gel electrophoresis (PFGE).
Molecular typing.
PFGE of SmaI-digested genomic DNA was performed according to the Harmony protocol (5) and analyzed using BioNumerics (Applied Maths NV, Belgium). A comparison of PFGE patterns in BioNumerics was performed using the Dice coefficient with the position tolerance set at 1.5% (change toward end of the fingerprint, 0.75%), and a dendrogram was constructed using the unweighted-pair group method using arithmetic averages (UPGMA). The isolates were grouped using two similarity cutoffs: 100% (indistinguishable patterns) and 80% (PFGE-100 and PFGE-80, respectively), as 80% is commonly used to define PFGE patterns that are likely to be epidemiologically related (6). Multilocus variable number of tandem repeats analysis (MLVA) was performed as reported previously (7). Each MLVA locus was amplified using single-primer-pair PCRs, and the amplification products were detected using gel electrophoresis with isolate pairs in adjacent lanes. The band sizes and matching bands between isolates were determined using BioNumerics and confirmed by visual inspection. MLVA patterns were considered to be different if the molecular weights of one or more loci varied by at least the size of one repeat for that locus. spa sequence typing was performed as described previously (8), and spa types were assigned using the SpaServer (Ridom Bioinformatics) (9). Three multiplex PCR/reverse line blot (mPCR/RLB) binary typing systems were employed (10, 11): (i) toxin gene profiling to target sea, seb, sec, sed, see, seg, seh, sei, eta, etb, etd, tst, and lukS-PV (Panton-Valentine leukocidin) genes (8), (ii) phage-derived open reading frame typing (PDORF) to examine 16 loci derived from integrated prophages (12), and (iii) staphylococcal cassette chromosome mec (SCCmec) subtyping to determine the mec class and ccr type and to interrogate 14 loci in the three junkyard regions (13). Reference strains that, in combination, were positive for each probe and a DNA-free control were used as the positive and negative controls, respectively.
Data analysis.
Stability was assessed by Kaplan-Meier survival analysis using SAS for Windows 9.3. For the purposes of survival analysis, an “event” was considered to have occurred when the members of the isolate pair had different results depending on the typing method under analysis. The time at which the “event” occurred was arbitrarily considered to be the midpoint between the collection times of the two isolates. An isolate pair was considered “censored” (at the time of collection of the second isolate) if the results for the typing method for the two isolates were indistinguishable, i.e., the time required for a change in molecular type was unknown for that patient, but it was longer than the period of observation. Survival analysis was used to estimate the probability that a typing method would remain unchanged for an isolate after 6 months. The log rank χ2 test was used to assess the differences in survival curves between methods, and P values were adjusted for multiple comparisons using the Šidák method (14). The initial isolate of each pair was used to calculate Simpson's index of diversity (2) of the typing method.
RESULTS
One hundred pairs of MRSA isolates were identified from the culture collection, and the time between isolate collections for each of the pairs varied between 1 month and 2.7 years. The members of 19 isolate pairs belonged to different MLST clonal complexes and therefore were excluded from further analysis.
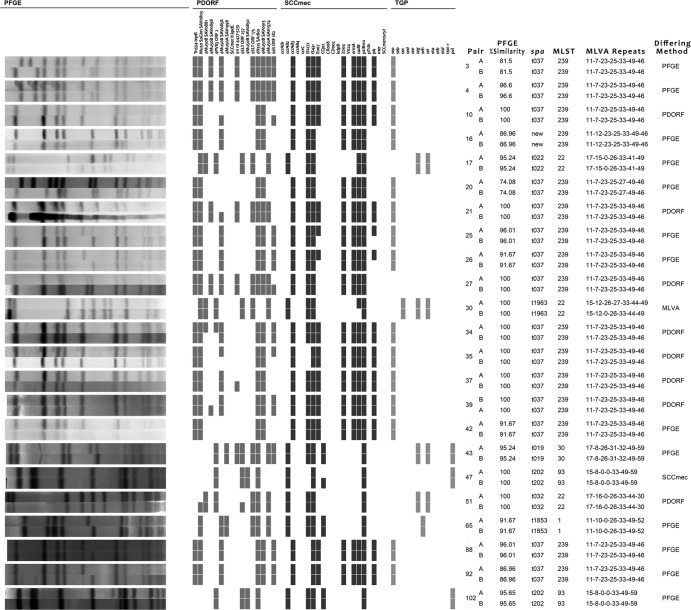
Of the remaining 81 isolate pairs, 37 had concordant results with all the methods (PFGE with 100% similarity [PFGE-100], PFGE with 80% similarity [PFGE-80], phage-derived open reading frame typing [PDORF], SCCmec subtyping, toxin gene profiling [TGP], spa sequence typing, and MLVA). Twenty-three isolate pairs differed by only 1 method (excluding PFGE-80): 13 for PFGE-100, 8 for PDORF, and 1 each for MLVA and SCCmec subtyping (Fig. 1). Fourteen isolate pairs differed by 2 methods, and the remaining 7 differed by 3 or more methods. Five isolate pairs differed by PFGE-80: one of these pairs was concordant for all non-PFGE methods, and the remaining four differed by 1, 2, 3, and 5 non-PFGE methods.
Fig 1.
Comparison of genotyping methods for 23 pairs, which differed by only one method (as indicated in the right-hand column). TGP, toxin gene profiling; PDORF, phage-derived open reading frame typing; MLVA, multilocus variable number of tandem repeats analysis. The MLVA repeats are for (from left to right) sspA, spa, sdrC, sdrD, sdrE, clfA, and clfB.
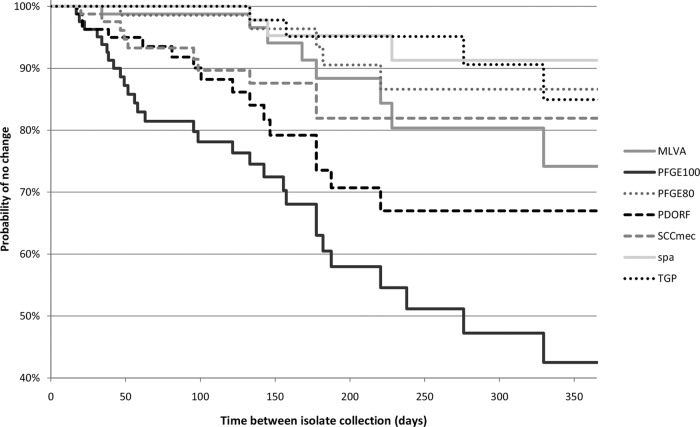
Kaplan-Meier survival curves show a clear separation of PFGE-100, PDORF, and SCCmec subtyping from the other more stable methods after as little as 3 months (Fig. 2). PFGE-100 was significantly less stable than all other methods tested (probability of no change at 6 months, 58%; 95% confidence interval [CI], 43 to 70%). spa typing was the most stable (probability of no change at 6 months, 95%; 95% CI, 82 to 99%), but this higher stability was significant only when compared with PFGE-100 and PDORF (P < 0.0001 and P = 0.03, respectively). PFGE-80 was significantly more stable but less discriminatory than PFGE-100. In general, there was an expected inverse relationship between stability and Simpson's index of diversity (Table 1). However, despite having a fractionally higher discriminatory power than PFGE-100 (Simpson's index of diversity, 0.89 versus 0.88), PDORF also had significantly higher stability, with a probability of no change at 6 months of 71% (95% CI, 55 to 82%) (Table 1).
Fig 2.
Kaplan-Meier survival curves for different genotyping methods. PFGE-100, PFGE-80: PFGE with similarity cutoffs of 100% and 80%, respectively. For other typing method abbreviations, see the Fig. 1 legend.
Table 1.
Stability and diversity measures of different genotyping methods
| Attribute | Value for indicated genotyping method |
||||||
|---|---|---|---|---|---|---|---|
| spa sequence typing | TGP | PFGE-80 | MLVA | SCCmec subtyping | PDORF | PFGE-100 | |
| No. (%) of pairs differing | 3 (4) | 5 (6) | 5 (6) | 9 (11) | 11 (14) | 17 (21) | 31 (38) |
| % probability of no change at 6 mo (95% CI) | 95 (82–99) | 95 (82–99) | 91 (76–96) | 88 (74–95) | 82 (68–90) | 71 (55–82) | 58 (43–70) |
| Simpson's index of diversity (95% CI) | 0.48 (0.35–0.62) | 0.47 (0.34–0.60) | 0.61 (0.50–0.73) | 0.70 (0.57–0.82) | 0.72 (0.64–0.79) | 0.89 (0.85–0.94) | 0.88 (0.81–0.94) |
| Log rank χ2 test statistic (P value) for comparison with PFGE-100 | 40.2 (<0.0001) | 34.8 (<0.0001) | 35.0 (<0.0001) | 25.1 (<0.0001) | 21.2 (<0.0001) | 10.3 (0.03) | |
DISCUSSION
Stability has an inverse relationship with discriminatory power and is a function of the molecular “clock speed” of the genetic loci being interrogated by a typing system. Housekeeping genes, for example, which are utilized for multilocus sequence typing (MLST), have a low molecular clock speed, resulting in high stability but low discriminatory power. Genetic loci on mobile genetic elements demonstrate a high molecular clock speed; utilizing these for typing may result in systems with low stability (possibly leading to misleading inferences about the relationships between isolates) but potentially high discriminatory power. The stability of a typing method has traditionally been measured by sampling strains at two time points and determining the fraction of strains that have the same results at each time point. Most commonly, sampling is performed before and after a given number of serial passages of the organism in the laboratory (in vitro stability); less commonly, it is performed by culturing an organism in its natural environment at different time points, e.g., conducting repeat cultures on a patient known to be colonized with the organism (in vivo stability). Measuring stability in vitro may lead to overestimation of the stability of a pathogen in its natural host environment, since the opportunities for genetic changes through transformation, the acquisition of mobile genetic elements, or in response to environmental conditions, such as antibiotic therapy or immune pressure, will be absent.
Our findings suggest that the consideration of stability of an MRSA typing system is especially important when one aims to measure the relatedness of MRSA isolates obtained in time intervals of more than 3 months. With studies examining shorter time periods (e.g., during a hospital outbreak), highly discriminatory but relatively unstable methods, such as PDORF or PFGE, will be suitable for examining the microepidemiology of strains which may otherwise be closely related. Studies examining longer time periods (macroepidemiology) benefit from more stable methods, such as spa sequence typing or PFGE, which have similarity cutoffs of 80%. These findings are consistent with those of a recent study that utilized next-generation sequencing to examine the microevolution of one geographically widespread MRSA clone (ST239) and which found an estimated molecular clock speed of one SNP mutation in the core genome every 6 weeks (15).
Simultaneous carriage of multiple MRSA strains, or loss of one strain and recolonization with another, are potentially confounding factors in studies of in vivo stability. Major changes in colonizing strains have been infrequent in other in vivo stability studies with MRSA (16–18), but the reported frequency of simultaneous carriage of multiple strains has varied in the literature (19). One report suggested that while simultaneous carriage of multiple methicillin-sensitive Staphylococcus aureus (MSSA) strains may be common, this is not the case for MRSA (20). Of 100 isolate pairs initially identified in our study, 19 differed by their MLST clonal complex and so were assumed to represent recolonization/infection with different strains.
The potential limitations of this study include the lack of serial sampling of isolates, which may have impaired the accuracy of the stability measures; more frequent sampling would require a prospective clinical study. While attempts were made to exclude reinfection/colonization by a different strain by excluding the isolate pairs belonging to different MLST clonal complexes, it is possible that some of the changes between isolate pairs represented reinfection with a different strain within the same clonal complex rather than the evolution of the initial isolate or perhaps the carriage of multiple strains at different body sites, since a combination of clinical and screening isolates was utilized for this study. This limitation may have led to an underestimate of stability, particularly that of methods with higher discriminatory power. However, the majority of discordant pairs differed by only one method, suggesting that reinfection with a different isolate was probably infrequent in this cohort. Despite the limitations, the estimates produced in this study should reliably reflect the relative stability of different genotyping methods.
In conclusion, PDORF offered greater stability than PFGE-100 over a 12-month period but had a similar ability to discriminate between apparently unrelated isolates from different individuals; this indicates that PDORF may be a suitable substitute for PFGE in the investigation of MRSA outbreaks and infection control surveillance. It also has additional advantages over PFGE, such as lower cost, faster turnaround time, higher portability of results, and easier interpretation. All other methods were relatively more stable but had correspondingly limited discriminatory power. Survival analysis techniques provide a useful quantitative measure of stability for the assessment of novel genotyping targets.
ACKNOWLEDGMENT
M.V.N.O. was supported by Australian National Health and Medical Research Council Postgraduate Scholarship no. 512029.
Footnotes
Published ahead of print 24 October 2012
REFERENCES
- 1.Struelens MJ. 1996. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin. Microbiol. Infect. 2:2–11 [DOI] [PubMed] [Google Scholar]
- 2.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J. Clin. Microbiol. 26:2465–2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrico JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H, Almeida JS, Ramirez M. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huygens F, Inman-Bamber J, Nimmo GR, Munckhof W, Schooneveldt J, Harrison B, McMahon JA, Giffard PM. 2006. Staphylococcus aureus genotyping using novel real-time PCR formats. J. Clin. Microbiol. 44:3712–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, Fussing V, Salmenlinna S, Vuopio-Varkila J, El Solh N, Cuny C, Witte W, Tassios PT, Legakis N, van Leeuwen W, van Belkum A, Vindel A, Laconcha I, Garaizar J, Haeggman S, Olsson-Liljequist B, Ransjo U, Coombes G, Cookson B. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sabat A, Krzyszton-Russjan J, Strzalka W, Filipek R, Kosowska K, Hryniewicz W, Travis J, Potempa J. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 41:1801–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai Y, Kong F, Wang Q, Tong Z, Sintchenko V, Zeng X, Gilbert GL. 2007. Comparison of single and multilocus sequence typing and toxin gene profiling for characterization of methicillin-resistant Staphylococcus aureus (MRSA). J. Clin. Microbiol. 45:3302–3308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kong F, Gilbert GL. 2006. Multiplex PCR-based reverse line blot hybridization assay (mPCR/RLB)—a practical epidemiological and diagnostic tool. Nat. Protoc. 1:2668–2680 [DOI] [PubMed] [Google Scholar]
- 11.O'Sullivan MV, Zhou F, Sintchenko V, Kong F, Gilbert GL. 2011. Multiplex PCR and reverse line blot hybridization assay (mPCR/RLB). J. Vis. Exp. (54):2781 doi:10.3791/2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O'Sullivan MV, Kong F, Sintchenko V, Gilbert GL. 2010. Rapid identification of methicillin-resistant Staphylococcus aureus transmission in hospitals by use of phage-derived open reading frame typing enhanced by multiplex PCR and reverse line blot assay. J. Clin. Microbiol. 48:2741–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai L, Kong F, Wang Q, Wang H, Xiao M, Sintchenko V, Gilbert GL. 2009. A new multiplex PCR-based reverse line-blot hybridization (mPCR/RLB) assay for rapid staphylococcal cassette chromosome mec (SCCmec) typing. J. Med. Microbiol. 58:1045–1057 [DOI] [PubMed] [Google Scholar]
- 14.Šidák Z. 1967. Rectangular confidence regions for the means of multivariate normal distributions. J. Am. Stat. Assoc. 62:626–633 [Google Scholar]
- 15.Harris SR, Feil EJ, Holden MTG, Quail MA, Nickerson EK, Chantratita N, Gardete S, Tavares A, Day N, Lindsay JA, Edgeworth JD, de Lencastre H, Parkhill J, Peacock SJ, Bentley SD. 2010. Evolution of MRSA during hospital transmission and intercontinental spread. Science 327:469–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartstein AI, Phelps CL, Kwok RY, Mulligan ME. 1995. In vivo stability and discriminatory power of methicillin-resistant Staphylococcus aureus typing by restriction endonuclease analysis of plasmid DNA compared with those of other molecular methods. J. Clin. Microbiol. 33:2022–2026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn G, Francioli P, Blanc DS. 2007. Double-locus sequence typing using clfB and spa, a fast and simple method for epidemiological typing of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 45:54–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakwinska O, Blanc DS, Lazor-Blanchet C, Moreillon M, Giddey M, Moreillon P. 2010. Ecological temporal stability of Staphylococcus aureus nasal carriage. J. Clin. Microbiol. 48:2724–2728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bloemendaal AL, Fluit AC, Jansen WT, Vriens MR, Ferry T, Amorim JM, Pascual A, Stefani S, Papaparaskevas J, Borel Rinkes IH, Verhoef J. 2009. Colonization with multiple Staphylococcus aureus strains among patients in European intensive care units. Infect. Control Hosp. Epidemiol. 30:918–920 [DOI] [PubMed] [Google Scholar]
- 20.O'Brien FG, Coombs GW, Pearman JW, Gracey M, Moss F, Christiansen KJ, Grubb WB. 2009. Population dynamics of methicillin-susceptible and -resistant Staphylococcus aureus in remote communities. J. Antimicrob. Chemother. 64:684–693 [DOI] [PMC free article] [PubMed] [Google Scholar]