Abstract
Self-collected foam nasal swabs (NS) obtained after instillation of saline spray were compared with nasal washes from immunocompetent subjects during 146 upper respiratory infections (URIs); the sensitivities for reverse transcription (RT)-PCR respiratory virus detection were 95% and 88%, respectively (P = 0.06). The sensitivities from NS collected with and without saline spray during 142 URIs from immunocompetent subjects were 96% and 86% (P = 0.004), respectively, and those from 140 URI samples from hematopoietic cell transplantation recipients were 88% and 85% (P = 0.56), respectively.
TEXT
A simple, sensitive, and noninvasive method for collection of respiratory samples is valuable for patient care and for studying respiratory virus epidemiology. Rapid diagnosis of respiratory viruses also enables implementation of potential treatment and infection prevention measures, essential for immunocompromised patients. Historically, the “gold standard” for respiratory virus detection has included nasal washes (NW) or nasopharyngeal swabs collected by medical personnel. Recent reports have shown that nasopharyngeal or nasal swabs (NS) collected from children compare favorably to NW/nasopharyngeal aspirates for PCR detection of most respiratory viruses (1–5). Furthermore, parent- or self-collected swabs are useful for community-based respiratory virus research (6–11). In these studies, cotton, Dacron, or nylon swabs were collected without nasal spray, stored in transport medium, and processed immediately or stored at 4°C.
We sought to optimize a simple and reliable method for self-collection of respiratory samples in immunocompetent subjects and allogeneic hematopoietic cell transplant (HCT) patients of all ages and specifically evaluated polyurethane foam swabs because of comfort and safety. We first compared self-collected foam NS obtained after nasal saline spray versus NW from immunocompetent subjects with upper respiratory infections (URIs). Subsequent comparisons between self-collected NS with and without nasal spray were conducted among immunocompetent subjects and HCT recipients to determine the importance of the saline spray.
Polyurethane foam swabs (Super Brush, LLC; catalog no. 71-4541) were used in immunocompetent volunteers. Before enrolling HCT recipients, we transitioned to sterile polyurethane foam swabs with a custom-shaped tip (Puritan Medical Products Co., LLC; no. 25-1805 1PF SC2 Arrow). For optimization, foam swabs were initially compared with flocked nylon swabs (Copan Diagnostics, Inc.; catalog no. 502CS01) and 3 transport media were evaluated: lysis buffer, universal transport medium (UTM) (Copan Diagnostics, Inc.), and isotonic saline. Using saline solution spiked with known concentrations of parainfluenza virus type 3 (PIV3) and influenza virus A, no inhibition of reverse transcription (RT)-PCR was seen with either swab type or transport media. Recovery rates of influenza virus A and PIV3 from foam swabs were similar for all 3 collection media at 1, 2, and 7 days and similar at room temperature and 4°C. Therefore, foam NS were collected and transported in dry tubes at room temperature for subsequent analyses.
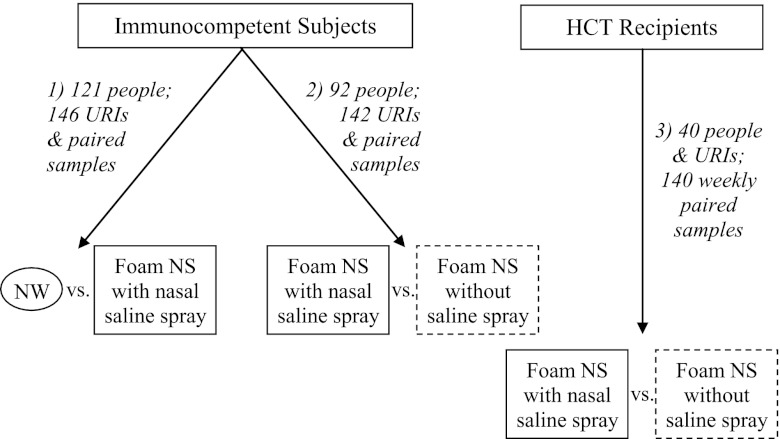
Immunocompetent volunteers with respiratory symptoms reported within 3 days were recruited for comparison of virus detection from self-collected foam NS and staff-collected NW (Fig. 1). Outpatient HCT recipients with documented viral URIs were recruited to provide weekly self-collected samples until viral testing was negative. Subjects were asked to blow their nose to remove mucus. For self-collection, 5 sprays (0.5 ml) of saline from a polyethylene metered spray bottle were instilled into the naris prior to swab insertion. The NS was rotated for 5 s, placed into a dry tube, and stored at room temperature. NW was collected from the opposite naris using 5 ml normal saline (2.5 ml for children) and transported at 4°C. Subsequently, study subjects (immunocompetent and immunocompromised) collected a “dry” NS by inserting a foam swab into the anterior naris, rotating it for 5 s, and placing the swab into an empty transport tube, followed by collection of a “wet” NS in the opposite naris after nasal saline spray (Fig. 1).
Fig 1.
Algorithm of respiratory sample collection and comparisons among immunocompetent subjects and hematopoietic cell transplant (HCT) recipients. URIs, upper respiratory infections; NW, nasal wash; NS, nasal swab. Study periods: 1, November 2006 to January 2009; 2, May 2008 to May 2012; 3, February 2010 to May 2012.
Subjects were allowed to participate during more than one URI if the URIs were >4 weeks apart. Paired collection of saline and dry foam swabs was performed 2 to 7 days after initial NW in 10 immunocompetent subjects. All subjects completed symptom surveys. The Fred Hutchinson Cancer Research Center and Seattle Children's Hospital Institutional Review Boards approved this study; written consent and assent forms were obtained.
Swabs from immunocompetent subjects were transported by study personnel; HCT recipients hand-carried weekly NS to the outpatient clinical laboratory. Before extraction, secretions in the saline NS were adjusted to 1 ml with addition of 0.5 ml Hank's balanced salt solution (HBSS); 1 ml was added to the dry-collected NS. Samples were vigorously vortexed, free liquid was pipetted off, and nucleic acid was extracted from 200 μl as previously described (12). Qualitative real-time RT-PCR assays were performed for 15 respiratory viruses by previously described methods and a newly developed in-house assay for human bocavirus (12–18).
Samples from incident URIs from immunocompetent subjects were treated as independent observations, but simultaneous virus detections were treated as repeated measures. Samples from HCT recipients were treated as dependent observations within the recipient. Each sample was considered true positive if either method was positive. To incorporate correlated data from multiple viruses per sample in an already-paired-sample data structure, a difference score was coded (i.e., for saline and dry swabs, −1 if saline negative/dry positive, 0 if concordant, and 1 if saline positive/dry negative), and analyzed via linear regression to compare method sensitivities. Sensitivities with 95% confidence intervals (95% CIs) were estimated via logistic regression models in true-positive samples. For all models, robust standard errors were calculated. P values were obtained from the Wald test without adjustment for multiple comparisons. Analyses were performed using SAS version 9.1 (SAS Institute, Inc., Cary, NC).
Paired samples were collected during 146 incident URIs from 121 immunocompetent subjects (including 3 parent-collected NS). Swab processing in the laboratory took place a median of 1 day after NS collection (range, 0 to 6 days). At least one sample had a respiratory virus detected by either collection method in 86/146 (59%) illness episodes (Table 1).
Table 1.
Comparison of detections of respiratory viruses by RT-PCR in nasal wash versus foam nasal swab samples collected with saline spraya
| Respiratory virusb | No. of virus detections |
Sensitivity, % (95% CI) |
P value | |||
|---|---|---|---|---|---|---|
| Both positive | NW positive, NS negative | NS positive, NW negative | NW | NS with saline spray | ||
| RSV | 3 | 0 | 0 | 100 | 100 | |
| Flu A | 7 | 0 | 0 | 100 | 100 | |
| Flu B | 2 | 0 | 1 | 67 | 100 | |
| HMPV | 2 | 0 | 0 | 100 | 100 | |
| PIV (types 1–4) | 6 | 0 | 1 | 86 | 100 | |
| HCoV (4 types) | 13 | 1 | 4 | 78 | 94 | |
| HRV | 39 | 2 | 4 | 91 | 96 | |
| AdV | 1 | 1 | 1 | 67 | 67 | |
| HBoV | 0 | 0 | 0 | |||
| Totalc | 73 | 4 | 11 | 88 (79–93) | 95 (88–98) | 0.06 |
Shown are results from 121 immunocompetent subjects during 146 incident upper respiratory infections.
RSV, respiratory syncytial virus; Flu, influenza virus; HMPV, human metapneumovirus; PIV, parainfluenza virus; HCoV, human coronaviruses (OC43, 229E, NL63, and HKU1); HRV, human rhinoviruses; AdV, adenovirus; and HBoV, human bocavirus.
A total of 60 swab pairs were negative in both samples; 86 pairs were positive in at least 1 swab, with 2 viruses detected in 2 swab pairs.
For comparison with and without saline spray, NS samples were collected during 142 URIs from 92 subjects (including 12 parent-collected NS). Swab processing took place a median of 1 day after collection (range, 1 to 6 days); 95% of NS were processed in ≤2 days. At least one sample had a respiratory virus detected by either method in 104/142 (73%) illness episodes (Table 2).
Table 2.
Comparison of detections of respiratory viruses by RT-PCR in foam nasal swabs collected with and without the use of nasal saline spraya
| Respiratory virusb | No. of virus detections |
Sensitivity, % (95% CI) |
P value | |||
|---|---|---|---|---|---|---|
| Both positive | Saline positive, dry negative | Dry positive, saline negative | NS with saline spray | Dry NS | ||
| Immunocompetent subjects | ||||||
| RSV | 7 | 0 | 0 | 100 | 100 | |
| Flu A | 12 | 1 | 0 | 100 | 92 | |
| Flu B | 2 | 0 | 0 | 100 | 100 | |
| HMPV | 2 | 0 | 0 | 100 | 100 | |
| PIV (types 1–4) | 6 | 2 | 0 | 100 | 75 | |
| HCoV (4 types) | 11 | 6 | 0 | 100 | 65 | |
| HRV | 52 | 2 | 4 | 93 | 97 | |
| AdV | 1 | 5 | 0 | 100 | 17 | |
| HBoV | 4 | 1 | 1 | 83 | 83 | |
| Totalc | 97 | 17 | 5 | 96 (90–98) | 86 (78–91) | 0.004 |
| Rhinorrhea | ||||||
| Positive (n = 117) | 84 | 11 | 4 | 96 (90–98) | 89 (81–94) | 0.06 |
| Negative (n = 25) | 13 | 6 | 1 | 95 (73–99) | 70 (50–84) | 0.007 |
| HCT recipients | ||||||
| RSV | 23 | 3 | 2 | 93 | 89 | |
| HMPV | 2 | 1 | 0 | 100 | 67 | |
| PIV (types 1–4) | 24 | 7 | 4 | 89 | 80 | |
| HCoV (4 types) | 5 | 3 | 1 | 89 | 67 | |
| HRV | 34 | 2 | 7 | 84 | 95 | |
| AdV | 0 | 2 | 0 | 100 | 0 | |
| Totald | 88 | 18 | 14 | 88 (78–94) | 85 (76–91) | 0.56 |
| Rhinorrhea | ||||||
| Positive (n = 84) | 62 | 8 | 7 | 91 (79–96) | 90 (80–95) | 0.80 |
| Negative (n = 54) | 26 | 9 | 7 | 83 (68–92) | 79 (62–89) | 0.66 |
Results are shown from 92 immunocompetent subjects with 142 incident upper respiratory infections and 40 HCT recipients with previously diagnosed respiratory virus infections with 140 weekly paired follow-up swabs.
RSV, respiratory syncytial virus; Flu, influenza virus; HMPV, human metapneumovirus; PIV, parainfluenza virus; HCoV, human coronaviruses (OC43, 229E, NL63, and HKU1); HRV, human rhinoviruses; AdV, adenovirus; and HBoV, human bocavirus.
A total of 38 swab pairs were negative in both samples; 104 pairs were positive in at least 1 swab, with 2 viruses detected in 9 swab pairs and 3 viruses detected in 3 swab pairs.
A total of 30 swab pairs were negative in both samples; 110 pairs were positive in at least 1 swab with 2 viruses detected in 10 swab pairs. No detections were made for influenza virus A, influenza virus B, and human bocavirus. Note that two patient symptom surveys were omitted because no data were reported for rhinorrhea.
Forty adult HCT patients with a clinical sample positive for a respiratory virus were followed longitudinally (32 patients were positive on first follow-up) for a median of 3 weeks (range, 1 to 9 weeks), providing 140 weekly paired NS samples. Swabs were processed a median of 1 day after collection (range, 1 to 8 days); 95% of swabs were processed in ≤2 days. Three patients had 2 viruses detected simultaneously; 2 patients had a second virus detected during follow-up. At least one sample had a respiratory virus detected by either method in 110/140 (79%) swab pairs (Table 2).
In general, swabs collected with saline spray performed better. This was most apparent in the immunocompetent group (Table 2). The improved performance of NS collected with saline was more pronounced in immunocompetent subjects without rhinorrhea than in those with rhinorrhea (P = 0.07 for interaction).
Seventy-five immunocompetent subjects completed an acceptability survey. Most strongly agreed or agreed that the process was comfortable (87%) and simple (96%); all reported willingness to participate in future studies. Sixty-one percent of 28 respondents who underwent NW comparison testing reported they strongly agreed or agreed they preferred self-collection over NW, 32% reported no preference, and 7% disagreed or strongly disagreed.
We report a comprehensive study of NS self-collection for diagnosis of respiratory virus infections in immunocompetent subjects and HCT recipients. We first examined optimal transport media and temperature, demonstrating that storage in dry tubes at room temperature was logistically feasible, with no apparent effect of time to processing on sensitivity of virus detection. We chose foam swabs because they are absorptive and potentially less likely than nylon, Dacron, or cotton to induce bleeding in HCT patients with mucositis or thrombocytopenia. Another study utilizing foam swabs documented better performance than nylon flocked swabs for rapid influenza virus antigen testing (19). Foam NS collected with saline nasal spray provided excellent results compared with NW, and saline spray improved performance compared to dry swabs alone. We hypothesize that saline spray may disrupt nasal respiratory epithelial cells, allowing for improved diagnostic yield.
Although we compared overall respiratory virus detections between collection methods, we lacked adequate numbers of subjects to make virus-specific comparisons. We also report results for a relatively small number of subjects without rhinorrhea. Collection of combined nasal and oropharyngeal samples could potentially increase sensitivity for respiratory virus detection (20–22). However, a small increase in sensitivity must be balanced by the cost of additional swabs, increased patient discomfort, and compliance issues. Our goal was to develop a comfortable, simple, and safe method for transplant patients with mucositis and nausea, and a throat swab in this situation might be undesirable. In the future, it would be of benefit to assess the added value of an oropharyngeal swab.
In conclusion, we have developed a well-accepted and sensitive method for self-collection of respiratory samples that is simple and safe for immunocompromised children and adults and does not require transport medium or refrigeration. This collection method will enable future study participants to collect samples at home that could be shipped to the laboratory, reducing infection control concerns while allowing for studies of outcomes or potential antiviral therapies. This method could potentially be applied to family members and health care workers caring for immunocompromised patients, or it could be employed in a pandemic situation (7), in which prompt diagnosis of viral disease is essential while minimizing patient exposure. Foam NS collected with saline spray increased sensitivity for virus detection in both immunocompetent subjects and HCT patients compared with dry swabs, and we recommend this approach as an alternative to NW for conducting respiratory virus surveillance.
(This work was presented in part at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy/46th Infectious Diseases Society of America Annual Meeting, Washington, DC, 2008, and the American Society of Bone Marrow Transplant/Center for International Blood and Marrow Transplant Research Tandem Meeting, San Diego, CA, 2012.)
ACKNOWLEDGMENTS
We thank Cheryl Callais, Victor Chow, Alex Weiss, Denise Della, Robert Farney, Karin Oesterich, Deborah Reyes, Srilata Remala, Noelle Gichohi, Gynevill Jolly, Jennifer Klein, and Christi Dahlgren for study coordination and assistance, Julia Emerson for statistical consultation, and Terry Stevens-Ayers, Tera Matson, Sam Chatterton-Kirchmeier, Abe Guerrero, Nancy Wright, Reggie Sampoleo, Rohit Shankar, and Anne Cent for laboratory expertise.
This work was supported by the National Institutes of Health (grants CA 18029, HL081595, K23HL091059, L40AI071572, and K24HL093294). A.P.C. also received support from the MedImmune Pediatric Fellowship Grant Award, the Pediatric Infectious Diseases Society Fellowship Award funded by MedImmune, and the Seattle Children's Center for Clinical and Translational Research and CTSA grant ULI RR025014.
M.B. received research support from Roche and GlaxoSmithKline and is a consultant for Gilead and GlaxoSmithKline. J.A.E. received research support from Novartis and Chimerix and is a consultant for Novavax and GlaxoSmithKline. All other authors have no potential conflicts of interests to disclose.
Footnotes
Published ahead of print 7 November 2012
REFERENCES
- 1. Chan KH, Peiris JS, Lim W, Nicholls JM, Chiu SS. 2008. Comparison of nasopharyngeal flocked swabs and aspirates for rapid diagnosis of respiratory viruses in children. J. Clin. Virol. 42:65–69 [DOI] [PubMed] [Google Scholar]
- 2. Lambert SB, Whiley DM, O'Neill NT, Andrews EC, Canavan FM, Bletchly C, Siebert DJ, Sloots TP, Nissen MD. 2008. Comparing nose-throat swabs and nasopharyngeal aspirates collected from children with symptoms for respiratory virus identification using real-time polymerase chain reaction. Pediatrics 122:e615–e620 [DOI] [PubMed] [Google Scholar]
- 3. Meerhoff TJ, Houben ML, Coenjaerts FE, Kimpen JL, Hofland RW, Schellevis F, Bont LJ. 2010. Detection of multiple respiratory pathogens during primary respiratory infection: nasal swab versus nasopharyngeal aspirate using real-time polymerase chain reaction. Eur. J. Clin. Microbiol. Infect. Dis. 29:365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Munywoki PK, Hamid F, Mutunga M, Welch S, Cane P, Nokes DJ. 2011. Improved detection of respiratory viruses in pediatric outpatients with acute respiratory illness by real-time PCR using nasopharyngeal flocked swabs. J. Clin. Microbiol. 49:3365–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sung RY, Chan PK, Choi KC, Yeung AC, Li AM, Tang JW, Ip M, Tsen T, Nelson EA. 2008. Comparative study of nasopharyngeal aspirate and nasal swab specimens for diagnosis of acute viral respiratory infection. J. Clin. Microbiol. 46:3073–3076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akmatov MK, Krebs S, Preusse M, Gatzemeier A, Frischmann U, Schughart K, Pessler F. 2011. E-mail-based symptomatic surveillance combined with self-collection of nasal swabs: a new tool for acute respiratory infection epidemiology. Int. J. Infect. Dis. 15:e799–e803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elliot AJ, Powers C, Thornton A, Obi C, Hill C, Simms I, Waight P, Maguire H, Foord D, Povey E, Wreghitt T, Goddard N, Ellis J, Bermingham A, Sebastianpillai P, Lackenby A, Zambon M, Brown D, Smith GE, Gill ON. 2009. Monitoring the emergence of community transmission of influenza A/H1N1 2009 in England: a cross sectional opportunistic survey of self sampled telephone callers to NHS Direct. BMJ 339:b3403 doi:10.1136/bmj.b3403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Esposito S, Molteni CG, Daleno C, Valzano A, Tagliabue C, Galeone C, Milani G, Fossali E, Marchisio P, Principi N. 2010. Collection by trained pediatricians or parents of mid-turbinate nasal flocked swabs for the detection of influenza viruses in childhood. Virol. J. 7:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lambert SB, Allen KM, Druce JD, Birch CJ, Mackay IM, Carlin JB, Carapetis JR, Sloots TP, Nissen MD, Nolan TM. 2007. Community epidemiology of human metapneumovirus, human coronavirus NL63, and other respiratory viruses in healthy preschool-aged children using parent-collected specimens. Pediatrics 120:e929–e937 [DOI] [PubMed] [Google Scholar]
- 10. Lambert SB, Allen KM, Nolan TM. 2008. Parent-collected respiratory specimens—a novel method for respiratory virus and vaccine efficacy research. Vaccine 26:1826–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Larios OE, Coleman BL, Drews SJ, Mazzulli T, Borgundvaag B, Green K, McGeer AJ. 2011. Self-collected mid-turbinate swabs for the detection of respiratory viruses in adults with acute respiratory illnesses. PLoS One 6:e21335 doi:10.1371/journal.pone.0021335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kuypers J, Wright N, Morrow R. 2004. Evaluation of quantitative and type-specific real-time RT-PCR assays for detection of respiratory syncytial virus in respiratory specimens from children. J. Clin. Virol. 31:123–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Foulongne V, Olejnik Y, Perez V, Elaerts S, Rodiere M, Segondy M. 2006. Human bocavirus in French children. Emerg. Infect. Dis. 12:1251–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuypers J, Martin ET, Heugel J, Wright N, Morrow R, Englund JA. 2007. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics 119:e70–e76 [DOI] [PubMed] [Google Scholar]
- 15. Kuypers J, Wright N, Corey L, Morrow R. 2005. Detection and quantification of human metapneumovirus in pediatric specimens by real-time RT-PCR. J. Clin. Virol. 33:299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuypers J, Wright N, Ferrenberg J, Huang ML, Cent A, Corey L, Morrow R. 2006. Comparison of real-time PCR assays with fluorescent-antibody assays for diagnosis of respiratory virus infections in children. J. Clin. Microbiol. 44:2382–2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lu X, Holloway B, Dare RK, Kuypers J, Yagi S, Williams JV, Hall CB, Erdman DD. 2008. Real-time reverse transcription-PCR assay for comprehensive detection of human rhinoviruses. J. Clin. Microbiol. 46:533–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Peck AJ, Englund JA, Kuypers J, Guthrie KA, Corey L, Morrow R, Hackman RC, Cent A, Boeckh M. 2007. Respiratory virus infection among hematopoietic cell transplant recipients: evidence for asymptomatic parainfluenza virus infection. Blood 110:1681–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scansen KA, Bonsu BK, Stoner E, Mack K, Salamon D, Leber A, Marcon MJ. 2010. Comparison of polyurethane foam to nylon flocked swabs for collection of secretions from the anterior nares in performance of a rapid influenza virus antigen test in a pediatric emergency department. J. Clin. Microbiol. 48:852–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hammitt LL, Kazungu S, Welch S, Bett A, Onyango CO, Gunson RN, Scott JA, Nokes DJ. 2011. Added value of an oropharyngeal swab in detection of viruses in children hospitalized with lower respiratory tract infection. J. Clin. Microbiol. 49:2318–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim C, Ahmed JA, Eidex RB, Nyoka R, Waiboci LW, Erdman D, Tepo A, Mahamud AS, Kabura W, Nguhi M, Muthoka P, Burton W, Breiman RF, Njenga MK, Katz MA. 2011. Comparison of nasopharyngeal and oropharyngeal swabs for the diagnosis of eight respiratory viruses by real-time reverse transcription-PCR assays. PLoS One 6:e21610 doi:10.1371/journal.pone.0021610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lieberman D, Shimoni A, Keren-Naus A, Steinberg R, Shemer-Avni Y. 2009. Identification of respiratory viruses in adults: nasopharyngeal versus oropharyngeal sampling. J. Clin. Microbiol. 47:3439–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]