Abstract
In a large prospective comparison, the illumigene test detected Clostridium difficile in 98% of toxin-positive and 58% of toxin-negative samples confirmed positive by other methods. The Xpert was uniformly sensitive. Most samples with discrepant results had C. difficile concentrations below the illumigene limit of detection. The significance of low-level C. difficile detection needs investigation.
TEXT
The incidence and severity of Clostridium difficile infection (CDI) have increased dramatically in North America and Europe over the last decade (1). In the United States, rates of CDI are now at an all-time high (2). With this change in incidence, the performance of C. difficile testing has been reexamined and there has been a movement toward tests that detect C. difficile antigens or DNA directly versus tests that detect C. difficile toxins (3). As a group, C. difficile detection tests (e.g., PCR, loop-mediated amplification [LAMP], glutamate dehydrogenase immunoassay) are more sensitive than toxin tests, but the reported sensitivities still vary for unclear reasons. For example, the published sensitivity of the illumigene C. difficile assay ranges from 81 to 98% (4–11), leaving laboratories with uncertainty about which test to use and what performance to expect at their own institution.
To address this issue, we evaluated the fecal C. difficile DNA load of positive samples as part of a large, prospective comparison of two FDA-approved nucleic acid amplification tests (NAATs) for C. difficile, the illumigene C. difficile test and the Xpert C. difficile/Epi test, with toxigenic culture. When the clinical results and fecal C. difficile load comparisons suggested a genuine difference in the sensitivity of these two NAATs, we evaluated the inoculum size and analytical limit of detection (LOD) of both assays to further investigate the causes and significance of discrepant results.
Study population and test methods.
Consecutive diarrheal stool samples submitted for C. difficile testing from adult inpatients ≥72 h after admission between January and October 2011 were included in the study; nonconforming stool samples were rejected. Each sample was tested for toxigenic C. difficile by three tests, (i) an illumigene C. difficile LAMP assay (Meridian Bioscience), (ii) an Xpert C. difficile/Epi real-time PCR assay (Cepheid), and (iii) a toxigenic culture. In addition, the fecal toxin status of each sample was determined by a toxin immunoassay (Premier C. difficile Toxins A & B; Meridian Bioscience) performed on all samples and a cytotoxicity assay (Wampole C. difficile Tox-B [TechLab]; MHRF cells [Diagnostic Hybrids]) performed on immunoassay-negative, C. difficile-positive samples from frozen aliquots when available (55/61; 90.2%) (12). All tests except cytotoxicity were performed daily on fresh stool by following the manufacturer's instructions. For toxigenic culture, 0.5 ml stool was mixed 1:1 with 95% EtOH for 10 min and a swab was used to inoculate a prereduced agar medium (cycloserine cefoxitin fructose agar supplemented with taurocholate [CCFA-ST]; Remel). Cultures were incubated anaerobically at 37°C for 3 days, and suspicious colonies were identified by colony and Gram stain morphology, odor, production of l-proline aminopeptidase, and fluorescence under long-range UV light. In vitro toxin production of isolates was confirmed by cytotoxicity testing of isolates grown in chopped meat broth (Remel) at 48 h (13). Samples that were negative with the initial EtOH shock culture but positive by either of the NAATs were cultured in enrichment broth (cycloserine cefoxitin mannitol broth with taurocholate lysozyme cysteine [CCMB-TAL]; Anaerobe Systems) from a frozen aliquot (n = 10) (14). The ability of the illumigene assay to detect toxigenic C. difficile isolates recovered from culture of illumigene-negative samples was tested directly using a 100-μl volume of a 4 McFarland suspension from fresh subculture.
Fecal load, inoculum size, and LOD methods.
The fecal C. difficile DNA load of positive samples was calculated from the Xpert tcdB gene PCR cycle number at the endpoint of detection using standard curves performed with each lot of test cartridges as previously described (12). The typical inoculum size of each test was evaluated by weighing five replicate samplings of one soft, one loose, and one watery stool sample using the sample collection brush (illumigene) or swab (Copan) by following the manufacturer's instructions. For the Xpert assay, our practice was to dip the swab fully and wipe any excess on the container wall. The LOD of each NAAT was determined by replicate testing of a 100-μl volume from serial dilutions (10-fold and 2-fold) of a 24-hour culture of C. difficile ATCC 43255 (VPI 10463), with the concentrations confirmed by the average of triplicate colony counts on brucella agar.
Data and statistical methods.
The first positive sample from each patient or first sample from negative patients was included; duplicate samples were excluded. Samples with ≥2 positive results (e.g., toxigenic culture, Xpert, illumigene, toxin by immunoassay or cytotoxicity) were considered to be confirmed as positive for toxigenic C. difficile. Sensitivity and specificity values and 95% confidence intervals (CIs) were calculated against the 2-test reference standard. Chi-square and Fisher's exact tests were used for categorical data.
Test results from clinical samples.
Of the 693 samples tested, 568 samples and patients were included in the analysis. One hundred (17.6%) samples, including 64 (64%) fecal-toxin-positive samples and 36 (36%) toxin-negative samples, had toxigenic C. difficile detected by ≥2 methods. The number of positives detected by each method (e.g., toxigenic culture, illumigene C. difficile, Xpert C. difficile/Epi) and the calculated sensitivities and specificities are shown in Table 1. Overall, both NAATs were highly sensitive (>98%) for C. difficile in fecal-toxin-positive samples, but the illumigene detected C. difficile in only 21/36 (58.3%) fecal-toxin-negative samples while the Xpert detected C. difficile in all 36 (100%) samples in this group (Table 1). The Xpert had 6 additional positive results that were not able to be confirmed by any other test. These were considered false positives.
Table 1.
Number of samples called positive by each test and overall sensitivity and specificity
| Reference classificationa | Total no. of samples | No. (%b) of samples testing positive with each test or % sensitivity/specificity (95% CI) |
||
|---|---|---|---|---|
| Toxigenic culture | illumigene C. difficile | Xpert C. difficile/Epi | ||
| Toxin positive/C. difficile positive | 64 | 62 (96.9) | 63 (98.4) | 64 (100.0) |
| Toxin negative/C. difficile positive | 36 | 32 (88.9) | 21 (58.3) | 36 (100.0) |
| C. difficile negative | 468 | 0 (0.0) | 0 (0.0) | 6 (1.3) |
| Sensitivity | 100 | 94 (89.4–98.7) | 84 (76.8–91.2) | 100 (95.4–100) |
| Specificity | 468 | 100 (99.0–100) | 100 (99.0–100) | 98.7 (97.7–99.7) |
≥2 tests positive.
Percentages are the number of samples positive by the test/the number of samples positive by the reference classification times 100.
Fecal C. difficile load, inoculum size, and LOD evaluations.
Comparison of the fecal DNA concentrations of samples with C. difficile detected by the Xpert but missed by the illumigene showed that virtually all (15/16; 93.8%) were stools with no fecal toxins detected and with a relatively low concentration of C. difficile DNA (Fig. 1). For direct comparison, the median concentration of C. difficile DNA for the 84 samples detected by illumigene was 6.64 (95% CI, 6.24 to 6.70) log10 C. difficile tcdB DNA copies/ml; the median concentration of the 16 illumigene-negative, Xpert-positive samples was 4.11 (95% CI, 3.82 to 4.60) log10 C. difficile tcdB DNA copies/ml. To better understand why the illumigene missed the C. difficile DNA in these low-concentration samples, we examined the inoculum size and LOD of both tests. The mean inoculum weights differed slightly between the two tests (ratio of sample weights between the Xpert swab and the illumigene brush, 0.7 to 1.6) but not to a degree that would explain the sensitivity difference among toxin-negative samples. In contrast, the analytical LOD of the two NAATs differed by 1.2 log10 DNA copies for the VPI 10463 strain we tested (Fig. 1). The illumigene detected 9/9 replicates at 32,750 CFU/ml (4.52 log10) and 5/10 replicates at 16,375 CFU/ml (4.21 log10), yielding an LOD of 4.52 log10 C. difficile tcdB DNA copies/ml. The Xpert detected 10/10 replicates at 2,047 CFU/ml (3.31 log10) and 0/5 replicates at 1,024 CFU/ml (3.01 log10), yielding an LOD of 3.31 log10 C. difficile tcdB DNA copies/ml. Using these LODs, 9/16 (56.3%) illumigene-negative, Xpert-positive samples had C. difficile DNA concentrations below the illumigene LOD; 7/16 (43.8%) had concentrations just above the LOD (range, 0.19 to 1.11 log10 above the LOD). To confirm that the illumigene assay can detect the C. difficile strains from samples with discrepant results, higher concentrations were tested directly from culture. Of the 15 available isolates, all were amplified and detected.
Fig 1.
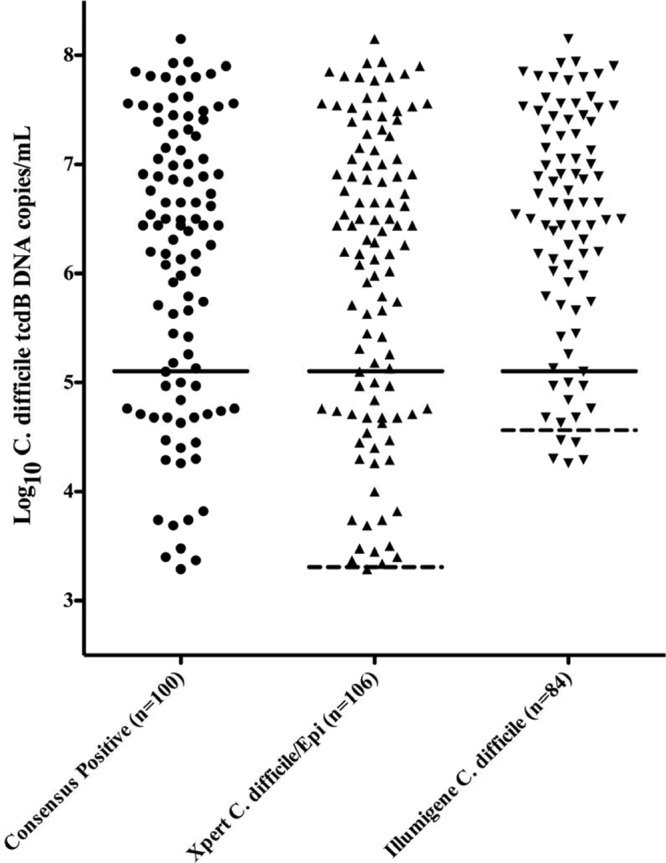
Fecal C. difficile concentrations of positive stool samples overall and detected by each test. Black circles (●) are consensus positive samples with toxigenic C. difficile detected by ≥2 tests. Upward-pointing triangles (▲) are all samples reported positive by the Xpert C. difficile/Epi test. Downward-pointing triangles (▼) are samples reported positive by the illumigene C. difficile test. Solid lines represent a 95% sensitivity cutoff for toxin detection (5.10 log10 C. difficile tcdB DNA copies/ml) from reference 12. Above this line, 61/73 (83.6%) of samples were toxin positive. Below this line, 24/27 (88.9%) samples were toxin negative. Dashed lines indicate the Xpert and illumigene C. difficile DNA LOD values discussed in the text (Xpert, 3.31 log10 C. difficile tcdB DNA copies/ml; illumigene, 4.52 log10 C. difficile tcdB DNA copies/ml).
Conclusions.
Overall, we found a 16% sensitivity difference between the illumigene C. difficile LAMP assay and the Xpert C. difficile/Epi real-time PCR test in a large-scale, prospective comparison with toxigenic culture. When the fecal toxin status of samples was considered, the illumigene and Xpert tests performed similarly and were both highly sensitive for toxin-positive samples, but the illumigene was much less sensitive with toxin-negative samples (58% for illumigene versus 100% for Xpert; P < 0.001). We suspected that this was due to the low concentration of C. difficile target DNA in these samples. To validate this hypothesis and our overall results, we compared the C. difficile concentrations of positive samples detected by both tests and positive samples missed by the illumigene and determined the LOD of both tests. These investigations confirmed that virtually all of the clinical samples with discrepant results had low C. difficile DNA concentrations that were near or below the illumigene LOD but above the Xpert LOD, as expected. The LOD thresholds we measured showed the illumigene LAMP assay to be less sensitive analytically than the Xpert real-time PCR test by about 1.2 log10 DNA copies/ml and were in agreement with the LOD data provided in the manufacturer's package insert for each test (15, 16). Conversely, when C. difficile isolates from samples originally called negative by the illumigene were retested at a higher concentration from culture, all were amplified and detected. Together, these findings demonstrate a clinical sensitivity difference between the illumigene C. difficile assay and the Xpert C. difficile/Epi test at low C. difficile concentrations that appears to be due to a difference in the analytical sensitivity of these two NAATs and not due to nonamplification due to DNA sequence polymorphisms. Careful review of the instructions for these two tests seems to suggest that the difference may be due to the additional sample dilution steps included in the preanalytical processing with the illumigene assay (15). Two other studies have reported sensitivities similar to ours for the illumigene and Xpert assays, but the underlying causes were not investigated (10, 11). Others have reported higher sensitivities for the illumigene assay in the range of 92 to 98% sensitivity, but it is possible that some of these were biased by use of a less-sensitive reference method or inclusion of large numbers of toxin-positive samples in the study (4–9).
We believe that these results show conclusively that the illumigene C. difficile LAMP assay and the Xpert C. difficile/Epi test differ in their analytical and clinical sensitivity, in particular in their ability to detect low-concentration, toxin-negative, C. difficile-positive samples. However, since the clinical significance of fecal samples with a low bacterial load of C. difficile is unknown, these results suggest that more questions need to be answered. If these patients are simply carriers without clinically significant CDI, then the illumigene assay may be more accurate. Alternatively, if there is no safe level of C. difficile for either CDI or transmission, then the more-sensitive Xpert assay may be preferable.
ACKNOWLEDGMENTS
This work was supported by grants from the National Center for Advancing Translational Sciences (8KL2TR000134-07) (C.R.P.).
All other authors declare no conflicts of interest.
Materials and reagents were partially provided by Cepheid, Meridian, and TechLab (C.R.P.).
We also thank the staff of the UC Davis Clinical Microbiology Laboratory for their help with this study and ongoing support.
Footnotes
Published ahead of print 10 October 2012
REFERENCES
- 1. Lessa FC, Gould CV, McDonald LC. 2012. Current status of Clostridium difficile infection epidemiology. Clin. Infect. Dis. 55(Suppl 2): S65–S70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McDonald LC, Lessa F, Sievert D, Wise M, Herrera R, Gould C, Malpiedi P, Dudeck M, Srinivasan A, Fridkin S, Cardo D. 2012. Vital signs: preventing Clostridium difficile infections. MMWR Morb. Mortal. Wkly. Rep. 61: 157–162 [PubMed] [Google Scholar]
- 3. Kufelnicka AM, Kirn TJ. 2011. Effective utilization of evolving methods for the laboratory diagnosis of Clostridium difficile infection. Clin. Infect. Dis. 52: 1451–1457 [DOI] [PubMed] [Google Scholar]
- 4. Boyanton BL, Jr, Sural P, Loomis CR, Pesta C, Gonzalez-Krellwitz L, Robinson-Dunn B, Riska P. 2012. Loop-mediated isothermal amplification compared to real-time PCR and enzyme immunoassay for toxigenic Clostridium difficile detection. J. Clin. Microbiol. 50: 640–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bruins MJ, Verbeek E, Wallinga JA, Bruijnesteijn van Coppenraet LE, Kuijper EJ, Bloembergen P. 2012. Evaluation of three enzyme immunoassays and a loop-mediated isothermal amplification test for the laboratory diagnosis of Clostridium difficile infection. Eur. J. Clin. Microbiol. Infect. Dis. 31: 3035–3039 [DOI] [PubMed] [Google Scholar]
- 6. Doing KM, Hintz MS. 2012. Prospective evaluation of the Meridian Illumigene loop-mediated amplification assay and the Gen Probe ProGastro Cd polymerase chain reaction assay for the direct detection of toxigenic Clostridium difficile from fecal samples. Diagn. Microbiol. Infect. Dis. 72: 8–13 [DOI] [PubMed] [Google Scholar]
- 7. Dubberke ER, Han Z, Bobo L, Hink T, Lawrence B, Copper S, Hoppe-Bauer J, Burnham CA, Dunne WM., Jr 2011. Impact of clinical symptoms on the interpretation of diagnostic assays for Clostridium difficile. J. Clin. Microbiol. 49: 2887–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lalande V, Barrault L, Wadel S, Eckert C, Petit JC, Barbut F. 2011. Evaluation of a loop-mediated isothermal amplification assay for diagnosis of Clostridium difficile infections. J. Clin. Microbiol. 49: 2714–2716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Noren T, Alriksson I, Andersson J, Akerlund T, Unemo M. 2011. Rapid and sensitive loop-mediated isothermal amplification test for Clostridium difficile detection challenges cytotoxin B cell test and culture as gold standard. J. Clin. Microbiol. 49: 710–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pancholi P, Kelly C, Raczkowski M, Balada-Llasat JM. 2012. Detection of toxigenic Clostridium difficile: comparison of the cell culture neutralization, Xpert C. difficile, Xpert C. difficile/Epi, and Illumigene C. difficile assays. J. Clin. Microbiol. 50: 1331–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Viala C, Le Monnier A, Maataoui N, Rousseau C, Collignon A, Poilane I. 2012. Comparison of commercial molecular assays for toxigenic Clostridium difficile detection in stools: BD GeneOhm Cdiff, XPert C. difficile and illumigene C. difficile. J. Microbiol. Methods 90: 83–85 [DOI] [PubMed] [Google Scholar]
- 12. Leslie JL, Cohen SH, Solnick JV, Polage CR. 20 July 2012, posting date Role of fecal Clostridium difficile load in discrepancies between toxin tests and PCR: is quantitation the next step in C. difficile testing? Eur. J. Clin. Microbiol. Infect. Dis. 31: 3295–3299 (Erratum, 4 August 2012, posting date doi:10.1007/s10096-012-1721-8.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reller ME, Lema CA, Perl TM, Cai M, Ross TL, Speck KA, Carroll KC. 2007. Yield of stool culture with isolate toxin testing versus a two-step algorithm including stool toxin testing for detection of toxigenic Clostridium difficile. J. Clin. Microbiol. 45: 3601–3605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tenover FC, Novak-Weekley S, Woods CW, Peterson LR, Davis T, Schreckenberger P, Fang FC, Dascal A, Gerding DN, Nomura JH, Goering RV, Akerlund T, Weissfeld AS, Baron EJ, Wong E, Marlowe EM, Whitmore J, Persing DH. 2010. Impact of strain type on detection of toxigenic Clostridium difficile: comparison of molecular diagnostic and enzyme immunoassay approaches. J. Clin. Microbiol. 48: 3719–3724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Meridian Bioscience, Inc 2011. illumigene C. difficile package insert. Meridian Bioscience, Inc., Cincinnati, OH [Google Scholar]
- 16. Cepheid 2011. Xpert C. difficile/Epi package insert. Cepheid, Sunnyvale, CA [Google Scholar]


