Abstract
From a collection of yeast isolates isolated from patients in Tunisian hospitals between September 2006 and July 2010, the yeast strain JEY63 (CBS 12513), isolated from a 50-year-old male that suffered from oral thrush, could not be identified to the species level using conventional methods used in clinical laboratories. These methods include matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), germ tube formation, and the use of CHROMagar Candida and metabolic galleries. Sequence analysis of the nuclear rRNA (18S rRNA, 5.8S rRNA, and 26S rRNA) and internal transcribed spacer regions (ITS1 and ITS2) indicated that the ribosomal DNA sequences of this species were not yet reported. Multiple gene phylogenic analyses suggested that this isolate clustered at the base of the Dipodascaceae (Saccharomycetales, Saccharomycetes, and Ascomycota). JEY63 was named Candida tunisiensis sp. nov. according to several phenotypic criteria and its geographical origin. C. tunisiensis was able to grow at 42°C and does not form chlamydospores and hyphae but could grow as yeast and pseudohyphal forms. C. tunisiensis exhibited most probably a haploid genome with an estimated size of 10 Mb on at least three chromosomes. Using European Committee for Antimicrobial Susceptibility Testing (EUCAST) and Clinical and Laboratory Standards Institute (CLSI) Candida albicans susceptibility breakpoints as a reference, C. tunisiensis was resistant to fluconazole (MIC = 8 μg/ml), voriconazole (MIC = 0.5 μg/ml), itraconazole (MIC = 16 μg/ml), and amphotericin B (MIC = 4 μg/ml) but still susceptible to posaconazole (MIC = 0.008 μg/ml) and caspofungin (MIC = 0.5 μg/ml). In conclusion, MALDI-TOF MS permitted the early selection of an unusual isolate, which was still unreported in molecular databases but could not be unambiguously classified based on phylogenetic approaches.
INTRODUCTION
The incidence of fungal infection has significantly increased during the past 2 decades. These infections mainly occur in susceptible patients afflicted with hematological malignancies or undergoing organ or bone marrow transplantation (1–5). Patients and treatment have changed, as well as fungi, which are involved in infections. Indeed, species that were previously unknown to the medical community are now emerging (4, 6). Invasive fungal infections are associated with high mortality rates, often related to a delayed diagnosis (1, 3), and novel fungal species isolated in human are being increasingly reported (6). Because fungal species show very different antifungal susceptibility profiles, it is important for clinicians to obtain accurate identifications in order to apply appropriate treatments (3, 7). Identification of yeast species is based on different criteria. Traditionally, fungi have been identified by phenotypic characters, including morphology, colony appearance, or pigmentation on chromogenic media (8, 9). However, molecular methods with high sensitivity and specificity have been now established. In particular, the D1/D2 domains of the nuclear large subunit ribosomal DNA (nucLSU rDNA) was found to be useful for identification of most ascomycetous yeasts (6, 10–12). Alternative methods using protein signatures with matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) have been developed with the aim of facilitating rapid yeast identification (13, 14). Introduction of this technique has facilitated the discovery of novel species (15, 16).
The purpose of the present study was to use the aforementioned morphological and molecular methods to characterize yeasts from a collection from different hospitals in Tunisia in order to identify them at the species level. Within this collection, an isolate with unusual antifungal susceptibility profile originating from a 50-year-old male suffering from oral thrush could not be identified by these methods. We consequently conducted phylogenetic multigene analyses to determine its placement within the Ascomycota.
MATERIALS AND METHODS
Strains and media.
Clinical yeast isolates were obtained from patients admitted at three public institutions between September 2006 and July 2010: Hospital Habib Thameur and Hospital La Rabta (Tunis) and Hospital Ibn El Jazzar of Kairouan in central Tunisia. The various clinical specimens were collected and analyzed by standard microbiological procedures with Sabouraud dextrose agar supplemented with chloramphenicol and gentamicin and incubated at 37°C for 24 at 48 h. Yeast isolates were identified on chromogenic medium CHROMagar Candida (CHROMagar Candida, Paris, France) based on the following criteria (C. albicans and C. dubliniensis, green colonies; C. tropicalis, metallic blue colonies; C. krusei, pink to light mauve colonies). Growth on YEPDA (1% Bacto peptone [Difco Laboratories, Basel, Switzerland]), 0.5% yeast extract [Difco], 2% glucose [Fluka, Buchs, Switzerland], 2% agar [Difco]) at 42°C differentiated between C. albicans (growth) and C. dubliniensis (no growth). The C. albicans ATCC 90028, C. dubliniensis CBS 7987, C. tropicalis ATCC 750, and C. krusei ATCC 6258 were used as control strains. For some isolates, ID 32C metabolic galleries (bioMérieux, Marcy l'Etoile, France) were used as described by the manufacturer.
Description of Candida tunisiensis J. Eddouzi, V. Hofstetter, M. Groenewald, M. Manai, D. Sanglard sp. nov. (JEY63).
Candida tunisiensis (tu.ni.si.en′sis, N.L. nom. fem. sing. adj., tunisiensis, referring to the region from which this yeast was isolated, Tunis, the capital of the Tunisia Republic). On YEPD agar, after 1 week of incubation at 25°C, the colonies are white in color, circular, rough, with convex elevation and invade agar. The cells are round to ovoid, single or in pairs, the budding is multipolar, and the cells are 3 to 4 μm by 5 to 6 μm. Pseudohyphal growth is dominant, but no transition to a hyphal form is observed. The formation of ascospores is not observed. The physiological characteristics are presented in Table S1 in the supplemental material. The optimal growth temperature is between 37 and 42°C. The type strain is JEY63 (CBS 12513), which was isolated from a 50-year-old male that suffered from oral thrush in a Tunisian hospital. The Mycobank accession number is 509092.
Formation of chlamydospores.
Yeasts were grown in complete YEPD medium (1% Bacto peptone [Difco]), 0.5% yeast extract [Difco], 2% glucose [Fluka]) at 30°C under constant shaking. The inoculum size was adjusted to 1.5 × 104 CFU/ml by measuring the absorbance in a spectrophotometer. A 50-μl of the third serial dilution was plated onto rice extract agar (rice extract at 0.7 g/liter and agar at 14.3 g/liter [Fluka], supplemented with 1% Tween 80) under a sterile glass coverslip to maintain a semianaerobic condition and grown in the dark for 7 days at 25°C. The rice extract is the sole source of nutrient in the medium. The lack of nutrients together with the oxygen-deficient culture conditions creates an environment that induces the formation of specific morphological forms in particular chlamydospores in some yeast. The test was performed in triplicate.
DNA isolation, amplification, and sequencing.
Genomic DNA was prepared from yeast cultures after overnight incubation in YEPD medium at 30°C under constant shaking as previously described (17). Six DNA regions of the fungal strain JEY63 were amplified and sequenced: four ribosomal gene regions (the small subunit [18S], the large subunit [26S], the 5.8S and flanking internal transcribed spacers 1 and 2 [ITS1-5.8S-ITS2], part of the mitochondrial small subunit [12S]), two protein-coding gene regions (part of the second largest subunit of the RNA polymerase II [subunit B150, regions 6 and 7; RPB2] and part of translation elongation factor 1α [TEF1α]). The primers used for amplification and/or sequencing are listed in Table 1. Amplification of nuclear ribosomal genes was performed in the presence of 200 μM concentrations of each deoxynucleoside triphosphate, 250 nM concentrations of each primer, 1.5 mM MgCl2, 2.6 U of Expand high-fidelity PCR System (Roche, Switzerland), and 5 ng of total genomic DNA. After enzyme activation at 94°C for 5 min, the reactions were subjected to 35 thermal cycles of 94 and 54°C for 60 s for each temperature, 72°C for 4 min, and a final elongation at 72°C for 10 min. Different regions of 12S, RPB2, and TEF1α were amplified and sequenced under the conditions and with the primers described by Zoller et al. (18), Matheny et al. (19), and Morehouse et al. (20), respectively. Sequencing was performed using reagents and conditions of the BigDye Terminator v1.1 cycle sequencing kit and on an ABI Prism 3130 XL DNA analyzer (Perkin-Elmer/Applied Biosystems, Foster City, CA). Final sequences were assembled and edited using the software package Sequencher 3.0 (Gene Codes Corp., USA). The GenBank accession numbers for ribosomal rRNA, mitSSU, and TEF1 are reported in Table S1 in the supplemental material. The GenBank accession number for RPB2 is JX104155.
Table 1.
Primers used in this study
| Analysis and primer | Sequence (5′–3′) |
|---|---|
| PCR of nuclear rRNA | |
| 18S-F | CTGGTTGATCCTGCCAGTAGT |
| 26Sext-R | ACTCCGTTGTACATCTAAGTCG |
| Sequencing of nuclear rRNA | |
| 18S-1 | CATGGTTTCAACGGGTAA |
| 18S-2 | CCTATTCTATTATTCCATGC |
| 18S-3 | TCCCTTCCAGAGGTCGGG |
| 18S-R | TCAGCCTTGCGACCATACTCC |
| L18S-F | GCTGGGGATAGAGCATTGCA |
| 26S-F | GTACAGTGATGGAAAGATGA |
| 26S-1 | GGTGAGTTGTTACACACTCC |
| 26S-2 | GCCATGGAAGTCGGAATCC |
| 26S-3 | AGAGCACTGGGCAGAAATCAC |
| 26S-4 | CTGACTGTCTAATTAAAACA |
| 26S-5 | AGCAGAACTGGCGATGCG |
| 26S-6 | TGCCGCGAAGCTACCATC |
| 26S-7 | TCCATTCATGCGCGTCAC |
| 26Sint-R | AGCATGGATTCTGACTTAGAGG |
| L28S-R | TCCAAACCGATGCTGGCC |
| ITS1 | TCCGTAGGTGAACCTGCGG |
| ITS2 | GCTGCGTTCTTCATCGAT |
| PCR and sequencing of mitSSU rDNA | |
| mrSSU-1 | AGCAGTGAGGAATATTGGTC |
| mrSSU-3R | ATGTGGCACGTCTATAGCCC |
| PCR and sequencing of RPB2 | |
| fRPB2-6F | TGGGGKWTGGTYTGYCCTGC |
| fRPB2-7cR | CCCATRGCTTGYTTRCCCAT |
| PCR and sequencing of TEF1 | |
| EF1a1F | TACAARTGYGGTGGTATYGACA |
| EF1a1R | ACNGACTTGACYTCAGTRGT |
Phylogenetic analyses.
The JEY63 sequences (18S, 26S, 12S, RPB2, and TEF1) were first added to the 6×-locus alignment for Ascomycota used by Schoch et al. (21; http://wasabi.lutzonilab.net/pub/alignments/download_alignments). Including JEY63, this alignment included 435 taxa. A second data set (4 loci/79 taxa) was built combining JEY63 sequences of the 18S, 26S, 12S, and TEF1 loci with sequence data of the same loci sampled in GenBank for 77 taxa representative of the major clades identified in the Saccharomycetes in previously published phylogenetic studies (22–24) and for an outgroup, Schizosaccharomyces pombe (see Table S1 in the supplemental material). Alignment of nucleotide sequences was performed manually using the MacClade 4.06 editor (25). Maximum-parsimony (MP) analyses were conducted in PAUP v40b10 (26). The search for the most parsimonious tree(s) used 500 heuristic searches of random addition sequence (RAS), with MAXTREE = 500 (for the 6-locus/435-taxon data set) or with MAXTREE = unlimited (for the 4-locus/79-taxon data set), with tree bisection and reconnection (TBR) branch swapping, the MULPARS option on, and gaps coded as missing characters. Branch robustness was estimated based on 100 (6-locus/435-taxona data set) and 500 (4-locus/79-taxon data set) MP bootstrap replicates, each of 100 RAS, and with the same settings as for the search for the most parsimonious tree(s). Branches that received MP bootstrap support (MPbs) ≥ 70% were considered significantly supported (27).
MALDI-TOF MS.
After incubation of strains for 24 h to 48 h at 35°C on YEPDA, sample preparation was performed on 4 to 5 yeast colonies as previously described by Pinto et al. (16). The analyzed of mass spectra were generated by the matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) MALDI-TOF mass spectrometry (MS) systems Microflex LT (Bruker Daltonics GmbH, Leipzig, Germany) with FlexControl (version 3.0) software (Bruker Daltonics). The data were processed by the associated software, and the sample spectra were compared to reference spectra in the provided database for microorganism identification. MALDI-TOF MS was performed on 11 identified reference strains (C. albicans ATCC 90028, C. albicans ATCC 24433, C. dubliniensis ATCC 2118, C. dubliniensis ATCC 2119, C. dubliniensis CBS 7987, C. glabrata ATCC 90030, C. krusei ATCC 6258, C. parapsilosis ATCC 22019, C. parapsilosis ATCC 90018, C. tropicalis ATCC 750, and Cryptococcus neoformans ATCC 90112) as a validation method. Each sample was tested in duplicate to ensure reproducibility of the spectra. A characterization score cutoff value was attributed to each sample and was interpreted as recommended by the manufacturer (scores of <1.7 indicate unreliable identification, scores from 1.7 to 1.999 indicate identification to the genus level; scores of 2 to 2.299 indicate probable species identification, and scores of ≥2.3 indicate highly probable species identification).
Susceptibility testing.
MIC values were determined by broth microdilution using the reference procedure of the Antifungal Susceptibility Testing Subcommittee of the European Committee for Antimicrobial Susceptibility Testing (EUCAST) for the testing of fermentative yeasts (28) with slight modification (5). The strains were cultivated overnight in YEPD at 30°C. Tests were performed in flat-bottom microdilution plates with RPMI 1640 medium supplemented with 2% of glucose, glutamine, and phenol red as pH indicator, but without bicarbonate. The inoculum sizes were adjusted to 0.5 × 105 to 2.5 × 105 CFU/ml by measuring the absorbance in a spectrophotometer. Drug dilutions were made for each in the corresponding solvents with concentration ranges for fluconazole (128 to 0.0625 mg/liter) and for itraconazole, voriconazole, posaconazole, amphotericin B, and caspofungin (16 to 0.0078 mg/liter). C. albicans ATCC 90028, C. glabrata ATCC 90030, and C. tropicalis ATCC 750 were included as control isolates. MIC values were determined with a spectrophotometer (at 530 nm) after 24 h of incubation as the lowest concentration of drug resulting in ≥50% (MIC50) and ≥90% (MIC90) growth inhibition, respectively.
Flow cytometry.
The estimation of the nuclear DNA content in yeast was achieved by flow cytometry with propidium iodide as fluorochrome staining. Yeast cells of three strains (C. albicans SC5314, C. glabrata CBS 138, and JEY63) were grown in YEPD broth overnight to the stationary phase of growth. Then, 10-ml samples of each culture were harvested, and the cells were fixed in 70% ethanol for 12 h at 4°C. The samples were washed once with 5 ml of 50 mM sodium citrate (pH 7.5) and resuspended in 1 ml of 50 mM sodium citrate. The cell concentration was diluted to 1.5 × 107 cells/ml. To each sample, 25 μl of RNase A (10 mg/ml) was added. After 1 h of incubation at 50°C, 100 μl of proteinase K (10 mg/ml) was added. The incubation was continued for an additional hour at 50°C, after 1 ml of 50 mM sodium citrate containing 16 μg of propidium iodide/ml was added. The samples were then incubated overnight at 4°C in the dark. Analyses were made by using a FACSCalibur flow cytometer (Becton Dickinson Biosciences, San Jose, CA) with a blue argon laser emitting at a 488-nm wavelength at 15 mW. For each histogram, 15,000 cells were analyzed.
Pulsed-field gel electrophoresis (PFGE).
Yeast cells were grown overnight in YEPD medium and washed twice with 5 ml of 50 mM EDTA (pH 9.0). Approximately 109 cells/ml were resuspended in 330 μl of 50 mM EDTA (pH 9.0). The volume was adjusted to 1 ml with 110 μl of solution A (1 M sorbitol, 10 mM EDTA, 100 mM sodium citrate, zymolase [100 U/ml], 5% [vol/vol] β-mercaptoethanol) and 560 μl of 1% (wt/vol) SeaKem GTG agarose in TE buffer (10 mM Tris-HCl, 1 mM EDTA [pH 8.0]) maintained at 50°C. After mixing, the solution was poured into the wells of plug molds and kept at room temperature for solidification. The plugs were transferred into tubes containing 3 ml of solution B (0.1 M Tris-HCl [pH 8.0], 0.45 M EDTA [pH 8.0], 7.5% β-mercaptoethanol) and incubated overnight at 37°C to obtain spheroplasts. The plugs were transferred into new tubes containing 5 ml of solution C (100 mM Tris-HCl [pH 8.0], 0.45 M EDTA [pH 8.0], 1% N-lauroylsarcosine, 0.15 mg of proteinase K/ml), followed by incubation for 6 h at 65°C. The agarose plugs were stored in 0.5 M EDTA (pH 9.0) at 4°C until use.
One-half of the plug was loaded into the wells of a 0.8% agarose gel (Bio-Rad). The gel was then placed into the electrophoresis chamber of a CHEF-DR III system (Bio-Rad) with running buffer (0.089 M Tris base, 0.089 M borate, 0.0025 M EDTA) diluted twice in water, and it was cooled to 14°C during recirculation. The best separation was achieved after 42 h (phase 1: pulse time of 120 to 300 s at 4.5 V, angle 120°, 24 h; phase 2: pulse time of 720 to 900 s at 2 V, angle 106°, 18 h). After electrophoresis, the gel was stained with ethidium bromide solution for 20 min and destained with distilled water.
RESULTS AND DISCUSSION
A collection of 423 clinical isolates was obtained from patients (blood, oral cavity, normally sterile body fluids, gastrointestinal tract, respiratory tract, biomedical devices, skin, and nail clippings) in three different hospitals in Tunisia between September 2006 and July 2010. The yeast isolates were first identified using the chromogenic medium CHROMagar Candida. This resulted in the rapid identification of C. albicans or C. dubliniensis, C. tropicalis, and C. krusei. C. dubliniensis was then specifically identified by its inability to grow at 42°C (29). Given the limitation of this medium for identifying all yeast species, the collection was systematically analyzed by MALDI-TOF MS. This method has been used successfully to determine the mass of proteins and peptides in addition to identifying previously unknown proteins. In recent years, MALDI-TOF MS has been implemented in routine laboratories and utilized as a completely new approach for the identification of bacteria, yeast, and filamentous fungi (16, 30–34).
From a collection containing 423 isolates, 413 exhibited MALDI-TOF analysis scores above the cutoff value of 2.0, which is empirically considered valid for correct yeast species identification. This demonstrates that the MALDI-TOF technology is an efficient tool for the rapid and reliable identification of clinically important identified yeasts. The MALDI-TOF test allowed us to relate the investigated fungal isolates to the following species: C. albicans (185 isolates), C. glabrata (70 isolates), C. parapsilosis (56 isolates), C. tropicalis (48 isolates), C. dubliniensis (21 isolates), C. krusei (8 isolates), C. metapsilosis (5 isolates), C. lusitaniae (4 isolates), C. kefyr (2 isolates), C. orthopsilosis (1 isolate), C. guilliermondii (1 isolate), C. utilis (1 isolate), C. intermedia (1 isolate), Saccharomyces cerevisiae (7 isolates), Trichosporon asahii (1 isolate), Trichosporon inkin (1 isolate), and Trichosporon mucoides (1 isolate) (see Table S3 in the supplemental material). The discriminatory power of MALDI-TOF MS could be demonstrated by its ability to distinguish three closely related species, including C. parapsilosis, C. metapsilosis, and C. orthopsilosis and others related species (34–36). There were no discrepancies between Chromagar and MALDI-TOF results (see Table S3 in the supplemental material). Ten remaining isolates could not be assigned to any reference isolate since the MALDI-TOF scores were below the discriminatory threshold value of 1.7 (see Fig. S1 [for spectra] and Table S3 [for scores] in the supplemental material). These isolates could not be identified at the species level by other methods such as metabolic galleries (see Table S3 in the supplemental material). The most likely explanation for the inability of MALDI-TOF to assign these isolates to specific species could be that the BioTyper instrument database is still not representative of the entire diversity of yeast species involved in human diseases. This conclusion was also drawn by Bizzini et al. (37) in a study on the identification of 410 clinical isolates that were not identified with conventional laboratory methods. Of 410 tested isolates, only 133 (32.4%) failed to be reliably identified (score < 1.7). Several other studies aiming to identify microbial species from clinical samples with MALDI-TOF attributed identification failures to the lack of reference spectra in the available instrument databases (33, 35, 36).
BLAST of ribosomal data.
In order to determine the species of the 10 remaining isolates, we undertook the sequencing of their D1/D2 and ITS1/ITS2 domains from the large subunit rDNA. This method resulted in the presumptive identification of 9 isolates to the following species (Table 2): Hanseniaspora opuntiae (3 isolates) Candida palmioleophila (2 isolates), Kodamea ohmeri (2 isolates), Debaryomyces hansenii (1 isolate), and Pichia caribbica (1 isolate). Only strain JEY63, for which the MALDI-TOF spectrum was unique (see Fig. S1 in the supplemental material) and not present in the MALDI-TOF instrument database, remained unknown after these steps, even including ITS1 or ITS2, which are considered the best loci for fungal barcodes (38).
Table 2.
Species assignment of isolates not identified by MALDI-TOF MS analysis
| Collection | Pairwise identity (%) of regions to existing database |
Species assignment | |
|---|---|---|---|
| D1/D2a region | ITS1/ITS2 region | ||
| JEY63 | 86.9 | 70.0 | |
| JEY182 | 100 | 98.7 | Kodamaea ohmeri |
| JEY234 | 100 | 99.7 | Kodamaea ohmeri |
| JEY258 | 100 | 99.5 | Hanseniaspora opuntiae |
| JEY267 | 100 | 99.8 | Pichia caribbica |
| JEY269 | 100 | 99.5 | Hanseniaspora opuntiae |
| JEY270 | 100 | 99.8 | Hanseniaspora opuntiae |
| JEY379 | 100 | 99.8 | Candida palmioleophila |
| JEY380 | 100 | 99.8 | Candida palmioleophila |
| JEY420 | 100 | 99.8 | Debaryomyces hansenii |
That is, domains D1 and D2 of the large subunit rDNA (LSU rDNA).
The GenBank accession numbers of JEY63 ribosomal sequences (18S, ITS1-5.8S-ITS2, and 26S) are reported in Table S1 in the supplemental material. The BLAST top scores of the 18S and 26S ribosomal regions were the most similar to members of the Saccharomycetes (Ascomycota) but with BLAST top scores that were too low to confirm JEY63's affiliation to this fungal class (BLAST top scores for 18S: Blastobotrys adeninivorans and several other Candida species, 95% sequence similarity; BLAST top scores for 26S: Sugiyamaella smithiae, Nakazawaea holstii, Spencermartinsiella europaea, Pichia xylosa, Scheffersomyces stipitis, and several other Candida species, 91% sequence similarity).
Clinical data from strain JEY63.
JEY63 was isolated from a 50-year-old man suffering from a pemphigus vulgaris disease. He was admitted to the University hospital of Habib Thameur in Tunis between February and May 2009. The diagnosis of pemphigus vulgaris was confirmed by histology and direct immunofluorescence. The patient showed extended mucocutaneous vesicles with clear or blood-stained contents. The treatment was controlled by corticosteroids (1.5 mg/kg/day). In March 2010, the patient was again admitted, and he was marked by the recurrence of extensive bullo-erosive lesions with involvement of the genital area. The second period of hospitalization lasted 4 months (11 March to 7 July 2010). During this period, three oral swabs were obtained from the patient each month. In these specimens, the same type of yeast was isolated, and JEY63 was the last sample. The patient also developed bacterial infections (Staphylococcus spp.). Therefore, he often received antibacterial agents such as bristopen, ciprofloxacin, and fucidin. During this period, no documented antifungal therapy was given.
Phenotypic and physiological characteristics.
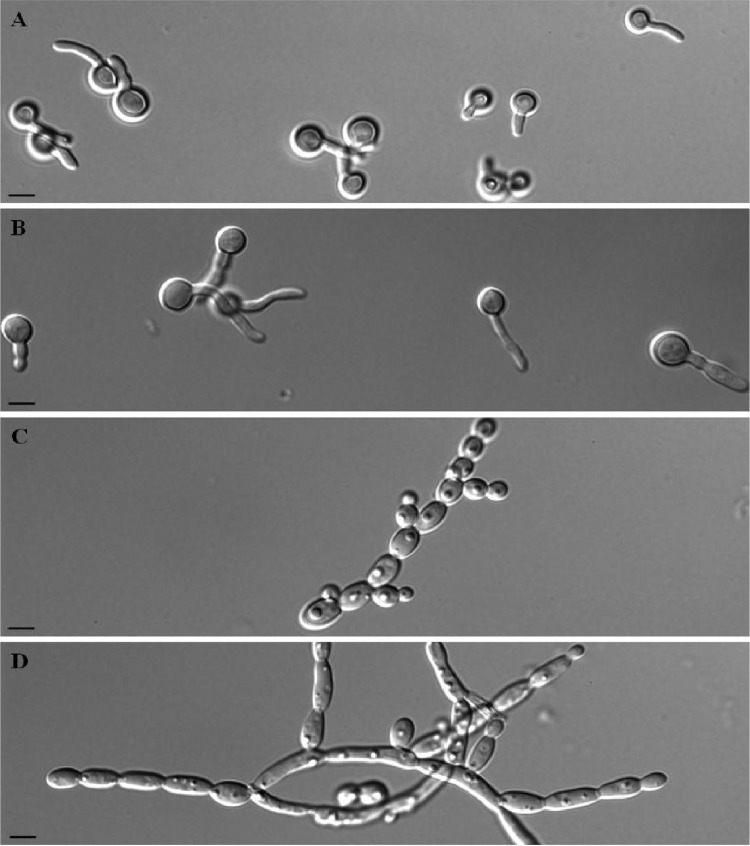
Microscopic observations showed the formation of germ tubes in C. albicans SC5314 in calf serum at 35°C, while only pseudohyphae were detected in JEY63 (Fig. 1A and C). When the incubation was continued for 3 h, the extension of gem tubes slowly elongated in C. albicans SC5314, but the dominant morphology was the development of pseudohyphae in JEY63 (Fig. 1B and D). The new strain did also produce pseudohyphae after overnight incubation in YEPD medium at 30°C under constant shaking (Fig. 2). Chlamydospore formation on rice extract agar plate was observed as expected in C. albicans SC5314; however, only pseudohyphae were observed in JEY63 (Fig. 3). The results from physiological characterization of JEY63 are shown in Table S2 in the supplemental material. According to the metabolic database available at the CBS Fungal Biodiversity Centre, the profile does not correspond to any deposited fungal isolate. One striking feature of this yeast species is its ability to grow at high temperature (42°C). It is also positive for glucose fermentation but does not exhibit a high metabolic capacity in metabolizing diverse sugars.
Fig 1.
Germ tube formation by C. albicans SC5314 and JEY63 after incubation of 106 cells/ml at 35°C in 50% calf serum. (A and B) C. albicans SC 5314 incubated for 1.5 and 3 h, respectively; (C and D): JEY63 incubated for 1.5 and 3 h, respectively. Bar, 10 μm.
Fig 2.
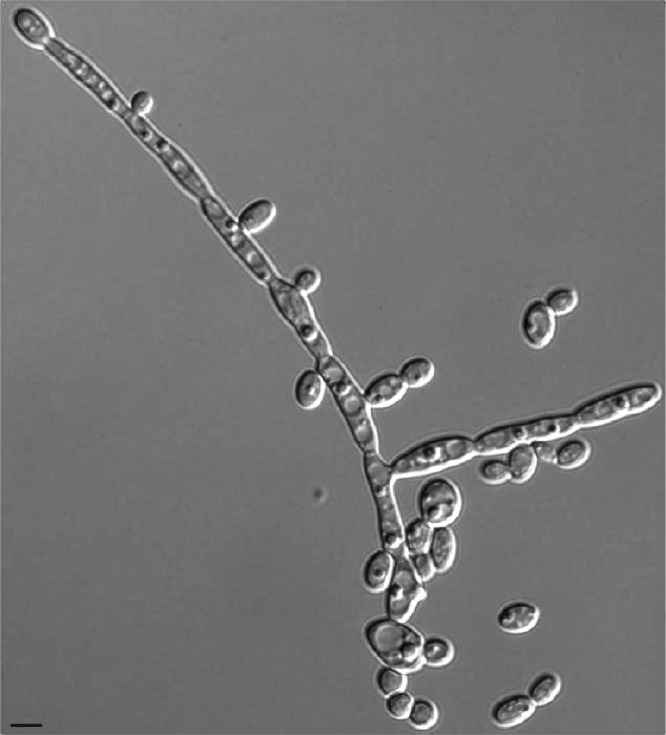
Morphology of yeast and pseudohyphal forms of JEY63 after overnight incubation in YEPD medium at 30°C with constant shaking. Bar, 10 μm.
Fig 3.
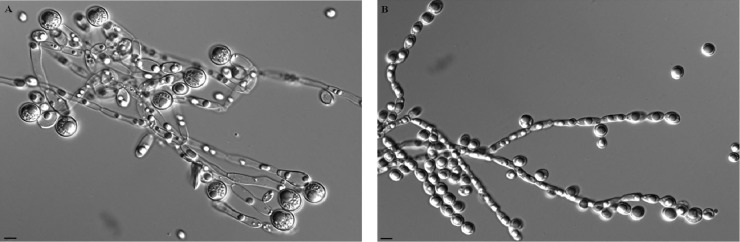
Production of chlamydospores by C. albicans SC5314 and JEY63 after incubation on rice extract agar plates supplemented with 1% (vol/vol) Tween 80 in the dark at 25°C for 7 days. (A) C. albicans SC5314; (B) JEY63. Bar, 10 μm.
Phylogenetic analyses.
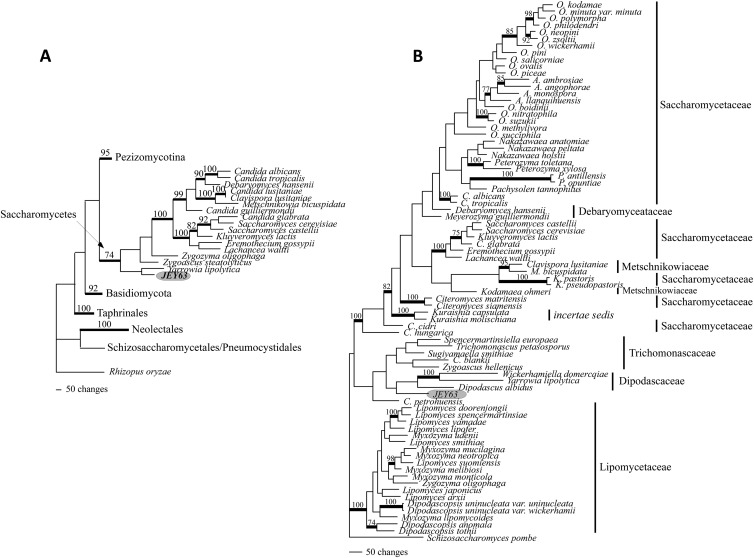
The GenBank accession numbers of JEY63 sequences (mitSSU and TEF1) are reported in Table S1 in the supplemental material. Since we were not able to determine the placement of JEY63 in the classification of Ascomycota using classical methods, including MALDI-TOF or BLAST, we first inferred the phylogenetic placement of JEY63 within the Ascomycota, adding this taxon to the alignment of Schoch et al. (21) and running MP analyses. This 6-locus/435-taxon alignment included 26,665 characters. After the exclusion of ambiguously aligned regions, 2,811 of the 5,128 characters that were kept for analyses were parsimony informative. Phylogenetic analyses of the 6-locus/435-taxon data set suggested with significant support (MPbs = 74%) that JEY63 was nested within the Saccharomycetes and resolved as a sister to Yarrowia lipolytica, though without support (Fig. 4A). Knowing that JEY63 belonged to the Saccharomycetes, we conducted a second MP analysis restricted to this class. The full length of this 4-locus/79-taxon alignment included 14,753 characters, 4,582 characters after the exclusion ambiguous regions with 1,202 of that were parsimony informative. MP analyses of the 4-locus/79-taxon data set (Fig. 4B) placed JEY63 basal to a clade including the members of Dipodascaceae (Dipodascus albidus, Yarrowia lipolytica, and Wickerhamiella domercqiae). Trichomonascaceae was resolved, without support, as the monophyletic and sister group of the Dipodascaceae (Fig. 4B). JEY63 appeared, however, clearly separated from the clade, including a paraphyletic Saccharomycetaceae (this clade also included the sampled Debaryomycetaceae and Metschnikowiaceae) with high support (MPbs = 100%) and from the Lipomycetaceae (MPbs = 100%). Overall, this 4-locus analysis exhibited low bootstrap support, which prevented us to discuss further internal relationships within the Saccharomycetes. The phylogenetic data place JEY63 alone in a branch at the base of Dipodascaceae family. We choose five species (Dipodascus aggregatus CBS 175.53, Galactomyces geotrichum CBS 772.71, Sporopachydermia lactativora CBS 6192, Wickerhamiella domercqiae CBS 4351, and Yarrowia lipolytica CBS 6124) belonging to this family for phenotypic and metabolic comparisons to JEY63. The comparisons revealed that JEY63 was distinguishable for its capacity to use complex carbohydrate (threalose, raffinose, sucrose, maltose, melezitose, and starch) as carbon sources (see Table S2 in the supplemental material). On the other hand, microscopic observations of the different strains revealed distinct morphologies and cell sizes compared to the other Dipodascaceae members (see Fig. S2 in the supplemental material). In particular, the formation of pseudohyphae was always observed in JEY63 but not in other isolates. Moreover, MALDI-TOF spectra of the chosen Dipodascaceae members yielded quite divergent spectra, suggesting a poor relationship between these isolates (see Fig. S3 in the supplemental material).
Fig 4.
Phylogenetic placement of JEY63. (A) One of the 500 most parsimonious trees inferred from the analysis of the 6-locus/435 taxon data set (length [L] = 82,021, consistency index [CI] = 0.083, retention index [RI] = 0.491, rescaled consistency index [RC] = 0.041, homoplasy index [HI] = 0.917). (B) One of the 12 most parsimonious trees inferred from the analysis of the 4-locus/79-taxon data set (L = 7,920, CI = 0292, RI = 0.592, RC = 0.173, HI = 0.708). Significant bootstrap values are reported along the branches, and supported internodes are highlighted in boldface. Genus abbreviations: A., Ambrosiozyma; C., Candida; K., Komagataella; M., Metschnikowia; O., Ogataea; P., Phaffomyces.
Taken together, the combined phylogenetic, metabolic, microscopic, and MALDI-TOF data suggest that JEY63 is quite distant from other members of the Dipodascaceae family and, in addition, do not form spores (no sexual cycle) as opposed to Dipodascaceae family members. We therefore proposed the name of Candida tunisiensis sp. nov. for this novel isolate to accommodate to the yeast taxonomic system.
Estimation of the genome size of C. tunisiensis and chromosome numbers.
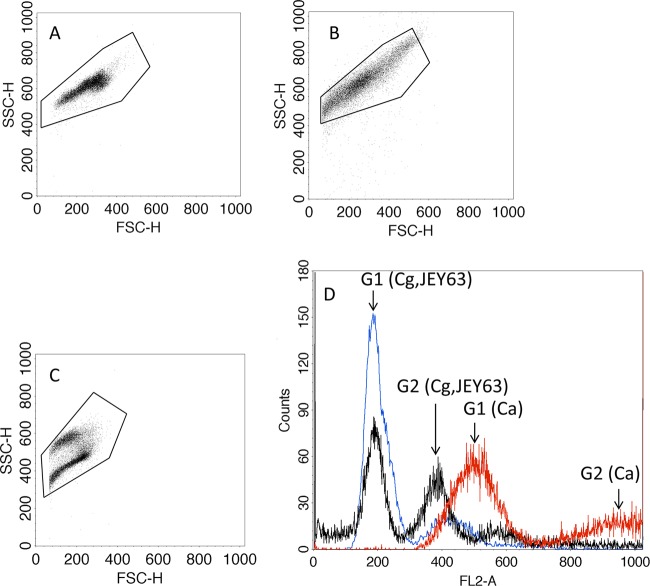
In the present study, flow cytometry was used as a tool to estimate the genome size of C. tunisiensis. Genome size estimation was based on the comparison of the amount of propidium iodide fluorescence with that was obtained from C. albicans SC5314 and C. glabrata CBS 138 as references for typical diploid and haploid genome sizes. The results show two peak intensities corresponding to the G1 and G2 phases (Fig. 5). A comparison of the peak positions of C. tunisiensis to those of the two external references indicates a good overlap with C. glabrata DNA content. This suggests that C. tunisiensis is haploid. The genome size of C. glabrata CBS 138 has been estimated to be ∼12.338 Mb on 13 chromosomes. PFGE was used to verify the number and sizes of the chromosomes of C. tunisiensis. C. albicans was included as a reference with known chromosome numbers and sizes. The separated chromosomal DNAs are shown in Fig. 6. We assumed that the migration distance of the chromosome bands depends on the chromosome size and that bands with increased UV staining within one lane probably contained two chromosomes with similar sizes. The karyotype patterns revealed that C. albicans SC5314 contained eight chromosomes; however, only three chromosomal bands were detected in C. tunisiensis. The slowest-migrating signal showed higher intensity than other chromosomes and thus suggests that probably two chromosomes are comigrating. The C. albicans chromosome sizes range from 0.949 to 3.188 Mb with a 14.284-Mb haploid genome size. According to these data and assuming sizes of 2 Mb, 2.26 Mb, and 2 × 3.18 Mb, we can estimate an approximate genome size of 10.62 Mb in C. tunisiensis.
Fig 5.
Flow cytometry analysis of nucleic DNA from overnight culture and stained with propidium iodide. (A) C. albicans SC5314; (B) JEY63 (C. tunisiensis CBS 12513); (C) C. glabrata CBS 138; (D) overlay histograms. The G1 and G2 phases of each species are indicated by arrows (Ca, C. albicans, red color; Cg, C. glabrata, blue color; JEY63, black). C. albicans SC5314 and C. glabrata CBS 138 were included as standards to facilitate estimations of DNA content of JEY63. The SSC-H and FSC-H axes indicate side-scatter and forward-scatter channel intensities, respectively.
Fig 6.
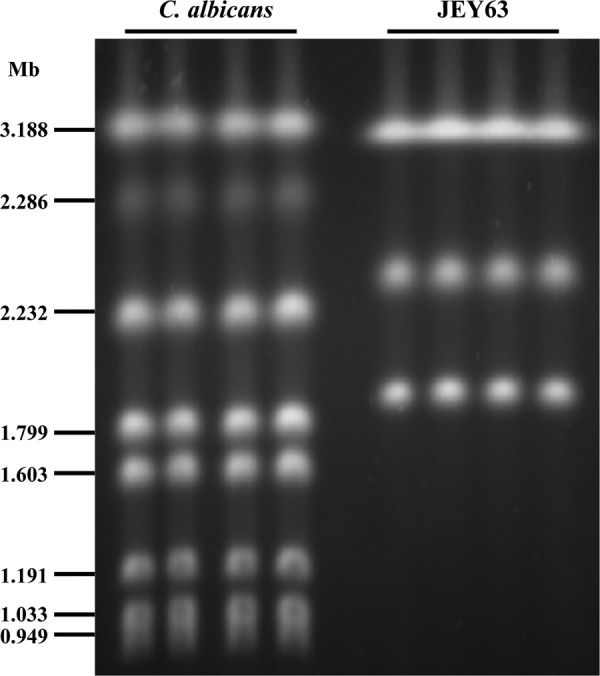
PFGE analysis of yeast chromosomes in C. albicans SC5314 and JEY63 (C. tunisiensis CBS 12513). The sizes of the C. albicans chromosomes are indicated on the left.
Antifungal susceptibility testing.
The MICs of fluconazole, itraconazole, posaconazole, caspofungin, and amphotericin B for C. tunisiensis were measured according to EUCAST protocols and are summarized in Table 3. Since we used here a unique clinical isolate, it was not possible to estimate epidemiological cutoff values for discriminating between antifungal susceptibility and resistance for this isolate. We arbitrarily took EUCAST and Clinical and Laboratory Standards Institute (CLSI) clinical breakpoints validated for C. albicans as a comparison. With these values, the data in Table 4 show that C. tunisiensis was resistant to amphotericin B (MIC = 4 μg/ml), fluconazole (MIC = 8 μg/ml), itraconazole (MIC = 16 μg/ml), and voriconazole (MIC = 0.5 μg/ml) (39–41). This isolate remained, however, susceptible to posaconazole (MIC = 0.008 μg/ml) and was still considered susceptible to caspofungin (MIC = 0.5 μg/ml). In these analyses, cross-resistance to all azoles was not observed in C. tunisiensis, and posaconazole in particular still exhibited high activity. Such patterns are not exceptional in yeast clinical isolates, and posaconazole is usually ranked as the most active azole (42). In conclusion, C. tunisiensis exhibits an unusual profile of antifungal susceptibility in presenting resistance to different agents. It is not possible to exactly determine whether the resistance profiles were acquired or not during potential antifungal therapy. Clinical data from this isolate indicate that the patient received probably only antibacterial agents therapy concomitantly with pemphigus vulgaris treatment. In the future, it is possible that other isolates from this species will be obtained, and thus it will be possible to discriminate between these possibilities.
Table 3.
In vitro susceptibility testing of amphotericin B, fluconazole, itraconazole, posaconazole, voriconazole, and caspofungin as determined by the EUCAST reference method
| Isolate | MIC (μg/ml) |
|||||
|---|---|---|---|---|---|---|
| Amphotericin B | Fluconazole | Itraconazole | Posaconazole | Voriconazole | Caspofungin | |
| C. albicans ATCC 90028 | 0.5 | 0.25 | <0.008 | <0.008 | <0.008 | 0.125 |
| C. glabrata ATCC 90030 | 1 | 8 | 0.25 | 0.25 | 0.125 | 0.25 |
| C. tropicalis ATCC 750 | 1 | 1 | 0.16 | <0.008 | <0.008 | 0.25 |
| JEY63 | 4 | 8 | 16 | 0.008 | 0.5 | 0.5 |
ACKNOWLEDGMENTS
We thank Françoise Ischer for technical assistance. We also thank Ridha Khelifa and Saïda Zouari (Service des Laboratoires, CHU Habib Thameur), Kacem Mahdouani (Laboratoire de Bactériologie, Hôpital Ibn El Jazzar de Kairouan), Dalenda El Euch (Service de Dermatologie, CHU La Rabta), and Emna Chaker (Laboratoire de Parasitologie-Mycologie, CHU La Rabta) for sampling yeast isolates. The MALDI-TOF analyses were assisted by the expertise of C. Durussel.
J.E. was the recipient of a grant from the Secrétariat d'Etat à l'Éducation et à la Recherche of the Swiss Confederation.
Footnotes
Published ahead of print 17 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01627-12.
REFERENCES
- 1. Menzin J, Meyers JL, Friedman M, Perfect JR, Langston AA, Danna RP, Papadopoulos G. 2009. Mortality, length of hospitalization, and costs associated with invasive fungal infections in high-risk patients. AJHP 66:1711–1717 [DOI] [PubMed] [Google Scholar]
- 2. Ohkubo T, Sugawara Y, Takayama T, Kokudo N, Makuuchi M. 2011. The risk factors of fungal infection in living-donor liver transplantations. J. Hepato-Biliary-Pancreatic Sci. 19:382–388 [DOI] [PubMed] [Google Scholar]
- 3. Pfaller MA, Diekema DJ. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36:1–53 [DOI] [PubMed] [Google Scholar]
- 4. Procop GW, Roberts GD. 2004. Emerging fungal diseases: the importance of the host. Clin. Lab. Med. 24:691–719 [DOI] [PubMed] [Google Scholar]
- 5. Slavin MA, Sorrell TC, Marriott D, Thursky KA, Nguyen Q, Ellis DH, Morrissey CO, Chen SC. 2010. Candidaemia in adult cancer patients: risks for fluconazole-resistant isolates and death. J. Antimicrob. Chemother. 65:1042–1051 [DOI] [PubMed] [Google Scholar]
- 6. Linton CJ, Borman AM, Cheung G, Holmes AD, Szekely A, Palmer MD, Bridge PD, Campbell CK, Johnson EM. 2007. Molecular identification of unusual pathogenic yeast isolates by large ribosomal subunit gene sequencing: 2 years of experience at the United Kingdom mycology reference laboratory. J. Clin. Microbiol. 45:1152–1158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cendejas-Bueno E, Gomez-Lopez A, Mellado E, Rodríguez-Tudela JL, Cuenca-Estrella M. 2010. Identification of pathogenic rare yeast species in clinical samples: comparison between phenotypic and molecular methods. J. Clin. Microbiol. 48:1895–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Odds FC, Bernaerts R. 1994. CHROMagar Candida, a new differential isolation medium for presumptive identification of clinically important Candida species. J. Clin. Microbiol. 32:1923–1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pfaller MA, Houston A, Coffmann S. 1996. Application of CHROMagar Candida for rapid screening of clinical specimens for Candida albicans, Candida tropicalis, Candida krusei, and Candida (Torulopsis) glabrata. J. Clin. Microbiol. 34:58–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daniel H-M, Meyer W. 2003. Evaluation of rRNA and actin gene sequences for the identification of ascomycetous yeasts. Int. J. Food Microbiol. 86:61–78 [DOI] [PubMed] [Google Scholar]
- 11. Kurtzman CP, Robnett CJ. 1998. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 73:331–371 [DOI] [PubMed] [Google Scholar]
- 12. Kurtzman CP, Robnett CJ. 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J. Clin. Microbiol. 35:1216–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Borman AM, Linton CJ, Oliver D, Palmer MD, Szekely A, Johnson EM. 2010. Rapid molecular identification of pathogenic yeasts by pyrosequencing analysis of 35 nucleotides of internal transcribed spacer 2. J. Clin. Microbiol. 48:3648–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Page BT, Shields CE, Merz WG, Kurtzman CP. 2006. Rapid identification of ascomycetous yeasts from clinical specimens by a molecular method based on flow cytometry and comparison with identifications from phenotypic assays. J. Clin. Microbiol. 44:3167–3171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, Theelen B, Groenewald M, Kostrzewa M, Cuenca-Estrella M, Gomez-Lopez A, Boekhout T. 2012. Reclassification of the Candida haemulonii complex: C. duobushaemulonii sp. nov. (C. haemulonii group II) and C. haemulonii var. vulnera var. nov., two multiresistant human pathogenic yeasts. J. Clin. Microbiol. 50:3641–3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pinto A, Halliday C, Zahra M, van Hal S, Olma T, Maszewska K, Iredell JR, Meyer W, Chen SC. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS One 6:e25712 doi:10.1371/journal.pone.0025712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanglard D, Ischer F, Calabrese D, Majcherczyk PA, Bille J. 1999. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 43:2753–2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zoller S, Lutzoni F, Scheidegger C. 1999. Genetic variation within and among populations of the threatened lichen Lobaria pulmonaria in Switzerland and implications for its conservation. Mol. Ecol. 8:2049–2059 [DOI] [PubMed] [Google Scholar]
- 19. Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, Hughes KW, Hofstetter V, Ammirati JF, Schoch CL, Langer E, Langer G, McLaughlin DJ, Wilson AW, Froslev T, Ge ZW, Kerrigan RW, Slot JC, Yang ZL, Baroni TJ, Fischer M, Hosaka K, Matsuura K, Seidl MT, Vauras J, Hibbett DS. 2007. Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Mol. Phylogenet. Evol. 43:430–451 [DOI] [PubMed] [Google Scholar]
- 20. Morehouse EA, James TY, Ganley AR, Vilgalys R, Berger L, Murphy PJ, Longcore JE. 2003. Multilocus sequence typing suggests the chytrid pathogen of amphibians is a recently emerged clone. Mol. Ecol. 12:395–403 [DOI] [PubMed] [Google Scholar]
- 21. Schoch CL, Sung GH, Lopez-Giraldez F, Townsend JP, Miadlikowska J, Hofstetter V, Robbertse B, Matheny PB, Kauff F, Wang Z, Gueidan C, Andrie RM, Trippe K, Ciufetti LM, Wynns A, Fraker E, Hodkinson BP, Bonito G, Groenewald JZ, Arzanlou M, de Hoog GS, Crous PW, Hewitt D, Pfister DH, Peterson K, Gryzenhout M, Wingfield MJ, Aptroot A, Suh SO, Blackwell M, Hillis DM, Griffith GW, Castlebury LA, Rossman AY, Lumbsch HT, Lucking R, Budel B, Rauhut A, Diederich P, Ertz D, Geiser DM, Hosaka K, Inderbitzin P, Kohlmeyer J, Volkmann-Kohlmeyer B, Mostert L, O'Donnell K, Sipman H, Rogers JD, Shoemaker RA, Sugiyama J, Summerbell RC, Untereiner W, Johnston PR, Stenroos S, Zuccaro A, Dyer PS, Crittenden PD, Cole MS, Hansen K, Trappe JM, Yahr R, Lutzoni F, Spatafora JW. 2009. The Ascomycota tree of life: a phylum-wide phylogeny clarifies the origin and evolution of fundamental reproductive and ecological traits. Syst. Biol. 58:224–239 [DOI] [PubMed] [Google Scholar]
- 22. Kurtzman CP, Robnett CJ. 2007. Multigene phylogenetic analysis of the Trichomonascus, Wickerhamiella, and Zygoascus yeast clades, and the proposal of Sugiyamaella gen. nov. and 14 new species combinations. FEMS Yeast Res. 7:141–151 [DOI] [PubMed] [Google Scholar]
- 23. Kurtzman CP, Robnett CJ. 2010. Systematics of methanol assimilating yeasts and neighboring taxa from multigene sequence analysis and the proposal of Peterozyma gen. nov., a new member of the Saccharomycetales. FEMS Yeast Res. 10:353–361 [DOI] [PubMed] [Google Scholar]
- 24. Peter G, Dlauchy D, Tornai-Lehoczki J, Suzuki M, Kurtzman CP. 2011. Spencermartinsiella europaea gen. nov., sp. nov., a new member of the family Trichomonascaceae. Int. J. Syst. Evol. Microbiol. 61:993–1000 [DOI] [PubMed] [Google Scholar]
- 25. Maddison WP, Maddison DR. 2003. MacClade: analysis of phylogeny and character evolution, version 4.06. Sinauer Associates, Sunderland, MA [Google Scholar]
- 26. Swofford DL. 2002. PAUP*: phylogenetic analysis using parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, MA [Google Scholar]
- 27. Mason-Gamer RJ, Kellogg EA. 1996. Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Syst. Biol. 45:524–545 [Google Scholar]
- 28. Subcommittee on Antifungal Susceptibility Testing of the ESCMID European Committee for Antimicrobial Susceptibility Testing 2008. EUCAST definitive document EDef 7.1: method for the determination of broth dilution MICs of antifungal agents for fermentative yeasts. Clin. Microbiol. Infect. 14:398–405 [DOI] [PubMed] [Google Scholar]
- 29. Pinjon E, Sullivan D, Salkin I, Shanley D, Coleman D. 1998. Simple, inexpensive, reliable method for differentiation of Candida dubliniensis from Candida albicans. J. Clin. Microbiol. 36:2093–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alanio A, Beretti JL, Dauphin B, Mellado E, Quesne G, Lacroix C, Amara A, Berche P, Nassif X, Bougnoux ME. 2011. Matrix-assisted laser desorption ionization time-of-flight mass spectrometry for fast and accurate identification of clinically relevant Aspergillus species. Clin. Microbiol. Infect. 17:750–755 [DOI] [PubMed] [Google Scholar]
- 31. Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. 2011. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 17:1359–1365 [DOI] [PubMed] [Google Scholar]
- 32. De Carolis E, Posteraro B, Lass-Florl C, Vella A, Florio AR, Torelli R, Girmenia C, Colozza C, Tortorano AM, Sanguinetti M, Fadda G. 2012. Species identification of Aspergillus, Fusarium, and Mucorales with direct surface analysis by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Microbiol. Infect. 18:475–484 [DOI] [PubMed] [Google Scholar]
- 33. Marklein G, Josten M, Klanke U, Muller E, Horre R, Maier T, Wenzel T, Kostrzewa M, Bierbaum G, Hoerauf A, Sahl HG. 2009. Matrix-assisted laser desorption ionization-time of flight mass spectrometry for fast and reliable identification of clinical yeast isolates. J. Clin. Microbiol. 47:2912–2917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quiles-Melero I, Garcia-Rodriguez J, Gomez-Lopez A, Mingorance J. 2012. Evaluation of matrix-assisted laser desorption/ionisation time-of-flight (MALDI-TOF) mass spectrometry for identification of Candida parapsilosis, C. orthopsilosis, and C. metapsilosis. Eur. J. Clin. Microbiol. Infect. Dis. 31:67–71 [DOI] [PubMed] [Google Scholar]
- 35. Jensen RH, Arendrup MC. 2011. Candida palmioleophila: characterization of a previously overlooked pathogen and its unique susceptibility profile in comparison with five related species. J. Clin. Microbiol. 49:549–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stevenson LG, Drake SK, Shea YR, Zelazny AM, Murray PR. 2010. Evaluation of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of clinically important yeast species. J. Clin. Microbiol. 48:3482–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bizzini A, Jaton K, Romo D, Bille J, Prod'hom G, Greub G. 2011. Matrix-assisted laser desorption ionization-time of flight mass spectrometry as an alternative to 16S rRNA gene sequencing for identification of difficult-to-identify bacterial strains. J. Clin. Microbiol. 49:693–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W. 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. U. S. A. 109:6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lass-Florl C, Arendrup MC, Rodriguez-Tudela JL, Cuenca-Estrella M, Donnelly P, Hope W. 2011. EUCAST technical note on amphotericin B. Clin. Microbiol. Infect. 17:E27–E29 [DOI] [PubMed] [Google Scholar]
- 40. Pfaller MA, Andes D, Arendrup MC, Diekema DJ, Espinel-Ingroff A, Alexander BD, Brown SD, Chaturvedi V, Fowler CL, Ghannoum MA, Johnson EM, Knapp CC, Motyl MR, Ostrosky-Zeichner L, Walsh TJ. 2011. Clinical breakpoints for voriconazole and Candida spp. revisited: review of microbiologic, molecular, pharmacodynamic, and clinical data as they pertain to the development of species-specific interpretive criteria. Diagn. Microbiol. Infect. Dis. 70:330–343 [DOI] [PubMed] [Google Scholar]
- 41. Pfaller MA, Andes D, Diekema DJ, Espinel-Ingroff A, Sheehan D. 2010. Wild-type MIC distributions, epidemiological cutoff values, and species-specific clinical breakpoints for fluconazole and Candida: time for harmonization of CLSI and EUCAST broth microdilution methods. Drug Resist. Updates 13:180–195 [DOI] [PubMed] [Google Scholar]
- 42. Pfaller MA. 2012. Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 125:S3–S13 [DOI] [PubMed] [Google Scholar]