Abstract
We describe using major outer membrane protein (MOMP) typing as a screen to compare the Campylobacter jejuni porA gene sequences of clinical outbreak strains from human stool with the porA sequences of dairy farm strains isolated during two milk-borne campylobacteriosis outbreak investigations in California. The genetic relatedness of clinical and environmental strains with identical or closely related porA sequences was confirmed by multilocus sequence typing and pulsed-field gel electrophoresis analysis. The first outbreak involved 1,644 C. jejuni infections at 11 state correctional facilities and was associated with consumption of pasteurized milk supplied by an on-site dairy (dairy A) at a prison in the central valley. The second outbreak involved eight confirmed and three suspect C. jejuni cases linked to consumption of commercial raw milk and raw chocolate colostrum at another central valley dairy (dairy B). Both dairies bottled fluid milk on the farm and distributed the finished product to off-site locations. Altogether, C. jejuni was isolated from 7 of 15 (46.7%) bovine fecal, 12 of 20 (60%) flush alley water, and 1 of 20 (5%) lagoon samples collected on dairy A. At dairy B, C. jejuni was cultured from 9 of 26 (34.6%) bovine fecal samples. Environmental strains indistinguishable from the clinical outbreak strains were found in five flush alley water samples (dairy A) and four bovine fecal samples (dairy B). The findings demonstrate that MOMP typing is a useful tool to triage environmental isolates prior to conducting more labor-intensive molecular typing methods.
INTRODUCTION
In the United States annually, an estimated 845,024 infections, 8,463 hospitalizations, and 76 deaths are caused by Campylobacter species (1). Preliminary data from the Foodborne Diseases Active Surveillance Network (FoodNet) indicate an overall incidence of 13 infections/100,000 population (2). Two species, Campylobacter jejuni and Campylobacter coli, cause >90% of reported human infections, although less-common species are considered emerging pathogens (3). Most patients experience a self-limiting gastrointestinal illness that resolves after 2 to 3 days. However, antecedent C. jejuni infection is the most common cause of Guillain-Barré syndrome, an acute inflammatory demyelinating polyneuropathy that leads to flaccid paralysis (4).
The majority of Campylobacter infections are zoonotic and linked to consumption of contaminated foods of animal origin or untreated water (3, 5, 6). Dairy products are the most common vehicle of transmission implicated in C. jejuni food-borne disease outbreaks, whereas sporadic infections are most frequently caused by poultry products (3, 7). An estimated 80% of dairy-related C. jejuni outbreaks are from consumption of raw (unpasteurized) milk, but outbreaks caused by failure in processing or postpasteurization contamination have been documented (3, 5, 6, 7, 8, 9, 10). Despite well-documented risks from raw dairy products, there continues to be consumer demand for access to these foods (11).
An increasing need exists to understand the mechanisms of dissemination of Campylobacter in the farm environment and to use this information for developing prevention strategies. Farm investigations can be challenging to conduct because of the lag time between the patient's exposure and identification of the food vehicle. Furthermore, the laboratory requirements to support source tracking from the farm can be expensive and labor-intensive. Given the ubiquitous presence of Campylobacter in livestock environments, finding a specific genotype or clinical outbreak strain on an implicated farm might be akin to searching for a needle in a haystack. In certain cases, analyzing numerous isolates from environmental samples by using pulsed-field gel electrophoresis (PFGE), the PulseNet standard for molecular subtyping of food-borne bacterial diseases (12, 13), is infeasible for a public health laboratory.
To address these concerns, we endeavored to identify an efficient molecular screening approach that allows triaging of a considerable number of Campylobacter isolates with the goal of source tracking the clinical outbreak strain back to the farm environment. We speculated that the DNA sequences of hypervariable regions of the porA gene (also referred to as cmp), encoding the major outer membrane protein (MOMP) of C. jejuni, might be exploited for this purpose (14, 15). As proof of concept, we developed a primer set that amplified a fragment of the porA gene (∼740 bp) and used phylogenetic analysis to compare the genetic relatedness of strains isolated from human stools with environmental strains isolated during two C. jejuni milk-borne outbreaks in California.
Outbreak 1.
In May 2006, a massive outbreak of C. jejuni infections involving 1,644 ill California prison inmates with 52 culture-confirmed cases was epidemiologically linked to consumption of pasteurized milk distributed to 11 state facilities (16). An on-site dairy (dairy A) and creamery at one of the prisons supplied the implicated milk to all affected facilities through a vocational program. No milk was distributed to the general public. Refrigerated, retained milk samples were phosphatase negative, indicating no global breach of pasteurization. C. jejuni was not recovered from the milk; however, the majority of samples had high aerobic standard plate (9.8 × 107 CFU/ml to 1.4 × 108 CFU/ml) and coliform (350 CFU/ml to >1.6 × 105 CFU/ml) counts. By regulatory standards for grade A pasteurized milk, standard plate counts should be <1.5 × 104 CFU/ml and there should be <10 CFU/ml coliforms. A quarantine order by the California Department of Agriculture was issued, and >2,900 cases of milk were returned.
Outbreak 2.
In November to December 2007, eight culture-confirmed and three suspect cases of C. jejuni infections were reported among consumers of commercially available raw milk and raw chocolate colostrum from a California licensed raw dairy (dairy B) with an on-site creamery (17). Cases from five geographically widespread counties had illness onsets clustered tightly within 11 days. Retained or leftover dairy product samples were unavailable for analysis. C. jejuni was not recovered from retail raw dairy products from other lots, but routine testing by the California Department of Food and Agriculture revealed a marked increase in aerobic standard plate counts from October (<2,500 CFU/ml) to November (>250,000 CFU/ml) preceding the illnesses. The aerobic standard plate count regulatory standard for grade A raw milk for human consumption was the same as that for pasteurized milk, but there was no coliform standard at the time. The short duration of the outbreak indicated that contaminated raw dairy products were available for only a brief time; thus, no quarantine order was issued.
MATERIALS AND METHODS
Study sites and sample collection.
The dairies were located in the central valley, a major inland milkshed in California. Unlike the majority of other central valley dairies, these dairies were relatively small (300 to 500 milking cows), and both bottled milk in a creamery on the property rather than shipping to an off-site processor. Dairy A was part of a state prison vocational program where inmates operated the dairy, grew forage on the property, and supplied milk to other state prisons in northern California. Dairy B was a state-licensed, commercial, organic, pasture-based raw-milk dairy that distributed raw dairy products to retail stores and farmers' markets statewide.
Environmental samples, including bovine feces, wastewater, and bulk tank raw milk (BTM), were collected at dairy A on 21 August 2006. Because of delays in gaining access to the prison dairy herd, our sampling occurred approximately 3 months after the outbreak was reported. Dairy A used a recycled wastewater system for manure management. We revisited dairy A on 21 March 2007 to conduct additional studies of the dairy wastewater system, including the flush alley water and lagoon (ponds).
We visited dairy B on 17 December 2007 to collect bovine feces, milking parlor discharge water, and BTM. Leftover or retained raw milk from the implicated lot was unavailable. Nine retail raw dairy samples with best-by dates between 25 December 2007 and 3 January 2008 produced by dairy B were collected from a health food store. Dairy B did not use a lagoon or flush alley recycled water system.
At dairy A, approximately 2 g of fresh feces collected from the pens was inoculated into C. jejuni semisolid aerobic transport medium (SAEM) prepared in 50-ml conical tubes as described previously (18). SAEM functions as both a transport and enrichment medium. BTM from dairy A was collected in 1-liter sterile bottles and divided into 2-ml samples in the laboratory and then inoculated into SAEM. Flush alley water samples were collected at the valve outlet in the stall barn by filling 1-liter sterile bottles and dividing the fluid into 2-ml samples in the laboratory. Lagoon water samples were collected similarly at different points from along the shore. At dairy B, approximately 10 g of fresh feces was collected with a sterile scoop and placed in a sterile Whirl-Pak bag (Wheatherby/Nasco, Inc., Fort Atkinson, WI). Raw milk and discharge water from the mobile milking parlor at dairy B were collected in 100-ml sterile containers. All samples were stored at 4°C in coolers and processed in <24 h.
Culture and identification.
Samples were cultured at the U.S. Department of Agriculture/Agricultural Research Service Western Regional Research Laboratory in Albany, CA, by using selective enrichment in Preston broth and plating onto blood-free Campylobacter charcoal differential agar (CCDA) medium (19). A 1:10 dilution of fresh fecal material was prepared by first emulsifying 1 g of feces in 10 ml of phosphate-buffered saline and processing for 30 s by vortex; a 1-ml aliquot of the liquid was pipetted into 9 ml of Preston broth poured into vented-cap cell culture flasks. A 1:5 dilution of unfiltered milk and water samples was prepared by inoculating 10 ml of liquid sample into 40 ml of Preston broth also in a cell culture flask. The flasks were placed on their sides in a plastic ziplock bag and grown under microaerobic conditions (5% O2, 10% CO2, and 85% N2) at 42°C with shaking (30 rpm) for 24 h. Enriched Preston broth was then spread-plated onto CCDA agar by pipetting 50 μl of liquid onto the plate. Plates were incubated for 48 h at 42°C under microaerobic conditions by using a plastic ziplock bag.
A subset of samples collected at dairy A were cultured at the University of California, Davis, by using selective enrichment in SAEM and plating on Campy-Cefex or CVA agar (BD, Franklin Lakes, NJ) as described previously (18). Approximately 2 g of feces was inoculated into SAEM prepared in a 50-ml conical tube. Milk and wastewater samples (lagoon and flush alley water) were processed by filtering 2 ml of liquid through a 0.22-μm-pore-size membrane and placing the filter in SAEM. Wastewater samples from dairy A were pipetted directly into SAEM without filtration. Conical tubes were mixed by inversion and incubated at 37°C for 24 h. The enriched SAEM was plated onto Campy-Cefex or CVA agar by using a 1-μl disposable loop and streaking for isolation. Plates were incubated for 48 h at 42°C in an anaerobic box containing three commercial CampyPak II gas-generating sachets (BBL Microbiology Systems, Cockeysville, MD).
Plates were checked for growth at 12, 24, 36, and 48 h. Presumptive identification of Campylobacter species was based on a Gram stain and examination of a wet mount by using phase-contrast microscopy to identify small, curved, or S-shaped bacteria with darting motility. Isolates were also characterized by hippurate hydrolysis, oxidase, and catalase tests. Multiple (8 to 12) pure colonies of presumptive C. jejuni were picked from each plate to increase the likelihood of finding the clinical outbreak subtype and stored on Microbank dry beads (Pro-Lab Diagnostics, Round Rock, TX) at −80°C.
C. jejuni was confirmed by a multiplex PCR, using the lipid A gene lpxA, with a modification by using only C. jejuni and C. coli primers (20). Additionally, suspect Campylobacter isolates from dairy B were identified to the species level by using matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (21). MALDI-TOF spectra from each isolate were compared with spectra from Campylobacter species validated previously by California's laboratory. In-house pattern-matching software was used for species identification by comparison of the presence of multiple ions in each spectrum, including certain ions previously identified as proteins encoded by annotated genes.
Quantification of indicator bacteria in dairy wastewater.
Quantification of total coliforms and Escherichia coli was determined for flush alley water and lagoon samples collected during the March 2007 visit to dairy A by using the Colilert QuantiTray 2000 (IDEXX Laboratories, Westbrook, ME) method, according to the manufacturer's recommendations (22). Briefly, duplicate samples (111 ml) of wastewater were diluted 1:10 and 1:100 in Butterfield's phosphate buffer. Colilert reagent containing 4-methylumbelliferyl-β-d-glucuronide (MUG) was added to 100 ml of each dilution, and samples were inoculated into 97-well trays and sealed. After 24 h of incubation at 35°C, wells were scored for yellow color from fermentation of O-nitrophenyl-β-d-galactoside; evidence of total coliforms, blue fluorescence from MUG (generic E. coli), and the most probable number (MPN) were calculated from the manufacturer's data tables (22). Four control strains (Pseudomonas aeruginosa ATCC 27853, Klebsiella pneumoniae ATCC 13882, E. coli ATCC 25922, and E. coli O157 ATCC 43888) were cultured and tested simultaneously for each assay.
DNA purification, amplification, and sequencing.
Genomic DNA for sequencing was prepared from single-colony Campylobacter isolates by using the DNeasy tissue kit (Qiagen USA, Inc., Valencia, CA) protocol according to the manufacturer's instructions (23). Alternately, DNA was extracted by boiling single Microbank bacterial storage beads from freezer stocks to resequence porA fragments (24). Genomic DNA concentrations were determined by using a NanoDrop microvolume spectrophotometer (Thermo Scientific, Wilmington, DE). PCRs were performed on an MJ Research Tetrad thermocycler with the following settings: 30 s at 94°C, 30 s at 53°C, and 2 min at 72°C (30 cycles). Each amplification mixture contained 50 ng genomic DNA, 1× PCR buffer, 1× PCR enhancer, 2.5 mM MgCl2, 250 μM (each) deoxynucleoside triphosphates, 50 pmol each primer, and 0.2 U Taq polymerase. Amplicons were purified on a BioRobot 8000 workstation (Qiagen USA, Valencia, CA).
Cycle-sequencing reactions were performed on an MJ Research Tetrad thermocycler by using the ABI Prism BigDye terminator cycle sequencing kit (version 3.0; Applied Biosystems, Carlsbad, CA) and standard protocols. All extension products were purified on DyeEx 96-well plates (Qiagen USA, Valencia, CA). DNA sequencing was performed on an ABI Prism 3130 genetic analyzer (Applied Biosystems, Carlsbad, CA) by using the POP-7 polymer, ABI Prism genetic analyzer data collection, and ABI Prism genetic analyzer sequencing analysis software.
MOMP, MLST, and PFGE typing.
Available clinical strains and up to 12 colonies from each positive environmental sample were subtyped. The MOMP primer sequences used for amplification and sequencing were designed from regions of the C. jejuni porA gene generating an ∼740-bp fragment (MOMP-L, 5′ CT AGT TAA AMT TAG TTT AGT WGC AGC 3′; MOMP-R, 5′ TCT TTW TYA CCR TAG TAT AAA CCA CC 3′). Strains were characterized further by extended multilocus sequence typing (MLST), as described previously (25), and 2-enzyme PFGE analyses, after digestion with SmaI and KpnI, according to the PulseNet protocol (26, 27). MOMP and MLST sequences were aligned by using ClustalW (European Bioinformatics Institute, Cambridge, United Kingdom); phylogenetic analysis of the aligned sequences was performed by using MEGA version 5 (Molecular Evolutionary Genetics Analysis; Tempe, AZ) (28). Sequences were queried against the Campylobacter PubMLST database (http://pubmlst.org/campylobacter/) to assign sequence types (ST). The porA allele of a subset of stored strains was resequenced using the Oxford oligonucleotide primers MOMP-1 and MOMP-2, published after our investigation (29); new porA allele sequences and trace files were submitted to the PubMLST for assignment of allele and MOMP peptide numbers.
Nucleotide sequence accession numbers.
The nucleotide sequences of porA (MOMP-L and MOMP-R) were deposited in the GenBank database under accession no. JX967537 to JX967548.
RESULTS
Human strains.
Twenty-six patient isolates associated with outbreak 1 were available to the state for PFGE analysis. All isolates had indistinguishable SmaI patterns (DBRS16.0206), which represented only 0.22% of isolates in the PulseNet national Campylobacter database. Twenty-four isolates were indistinguishable by using the secondary enzyme, KpnI (DBRK02.0201); two isolates demonstrated a two-band difference from the dominant KpnI pattern. Three patient isolates (RM5846, RM5847, and RM6591) with the dominant SmaI/KpnI pattern were saved by the state and made available for this study for MOMP and MLST typing. Because of the time lag (∼3 months) between the original laboratory investigation and this study, the other patient isolates already had been discarded. All three available strains belonged to ST-21, a common subtype in the Campylobacter PubMLST database.
Only one of the eight laboratory-confirmed isolates (RM6863) from outbreak 2 was available to us. The other isolates were discarded by clinical laboratories before county and state public health officials recognized the outbreak, which is routine because Campylobacter strains typically are not submitted to PulseNet for cluster detection. The SmaI (DBRS16.0006) and KpnI (DBRK02.0011) patterns of this single-patient isolate had not been recorded previously in the national PulseNet Campylobacter database. The strain also belonged to a rare MLST subtype, ST-1244. At the time of this study in 2007, only one human ST-1244 isolate was present in the PubMLST database, although not all researchers deposit their sequences.
Environmental investigation.
C. jejuni was isolated from 7 of 15 (46.7%) bovine fecal samples, 10 of 10 (100%) flush alley water samples, and none of the BTM samples collected at dairy A (outbreak 1) in August 2006. C. coli was also isolated from 4 (26.7%) of the bovine fecal samples but none of the flush alley water samples. During the follow-up visit in March 2007 to study the dairy wastewater system, C. jejuni was isolated from 2 of 10 (20%) flush alley water and 1 of 20 (5%) lagoon samples. In contrast, C. coli was cultured from 9 of 10 (90%) flush alley water and 20 of 20 (100%) lagoon samples during the second visit. The coliform and E. coli counts (MPN) for the flush alley water samples were 683,000/g and 546,000/g, respectively. The coliform and E. coli counts (MPN) for the lagoon water were 579,400/g and 327,000/g, respectively.
C. jejuni was isolated from 9 of 26 (34.6%) bovine fecal samples at dairy B (outbreak 2) collected in December 2007. C. jejuni was not isolated from BTM, retail raw milk, or milking parlor discharge water. Other Campylobacter species were also cultured from the bovine fecal samples, including C. coli, C. fetus, C. hyointestinalis, and C. lari. C. coli was also cultured from milking parlor discharge water.
Identification of the clinical outbreak strain in farm samples.
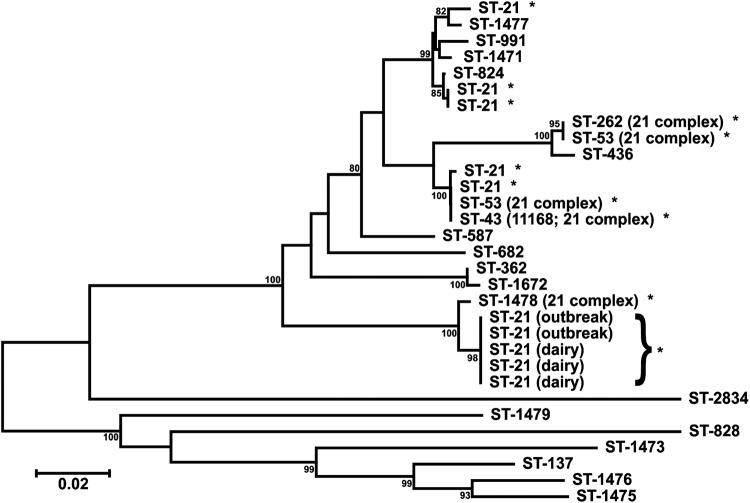
Because ST-21 (outbreak 1) was a common sequence type in the PubMLST database, we initially evaluated the uniqueness of the dairy A-related clinical outbreak porA sequence by comparing it with the porA sequences of 26 representative C. jejuni ST-21, clonal complex-21, and related isolates in our strain collection. Sufficient genetic diversity was observed to proceed with screening the farm isolates (Fig. 1).
Fig 1.
Phylogenetic tree comparing the porA sequence of the Campylobacter jejuni clinical outbreak strain (sequence type 21) from dairy A with those of related isolates from our strain collection. The sequences of two clinical outbreak isolates (outbreak) and three flush alley water isolates (dairy) identified through major outer membrane protein screening are displayed. *, sequence and clonal type 21 strains. The scale bar represents the number of nucleotide changes per site.
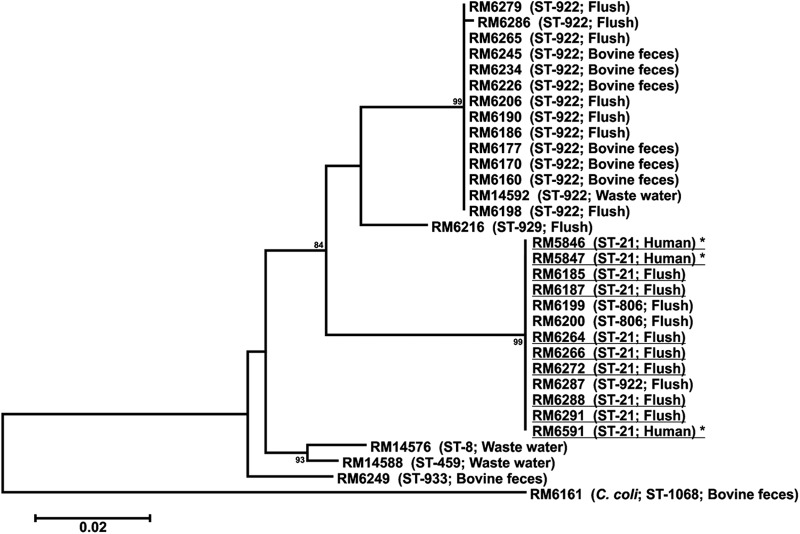
Phylogenetic comparison of 70 C. jejuni MOMP sequences from 17 dairy A samples revealed 10 strains recovered from five flush alley water samples with the same porA sequence as the clinical outbreak strain (Fig. 2, MOMP group A3). Of these 10 candidate strains, 8 had indistinguishable SmaI (Table 1, pulsotype 3) and KpnI patterns, compared with the clinical outbreak strain, and belonged to the same sequence type (ST-21). Two strains (RM6199 and RM6200) differed from the clinical outbreak strain by a single band (Table 1, pulsotype 4) and belonged to ST-806, a sequence type closely related to ST-21. The outbreak strain was not identified in 30 lagoon and flush alley water samples during the follow-up visit in March 2007; however, four wastewater isolates belonged to ST-8, ST-459, and ST-922 (Table 1; Fig. 2).
Fig 2.
Phylogenetic tree comparing the porA sequences of representative Campylobacter jejuni and Campylobacter coli strains isolated from dairy A environmental samples with that of the clinical outbreak strain (outbreak 1, 2006 to 2007). *, clinical outbreak strain. Dairy strains with the identical porA and multilocus sequence typing alleles and indistinguishable SmaI and KpnI patterns are underlined. The scale bar represents the number of nucleotide changes per site.
Table 1.
Genotypes of Campylobacter jejuni strains cultured from environmental samplesa
| Dairy | Sourceb | No. of isolatesd | MOMP group | Oxford porA/MOMPe | Pulsotype (SmaI) | MLST ST | No. of MLST alleles |
MLST clonal complex | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| atpA | aspA | glnA | gltA | glyA | pgm | tkt | ||||||||
| A | BF, FWc | 51 | A1 | 747/692 | 1 | 922 | 1 | 6 | 1 | 2 | 83 | 2 | 3 | Unassigned |
| FW | 3 | A1 | 1524/1398 | 1 | 922 | 1 | 6 | 1 | 2 | 83 | 2 | 3 | Unassigned | |
| FW | 1 | A2 | 749/694 | 2 | 929 | 9 | 17 | 2 | 4 | 62 | 4 | 5 | 257 | |
| FW, H | 10 | A3 | 1533/1377 | 3 | 21 | 2 | 5 | 1 | 1 | 3 | 2 | 1 | 21 | |
| FW | 2 | A3 | 1500/1377 | 4 | 806 | 2 | 5 | 1 | 1 | 3 | 140 | 3 | 21 | |
| FWc | 1 | A4 | 745/690 | 5 | 8 | 2 | 6 | 1 | 1 | 3 | 2 | 1 | 21 | |
| Lc | 2 | A5 | 44/41 | 6 | 459 | 1 | 3 | 2 | 3 | 3 | 5 | 9 | 42 | |
| BF | 8 | A6 | 1523/1397 | 1, 7, 8, 9 | 933 | 10 | 7 | 1 | 59 | 19 | 10 | 5 | 403 | |
| B | BF, H | 18 | B1 | 1499/1376 | 10, 11 | 1244 | 1 | 17 | 1 | 2 | 2 | 225 | 3 | 61 |
| BF | 2 | B1 | 1526/1400 | 10 | 1244 | 1 | 17 | 1 | 2 | 2 | 225 | 3 | 61 | |
| BF | 8 | B2 | 48/44 | 12 | 22 | 1 | 3 | 3 | 6 | 4 | 3 | 3 | 22 | |
| BF | 3 | B3 | 1529/1403 | 13 | 922 | 1 | 6 | 1 | 2 | 83 | 2 | 3 | Unassigned | |
| BF | 3 | B4 | 1528/1402 | 14 | 45 | 4 | 1 | 7 | 10 | 4 | 1 | 7 | 45 | |
Genotypes of Campylobacter jejuni strains cultured from environmental samples collected during pasteurized (dairy A) and raw (dairy B) milk outbreak investigations, California, 2006 to 2007. MLST, multilocus sequence typing; MOMP, major outer membrane protein; ST, sequence type.
BF, bovine feces; FW, flush alley water; H, human stool; L, lagoon.
ST-8 (MOMP group A4; n = 1), ST-459 (MOMP group A5; n = 2), and ST-922 (MOMP group A1; n = 1) strains were from samples collected in March 2007.
Includes multiple colony picks from individual samples (n = 112).
Representative strains from each MOMP group were subtyped by using the Oxford porA/MOMP method (n = 86). porA allele and MOMP peptide numbers were assigned by PubMLST (http://pubmlst.org/campylobacter/).
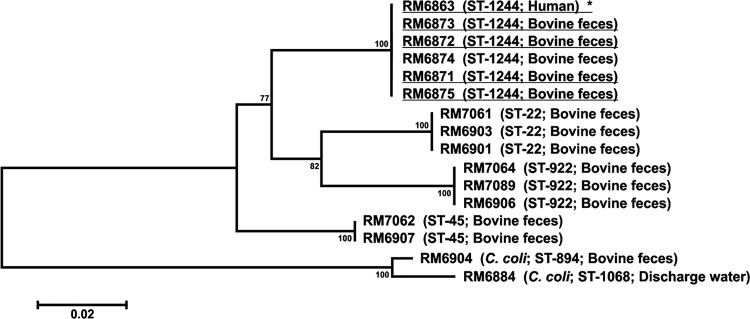
In December 2007, we used the MOMP screen again to compare isolates from dairy B (outbreak 2) samples with the one clinical outbreak strain available to us. Among 34 C. jejuni isolates recovered from 9 bovine fecal samples at dairy B, we identified 20 strains from four samples with porA sequences identical to that of the patient strain (Fig. 3, group B1). Of the 20 candidate strains, 17 had SmaI and KpnI patterns indistinguishable from those of the clinical outbreak strain (Table 1, pulsotype 10). Three strains, represented by RM6874 in Fig. 3, indicated a two-band difference from the outbreak strain by SmaI PFGE analysis. The MLST results revealed that all 20 MOMP group B1 strains were ST-1244, including those with slightly different pulsotypes (Table 1).
Fig 3.
Phylogenetic tree comparing the porA sequences of representative Campylobacter jejuni and Campylobacter coli strains isolated from dairy B environmental samples with that of the clinical outbreak strain (outbreak 2, 2007). *, clinical outbreak strain. Dairy strains with the identical porA and multilocus sequence typing alleles and indistinguishable SmaI and KpnI patterns are underlined. The scale bar represents the number of nucleotide changes per site.
Genetic diversity of C. jejuni and C. coli from the dairy environmental samples.
Overall, 10 unique MOMP types using our primer set and 15 unique MOMP-MLST-PFGE groups were identified among the 108 C. jejuni isolates subtyped in 2006 and 2007 (Table 1). In comparison, resequencing the porA fragment of 86 strains by using the Oxford primers (MOMP-1, MOMP-2) revealed 13 MOMP subtypes and a combined 17 MOMP-MLST-PFGE subtypes. Analysis of multiple colony picks resulted in recovery of up to four unique subtypes per sample. The most common MLST sequence type was ST-922, which was also the only sequence type identified for both dairies (Table 1). Fourteen SmaI pulsotypes were identified with ST-933 strains from dairy A showing the most diversity (four pulsotypes; Table 1).
In contrast, the C. coli isolates from bovine feces and wastewater samples collected at dairy A during the August 2006 (n = 42) and March 2007 (n = 33) visits were remarkably clonal; all strains belonged to ST-1068 and had identical porA sequences (Fig. 2). Similarly, the C. coli strains (n = 23) from bovine feces and discharge water collected at dairy B belonged to ST-894 and ST-1068, respectively, and had closely related porA sequences (Fig. 3).
DISCUSSION
We used MOMP typing to screen a substantial number of farm isolates collected after two milk-borne C. jejuni outbreaks and successfully identified the clinical outbreak strains. This study demonstrates that sequencing the C. jejuni porA allele is a promising target for rapid environmental screening to identify specific clones prior to conducting labor-intensive molecular typing methods following outbreaks.
By implementing a single-allele screening approach, public health laboratories could decrease time and labor costs associated with C. jejuni molecular epidemiologic studies in the food supply continuum. Specifically, sequencing of the porA gene and construction of a phylogenetic tree can be accomplished in under 24 h compared with MLST and 2-enzyme PFGE confirmation, which may take at least a week to complete in public health laboratories with limited staff and resources. The screen is also amendable to retrospective high-throughput sequencing of strains by boiling single MicroBank freezer stocks to extract DNA directly from the bead, thus eliminating the need to regrow isolates by culture (24). We used the bead extraction technique to rapidly resequence porA alleles using the Oxford primer set published subsequently to our original analysis as described below (29). Laboratories could further streamline the screen by developing an allelic discrimination real-time PCR assay to detect single nucleotide polymorphisms in the porA sequence specific to the clone (clinical outbreak strain) of interest (30). This approach provides a method for screening a large number of clinical and, possibly, environmental isolates in investigations for rapid bacterial source tracking and selection of strains for next-generation whole-genome sequencing for high-resolution differentiation (31).
In 2008, an extended 8-locus MLST-porA typing scheme was published by researchers in Oxford, England (29). The Oxford porA product (570 bp, minimum; 738 bp, maximum) overlaps with the fragment we analyzed in this study. Cody et al. (2009) used the Oxford primer set to type a C. jejuni strain collection and demonstrated that the porA allele has sufficient genetic diversity and stability for use as a molecular epidemiologic tool (32). Results from our study are comparable and show that the Oxford typing scheme discriminated an additional three porA/MOMP types.
This study has some limitations that indicate the need for further investigation. For example, and unfortunately for effective trace-back investigations, many clinical laboratories often discard C. jejuni isolates after confirmation of species, leaving few human isolates available for analysis. Even in this very large outbreak, isolates were not saved, thus limiting our ability to evaluate the robustness of MOMP typing in discriminating clinical MLST and PFGE subtypes. Our analysis also emphasizes the importance of selecting multiple pure colonies from individual samples during C. jejuni outbreak investigations for successful molecular subtyping, as described previously in the human and veterinary literature (33, 34). Indeed, our results confirmed that multiple C. jejuni subtypes in individual bovine fecal and wastewater samples are not uncommon (Table 1). However, the regional prevalence of the C. jejuni outbreak strains at dairy A (ST-21) and dairy B (ST-1244) was unknown at the time of the investigations. Follow-up studies of multiple dairies in the central valley indicate that ST-1244 is relatively common from bovine sources, including bulk tank milk (M. T. Jay-Russell, unpublished data). Noteworthy is a report by Kwan et al. (2008) of ST-1244 as predictive potentially of strains that are bovine adapted, although further studies will be required to determine whether this is a marker for host-adapted C. jejuni (35).
Public health implications.
An important goal of this study was to show the utility of MOMP typing to source-track genetically related strains in the context of an outbreak and identify potential on-farm risk factors and prevention strategies. Although C. jejuni was not isolated from milk samples during either outbreak investigation, our molecular typing results support strongly the epidemiologic findings implicating milk products produced by the dairies as the sources of both outbreaks (16, 17). Campylobacter is notoriously difficult to isolate from milk following outbreaks, possibly due to die-off during the lag time from consumption to recognition of the outbreak, uneven distribution of the bacteria in bulk tank milk, or inadequate culture and isolation methods for milk.
The exact mechanism of contamination at dairy A remains unclear, but it might have been the result of cross-contamination after pasteurization or heavy loads of Campylobacter before pasteurization. Based on the ease of recovery of C. jejuni from flush alley water, we speculate it is possible that cross-contamination with environmental dairy wastewater during milk processing occurred. Notably, the dairy A pasteurization room was located adjacent to the milking parlor and ∼100 m from the dairy lagoon. Moreover, the recovery of C. jejuni from flush alley water and lagoon samples at dairy A, approximately 3 months after the outbreak, suggests that the outbreak strain was predominant at this dairy and the lagoon could be an important environmental reservoir. The high numbers of indicator bacteria (coliforms and E. coli) identified in lagoon and flush alley water are consistent with the pathogen prevalence. Additional research is needed to define the spread of Campylobacter in recycled wastewater systems and strategies to minimize contamination at dairies that use these systems for manure management.
Similar to the results at dairy A, the clinical outbreak strain was cultured from bovine feces during the environmental investigation of the dairy B outbreak linked to organic raw milk and raw chocolate colostrum. Dairy B also had an on-site creamery adjacent to cattle areas. Raw dairy products are not subjected to a “kill step,” so it is plausible to suggest that viable C. jejuni from bovine fecal material contaminated the milk/colostrum sometime during production or processing. Cross-contamination with environmental waters represents another possible mechanism of contamination. Indeed, a different pathogen, C. coli, was isolated from milking parlor discharge water, suggesting the possibility of cross-contamination.
The results obtained by MOMP typing the environmental isolates ultimately facilitated more intensive investigation into on-farm food safety practices at the implicated dairies and were used by state agriculture and public health officials to reinforce the importance of sanitation and education of dairy workers about best practices in dairy production (16, 17). Additionally, the California state legislature passed a bill that became effective on 1 January 2008 requiring no more than 10 coliform bacteria per milliliter in grade A raw milk for human consumption, the same coliform limit required for pasteurized milk (36).
In summary, our study highlights the potential for ubiquitous dissemination of C. jejuni in the dairy farm environment and the utility of using MOMP typing as an epidemiologic tool for screening C. jejuni isolates to identify specific strains during dairy-related outbreak investigations to support implementation of on-farm prevention strategies. The approach could be applied during investigation of other potential food and water sources where there is a need to prioritize a large number of C. jejuni environmental isolates before confirmation by other molecular methods such as MLST, PFGE, or whole-genome sequencing analyses.
ACKNOWLEDGMENTS
We are grateful for technical assistance provided by Leslie Harden and Feli Bautista, U.S. Department of Agriculture/Agricultural Research Service (USDA/ARS) Western Regional Research Center. We also thank Leta Crawford-Miksza and Linda Guthertz at the Food and Drug Laboratory Branch, California Department of Public Health, for technical assistance and Jennifer Schneider, Jeffrey Higa, and James Sigl from the California Department of Public Health for work on the epidemiologic and farm investigations. We also thank Stephen Beam with the California Department of Food and Agriculture for sharing findings from their investigation.
This work was supported by funds from USDA/ARS Current Research Information System (CRIS) project no. 5325-42000-045-00D.
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print 31 October 2012
REFERENCES
- 1.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson MA, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention 2010. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food—10 states, 2009. MMWR Morb. Mortal. Wkly. Rep. 59:418–422 [PubMed] [Google Scholar]
- 3.Miller WG, Mandrell RE. 2005. Prevalence of Campylobacter in the food and water supply: incidence, outbreaks, isolation and detection, p 101–163. In Ketley JM, Konkel ME. (ed), Campylobacter. Molecular and cellular biology. Horizon Bioscience, Norfolk, United Kingdom [Google Scholar]
- 4.Yan SS, Pendrak ML, Foley SL, Powers JH. 2005. Campylobacter infection and Guillain-Barré syndrome: public health concerns from a microbial food safety perspective. Clin. Appl. Immunol. Rev. 5:285–305 [Google Scholar]
- 5.Altekruse SF, Stern NJ, Fields PI, Swerdlow DL. 1999. Campylobacter jejuni—an emerging foodborne pathogen. Emerg. Infect. Dis. 5:28–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oliver SP, Boor KJ, Murphy SC, Murinda SE. 2009. Food safety hazards associated with consumption of raw milk. Foodborne Pathog. Dis. 6:793–806 [DOI] [PubMed] [Google Scholar]
- 7.Taylor EV, Herman KM, Ailes EC, Fitzgerald C, Yoder JS, Mahon BE, Tauxe RV. 2012. Common source outbreaks of Campylobacter infection in the USA, 1997–2008. Epidemiol. Infect. 15:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Birkhead G, Vogt RL, Heun E, Evelti CM, Patton CM. 1988. A multiple-strain outbreak of Campylobacter enteritis due to consumption of inadequately pasteurized milk. J. Infect. Dis. 157:1095–1097 [DOI] [PubMed] [Google Scholar]
- 9.Cronquist AB, Haubert NA, Scott A, Meyer K, Peden JR, Pilonetti-Hall T, Klug P. 2005. Outbreak of Campylobacter jejuni enteritis associated with consumption of pasteurized milk processed at a prison dairy, Colorado 2005, abstr P-65, p 55. Abstr. Int. Conf. Emerg. Infect. Dis. 2006, Atlanta, Georgia [Google Scholar]
- 10.Langer AJ, Ayers T, Grass J, Lynch M, Angulo FJ, Mahon BE. 2012. Nonpasteurized dairy products, disease outbreaks, and state laws—United States, 1993–2006. Emerg. Infect. Dis. 18:385–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jay-Russell MT. 2010. Raw (unpasteurized) milk: are health-conscious consumers making an unhealthy choice? Clin. Infect. Dis. 51:1418–1419 [DOI] [PubMed] [Google Scholar]
- 12.Gerner-Smidt P, Stroika SG, Fitzgerald C. 2008. National molecular subtyping network for food-borne bacterial disease surveillance in the United States, p 277–285. In Nachamkin I, Szymanski CM, Blaser MJ. (ed), Campylobacter, 3rd ed. ASM Press, Washington DC [Google Scholar]
- 13.Swaminathan B, Barrett TJ, Hunter SB, Tauxe RV. 2001. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerg. Infect. Dis. 7:382–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang S, Luangtongkum T, Morishita TY, Zhang Q. 2005. Molecular typing of Campylobacter strains using the cmp gene encoding the major outer membrane protein. Foodborne Pathog. Dis. 2:12–23 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Q, Meitzler JC, Huang S, Morishita T. 2000. Sequence polymorphism, predicted secondary structures, and surface-exposed conformational epitopes of Campylobacter major outer membrane protein. Infect. Immun. 68:5679–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.California Department of Public Health 2007. Campylobacteriosis outbreak—May 2006 California Department of Public Health, Richmond, CA [Google Scholar]
- 17.California Department of Public Health 2008. Cluster of Campylobacter infections possibly associated with raw dairy products. California Department of Public Health, Gardena, CA [Google Scholar]
- 18.Jeffrey JS, Hunter A, Atwill ER. 2000. A field-suitable, semisolid aerobic enrichment medium for isolation of Campylobacter jejuni in small numbers. J. Clin. Microbiol. 38:1668–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Uyttendaele M, Debevere J. 1996. Evaluation of Preston medium for detection of Campylobacter jejuni in vitro and in artificially and naturally contaminated poultry products. Food Microbiol. 13:115–122 [Google Scholar]
- 20.Klena JD, Parker CT, Knibb K, Ibbitt JC, Devane PM, Horn ST, Miller WG, Konkel ME. 2004. Differentiation of Campylobacter coli, Campylobacter jejuni, Campylobacter lari, and Campylobacter upsaliensis by a multiplex PCR developed from the nucleotide sequence of the lipid A gene lpxA. J. Clin. Microbiol. 42:5549–5557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mandrell RE, Harden LA, Bates A, Miller WG, Haddon WF, Fagerquist CK. 2005. Speciation of Campylobacter coli, C. jejuni, C. helveticus, C. lari, C. sputorum, and C. upsaliensis by matrix-assisted laser desorption ionization–time of flight mass spectrometry. Appl. Environ. Microbiol. 71:6292–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.IDEXX 2007. Colilert® QuantiTray 2000 instruction manual. IDEXX Laboratories, Westbrook, ME [Google Scholar]
- 23.Qiagen USA 2006. DNeasy® blood & tissue handbook. Qiagen, Valencia, CA [Google Scholar]
- 24.Miller WG, Parker CT, Heath S, Lastovica AJ. 2007. Identification of genomic differences between Campylobacter jejuni subsp jejuni and C. jejuni subsp doylei at the nap locus leads to the development of a C. jejuni subspeciation multiplex PCR method. BMC Microbiol. 7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller WG, On SL, Wang G, Fontanoz S, Lastovica AJ, Mandrell RE. 2005. Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus. J. Clin. Microbiol. 43:2315–2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention 2000. Standardized molecular subtyping of foodborne bacterial pathogens by pulsed-field gel electrophoresis: training manual. U.S. Department of Health and Human Services, CDC, Atlanta, GA [Google Scholar]
- 27.Ribot EM, Fitzgerald C, Kubota K, Swaminathan B, Barrett TJ. 2001. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J. Clin. Microbiol. 39:1889–1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dingle KE, McCarthy ND, Cody AJ, Peto TE, Maiden MC. 2008. Extended sequence typing of Campylobacter spp., United Kingdom. Emerg. Infect. Dis. 14:1620–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Best EL, Fox AJ, Frost JA, Bolton FJ. 2004. Identification of Campylobacter jejuni multilocus sequence type ST-21 clonal complex by single-nucleotide polymorphism analysis. J. Clin. Microbiol. 42:2836–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuehn BM. 2012. Database of food-borne pathogen genomes created. JAMA 8:557. [DOI] [PubMed] [Google Scholar]
- 32.Cody AJ, Maiden MJ, Dingle KE. 2009. Genetic diversity and stability of the porA allele as a genetic marker in human Campylobacter infection. Microbiology 155:4145–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardner TJ, Fitzgerald C, Xavier C, Klein R, Pruckler J, Stroika S, McLaughlin JB. 2011. Outbreak of campylobacteriosis associated with consumption of raw peas. Clin. Infect. Dis. 53:26–32 [DOI] [PubMed] [Google Scholar]
- 34.Sahin O, Plummer PJ, Jordan DM, Sulaj K, Pereira S, Robbe-Austerman S, Wang L, Yaeger MJ, Hoffman LJ, Zhang Q. 2008. Emergence of a tetracycline-resistant Campylobacter jejuni clone associated with outbreaks of ovine abortion in the United States. J. Clin. Microbiol. 46:1663–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwan PS, Birtles A, Bolton FJ, French NP, Robinson SE, Newbold LS, Upton M, Fox AJ. 2008. Longitudinal study of the molecular epidemiology of Campylobacter jejuni in cattle on dairy farms. Appl. Environ. Microbiol. 74:3626–3633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.California Department of Food and Agriculture 2007. New coliform bacteria standard for California raw milk producers. http://www.cdfa.ca.gov/egov/Press_Releases/Press_Release.asp?PRnum=07-090