Abstract
A total of 120 Burkholderia cepacia complex isolates collected during 2004–2010 from 66 patients in two cystic fibrosis reference centers in Argentina were analyzed. Burkholderia contaminans was the species most frequently recovered (57.6%), followed by Burkholderia cenocepacia (15%), a species distribution not reported so far. The recA-PCR-based techniques applied to the B. contaminans isolates revealed that 85% of the population carried the recA-ST-71 allele. Our results showed the utility of BOX-PCR genotyping in analyzing B. contaminans diversity. This approach allowed us to address clonal transmission during an outbreak and the genetic changes occurring in infecting bacteria over the course of chronic infection.
TEXT
Burkholderia cepacia complex species are capable of causing chronic and often severe respiratory tract infections in cystic fibrosis (CF) patients and other types of infections in immunocompromised patients. Although many CF patients remain infected by these bacteria and yet stay relatively healthy for prolonged periods, others either have a severe decline in their pulmonary status or die shortly after the initial colonization (1). In Argentina, the first reports of B. cepacia complex bacteria infecting CF patients started around 1990. At that time, Burkholderia spp. were recovered sporadically and with a very low prevalence (<0.1%). The prevalence increased over the last decade from 0.2 to 3.6%, depending on the medical center. In early 2004, an outbreak involving main Argentine CF care centers occurred, and the proportion of patients from whom B. cepacia complex species were recovered ranged from 19 to 36% (2–4). Currently, through strict infection control procedures, the prevalence of Burkholderia spp. in local patients has decreased to approximately 10%. Recent worldwide surveillance studies concerning the distribution of B. cepacia complex species in CF patients have cited B. multivorans and B. cenocepacia as the most frequently recovered species. These species account for approximately 80% of the infected patients, and the prevalence of one or the other is geographically and temporally dependent (5–7). Unlike this worldwide situation, in Argentina a relatively high prevalence of B. contaminans followed by B. cenocepacia has been observed (3, 4). To our knowledge, no other geographical region has been reported in the literature to have such a high occurrence of B. contaminans infecting CF patients. In view of this particular scenario, coupled with the lack of biodiversity information available for B. contaminans species, we decided to study this local population by recA-PCR-based techniques and by repetitive element sequence-based PCR (rep-PCR) approaches. These methods allowed us to address the genetic diversity of B. contaminans isolates recovered during that outbreak along with the genetic changes occurring in the infecting bacteria over the course of a chronic infection.
Distribution of Burkholderia cepacia complex species in CF patients.
This study involved the analysis of a total of 120 clinical Burkholderia cepacia complex isolates recovered from 66 CF patients from 2004 through 2010 and 13 reference strains (see Table S1 in the supplemental material). The isolates were recovered from sputum samples of CF patients treated at two main CF reference centers of Argentina (Hospital de Niños Sor María Ludovica, La Plata [HNLP], and Hospital Santísima Trinidad, Córdoba [HST], located 800 km north of La Plata) plus one hospital (Hospital Dr. Pedro Moguillanski, Cipoletti, Río Negro [HNRN], 1,200 km south of La Plata). Twenty-two isolates (indicated with a “v” in Table S1 in the supplemental material) were withdrawn from patients infected during the B. cepacia complex outbreak, and a cohort of 51 isolates was recovered from 21 patients chronically infected with B. cepacia complex bacteria. Patients were considered chronically infected when three positive cultures of B. cepacia complex strains were isolated within a 6-month period (8). Bacterial isolation was performed on B. cepacia selective agar medium (BCSA; Laboratorios Britania S.A., Argentina) according to clinical microbiology practice recommendations for respiratory tract specimens from CF patients (9, 10), and the strains recovered were phenotypically characterized by means of conventional biochemical techniques (9–12). The species-level identification was achieved by recA-PCR-based techniques. The recA gene (1,040 bp) was amplified with BCR1 and BCR2 primers (13), and the amplified products were subjected to restriction fragment length polymorphism (RFLP) analysis with the restriction enzyme HaeIII (Promega, Inc.) (13). The restriction PCR patterns obtained were analyzed and compared with those reported in the literature (13–16). The amplified products were purified with a QIAquick PCR purification kit (Qiagen Inc., CA) and sequenced on both strands with the BigDye terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA) at Macrogen, Inc. (Seoul, South Korea). A final identification was achieved by recA DNA sequence analysis and by gyrB DNA fragment (1,900 bp) sequencing when an identification remained ambiguous. The recA sequence type (recA-ST) within each species was obtained by comparing the sequence of a 393-bp recA fragment against the multilocus sequence typing (MLST) database (http://pubmlst.org/bcc/) (1).
The analysis of species distribution among the patients studied showed that 8 of the 17 species described for the B. cepacia complex had infected the local CF population, with a remarkably high representation of B. contaminans, i.e., 57.6% of the infected patients. The data on the species of the first isolate available from each patient indicated that B. cenocepacia was the second most frequent species, it encountered at a frequency of almost 15% of the patients, followed by B. cepacia at 7.5%, B. multivorans at 6.0%, B. stabilis and B. vietnamiensis at 5%, B. seminalis at 3.0%, and B. ambifaria at 1.5%.
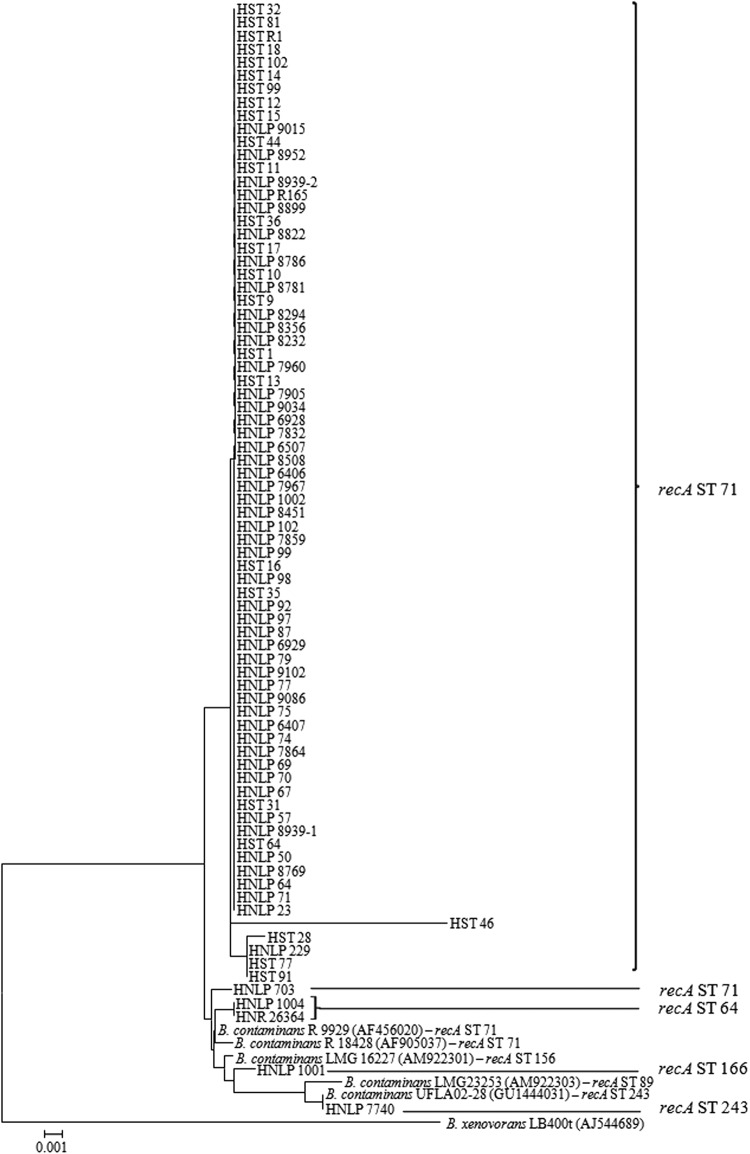
In order to examine the genetic diversity among the B. contaminans isolates, sequence analysis of a 900-bp fragment of the recA gene was performed for the 79 B. contaminans isolates (see Table S1 in the supplemental material). The phylogenetic relationship among them was investigated by using MEGA version 4.0 software (http://www.megasoftware.net) (Fig. 1). Isolates were clearly separated into one large homogenous cluster that grouped 85% of the B. contaminans population under study together, along with four small clusters containing only 5 isolates in total. When the sequence of the 393-bp DNA fragment from the recA gene was compared against the MLST database (1), all the bacteria included in the main cluster were found to belong to the recA-ST-71 allele, while 4 of the isolates grouping outside this cluster displayed different recA-ST alleles (ST-64, -166, and -243). The isolates grouped in the main cluster had a recA-HaeIII restriction pattern corresponding to the K type, whereas the isolates outside this group gave profiles of K, AT, and J (see Table S1 in the supplemental material). Interestingly, one of the isolates (HNLP 1001) exhibited the J recA-RFLP pattern, which was reported to be representative of the species B. cenocepacia, B. stabilis, B. lata, B. seminalis, and B. arboris (13, 17, 18) but has not been documented so far for B. contaminans. It is notable that none of the recA sequences corresponding to isolates that grouped in the main cluster was represented by the recA reference sequences available at GenBank. It should also be remarked that two reference sequences displaying the recA-ST-71 allele, those belonging to two isolates recovered from CF patients in Brazil (B. contaminans R18428 and R9929), clustered at a short distance from the homogenous cluster (Fig. 1). A further exploration of a possible regional incidence of B. contaminans among isolates circulating in Brazil should prove interesting. In this regard, the incidence of RFLP K pattern strains in CF patients seen at two different hospitals in Brazil (15, 19) might also indicate the existence of regional spreading of B. contaminans between bordering countries.
Fig 1.
Phylogenetic tree of the 79 B. contaminans CF isolates (see Table S1 in the supplemental material) based on the analysis of a 900-bp recA fragment sequence using the neighbor-joining method. Bootstrap values greater than 70% are shown for 1,500 replicates. Five B. contaminans reference sequences obtained from the GenBank sequence database were included in the analysis (accession numbers in parentheses). recA sequence types (recA-STs) were obtained by comparing the sequence of a 393-bp recA fragment against the public MLST database (http://pubmlst.org/bcc/). Burkholderia xenovorans was used as the outgroup.
Genetic diversity among B. contaminans isolates with the recA-ST-71 allele as investigated by rep-PCR DNA fingerprinting analysis.
The low level of polymorphism in the recA gene encountered in the B. contaminans population studied here prompted us to explore the genetic diversity among the isolates carrying the recA-ST-71 allele by rep-PCR (20). This subset of 75 B. contaminans isolates was analyzed by repetitive extragenic palindromic (REP), enterobacterial repetitive intergenic consensus (ERIC), and BOX-PCRs. Amplification reactions were performed in a MyCycler thermal cycler (Bio-Rad) with the primers REP1R-I, REP2-I, ERIC1R-I, and BOX-A1R (20). The amplicons generated were electrophoresed in agarose gels, and the banding patterns obtained were analyzed with the GelCompar II version 2.1 software package (Applied Maths, Kortrijk, Belgium). The profiles yielding a band similarity of 85% or more were considered related clones for all assays. According to this criterion, both REP and ERIC banding patterns displayed a very limited diversity. With the REP primers, the isolates showed only four different banding patterns, with one being observed in 92% of the isolates. Similarly, when ERIC-PCR primers were used, 5 different patterns were found, with one subtype including 89% of the isolates (data not shown). The highest discrimination power was obtained when BOX-PCR was performed. This fingerprint typing yielded 10 distinct amplification profiles (B1 to B10) (Fig. 2). The distribution of the isolates' amplification profiles was as follows: subtype B8, 50%; subtype B1, 22.9%; subtypes B6 and B7, 6.75%; subtype B9, 5.4%; and subtypes B2, B3, B7, B5, and B10, 1.35% (these comprised only 1 isolate each).
Fig 2.
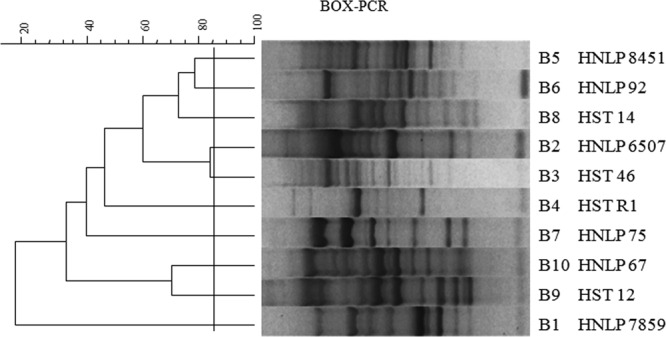
Dendrogram showing the relatedness among representative BOX-PCR fingerprinting patterns obtained from Burkholderia contaminans recA-ST-71 clinical isolates. The dendrogram was produced by the UPGMA (unweighted pair group method with arithmetic average) method. Clusters were delineated with an 85%-similarity cutoff value, as indicated by the vertical line. One isolate representative of each pattern is indicated.
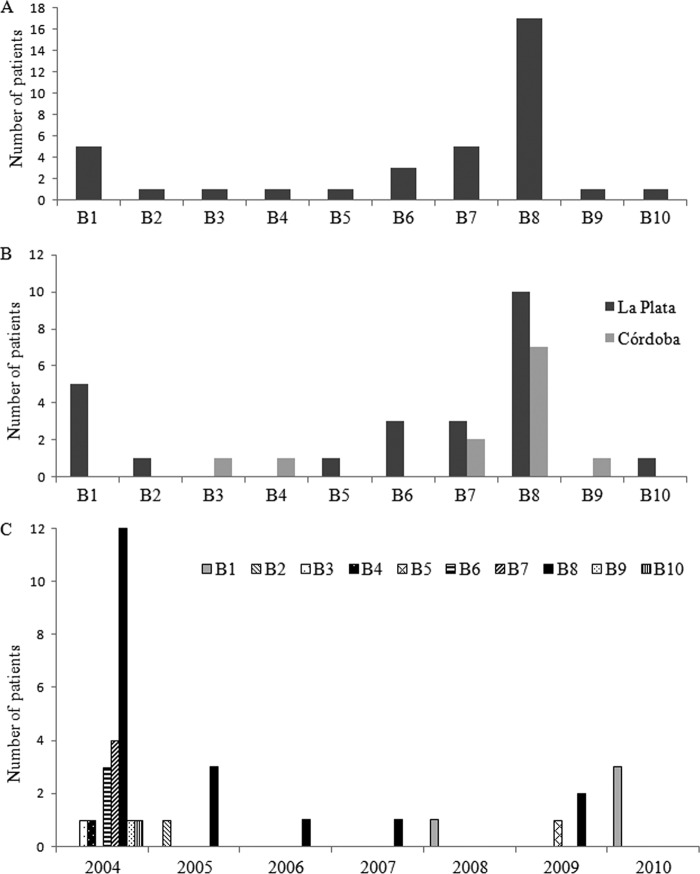
The first B. contaminans isolate recovered from each patient over the period 2004–2010 was used to determine the incidence of each BOX subtype. Three subtypes, B1, B7, and B8, were the most abundant patterns found, altogether accounting for 75% of the isolates. The B6 subtype was observed in 3 patients, while the remaining 6 subtypes were unique fingerprints observed in only one patient each (Fig. 3A). With respect to geographical distribution, whereas the isolates with B7 and B8 subtypes were found at both reference centers at a remarkably high incidence, all the other BOX subtypes were present at only one of the two locations (Fig. 3B). Thus, although the interpatient transmission of B. contaminans with the B7 and B8 BOX subtypes could very likely have been a dominant occurrence at both CF centers, other sources of new infections were clearly present. Regarding the prevalence of each BOX subtype over time, during the period of the outbreak (2004–2005), the CF patients were in most instances infected with the B8 (55%), B7 (15%), and B6 (11%) subtypes but were also infected, in a much lower proportion, with isolates belonging to other genotypes (B2, B3, B4, B9, and B1) (Fig. 3C). We therefore infer that the isolates belonging to the B8 genotype (and to a lesser extent to B7) were efficiently transmitted at the outbreak period to produce a clonal spreading among the patients (Fig. 3B and C). Nevertheless, the fact that other unique BOX subtypes (e.g., the B2, B3, B4, B9, and B10 subtypes) were also present should be stressed (Fig. 3C). Hence, as previously reported for different Burkholderia sp. outbreaks (21), although a high number of infections could have been the result of particular BOX subtype patient transmission, other sources of infection—such as the environment or industrial products—might have accounted for the infections in CF patients within that period. Additionally, it should be noted that since 2005, isolates belonging to BOX subtypes B1 and B8 most notably seem to be responsible for new infections in CF patients. The PCR fingerprinting techniques have been successfully used for B. cenocepacia, B. cepacia, B. multivorans, B. dolosa, and B. pseudomallei clinical isolate discrimination (21, 22). In fact, ERIC- and BOX-PCR have proven to be valid technical alternatives to pulsed-field gel electrophoresis (22–24). In the present work, we were able to demonstrate that BOX-PCR is also a reliable typing method to examine the genetic relatedness among B. contaminans isolates so as to enable the identification of spreading of two different genotypes within a very closely related group of isolates.
Fig 3.
Distribution of BOX-PCR subtypes (B1 to B10) of B. contaminans isolates with the recA-ST-71 allele obtained over a 7-year period, with only the first isolate recovered from each patient being considered. (A) Incidence; (B) geographical distribution; (C) temporal distribution.
B. contaminans diversity in chronically infected patients.
CF patients are known to be susceptible to respiratory-tract chronic infection by certain Burkholderia species (25). Both the persistence of the same strain and the colonization with a different strain during the course of long-term infection have been reported (25–27). In 8 of the 21 chronically infected patients analyzed during the 7-year period of this work, more than one Burkholderia species was recovered during the course of infection (Table 1), either as a consequence of coinfection or through subsequent colonization by different species (so-called “species replacement”). In contrast, in the remaining 13 chronically infected patients, only one species (B. contaminans) was obtained (Table 1). Upon consideration of the BOX subtypes of serially recovered isolates during the infection of each of these 13 patients, we found that 5 patients remained infected with isolates carrying the same BOX subtype, whereas the other 8 evidenced a change in their isolate's BOX subtype during chronic infection. The latter result again could have occurred either as a consequence of a strain replacement event or through coinfection, with different isolates being collected—completely by chance—at the different samplings (28). In addition, the BOX-PCR diversity among the isolates recovered throughout the course of prolonged infection demonstrated that those clones isolated during the first year or years of infection belonged to different BOX subtypes (B6 to B8), with the B8 subtype being predominant—probably since the sample collection overlapped with the outbreak period (2004–2005). In contrast, after a long-lasting infection (more then 3 years of being colonized by B. contaminans), either the B1 or B8 BOX subtype became predominant, and the two were likely coexistent (28). Therefore, we conclude that the BOX-subtype diversity of the infecting strains responsible for the chronic cases investigated seemed to decrease over the time of infection, with B1 and B8 subtypes being the ones that mostly persisted and, apparently, replacing the initially infecting strains. This type of occurrence was found to be common in P. aeruginosa infections, where after a period of recurrent colonization, the CF patients became colonized permanently with a single lineage (29, 30). Once this P. aeruginosa lineage became adapted to the hostile environment of the CF lung (i.e., as the “adapted dominant epidemic strain”), that bacterium could persist for several decades so as to overcome both the host defense mechanisms and any intensive antibiotic therapy (29, 31–33).
Table 1.
Burkholderia cepacia complex species recovered from 21 patients over the period of chronic infection
|
Burkholderia cepacia complex species recovered from: |
No. of patients | |
|---|---|---|
| Initial samplea | Subsequent samples | |
| B. contaminans | B. cenocepacia | 4 |
| B. cepacia | B. vietnamiensis | 2 |
| B. cenocepacia | B. contaminans,B. cenocepacia | 2b |
| B. contaminans | B. contaminans | 5c |
| B. contaminans | B. contaminans | 8d |
Identification of the first available clinical isolates recovered from the patients with chronic infections.
A species different from the initial one was detected, and then, in later cultures, the initial species was found again, indicating probable species coinfection.
The same BOX subtype was detected in subsequent cultures.
Different BOX subtype patterns were detected in subsequent sputum cultures.
In conclusion, the species B. contaminans was indeed able to develop chronic colonization in CF patients. In particular, isolates belonging to BOX subtypes B8 and B1 persisted during the 7 years of our surveillance, either infecting CF patients for the first time (Fig. 3C) or colonizing CF patients chronically. We therefore infer that the aforementioned genotypes can survive in certain ecological niches or industrial products, spread among patients, and most readily adapt to the lungs of CF patients. This research has enabled a deeper understanding of the diversity of the B. contaminans isolates recovered from CF patients treated in Argentina. Since a knowledge of currently infectious strains is critical in prescribing patient treatment and making prognoses, isolates characterized by the B1 and B8 subtypes have now become the object of ongoing investigations in our laboratory. This and the forthcoming information will provide deeper insights into the strategies employed by B. contaminans to adapt and persist within CF patients.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Agencia Nacional de Promoción Científica y Tecnológica (grant BID 1728/OC-AR-PICT 34836) and Proyecto Extensión Fac. Cs. Exactas, UNLP. A. Bosch is a member CIC PBA, and P. Martina holds a CONICET fellowship.
We thank Julio Figari for his technical assistance. We are grateful to Laura Galanternik for providing the B. cepacia complex reference strains and to Donald F. Haggerty for editing the final version of the manuscript.
Footnotes
Published ahead of print 7 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02500-12.
REFERENCES
- 1.Baldwin A, Mahenthiralingam E, Thickett KM, Honeybourne D, Maiden MCJ, Govan JR, Speert DP, LiPuma JJ, Vandamme P, Dowson CG. 2005. Multilocus sequence typing scheme that provides both species and strain differentiation for the Burkholderia cepacia complex. J. Clin. Microbiol. 43:4665–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bosch A, Miñán A, Vescina C, Degrossi J, Gatti B, Montanaro P, Messina M, Franco M, Vay C, Schmitt J, Naumann D, Yantorno O. 2008. Fourier transform infrared spectroscopy for rapid identification of nonfermenting gram-negative bacteria isolated from sputum samples from cystic fibrosis patients. J. Clin. Microbiol. 46:2535–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jordá-Vargas L, Degrossi J, Castañeda NC, D'Aquino M, Valvano MA, Procopio A, Galanternik L, Centrón D. 2008. Prevalence of indeterminate genetic species of Burkholderia cepacia complex in a cystic fibrosis center in Argentina. J. Clin. Microbiol. 46:1151–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miñán A, Bosch A, Lasch P, Stämmler M, Serra DO, Degrossi J, Gatti B, Vay C, D'aquino M, Yantorno O, Naumann D. 2009. Rapid identification of Burkholderia cepacia complex species including strains of the novel Taxon K, recovered from cystic fibrosis patients by intact cell MALDI-ToF mass spectrometry. Analyst. 134:1138–1148 [DOI] [PubMed] [Google Scholar]
- 5.Drevinek P, Mahenthiralingam E. 2010. Burkholderia cenocepacia in cystic fibrosis: epidemiology and molecular mechanisms of virulence. Clin. Microbiol. Infect. 16:821–830 [DOI] [PubMed] [Google Scholar]
- 6.Hauser AR, Jain M, Bar-Meir M, McColley SA. 2011. Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin. Microbiol. Rev. 24:29–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LiPuma JJ. 2010. The changing microbial epidemiology in cystic fibrosis. Clin. Microbiol. Rev. 23:299–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nørskov-Lauritsen N, Johansen HK, Mette G, Nielsen XC, Pressler T, Hanne V, Høiby N, Fenger MG, Olesen HV. 2010. Unusual distribution of Burkholderia cepacia complex species in Danish cystic fibrosis clinics may stem from restricted transmission between patients. J. Clin. Microbiol. 48:2981–2983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coenye T, Vandamme P, Govan JRW, LiPuma JJ. 2001. Taxonomy and identification of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:3427–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henry DA, Mahenthiralingam E, Vandamme P, Coenye T, Speert DP. 2001. Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:1073–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.NCCLS 1999. Performance standards for antimicrobial disk and dilution susceptibility tests for bacteria isolated from animals. Approved standard M31-A. NCCLS, Wayne, PA [Google Scholar]
- 12.Zhou J, Garber E, Desai M, Saiman L. 2006. Compliance of clinical microbiology laboratories in the United States with current recommendations for processing respiratory tract specimens from patients with cystic fibrosis. J. Clin. Microbiol. 44:1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahenthiralingam E, Bischof J, Byrne SK, Radomski C, Davies JE, Av-Gay Y, Vandamme P. 2000. DNA-based diagnostic approaches for identification of Burkholderia cepacia complex, Burkholderia vietnamiensis, Burkholderia multivorans, Burkholderia stabilis, and Burkholderia cepacia genomovars I and III. J. Clin. Microbiol. 38:3165–3173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalmastri C, Fiore A, Alisi C, Bevivino A, Tabacchioni S, Giuliano G, Sprocati AR, Segre L, Mahenthiralingam E, Chiarini L, Vandamme P. 2003. A rhizospheric Burkholderia cepacia complex population: genotypic and phenotypic diversity of Burkholderia cenocepacia and Burkholderia ambifaria. FEMS Microbiol. Ecol. 46:179–187 [DOI] [PubMed] [Google Scholar]
- 15.Detsika MG, Corkill JE, Magalha M, Glendinning KJ, Hart CA, Winstanley C. 2003. Molecular typing of, and distribution of genetic markers among, Burkholderia cepacia complex isolates from Brazil. J. Clin. Microbiol. 41:4148–4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermis K, Coenye T, Mahenthiralingam E, Nelis HJ, Vandamme P. 2002. Evaluation of species-specific recA-based PCR tests for genomovar level identification within the Burkholderia cepacia complex. J. Med. Microbiol. 51:937–940 [DOI] [PubMed] [Google Scholar]
- 17.Vanlaere E, Baldwin A, Gevers D, Henry D, De Brandt E, LiPuma JJ, Mahenthiralingam E, Speert DP, Dowson C, Vandamme P. 2009. Taxon K, a complex within the Burkholderia cepacia complex, comprises at least two novel species, Burkholderia contaminans sp. nov. and Burkholderia lata sp. nov. Int. J. Syst. Evol. Microbiol. 59:102–111 [DOI] [PubMed] [Google Scholar]
- 18.Vanlaere E, LiPuma JJ, Baldwin A, Henry D, De Brandt E, Mahenthiralingam E, Speert D, Dowson C, Vandamme P. 2008. Burkholderia latens sp. nov., Burkholderia diffusa sp. nov., Burkholderia arboris sp. nov., Burkholderia seminalis sp. nov. and Burkholderia metallica sp. nov., novel species within the Burkholderia cepacia complex. Int. J. Syst. Evol. Microbiol. 58:1580–1590 [DOI] [PubMed] [Google Scholar]
- 19.Assaad W, Magalhães M, Plesa M, Hart CA, Cornelis P, Winstanley C. 2006. Identical Burkholderia cepacia complex strain types isolated from multiple patients attending a hospital in Brazil. J. Med. Microbiol. 55:247–249 [DOI] [PubMed] [Google Scholar]
- 20.Versalovic J, Koeuth T, Lupski JR. 1991. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 19:6823–6831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Biddick R, Spilker T, Martin A, LiPuma JJ. 2003. Evidence of transmission of Burkholderia cepacia, Burkholderia multivorans and Burkholderia dolosa among persons with cystic fibrosis. FEMS Microbiol. Lett. 228:57–62 [DOI] [PubMed] [Google Scholar]
- 22.Coenye T, Spilker T, Martin A, LiPuma JJ. 2002. Comparative assessment of genotyping methods for epidemiologic study of Burkholderia cepacia genomovar III. J. Clin. Microbiol. 40:3300–3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campana S, Taccetti G, Ravenni N, Favari F, Cariani L, Sciacca A, Savoia D, Collura A, Fiscarelli E, Intinis GD, Busetti M, Cipolloni A, Aprile A, Provenzano E, Collebrusco I, Frontini P, Stassi G, Trancassini M, Tovagliari D, Lavitola A, Doherty CJ, Coenye T, Govan JRW, Vandamme P. 2005. Transmission of Burkholderia cepacia complex: evidence for new epidemic clones infecting cystic fibrosis patients in Italy. J. Clin. Microbiol. 43:5136–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Currie BJ, Gal D, Mayo M, Ward L, Godoy D, Spratt BG, LiPuma JJ. 2007. Using BOX-PCR to exclude a clonal outbreak of melioidosis. BMC Infect. Dis. 7:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernhardt S, Spilker T, Coffey T, LiPuma JJ. 2003. Burkholderia cepacia complex in cystic fibrosis: frequency of strain replacement during chronic infection. Clin. Infect. Dis. 37:780–785 [DOI] [PubMed] [Google Scholar]
- 26.Chen JS, Ka Witzmann Spilker T, Fink RJ, LiPuma JJ. 2001. Endemicity and inter-city spread of Burkholderia cepacia genomovar III in cystic fibrosis. J. Pediatr. 139:643–649 [DOI] [PubMed] [Google Scholar]
- 27.Mahenthiralingam E, Vandamme P, Campbell ME, Henry DA, Gravelle AM, Wong LT, Davidson AG, Wilcox PG, Nakielna B, Speert DP. 2001. Infection with Burkholderia cepacia complex genomovars in patients with cystic fibrosis: virulent transmissible strains of genomovar III can replace Burkholderia multivorans. Clin. Infect. Dis. 33:1469–1475 [DOI] [PubMed] [Google Scholar]
- 28.Yang JH, Spilker T, LiPuma JJ. 2006. Simultaneous coinfection by multiple strains during Burkholderia cepacia complex infection in cystic fibrosis. Diagn. Microbiol. Infect. Dis. 54:95–98 [DOI] [PubMed] [Google Scholar]
- 29.Jelsbak L, Johansen HK, Frost A- L, Thøgersen R, Thomsen LE, Ciofu O, Yang L, Ja Haagensen J, Høiby N, Molin S. 2007. Molecular epidemiology and dynamics of Pseudomonas aeruginosa populations in lungs of cystic fibrosis patients. Infect. Immun. 75:2214–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rau MH, Hansen SK, Johansen HK, Thomsen LE, Workman CT, Nielsen KF, Jelsbak L, Høiby N, Yang L, Molin S. 2010. Early adaptive developments of Pseudomonas aeruginosa after the transition from life in the environment to persistent colonization in the airways of human cystic fibrosis hosts. Environ. Microbiol. 12:1643–1658 [DOI] [PubMed] [Google Scholar]
- 31.Feliziani S, Luján AM, Moyano AJ, Sola C, Bocco JL, Montanaro P, Canigia LF, Argaraña CE, Smania AM. 2010. Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS One 5:e12669 doi:10.1371/journal.pone.0012669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc. Natl. Acad. Sci. U. S. A. 103:8487–8492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang L, Jelsbak L, Marvig RL, Damkiær S, Workman CT, Rau MH, Hansen SK, Folkesson A, Johansen HK, Ciofu O, Høiby N, Sommer MO, Molin S. 2011. Evolutionary dynamics of bacteria in a human host environment. Proc. Natl. Acad. Sci. U. S. A. 108:7481–7486 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.