Abstract
The anterior nares are the site of choice for the Veterans Administration methicillin-resistant Staphylococcus aureus (MRSA) surveillance program; however, a correlation between nares colonization and concomitant wound infections has not been well established. The purpose of this study was 3-fold: to determine the relatedness of MRSA isolates from 40 paired wound and nares specimens by four different strain typing methods, to determine concordance of typing methods, and to establish a baseline of MRSA types at this medical center. Isolates were typed by repetitive PCR (rep-PCR) (DiversiLab System; DL) and SpectraCell Raman analysis (SCRA) (commercially available methods that can be performed within a clinical lab), pulsed-field gel electrophoresis (PFGE), and an antibiotic susceptibility profile (AB). Whole-genome optical mapping (WGM) (OpGen, Inc.) was performed on selected isolates. All methods agreed that 26 pairs were indistinguishable and four pairs were different. Discrepant results were as follows: 4 where only SCRA was discordant, 3 where only AB was discordant, 2 where both DL and AB were discordant, and 1 where both DL and SCRA were discordant. All WGM agreed with PFGE. After discrepancy resolution, 80% of the pairs were indistinguishable and 20% were different. A total of 56% of nares results were nonpredictive if negative nares and positive wound cultures are included. Methods agreed 85 to 93% of the time; however, congruence of isolates to a clade was lower. Baseline analysis of types showed that 15 pairs were unique to single patients (30 strains, 38%; 47% of the matching pairs). Twenty-five strains (30%) represented a single clade identical by PFGE, SCRA, and DL, decreasing specificity. Typing method and institutional type frequency are important in assessing MRSA strain relatedness.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is a major cause of hospital-associated and community-associated infection in the Veteran's Affairs (VA) healthcare system, with the incidence of MRSA infection increasing significantly in the first part of the millennium. Skin and soft tissue infections (SSTI) were the most commonly reported type of infection (1), and 65% of SSTI were caused by MRSA, as reported by one VA hospital (2). As a result, in 2007, the VA system instituted a nationwide acute-care surveillance program in their 153 medical centers with the aim of decreasing both carriage and disease due to MRSA (3). A 2011 report notes that implementation of the “MRSA bundle,” a multifaceted program including universal surveillance, contact precautions, hand hygiene, and institutional culture change, was associated with but not necessarily the cause of a decrease in health care-associated infections with MRSA across the large hospital system (3).
The correlation between nasal carriage and bloodstream infection has been previously established (4), and the rate of bloodstream infection is a recognized surrogate marker to assess changes in the overall rate of infection (3). However, the correlation between nasal carriage and wound infection has not been clearly established. In addition, the evaluation of nosocomial transmission is more complex than can be reflected by a simple tabulation of MRSA surveillance culture results. Accurate strain typing methods are essential if we are to adequately evaluate acquisition of and risk of infection by MRSA.
There have been numerous studies to compare molecular typing methods to determine MRSA strain relatedness (5, 6). Pulsed-field gel electrophoresis (PFGE) remains the gold standard for molecular strain typing of MRSA (7). The major advantages of PFGE are standardized protocols and its high discriminatory power. Other methods, such as multilocus sequence typing (MLST), staphylococcal cassette chromosome (SCCmec) typing, and spa typing, are widely used methods for distinguishing genetic relationships between groups of MRSA strains (8–10). Other less-used typing systems, including multilocus variable-number tandem-repeat analysis (MLVA) (11), repetitive PCR (rep-PCR) with the DiversiLab system (DL) (bioMeriuex, Durham, NC) (12), and the SpectraCellRA Raman analysis typing (SCRA) system (River Diagnostics, Rotterdam, Netherlands) (13), have been compared to PFGE and other methods in terms of discriminatory power, reproducibility, ease of use, and cost. DL and SCRA are integrated microbial typing systems that can be purchased and performed by routine clinical laboratories.
Antibiotic susceptibility testing is widely used in the clinical microbiology laboratory to suggest the relatedness of multiple isolates recovered from the same patient's specimen or from multiple specimens collected around the same time. The value of the isolate's overall antibiotic susceptibility profile, or antibiogram (AB), lies in its ability to rapidly show preliminary discrimination between multiple isolates that may otherwise appear phenotypically equivalent; however, this method lacks stability and discriminatory power for general epidemiological studies (14).
The initial objective for this study was to answer a practical question: for a single patient, is the MRSA strain isolated from the nares the same as the strain isolated from the wound at about the same time? We also measured concurrence and the relationship between the following four different typing methods: rep-PCR, pulsed-field gel electrophoresis, Raman spectroscopy, and the antibiotic susceptibility profile. We selectively utilized whole-genome mapping (WGM) (OpGen, Gaithersburg, MD) to resolve discrepancies between the methods. Since our surveillance is an ongoing activity, we established a baseline for the frequency and variability of MRSA types at this medical center.
(These data were presented in part at the 112th General Meeting of the American Society for Microbiology, San Francisco, CA, 2012, the 109th General Meeting of the American Society for Microbiology, Philadelphia, PA, 2009, and the 20th European Conference for Clinical Microbiology and Infectious Disease, Vienna, Austria, 2010.)
MATERIALS AND METHODS
MRSA strains.
At the VA Puget Sound Health Care System in Seattle, WA, we track all MRSA nares cultures and all wounds positive for MRSA from routine clinical and surveillance specimens. From these, we identified 40 consecutive patients for whom both nares and wound specimens were positive for MRSA and the nares isolate was obtained prior to, at the same time as, or within 48 h of the wound isolate. MRSA strains were identified by culture (after broth enrichment for nares specimens) on chromogenic MRSASelect agar (Bio-Rad, France) (15). Wound specimens included abscess aspirate, joint fluid, pleural fluid, superficial and deep wound, bone, tissue, and other material from surgical sites. The patients were deidentified and assigned numbers from 1 to 40, with associated isolates randomly assigned numbers from 1 to 80. Patient identifier and isolate numbers were relinked only after all analyses were completed.
Rep-PCR.
Rep-PCR was performed using a standardized commercial system called DiversiLab (DL) (bioMérieux, Durham, NC). The DNA from the 80 MRSA isolates was extracted using the UltraClean microbial DNA isolation kit (MoBio Laboratories, Carlsbad, CA), semiquantitated using DiversiLab DNA gel standards, and visualized by agarose gel electrophoresis. Rep-PCR was performed using the DiversiLab Staphylococcus fingerprinting kit, including master mix and primers as described by the manufacturer. Thermal cycling was performed using a GeneAmp PCR System 9700 (Applied Biosystems, Foster City, CA). Parameters were as follows: initial denaturation at 94°C for 120 s, denaturation at 94°C for 30 s, annealing at 50°C for 30 s, extension at 70°C for 90 s, and final extension at 70°C for 180 s. Analysis of the rep-PCR product was performed using a DiversiLab microfluidic chip and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Digital virtual gel banding patterns were created and visualized by the DiversiLab software. Relatedness of isolates was determined by DiversiLab software and by the analysis of the raw electropherograms. Strains with <95% similarity and 1 to 2 band differences were considered to be unrelated. Each of the seven virtual gel patterns was given a unique letter value of A through H (F was not found to be unique on closer analysis).
Pulsed-field gel electrophoresis.
PFGE analyses were done as previously described (7, 16). The DNA blocks were prepared, digested separately with SmaI (Fermentas Inc., Glen Burnie, MD) for 6 h at 28°C, and subjected to electrophoresis in a contour-clamped homogeneous electric field (CHEF DR II system; Bio-Rad Laboratories Inc., Hercules, CA) for 21 h at 14°C with switch times of 5 s (initial) and 40 s (final) at 6 V cm−1 as described previously (7). The gels were stained with ethidium bromide, destained in distilled water, and photographed under UV transillumination. Isolates were considered to have the same PFGE pattern if they had indistinguishable patterns. Because we were not using the patterns for epidemiologic evaluations, we did not use a system in which those that differed by ≤3 bands were considered the same, as previously described (7). Each PFGE pattern was given a unique number value; in our data, these are represented as numbers 1 through 21. Only after assessing all gels did we break the code to reveal patient identification.
Raman spectroscopy.
Raman spectroscopy was performed using a standardized commercial system from River Diagnostics called SpectraCellRA (SCRA). Before Raman measurements were performed, the MRSA isolates were washed and transferred to a fused silica glass slide and air dried, resulting in small pellets of biomass. Strains with <99.9% similarity were considered to be unrelated. All SCRA patterns that clustered above this similarity cutoff were given a unique number value. Nine SCRA patterns were distinguished (numbers 1, 2, 3, 4, 6, 7, 8, 9, and 10).
Antibiogram analysis.
Antimicrobial susceptibility data were generated using customized frozen Sensititre plates from Trek Diagnostics (Cleveland, OH). MICs were determined for the following drugs: erythromycin (ERY), clindamycin (CLI), daptomycin (DAP), gentamicin (GEN), linezolid (LZD), rifampin (RIF), vancomycin (VAN), trimethoprim-sulfamethoxazole (SXT), levofloxacin (LVX), quinupristin-dalfopristin (Q-D), tigecycline (TGC), moxifloxacin (MXF), fusidic acid (FUS), mupirocin (MUP), and retapamulin (RET). The MIC values for each isolate were put in an Excel spreadsheet, and an overall antibiogram (AB) for each organism was created. A pattern was considered unique if there was a ≥3-fold difference in at least one antibiotic or a ≥2-fold difference in at least 3 antibiotics. Each unique pattern was assigned a letter, with 11 distinct patterns assigned 11 random letters.
Whole-genome mapping.
Analysis was performed by OpGen, Inc., Gaithersburg, MD. Approximately 107 cells of each MRSA strain were extracted and evaluated for quality and concentration through a standard protocol. Single-molecule MapSets were collected on the Argus system. DNA molecules were captured using a microfluidic device, MapCard, composed of a charged glass surface and polymer overlay with nanometer grooves. Molecules were subjected to restriction enzyme digestion with the NcoI enzyme. After digestion, the molecules were stained with a fluorescent DNA intercalating dye. Once processed, the card was imaged through fluorescence microscopy for image capture and single-molecule markup. Pixelated images of the stained, restricted DNA fragments were converted into the kbp size of the restricted molecules generated. An automated image acquisition collected all single-molecule restriction maps fulfilling size expectations. Single-molecule maps were assembled to create a circular map spanning the entire genome with coverage of greater than 30-fold. MapSolver software was used to create dendrograms to cluster strains using the unweighted-pair group method using average linkages (UPGMA). Further comparative genomics revealed genetic rearrangements occurring between strains.
Analysis of microbial strain typing results.
The congruence of the strain typing results generated by PFGE, DL, and SCRA was determined by calculating the adjusted Rand and Wallace coefficients as described by Carrico et al. (17) and Pinto et al. (18) using the Comparing Partitions website (http://darwin.phyloviz.net/ComparingPartitions/index.php?link=Tool). Pairwise comparisons were made on data sets. Simpson's index of diversity was also determined for the three typing methods.
Human subjects.
This study was approved by the VA Puget Sound Health Care System's Institutional Review Board.
RESULTS
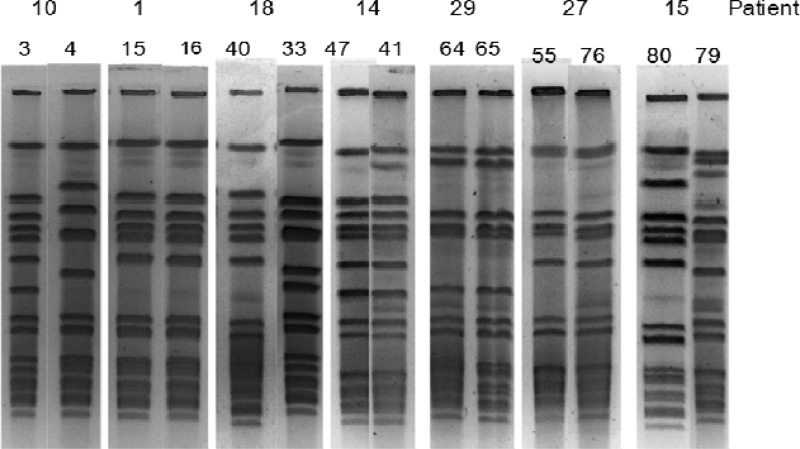
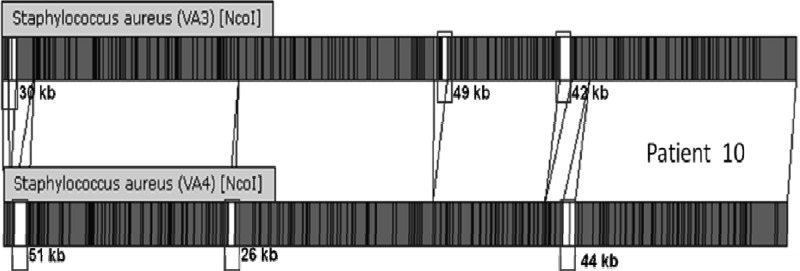
The designated strain types for the 80 strains for each of the four methods used are shown in Table 1. PFGE, SCRA, DL, and AB provided 21, 9, 7, and 11 unique groups, respectively. There were 26 patients for whom the pairs were indistinguishable and four patients from whom we isolated clearly different strains. There were 10 patients for whom there were discrepant results (Table 2). WGM was performed on four of the discrepant pairs and two control pairs. Discrepant pairs included 4 pairs with only the SCRA result discordant, 3 pairs with only the AB result discordant, 2 pairs with both DL and AB results discordant, and 1 pair with DL and SCRA discordant (Table 2). In each case, the WGM results agreed with the PFGE designation. After resolution, 80% (32/40) of the MRSA paired isolates from nares and wound were indistinguishable and 20% were different. The gels from discrepant results are shown in Fig. 1. Patients 1, 14, and 29 were considered to be the same by PFGE, while patients 10 and 18 are different. The WGM comparison of the nares and wound strains from patient 10 (Fig. 2) confirms that the two strains are genetically distinct. The agreement between methods was 92.5% for PFGE and DL, 87.5% for PFGE and SCRA, and 85% for DL and SCRA.
Table 1.
Detailed comparison of typing methods
| Patient no. | Strain no.a | Type of specimen | PFGE type | SCRA type | DL type | Antibiotic profile |
|---|---|---|---|---|---|---|
| 1 | 15* | Nares (admit) | 1 | 10 | C | B |
| 16* | Skin | 1 | 1 | C | B | |
| 2 | 7 | Nares (admit) | 2 | 10 | C | B |
| 6 | Surgical wound | 2 | 10 | C | B | |
| 3 | 18 | Nares (admit) | 13 | 6 | H | C |
| 17 | Abscess | 13 | 6 | H | C | |
| 4 | 2 | Nares | 5 | 7 | G | C |
| 1 | Other tissue | 5 | 7 | G | C | |
| 5 | 8 | Nares (admit) | 10 | 10 | E | B |
| 9 | Abscess | 10 | 10 | E | B | |
| 6 | 21 | Nares | 1 | 10 | C | D |
| 20 | Abscess | 1 | 10 | C | D | |
| 7 | 23* | Nares (admit) | 1 | 7 | C | D |
| 24* | Abscess | 1 | 10 | C | D | |
| 8 | 19 | Nares | 1 | 10 | C | B |
| 22 | Abscess | 1 | 10 | C | B | |
| 9 | 5 | Nares | 1 | 10 | C | B |
| 14 | Abscess | 1 | 10 | C | B | |
| 10 | 4* | Nares (admit) | 15 | 7 | C | B |
| 3* | Superficial | 14 | 10 | C | B | |
| 11 | 10 | Nares (admit) | 12 | 6 | H | C |
| 11 | Other wound | 2 | 10 | C | D | |
| 12 | 13 | Nares (admit) | 10 | 10 | E | B |
| 12 | Surgical wound | 10 | 10 | E | B | |
| 13 | 38 | Nares (admit) | 1 | 10 | C | B |
| 39 | Abscess | 1 | 10 | C | B | |
| 14 | 41* | Nares (admit) | 16 | 6 | G | C |
| 47* | Other tissue | 16 | 6 | G | N | |
| 15 | 79 | Nares (admit) | 12 | 7 | H | K |
| 80 | Other tissue | 10 | 10 | E | R | |
| 16 | 26 | Nares | 5 | 2 | G | O |
| 25 | Wound | 1 | 10 | C | B | |
| 17 | 44 | Nares (admit) | 8 | 6 | G | L |
| 35 | Surgical wound | 8 | 6 | G | L | |
| 18 | 33* | Nares (admit) | 4 | 10 | C | W |
| 40* | Abscess | 1 | 10 | C | D | |
| 19 | 43 | Nares | 1 | 10 | C | D |
| 42 | Abscess | 1 | 10 | C | D | |
| 20 | 36 | Nares | 18 | 10 | C | D |
| 31 | Abscess | 18 | 10 | C | D | |
| 21 | 32 | Nares | 5 | 7 | G | C |
| 45 | Other wound | 5 | 7 | G | C | |
| 22 | 37 | Nares (admit) | 21 | 6 | G | C |
| 46 | Surgical wound | 5 | 7 | G | C | |
| 23 | 27 | Nares (admit) | 1 | 10 | C | D |
| 28 | Abscess | 1 | 10 | C | D | |
| 24 | 48 | Nares | 3 | 10 | C | Q |
| 34 | Other wound | 3 | 10 | C | Q | |
| 25 | 30 | Nares (admit) | 5 | 3 | G | O |
| 29 | Other wound | 4 | 7 | A | A | |
| 26 | 70 | Nares (admit) | 1 | 10 | C | B |
| 69 | Abscess | 1 | 10 | C | R | |
| 27 | 55 | Nares (admit) | 19 | 4 | G | C |
| 76 | Ulcer | 19 | 4 | G | C | |
| 28 | 78 | Nares (admit) | 20 | 10 | D | A |
| 52 | Other wound | 20 | 10 | D | A | |
| 29 | 64 | Nares (discharge) | 7 | 7 | G | C |
| 65 | Other wound | 7 | 9 | G | C | |
| 30 | 75* | Nares (discharge) | 1 | 10 | C | L |
| 54* | Superficial | 9 | 10 | E | B | |
| 31 | 59 | Nares | 9 | 10 | A | D |
| 58 | Other wound | 9 | 10 | A | D | |
| 32 | 66 | Nares (admit) | 1 | 8 | C | D |
| 67 | Abscess | 1 | 8 | C | D | |
| 33 | 71 | Nares | 1 | 10 | C | B |
| 61 | Abscess | 1 | 10 | C | B | |
| 34 | 68 | Nares (admit) | 1 | 10 | C | B |
| 49 | Other wound | 1 | 10 | C | B | |
| 35 | 74* | Nares | 1 | 10 | C | W |
| 73* | Abscess | 1 | 10 | C | L | |
| 36 | 72 | Nares (admit) | 9 | 10 | E | B |
| 77 | Other wound | 9 | 10 | E | B | |
| 37 | 57 | Nares (admit) | 6 | 4 | G | L |
| 60 | Abscess | 6 | 4 | G | L | |
| 38 | 63 | Nares (discharge) | 4 | 7 | C | B |
| 62 | Other wound | 4 | 7 | C | L | |
| 39 | 56 | Nares | 12 | 6 | H | L |
| 53 | Other wound | 12 | 6 | H | L | |
| 40 | 51 | Nares (discharge) | 11 | 6 | B | G |
| 50 | Wound | 11 | 6 | B | G |
Asterisks indicate pairs that are not in the same groups for all typing methods.
Table 2.
Analysis of discrepant resultsa
| Category and patient no. | PFGE | SCRA | DL | Antibiotic profile | OpGen | Final assessment |
|---|---|---|---|---|---|---|
| Discrepant results | ||||||
| 1 | S | D | S | S | S | |
| 7 | S | D | S | S | S | S |
| 10 | D | D | S | S | D | D |
| 14 | S | S | S | Db | S | |
| 18 | D | S | S | D | D | D |
| 22 | D | D | S | S | D | D |
| 26 | S | S | S | Dc | S | |
| 29 | S | D | S | S | S | |
| 30 | D | S | D | D | D | |
| 35 | S | S | S | Dd | S | |
| Control | ||||||
| 27 | S | S | S | S | S | S |
| 36 | S | S | S | S | S | S |
S, strains from the patient are in the same group; D, strains from the patient are in different groups.
Only different for mupirocin.
Only different for erythromycin.
Only different for trimethoprim-sulfamethoxazole.
Fig 1.
Visual comparison of PFGE gels from selected pairs. Analysis of selected pairs with the discrepant results. The final assessment was that the strains were different for patients 10 and 18 and the same for patients 1, 14, and 29. Strains from patient 27 and 15 were not discrepant and are included as controls to illustrate similarity of strains (patient 27) and differences (patient 15); all testing methods agreed that both strains from patient 27 were the same and both strains from patient 15 were different.
Fig 2.
Whole-genome map comparison of the naris and wound strains from patient 10. Comparison of the high-resolution, ordered restriction maps of two isolates from the same patient demonstrates significant genomic differences, indicated by white spaces and noted as the number of kilobases.
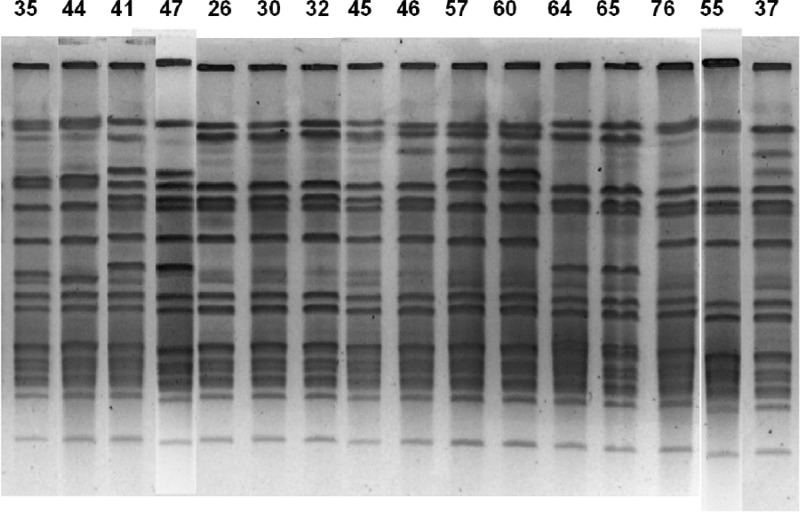
Figure 3 demonstrates increased discriminatory power for PFGE compared to that of DL. All 16 of the shown strains were group G by DL. However, when compared pairwise, the strains of patients 17 (strains 35 and 44), 14 (strains 41 and 47), 37 (strains 57 and 60), 29 (strains 64 and 65), and 27 (strains 55 and 76) have clearly distinguishable patterns by PFGE. The lack of discrimination for many isolates by DL is further shown in that the two largest clonal groups, C and G, comprise 61% of isolates. The four typing results in conjunction found 15 unique pairs (30 strains, 38%) of two strains that were both identical to each other and unrelated to any other isolate (patients 2, 3, 14, 17, 20, 24, 27, 28, 29, 31, 32, 37, 38, 39, and 40). By summarizing data for PFGE, DL, and SCRA from Table 1, there were 30 unique groups. The 25 isolates (31%) in the largest unique group were all PFGE 1, SCRA 10, and DL C types. Although these isolates were not clustered by time or ward at the time of specimen collection, they have not been evaluated for longitudinal or less-obvious connections.
Fig 3.
Greater discrimination by PFGE. Each isolate was designated group G by DL. However, the strains of patients 17 (strains 35 and 44), 14 (strains 41 and 47), 37 (strains 57 and 60), 29 (strains 64 and 65), and 27 (strains 55 and 76) have distinct patterns by PFGE when evaluated for a single band difference. Other methods of evaluating gel patterns can allow up to 3 band differences and still consider the strains to be in the same group.
Analysis of the typing data by the four methods is shown by the adjusted Rand coefficients (Table 3). The adjusted Rand coefficients for the pairwise comparisons of typing results ranged from 0.154 to 0.562, where values close to 1.0 indicate high congruence of results between methods. PFGE and DL had the highest congruence. The contingency table (Table 4) shows the relationship between PFGE and DL. Outside the large group of 29 isolates which share the PFGE 1/DL C signature, the combination of the two methods provides greater discrimination. For example, using Tables 1 and 4, we see that the only person to have a strain with a PFGE 9/DL A signature is patient 31. If a subsequent patient were to have a strain with that signature, it would be strong evidence to pursue the possibility of strain transmission between patient 31 and the subsequent patient, whereas finding a strain with a PFGE 1/DL C signature would suggest many possible sources of transmission. All the cells in Table 4 with only two strains in it are from a single patient.
Table 3.
Congruence of strain typing results as indicated by adjusted Rand coefficientsa
| First method | Congruence between first method and: |
||
|---|---|---|---|
| PFGE | SCRA | DL | |
| PFGE | |||
| SCRA | 0.241 (0.099–0.389) | ||
| DL | 0.562 (0.396–0.739) | 0.377 (0.221–0.542) | |
| AB | 0.154 (0.058–0.253) | 0.166 (0.040–0.295) | 0.251 (0.121–0.379) |
Values are adjusted Rand and jackknife pseudovalues (95% confidence intervals). Data were generated using the Comparing Partitions website (http://darwin.phyloviz.net/ComparingPartitions/index.php?link=Tool). Although PFGE and DL agreed 93% in their assessment of the patient pairs, the congruence value of 0.563, while highest among our testing methods, is not high. This is because PFGE is more discriminatory, as shown in Fig. 2 and in the contingency table (Table 4). The congruence of AB typing is the lowest with all methods.
Table 4.
Contingency tablea
| PFGE type | No. of strains with each DL type that have the indicated PFGE type |
||||||
|---|---|---|---|---|---|---|---|
| C | G | E | H | A | D | Total | |
| 21 | 1 | 1 | |||||
| 14 | 1 | 1 | |||||
| 15 | 1 | 1 | |||||
| 8 | 2 | 2 | |||||
| 20 | 2 | 2 | |||||
| 19 | 2 | 2 | |||||
| 7 | 2 | 2 | |||||
| 16 | 2 | 2 | |||||
| 12 | 2 | 2 | |||||
| 4 | 1 | 1 | 2 | ||||
| 3 | 2 | 2 | |||||
| 18 | 2 | 2 | |||||
| 13 | 2 | 2 | |||||
| 2 | 3 | 3 | |||||
| 9 | 3 | 2 | 5 | ||||
| 10 | 5 | 5 | |||||
| 5 | 7 | 7 | |||||
| 1 | 29 | 29 | |||||
| Total | 39 | 16 | 8 | 4 | 3 | 2 | 72 |
This table, generated by the Comparing Partitions website (http://darwin.phyloviz.net/ComparingPartitions/index.php?link=Tool), shows the relationship between PFGE and DL. The cells with only a “2” in them represent unique pairs from a single patient. For example, using Tables 1 and 4, it is shown that the only person with the PFGE 9/DL A signature is patient 31.
Although antibiotic susceptibility pattern comparison in general is not considered an accurate typing method for unrelated strains, in this study, where one strain was isolated within 48 h of its pair, the results correlated well with those of the other methods of strain typing. The AB profiles showed 11 distinct patterns. The four pairs that were different by all methods showed extreme variation in the AB profile, with >4 antibiotic resistance differences. However, for the three pairs (patient numbers 14, 26, and 35) that were indistinguishable by all methods except the AB profile, there was only a one-drug difference (erythromycin, trimethoprim-sulfamethoxazole, or mupirocin).
DISCUSSION
The published molecular strain type correlation between MRSA nares colonization and concomitant wound infections has varied greatly; Kazakova et al. found no evidence of MRSA nasal carriage as the source for MRSA infection in an outbreak among professional football players (19). Chen et al. and Berthelot et al. found that ∼50 to 60% of available nares and wound isolate pairs were indistinguishable in pediatric patients or patients undergoing orthopedic surgery (20, 21). Frazee et al. and Reighard et al. found a much tighter correlation, with ∼80 to 100% of available isolate pairs indistinguishable in patients evaluated in the emergency department of an urban teaching hospital or a university hospital burn trauma unit (22, 23). In addition to patient populations evaluated, studies vary in their interpretation criteria and assessment of discriminatory power of strain typing methods (20, 22, 24). It is clear that the overall categorical designation of strain types (indistinguishable versus unrelated) relies on the methodology and interpretation criteria used in the study and that not all analyses are equivalent (12, 25–27).
We used three molecular strain typing methods and the AB profile to determine the relatedness of 80 paired wound and nares isolates and to examine the concordance of two commercially integrated systems with PFGE and the AB profile. We found that the paired isolates from the nares and the wound site from the same person at the same time were indistinguishable for 80% and different for 20% of the patients. Thus, for the majority of the cases, when both wound and nares isolates were available, the nares isolate predicted the wound isolate. These data are comparable to those that have been established for bloodstream infection (4). Within the same time frame that we collected the 40 pairs of MRSA isolates, we also found 33 patients that had a wound positive for MRSA while the nares swab obtained within 48 h was negative for MRSA. Thus, we identified 73 patients with paired nares and wound cultures. If we consider the 33 patients with negative nares cultures and 8 patients with a different strain isolated, a total of 56% (41/73) of patients' nares results were nonpredictive of wound isolate type or result. This is consistent with other findings in the literature, in which ∼60% of nares cultures are negative in the setting of a concomitant MRSA wound infection (8, 20–22). These data are striking considering the fact that MRSA surveillance programs are founded on the presumed increased risk of infection due to or correlated with nares colonization. Our data, along with those of others, suggest that the source for MRSA wound infection, if endogenous, may also be extranasal. However, it is possible that there was more than one MRSA strain in the nares, as we were not able to distinguish different strains by colony appearance on the chromogenic MRSASelect plates that we used.
Currently, strain typing of MRSA isolates is not commonly performed in a routine clinical laboratory. This is, in part, because many methods are labor-intensive, cost prohibitive, or technically difficult to perform. We therefore chose to evaluate two methods that are technically straightforward to perform and commercially available for a clinical laboratory. We included PFGE as the standard and the AB profile because it can be performed without any additional materials. In our comparison of methods, DL and PFGE were concordant when assessing whether the two patient isolates were indistinguishable at 92.5% agreement (Table 3), a percentage which was comparable to those of other published studies (28, 29). Although agreement between PFGE and DL was high, as shown by the adjusted Rand analysis, the congruence value of 0.562 (Table 4), while highest among our testing methods, is not as high as in a recent publication (30); however, because we were evaluating matched pairs, our study included fewer strains and may not have allowed as robust of an analysis. In our study, PFGE was the most discriminatory strain typing method, as shown in Fig. 3 and Table 4. The congruence of AB typing was the lowest of all methods; however, we found that the antibiotic profile was useful in suggesting nonidentity between the paired strains from a single individual if single antibiotic changes were not counted. This rapid and inexpensive method, although limited, is available to almost all microbiology laboratories.
Our evaluation revealed a major clone comprising about 31% of the total strains. While problematic for use in attributing a particular incident, health care professional, or copatient to transmission of MRSA, the finding that a preponderance of surveillance isolates belong to just one or two strain types is reported elsewhere (30, 31). Since it is known that S. aureus demonstrates limited clonal diversity within given regional and patient demographics (12), it was not unexpected that PFGE, SCRA, and DL each defined a major clade in which approximately one-third to one-half of the isolates were indistinguishable. Since many of our veterans have an extensive long-term connection to our medical center, it may be that this clade is common in our geographic area or medical center and does not indicate recent person-to-person passage. Baseline data can reveal preexisting large clonal groups that are common in an environment, findings which can skew the interpretation of possible nosocomial transmission of MRSA. In addition, baseline data can also highlight strains that are rare. Although the methods examined in this study may potentially be effective in tracking transmission in an outbreak setting with a unique strain, if there are common strains within the population, the discrete patient-to-patient transmission is difficult to assess. Other techniques, such as spa typing, MLST, or binary typing, may provide additional discrimination (25, 26, 30, 32, 33).
Even though the importance of integrating molecular strain typing into an infection control program has been suggested, its performance by the clinical microbiology laboratory is not standardized or routine. The Veterans Administration MRSA surveillance program as a whole has been considered a success because of a decrease in the rate of MRSA bloodstream infections; however, strain typing was not performed and the overall causality of the decrease has been reevaluated (34). Since 80% of our nares-wound pairs in our sample population were concordant and 47% of the concordant pairs were also unique to that single patient, the data suggest that many patients' wound infections are endogenous and that infection control measures designed only to prevent interpersonal spread of MRSA may not be the most efficient or efficacious approach. Rather, decolonization, hand hygiene compliance, and catheter hygiene might be emphasized. If hospitals are to be held accountable for the consequences of presumed nosocomial MRSA infections, in order to establish causality in hospital-associated transmission, typing method and institutional type frequency of MRSA must be considered.
ACKNOWLEDGMENTS
We thank OpGen, Inc., for providing resources and especially Erin Newburn and Emily Zentz for their work and consultation regarding the whole-genome mapping studies. We thank GlaxoSmithKline for providing resources and especially Rhibi Shawar for consultation. We thank Jennifer Black for her technical expertise regarding the antibiotic susceptibility testing and Diana Willemse for technical assistance in performing the Raman spectroscopy measurements. We thank the Seattle VA Infection Control team and the hospital staff for their contributions to the MRSA surveillance program. Most of all, we thank the Seattle VA microbiology section members, who have the highest IQ per grade, for their excellent support.
This work was supported in part by resources from the VA Puget Sound Health Care System, Seattle, WA.
Footnotes
Published ahead of print 7 November 2012
REFERENCES
- 1. Caffrey AR, Laplante KL. 2012. Changing epidemiology of methicillin-resistant Staphylococcus aureus in the Veterans Affairs Healthcare System, 2002-2009. Infection 40:291–297 [DOI] [PubMed] [Google Scholar]
- 2. Tracy LA, Furuno JP, Harris AD, Singer M, Langenberg P, Roghmann MC. 2011. Staphylococcus aureus infections in US veterans, Maryland, USA, 1999-2008. Emerg. Infect. Dis. 17:441–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jain R, Kralovic SM, Evans ME, Ambrose M, Simbartl LA, Obrosky DS, Render ML, Freyberg RW, Jernigan JA, Muder RR, Miller LJ, Roselle GA. 2011. Veterans Affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections. N. Engl. J. Med. 364:1419–1430 [DOI] [PubMed] [Google Scholar]
- 4. von Eiff C, Becker K, Machka K, Stammer H, Peters G. 2001. Nasal carriage as a source of Staphylococcus aureus bacteremia. N. Engl. J. Med. 344:11–16 [DOI] [PubMed] [Google Scholar]
- 5. van Belkum A. 2007. Tracing isolates of bacterial species by multilocus variable number of tandem repeat analysis (MLVA). FEMS Immunol. Med. Microbiol. 49:22–27 [DOI] [PubMed] [Google Scholar]
- 6. van Leeuwen WB, Snoeijers S, van der Werken-Libregts C, Tuip A, van der Zee A, Egberink D, de Proost M, Bik E, Lunter B, Kluytmans J, Gits E, van Duyn I, Heck M, van der Zwaluw K, Wannet W, Noordhoek GT, Mulder S, Renders N, Boers M, Zaat S, van der Riet D, Kooistra M, Talens A, Dijkshoorn L, van der Reyden T, Veenendaal D, Bakker N, Cookson B, Lynch A, Witte W, Cuny C, Blanc D, Vernez I, Hryniewicz W, Fiett J, Struelens M, Deplano A, Landegent J, Verbrugh HA, van Belkum A. 2002. Intercenter reproducibility of binary typing for Staphylococcus aureus. J. Microbiol. Methods 51:19–28 [DOI] [PubMed] [Google Scholar]
- 7. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. 2003. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J. Clin. Microbiol. 41:5113–5120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC) 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob. Agents Chemother. 53:4961–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Belkum A, Tassios PT, Dijkshoorn L, Haeggman S, Cookson B, Fry NK, Fussing V, Green J, Feil E, Gerner-Smidt P, Brisse S, Struelens M. 2007. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin. Microbiol. Infect. 13(Suppl 3):1–46 [DOI] [PubMed] [Google Scholar]
- 11. Lindstedt BA. 2005. Multiple-locus variable number tandem repeats analysis for genetic fingerprinting of pathogenic bacteria. Electrophoresis 26:2567–2582 [DOI] [PubMed] [Google Scholar]
- 12. Tenover FC, Gay EA, Frye S, Eells SJ, Healy M, McGowan JE., Jr 2009. Comparison of typing results obtained for methicillin-resistant Staphylococcus aureus isolates with the DiversiLab system and pulsed-field gel electrophoresis. J. Clin. Microbiol. 47:2452–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Willemse-Erix DF, Scholtes-Timmerman MJ, Jachtenberg JW, van Leeuwen WB, Horst-Kreft D, Bakker Schut TC, Deurenberg RH, Puppels GJ, van Belkum A, Vos MC, Maquelin K. 2009. Optical fingerprinting in bacterial epidemiology: Raman spectroscopy as a real-time typing method. J. Clin. Microbiol. 47:652–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shopsin B, Kreiswirth BN. 2001. Molecular epidemiology of methicillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 7:323–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Harrington AT, Mahlen SD, Clarridge JE., III 2010. Significantly larger numbers of methicillin-resistant Staphylococcus aureus bacteria are recovered from polymicrobial respiratory and wound sites by use of chromogenic primary media than by use of conventional culture. J. Clin. Microbiol. 48:1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Roberts MC, Soge OO, No D, Beck NK, Meschke JS. 2011. Isolation and characterization of methicillin-resistant Staphylococcus aureus from fire stations in two northwest fire districts. Am. J. Infect. Control 39:382–389 [DOI] [PubMed] [Google Scholar]
- 17. Carrico JA, Silva-Costa C, Melo-Cristino J, Pinto FR, de Lencastre H Almeida JS, Ramirez M. 2006. Illustration of a common framework for relating multiple typing methods by application to macrolide-resistant Streptococcus pyogenes. J. Clin. Microbiol. 44:2524–2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pinto FR, Melo-Cristino J, Ramirez M. 2008. A confidence interval for the wallace coefficient of concordance and its application to microbial typing methods. PLoS One 3:e3696 doi:10.1371/journal.pone.0003696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kazakova SV, Hageman JC, Matava M, Srinivasan A, Phelan L, Garfinkel B, Boo T, McAllister S, Anderson J, Jensen B, Dodson D, Lonsway D, McDougal LK, Arduino M, Fraser VJ, Killgore G, Tenover FC, Cody S, Jernigan DB. 2005. A clone of methicillin-resistant Staphylococcus aureus among professional football players. N. Engl. J. Med. 352:468–475 [DOI] [PubMed] [Google Scholar]
- 20. Berthelot P, Grattard F, Cazorla C, Passot JP, Fayard JP, Meley R, Bejuy J, Farizon F, Pozzetto B, Lucht F. 2010. Is nasal carriage of Staphylococcus aureus the main acquisition pathway for surgical-site infection in orthopaedic surgery? Eur. J. Clin. Microbiol. Infect. Dis. 29:373–382 [DOI] [PubMed] [Google Scholar]
- 21. Chen AE, Cantey JB, Carroll KC, Ross T, Speser S, Siberry GK. 2009. Discordance between Staphylococcus aureus nasal colonization and skin infections in children. Pediatr. Infect. Dis. J. 28:244–246 [DOI] [PubMed] [Google Scholar]
- 22. Frazee BW, Lynn J, Charlebois ED, Lambert L, Lowery D, Perdreau-Remington F. 2005. High prevalence of methicillin-resistant Staphylococcus aureus in emergency department skin and soft tissue infections. Ann. Emerg. Med. 45:311–320 [DOI] [PubMed] [Google Scholar]
- 23. Reighard A, Diekema D, Wibbenmeyer L, Ward M, Herwaldt L. 2009. Staphylococcus aureus nasal colonization and colonization or infection at other body sites in patients on a burn trauma unit. Infect. Control Hosp. Epidemiol. 30:721–726 [DOI] [PubMed] [Google Scholar]
- 24. Tenover FC, Tickler IA, Goering RV, Kreiswirth BN, Mediavilla JR, Persing DH. 2012. Characterization of nasal and blood culture isolates of methicillin-resistant Staphylococcus aureus from patients in United States hospitals. Antimicrob. Agents Chemother. 56:1324–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Babouee B, Frei R, Schultheiss E, Widmer AF, Goldenberger D. 2011. Comparison of the DiversiLab repetitive element PCR system with spa typing and pulsed-field gel electrophoresis for clonal characterization of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 49:1549–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Church DL, Chow BL, Lloyd T, Gregson DB. 2011. Comparison of automated repetitive-sequence-based polymerase chain reaction and spa typing versus pulsed-field gel electrophoresis for molecular typing of methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 69:30–37 [DOI] [PubMed] [Google Scholar]
- 27. Cookson BD, Robinson DA, Monk AB, Murchan S, Deplano A, de Ryck R, Struelens MJ, Scheel C, Fussing V, Salmenlinna S, Vuopio-Varkila J, Cuny C, Witte W, Tassios PT, Legakis NJ, van Leeuwen W, van Belkum A, Vindel A, Garaizar J, Haeggman S, Olsson-Liljequist B, Ransjo U, Muller-Premru M, Hryniewicz W, Rossney A, O'Connell B, Short BD, Thomas J, O'Hanlon S, Enright MC. 2007. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. J. Clin. Microbiol. 45:1830–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Deplano A, Schuermans A, Van Eldere J, Witte W, Meugnier H, Etienne J, Grundmann H, Jonas D, Noordhoek GT, Dijkstra J, van Belkum A, van Leeuwen W, Tassios PT, Legakis NJ, van der Zee A, Bergmans A, Blanc DS, Tenover FC, Cookson BC, O'Neil G, Struelens MJ, The European Study Group on Epidemiological Markers of the ESCMID. 2000. Multicenter evaluation of epidemiological typing of methicillin-resistant Staphylococcus aureus strains by repetitive-element PCR analysis. J. Clin. Microbiol. 38:3527–3533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ross TL, Merz WG, Farkosh M, Carroll KC. 2005. Comparison of an automated repetitive sequence-based PCR microbial typing system to pulsed-field gel electrophoresis for analysis of outbreaks of methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 43:5642–5647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Sullivan MV, Zhou F, Sintchenko V, Gilbert GL. 2012. Prospective genotyping of hospital-acquired methicillin-resistant Staphylococcus aureus using a novel, highly discriminatory binary typing system. J. Clin. Microbiol. 50:3513–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Luo L, Xie Y, He C, Qiao F, Zhuang H, Guo L, Yin W, Kang M, Wang L. 3 August 2012, posting date. Molecular epidemiological analysis of methicillin-resistant Staphylococcus aureus isolates from a medical intensive care unit: a comparison of nasal and clinical isolates. Am. J. Med. Sci. doi:10.1097/MAJ.0b013e31825b5443 [DOI] [PubMed] [Google Scholar]
- 32. Melles DC, Schouls L, Francois P, Herzig S, Verbrugh HA, van Belkum A, Schrenzel J. 2009. High-throughput typing of Staphylococcus aureus by amplified fragment length polymorphism (AFLP) or multi-locus variable number of tandem repeat analysis (MLVA) reveals consistent strain relatedness. Eur. J. Clin. Microbiol. Infect. Dis. 28:39–45 [DOI] [PubMed] [Google Scholar]
- 33. Udo EE, Aly NY, Sarkhoo E, Al-Sawan R, Al-Asar AS. 2011. Detection and characterization of an ST97-SCCmec-V community-associated meticillin-resistant Staphylococcus aureus clone in a neonatal intensive care unit and special care baby unit. J. Med. Microbiol. 60:600–604 [DOI] [PubMed] [Google Scholar]
- 34. Gurieva T, Bootsma MC, Bonten MJ. 2012. Successful Veterans Affairs initiative to prevent methicillin-resistant Staphylococcus aureus infections revisited. Clin. Infect. Dis. 54:1618–1620 [DOI] [PubMed] [Google Scholar]