Abstract
In recent years, Geosmithia argillacea has been increasingly reported in humans and animals and can be considered an emerging pathogen. The taxonomy of Geosmithia was recently studied, and Geosmithia argillacea and related species were transferred to the new genus Rasamsonia. The diversity among a set of Rasamsonia argillacea strains, including 28 clinical strains, was studied, and antifungal susceptibility profiles were generated. Data obtained from morphological studies and from phylogenetic analyses of internal transcribed spacer (ITS) and partial β-tubulin and calmodulin sequences revealed the presence of four species in the Rasamsonia argillacea complex, two of which are newly described here: R. piperina sp. nov. and R. aegroticola sp. nov. In contrast to other related genera, all Rasamsonia species can be identified with ITS sequences. A retrospective identification was performed on recently reported clinical isolates from animal or human patients. Susceptibility tests showed that the antifungal susceptibility profiles of the four members of the R. argillacea complex are similar, and caspofungin showed significant activity in vitro, followed by amphotericin B and posaconazole. Voriconazole was the least active of the antifungals tested. The phenotypically similar species R. brevistipitata and R. cylindrospora had different antifungal susceptibility profiles, and this indicates that correct species identification is important to help guide appropriate antifungal therapy.
INTRODUCTION
Phylogenetic analyses showed that Geosmithia is polyphyletic and forms lineages in the Hypocreales and Eurotiales (1, 2). Recently, the eurotialean Geosmithia species G. argillacea, G. cylindrospora, and G. eburnea (teleomorph, Talaromyces eburneus) were transferred to the genus Rasamsonia, and new combinations for Talaromyces emersonii and T. byssochlamydoides were made in this genus. Currently, this genus consists of seven species: R. argillacea, R. brevistipitata, R. byssochlamydoides, R. composticola, R. cylindrospora, R. eburnea, and R. emersonii (3, 4). Rasamsonia phenotypically resembles Paecilomyces, and both genera contain thermotolerant species, produce olive brown conidia, and form ascomata with no or scarce ascomatal covering, but Rasamsonia differs from Paecilomyces in having more regularly branched conidiophores with distinct rough-walled structures (3).
Rasamsonia argillacea is a thermotolerant fungus and has previously been isolated from hot environments, such as mine tips with a very high surface temperature, and from (indoor) air and clinical specimens. This species is considered to be a rare pathogen; however, R. argillacea is increasingly being reported in the literature as the causal agent of invasive mycosis. The first case of an invasive infection was reported in 2009 in a German shepherd dog (5), and more recently, two case series covering nine patients with chronic granulomatous disease (CGD) were documented (6, 7), and Valentin et al. (8) reported this species in a patient with graft-versus-host disease (GvHD). This emerging pathogen also gained attention as a chronic colonizer of airways in patients with cystic fibrosis (CF) (9, 10). Despite chronic colonization in these patients, there was no correlation between pulmonary deterioration and R. argillacea isolation, nor did the fungus seem to present a problem in patients undergoing lung transplantation. Rasamsonia argillacea might be more common than reported in the literature. A retrospective identification showed that all stored Paecilomyces variotii strains from patients with CGD were R. argillacea, and similar misidentifications also occurred for non-CGD patients (6, 11).
In this study, we have examined a set of isolates belonging to the R. argillacea species complex (R. argillacea sensu lato) originating from various substrates, with emphasis on strains obtained from clinical sources. In order to specify the taxonomy of this species, we analyzed three different loci (internal transcribed spacer [ITS] regions and partial β-tubulin and calmodulin genes) combined with macro- and micromorphological characteristics. Furthermore, antifungal susceptibility profiles were determined for these isolates, and a retrospective identification was performed for recently reported clinical isolates from animals or human patients.
MATERIALS AND METHODS
Isolates.
The strains examined are listed in Table 1. Isolates from both environmental and clinical origins were included, with emphasis on strains obtained from clinical sources. Only isolates identified as R. argillacea and R. eburnea were included in the taxonomical part of this study, because other members of the genus Rasamsonia are phylogenetically more distant and less frequently isolated from clinical specimens (3).
Table 1.
Overview of examined isolatesa
| Species | Designation(s) in other collections | Origin (isolation date) | Reference(s) |
|---|---|---|---|
| R. aegroticola | CBS 132819T = DTO 137A8 = IHEM 22641 | Respiratory secretions from CF patient D, France (August 2005) | 10 |
| R. aegroticola | DTO 049D4* = NCPF 2801 | Sputum of cystic fibrosis patient, UK (1991) | 9, 11 |
| R. aegroticola | DTO 137A9* = IHEM 22642 | Respiratory secretions from CF patient D, France (November 2007) | 10 |
| R. aegroticola | DTO 137B5* = IHEM 22928 | Respiratory secretions from CF patient F, France (October 2008) | 10 |
| R. aegroticola | DTO 137B6* = IHEM 22647 | Respiratory secretions from CF patient G, France (April 2007) | 10 |
| R. aegroticola | DTO 137B7 = IHEM 22685 | Respiratory secretions from CF patient G, France (June 2007) | 10 |
| R. aegroticola | DTO 137B8 = IHEM 22648 | Respiratory secretions from CF patient G, France (June 2007) | 10 |
| R. aegroticola | DTO 137B9* = IHEM 23431 | Respiratory secretions from CF patient G, France (November 2007) | 10 |
| R. aegroticola | DTO 137C2* = IHEM 14262 | Respiratory secretions from CF patient I, France (September 1997) | 10 |
| R. aegroticola | DTO 137C3 = IHEM 23429 | Respiratory secretions from CF patient I, France (December 1997) | 10 |
| R. argillacea | CBS 101.69T* = DTO 097E4 = IBT 31199 | Type; mine tip with a very high surface temp, Staffordshire, UK | |
| R. argillacea | CBS 102.69* = DTO 097E5 = IBT 31200 | Air, UK | |
| R. argillacea | CBS 128787* = DTO 073F3 | Heat-treated fruit concentrate, imported into the Netherlands | |
| R. argillacea | CBS 907.70* = DTO 097E7 | Unknown source, Reading, UK | |
| R. argillacea | DTO 137A2 = IHEM 22636 | Respiratory secretions from CF patient A, France (September 2007) | 10 |
| R. argillacea | DTO 137A4 = IHEM 22929 | Respiratory secretions from CF patient A, France (February 2008) | 10 |
| R. argillacea | DTO 137A5* = IHEM 22894 | Respiratory secretions from CF patient A, France (September 2008) | 10 |
| R. argillacea | DTO 137A6* = IHEM 22640 | Respiratory secretions from CF patient B, France (December 2006); ID based on ITS sequence deposited in GenBank | 10 |
| R. argillacea | DTO 137C1 = IHEM 22033 | Respiratory secretions from CF patient H, France (February 2009) | 10 |
| R. brevistipitata | CBS 128785T** = DTO 25H2 = IBT 31187 | Indoor environment of school, from cork with bitumen, Germany | |
| R. brevistipitata | CBS 128786** = DTO 26B1 = IBT 31187 | Indoor environment of school, from cork with bitumen, Germany | |
| R. cylindrospora | CBS 275.58NT** = DTO 138F8 = IBT 31202 | Culture contaminant, Berkshire, England, UK | |
| R. cylindrospora | CBS 432.62** = DTO 138F7 = IBT 31201 | Human, sputum, the Netherlands | |
| R. eburnea | CBS 100538T* = DTO 105D6 = IBT 17519 | Type; soil, Taipei, Taiwan | |
| R. eburnea | CBS 102881* = DTO 097E9 = IBT 31195 = UAMH 9714 | Human, bronchial wash specimen (apparently etiologic), Toronto, Ontario, Canada | |
| R. eburnea | CBS 124445* = DTO 049D7 = IBT 31193 = NCPF 7594 | Blood culture, patient with peritonitis, UK; same patient as CBS 124446 (2002) | 9, 11 |
| R. eburnea | CBS 124446* = DTO 049D9 = IBT 31192 = NCPF 7596 | Peritoneal dialysis fluid, UK; same patient as CBS 124445 (2002) | 9, 11 |
| R. eburnea | CBS 124447* = DTO 045I3 = IBT 31191 | Contaminant of blood culture (pseudo-outbreak), UK | 11 |
| R. piperina | CBS 104.69* = DTO 097E6 | Wood chips of Picea abies and Pinus sylvestris, Sweden | |
| R. piperina | CBS 105.69* = DTO 138G1 | Wood chips of Picea abies and Pinus sylvestris, Sweden | |
| R. piperina | CBS 106.69* = DTO 138G2 | Wood chips of Picea abies and Pinus sylvestris, Sweden | |
| R. piperina | CBS 128034* = DTO 138F5 = UAMH 10933 | Necropsy thoracic vertebra, German shepherd dog, USA | 5 |
| R. piperina | CBS 187.90* = DTO 138F9 | Air, Stuttgart, Germany | |
| R. piperina | CBS 406.73* = DTO 076F1 = IJFM 1073 | Seed of Piper nigrum, Spain | |
| R. piperina | CBS 407.73* = DTO 097E8 = IJFM 1405 | Seed of Piper nigrum, Spain | |
| R. piperina | CBS 408.73T* = DTO 138G3 = IJFM 1326 | Seed of Piper nigrum, Spain | |
| R. piperina | DTO 137A7* = IHEM 16291 | Respiratory secretions from CF patient C, France (September 1999) | 10 |
| R. piperina | DTO 137B1 = IHEM 22643 | Respiratory secretions from CF patient E, France (August 2007) | 10 |
| R. piperina | DTO 137B2 = IHEM 22644 | Respiratory secretions from CF patient E, France (October 2007) | 10 |
| R. piperina | DTO 137B3* = IHEM 22645 | Respiratory secretions from CF patient E, France (October 2007) | 10 |
| R. piperina | DTO 137B4 = IHEM 23433 | Respiratory secretions from CF patient E, France (February 2008) | 10 |
| R. piperina | DTO 138F6* = UAMH 10933 | Necropsy of thoracic vertebra, German shepherd dog, USA | 5 |
| R. piperina | DTO 139A8* = UAMH 10932 | Urine sample of German shepherd dog, USA | 5 |
Strains indicated with an asterisk were included in the susceptibility tests; strains labeled with two asterisks were included only in the susceptibility tests and not in the phylogenetic analysis. CBS, culture collection of the CBS-Fungal Biodiversity Centre, Utrecht, the Netherlands; IHEM, culture collection of the Scientific Institute of Public Health, Mycology Section, Brussels, Belgium; DTO, internal culture collection of the CBS-Fungal Biodiversity Centre; IBT, culture collection of Center for Microbial Biotechnology (CMB) at the Department of Systems Biology, Technical University of Denmark; UAMH, University of Alberta Microfungus Collection and Herbarium, Alberta, Canada; NCPF, National Collection of Pathogenic Fungi, Mycology Reference Laboratory, Bristol, United Kingdom.
Phenotypic examination and extrolite analysis.
Macroscopic characteristics were studied by using the following agar media: Czapek yeast extract agar (CYA), yeast extract sucrose agar (YES), creatine sucrose agar (CREA), dichloran–18% glycerol agar (DG18), oatmeal agar (OA), and malt extract agar (MEA) (Oxoid). The isolates were inoculated at three points on 90-mm petri dishes and incubated for 7 days at 37°C in darkness. All media were prepared as described previously by Samson et al. (12). Fungal material was examined by light microscopy (Olympus BH2 and Zeiss Axioskop 2 Plus). Microscopic mounts were made in lactic acid from MEA or OA, and a drop of alcohol was added to remove air bubbles and excess conidia. Extrolite analysis was performed according to methods described previously by Houbraken et al. (3).
DNA extraction, PCR, and DNA sequencing.
Isolates were grown on MEA for 7 to 14 days at 37°C prior to DNA extraction. Genomic DNA was extracted by using the Ultraclean microbial DNA isolation kit (MoBio), according to the manufacturer's instructions. The extracted DNA was stored at −20°C until use. The ITS regions (including 5.8S rRNA genes) and part of the β-tubulin and calmodulin genes were amplified and sequenced. An overview of primers and PCR conditions for the amplification of these loci was reported previously by Houbraken et al. (3). The obtained sequences were aligned by using the Muscle program in MEGA5 (13, 14). The individual data sets were analyzed by maximum likelihood (ML) analysis using MEGA5. One thousand rapid bootstrap inferences were executed, and thereafter, a thorough ML search was performed. The combined data set was analyzed as two distinct data partitions by using RAxML (Randomized Axelerated Maximum Likelihood) software (15). Rasamsonia emersonii CBS 396.64 was used as an outgroup.
Antifungal susceptibility testing.
Antifungal susceptibility testing was performed by the broth microdilution reference method of the Clinical and Laboratory Standards Institute (CLSI), as described in document M38-A2 (16). Briefly, isolates were inoculated onto Sabouraud dextrose agar at 35°C. After 48 h of incubation, five colonies greater than 1 mm in diameter were selected, suspended in a 0.85% saline solution, and adjusted to a final concentration of 0.5 × 103 to 2.5 × 103 conidia/ml in RPMI 1640 medium (equivalent to a 0.5 McFarland standard) buffered at pH 7.0 with 0.165 M morpholinepropanesulfonic acid (MOPS; Sigma). Five antifungal drugs were tested: amphotericin B (AMB; Sigma Chemical Co., St. Louis, MO), itraconazole (ITZ; Janssen Research Foundation, Beerse, Belgium), voriconazole (VCZ; Pfizer Central Research, Sandwich, United Kingdom), posaconazole (PCZ; Schering-Plough, Kenilworth, NJ), and caspofungin (CAS; MSD, Haarlem, the Netherlands). All antifungals were dissolved in dimethyl sulfoxide, except CAS, which was solubilized in sterile distilled water. The drugs were prepared at the following concentrations: 1,250 μg/ml for FCZ and 640 μl/ml for the others. The solutions were diluted in RPMI medium, and final drug concentrations ranged from 16 to 0.03 μg/ml for ITZ, VCZ, PCZ, and AMB and from 8 to 0.015 μg/ml for CAS. After 48 h of incubation at 35°C, MICs were determined visually by comparison with the drug-free growth control. For AMB, ITZ, PCZ, and VCZ, MIC values were defined as the lower drug concentration which resulted in a reduction in the turbidity of 100%. For CAS, minimum effective concentration (MEC) values corresponding to the lowest concentration of CAS causing abnormal hyphal growth with short abundant branching were determined by microscopic examination of the microdilution plates after 48 h of incubation (16). For those isolates that were not inhibited by the highest drug concentration, the next-highest concentration was used to calculate the geometric mean (GM) MIC. The susceptibility profile of a selected set of isolates was determined, and these isolates are marked in Table 1 with one or two asterisks.
Nucleotide sequence accession numbers.
Newly obtained ITS, β-tubulin, and calmodulin sequences were deposited in the GenBank nucleotide sequence database under accession numbers JX272931 to JX273026.
RESULTS
Phenotypic characterization.
Based on phenotypic characteristics, four species could be distinguished in the Rasamsonia argillacea species complex: R. argillacea, R. eburnea, and two newly described species. We propose the names R. piperina sp. nov. and R. aegroticola sp. nov. for the newly described species. The results of this phenotypic comparison are listed in Table 2. Rasamsonia argillacea, R. eburnea, R. piperina, and R. aegroticola share various features typical of the genus Rasamsonia, such as good growth at 40°C; the production of olive brown conidia on MEA, CYA, OA, and/or YES; distinct rough-walled conidiophores, metulae, and phialides; and phialides consisting of a narrow cylindrical base, tapering more or less abruptly to a narrowed conidium-bearing tube. Several characters were useful for species recognition. Among the studied strains, R. piperina isolates appeared to grow more restricted, were less dense, and sporulated less abundantly on CYA and YES incubated at 37°C than other members of the R. argillacea species complex. Furthermore, R. eburnea isolates were unique in having a dark brown or blackish brown reverse on MEA, while the reverse of other species were at most grayish brown. The formation of a dark brown reverse is shared with R. cylindrospora, a species outside the R. argillacea complex. The shape and size of the conidia varied within and among strains of the same species. However, there were differences observed: the majority of the conidia produced by R. argillacea were cylindrical, with a length-to-width ratio above 1.8 (average, 2.0), while the conidia of the other three species were (broadly) ellipsoidal or cylindrical, with a length-to-width ratio that was always below 1.7 (average, 1.4).
Table 2.
Differential characteristics of Rasamsonia speciesa
| Species | Colony diam (mm), growth on CYA (7 days, 37°C) | Shape and size of conidia | Length/width ratio of conidia | Reverse color on MEA |
|---|---|---|---|---|
| R. aegroticola | (20–) 25–40, good growth | Variable; predominantly cylindrical or ellipsoid, 2.5–3.5 × 1.8–2.5 μm | 1.3–1.6:1 | Grayish brown |
| R. argillacea | 30–40, good growth | Cylindrical or ovoid, (3–) 3.5–4.5 (–5.0) × 1.5–2.0 (–2.3) μm | 1.8–2.3:1 | (Light) brown |
| R. brevistipitata | 11–17, good growth | Ellipsoidal or ovoid, (2.0–) 2.5–3.0 (–3.5) × 1.7–2.1 μm | 1.3–1.5:1 | (Light) brown |
| R. cylindrospora | 5–10, good growth | Cylindrical, 4.0–5.0 × 1.6–2.1 μm | 2.1–2.5:1 | Blackish brown |
| R. eburnea | 30–40, good growth | Variable, predominantly ellipsoidal or ovoid but also cylindrical, 2.5–3.5 (–4) × 1.8–2.5 μm | 1.1–1.4:1 | Dark brown or blackish brown |
| R. piperina | CYA (10–)15–25, moderate growth | Ellipsoidal or cylindrical, 2.0–3.5 × 1.7–2.5 μm | 1.3–1.7:1 | Brown or grayish brown |
The phenotypically similar species R. cylindrospora and R. brevistipitata are included, while the ascoma- and ascospore-producing species R. byssochlamydoides, R. composticola, and R. emersonii are excluded.
Phylogenetic analysis.
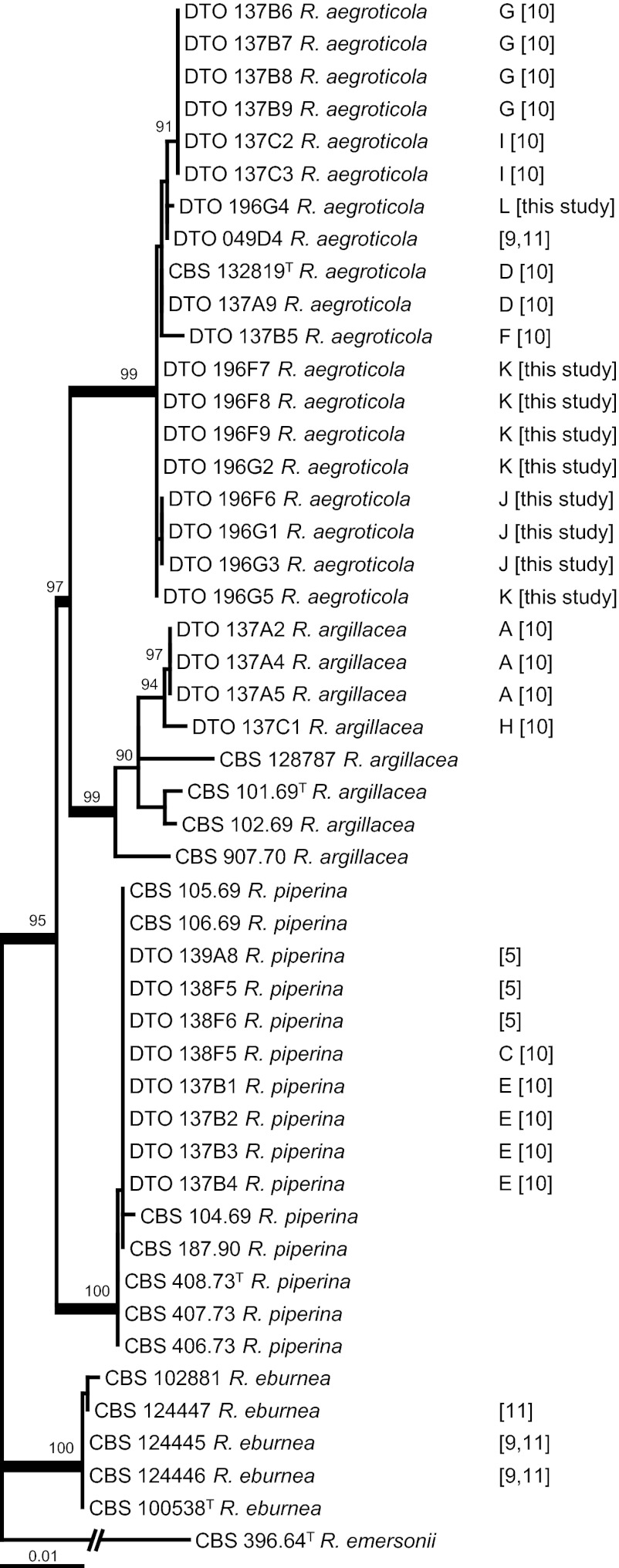
The total lengths of the individual calmodulin, ITS, and β-tubulin data sets were 525, 680, and 481 bp, respectively, resulting in a total length of the combined data set of 1,686 bp. The combined data set had 176 distinct alignment patterns (64 for calmodulin, 71 for ITS, and 41 for β-tubulin), and the proportion of gaps and completely undetermined characters in this alignment was 5.64%. The result of the phylogenetic analysis is shown Fig. 1, and the phylograms of the individual loci are presented in Fig. S1 in the supplemental material. Four highly supported clades were observed for the R. argillacea complex. One clade was centered on the type strain of R. argillacea, CBS 101.69T, and another clade was centered on the type strain of R. eburnea, CBS 100538T. The other two clades were centered on the two newly described species, R. piperina and R. aegroticola. A high level of sequence variation was detected in R. argillacea, and this may imply that this species comprises other cryptic species. More strains need to be analyzed to test this hypothesis. The phylogram based on the individual ITS data set was poorly supported, and two well-supported clades could be recognized (R. aegroticola and R. eburnea). Three well-supported clades could be recognized in the β-tubulin phylogram (all except R. argillacea), and the analysis of the calmodulin data set resulted in four well-supported clades. The deeper nodes were also best supported in the calmodulin phylogram; however, the result of this analysis was contradictory to that of the combined analysis. Rasamsonia aegroticola and R. piperina are sister species on a well-supported branch in the calmodulin data set (96% bootstrap support), while R. argillacea and R. aegroticola were related in the combined analysis (97% bootstrap support). Although each data set has a different phylogenetic signal, all three studied genes exhibited sufficient interspecific variation for identification purposes.
Fig 1.
Best-scoring maximum likelihood tree using RAxML based on a combined data set of partial β-tubulin, calmodulin, and ITS sequences and showing the close relationship of Rasamsonia aegroticola, R. argillacea, and R. piperina (95% bootstrap support). The numbers in brackets to the right of the species names are reference numbers, and the letters refer to the different cases. The sequences obtained from clinical isolates in this study were all obtained from CF patients. Well-supported branches (>95% bootstrap support) are in boldface type, and values with less than 70% bootstrap support are not shown. Rasamsonia emersonii CBS 396.64 was used as an outgroup.
Antifungal susceptibility testing.
Susceptibility data, i.e., the geometric mean (GM) of the MIC and the range, are shown in Table 3. Due to the limited number of isolates (2 to 11 strains per species), MIC50 and MIC90 values could not be obtained. The phenotypically similar species R. cylindrospora and R. brevistipitata were included in the susceptibility test. The susceptibility profiles of most R. argillacea, R. eburnea, R. aegroticola, and R. piperina isolates were very similar and differed from those of R. cylindrospora and R. brevistipitata. Little intraspecific variation in antifungal susceptibility was noted. Caspofungin showed the lowest MICs, as all isolates were inhibited by a concentration of 0.5 mg/liter. Amphotericin B also showed significant activity in vitro against the clinical isolates, followed by posaconazole. Voriconazole was not active, and with exception of the two tested R. brevistipitata isolates, the majority of the strains were not inhibited at a concentration of 16 mg/liter. With the exception of R. argillacea, all species had similar MICs of itraconazole (0.06 to 2 mg/liter). In R. argillacea, three strains were insensitive to itraconazole (16 to 32 mg/liter), while three other strains were inhibited by a concentration of 1 mg/liter.
Table 3.
Susceptibility results of Rasamsonia species, including GM and MIC distributions by species and antifungal agenta
| Species | No. of isolates | GM MIC (mg/liter) (range) | ||||
|---|---|---|---|---|---|---|
| AMB | ITZ | PCZ | CAS | VCZ | ||
| R. aegroticola | 5 | 2.00 (0.5–2) | 1.32 (1–2) | 1.74 (1–4) | 0.19 (0.06–0.5) | >16 |
| R. argillacea | 6 | 2.00 (0.125–2) | 5.04 (1–32) | 3.17 (1–4) | 0.28 (0.125–0.5) | >16 |
| R. brevistipitata | 2 | 0.09 (0.03–0.25) | 0.12 (0.06–0.25) | 0.06 (0.03–0.12) | 0.35 (0.25–0.5) | 0.12 (0.06–0.25) |
| R. cylindrospora | 2 | 1.41 (1–2) | 1.41 (1–2) | 2.83 (1–8) | 0.13 | >16 |
| R. eburnea | 4 | 2.00 (1–4) | 2.38 (1–4) | 2.00 (1–4) | 0.35 (0.125–0.5) | >16 |
| R. piperina | 11 | 1.22 (0.25–2) | 1.00 (0.5–1) | 1.41 (0.06–2) | 0.25 (0.03–0.5) | 19.33 (8–>16) |
The phenotypically similar species R. cylindrospora and R. brevistipitata are included in this analysis. Given are the geometric mean MICs, and the range in MICs, if observed, is given in parentheses. AMB, amphotericin B; ITZ, itraconazole; PCZ, posaconazole; CAS, caspofungin; VCZ, voriconazole.
TAXONOMY
Rasamsonia aegroticola Houbraken, Giraud, and Samson sp. nov.
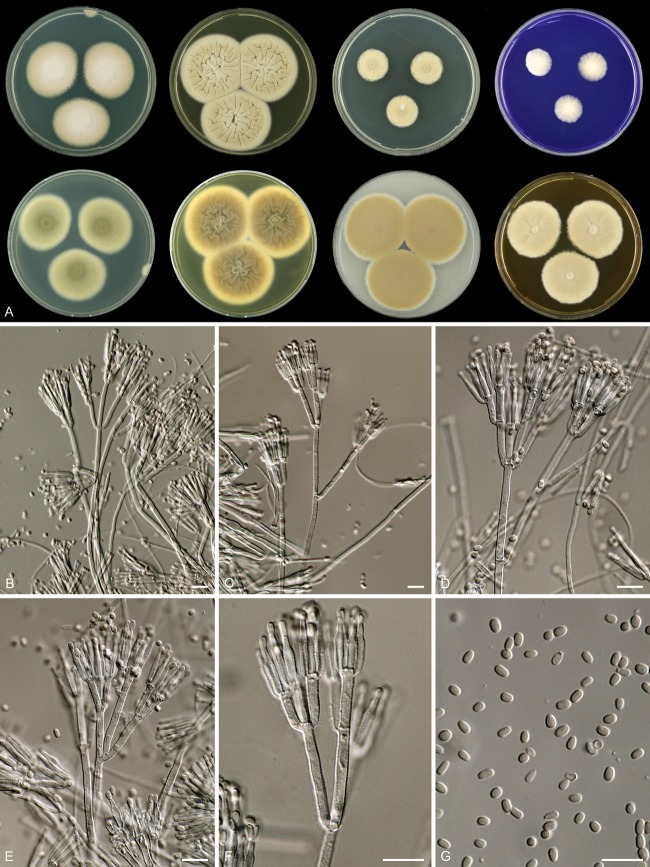
Mycobank accession number MB801150 (Fig. 2).
Fig 2.
Rasamsonia aegroticola DTO 196-G4. (A) Seven-day-old cultures at 37°C. From left to right, first row, all obverse, CYA, YES, DG18, and CREA; second row, CYA reverse, YES reverse, OA obverse, and MEA obverse. (B to F) Conidiophores. (G) Conidia. Scale bars = 10 μm.
(i) Etymology.
This species is isolated mainly from CF patients, and therefore, the name aegroticola is chosen, derived from aegrotus and meaning patient in Latin.
(ii) Typus.
France: isolated from respiratory secretions from a CF patient, August 2005, J. P. Bouchara (holotype CBS H-21031; culture ex-type CBS 132819 = DTO 137-A8 = IHEM 22641).
(iii) Diagnosis.
Rasamsonia aegroticola isolates are thermotolerant with good growth on CYA ([20–] 25 to 40 mm) and YES (35 to 55 mm) at 37°C; reverse on MEA is grayish brown; length/width ratio of conidia is 1.3:1 to 1.5:1.
(iv) Description.
Colony diameters at 7 days at 37°C are (20–) 25 to 40 mm on CYA, 25 to 45 mm on MEA, 35 to 55 mm on YES, 15 to 25 mm on DG18, (30–) 35 to 50 mm on OA, and (10–) 15 to 25 mm on creatine sucrose agar, with poor to weak growth, moderate to good sporulation, and no acid and no base production.
Colonies on CYA at 37°C are spreading, raised at center, and weakly wrinkled or radiate sulcate; margins are small (0 to 2 mm), entire; mycelium is white; conidiogenesis is variable (absent to dense); conidia are pale olive brown to olive brown en masse; texture is velvety; exudate and soluble pigments are absent; reverse is greenish brown, often with a shade of orange. Conidiogenesis is moderately dense on YES at 37°C; conidia are pale olive brown en masse; mycelium is white; exudate and soluble pigment are absent; reverse is dark brown in center with an orange or orange-brown edge. Colonies on MEA are raised in the center, entire; texture is velvety, occasionally with white aerial mycelium in center; exudate and soluble pigment are absent; conidia are olive brown en masse; reverse is brown to dark brown with a shade of gray. Colonies are restricted on DG18, entire; conidia are olive brown, occasionally with pale olive brown sectors; reverse is dark brown or brown in center with yellow-brown edge. Ehrlich reaction is negative.
Sclerotia and ascomata are absent. Conidiophores are biverticillate and appressed, often with one or two subterminal branches present at one stage, resulting in “double” or “triple” biverticillate conidiophores. Stipes are smooth walled when young, coarsely roughened in older parts of the colony, and 75 to 250 by 2.5 to 3.5 μm. Branches are 18 to 30 μm. Metulae are in terminal whorls of 2 to 5, somewhat appressed, nonvesiculate, roughened in older parts, and 11 to 15 by 2.0 to 3.0 μm. Phialides are roughened, consisting of a narrow cylindrical base, tapering more or less abruptly to a narrowed conidium-bearing tube, typically occurring in dense clusters of 4 to 12, but are smooth walled in reduced conidiophores and 11 to 15 by 2.0 to 2.5 μm; conidia are smooth walled, variable, mostly cylindrical or ellipsoid, with a small portion ovoid, and 2.5 to 3.5 by 1.8 to 2.5 μm; the mean length/width ratio is between 1.3:1 and 1.6:1.
(v) Extrolites.
There are several uncharacterized extrolites from different chromophore families, and griseofulvin is detected in 3 of 6 examined strains (CBS 132819, DTO 137-B6, and DTO 137-C2).
(vi) Distribution and ecology.
Distribution is in France, the Netherlands, the United Kingdom, and the United States. The species has been isolated from respiratory secretions of CF patients and patients with CGD.
Rasamsonia piperina Houbraken, Giraud, and Samson sp. nov.
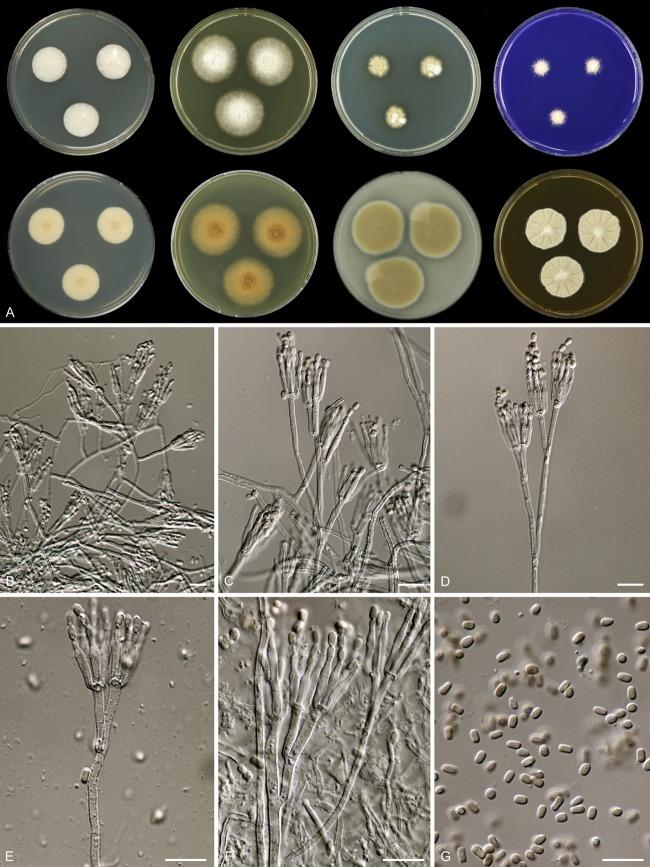
Mycobank accession number MB801151 (Fig. 3).
Fig 3.
Rasamsonia piperina CBS 408.73T. (A) Seven-day-old cultures at 37°C. From left to right, first row, all obverse, CYA, YES, DG18, and CREA; second row, CYA reverse, YES reverse, OA obverse, and MEA obverse. (B to F) Conidiophores. (G) Conidia. Scale bars = 10 μm.
(i) Etymology.
Etymology refers to pepper, the substrate from which the type strain was isolated.
(ii) Typus.
Spain: seed of Piper nigrum, C. Ramírez (holotype CBS H-21030; culture ex-type CBS 408.73 = DTO 138G3 = IJFM 1326).
(iii) Diagnosis.
Rasamsonia piperina isolates are thermotolerant, with moderate growth on CYA ([10–] 15 to 25 mm) and YES ([10–] 20 to 30 mm) at 37°C; reverse on MEA is brown or grayish brown; the length/width ratio of conidia is 1.3:1 to 1.7:1.
(iv) Description.
Colony diameters at 7 days at 37°C are (10–) 15 to 25 mm on CYA, 20 to 35 mm on MEA, (10–) 20 to 30 mm on YES, 5 to 20 mm on DG18, (10–) 20 to 45 mm on OA, and 10 to 25 mm on creatine sucrose agar, with weak to moderate growth, absent or weak sporulation, and no acid and no base production.
Colonies on CYA at 37°C are restricted, with moderate growth; margins are entire, and sulcations are absent; mycelium is white; conidiogenesis is absent; exudate and soluble pigments are absent; reverse is unaffected, in some strains with a pale brown center. Conidiogenesis is absent on YES at 37°C; growth is moderate to good; mycelium is white or light crème; exudate and soluble pigment are absent; reverse is unaffected or pale brown with a brown center. Colonies on MEA are raised in the center, entire; mycelium is white; texture is velvety or floccose, occasionally in the center with funicules similarly to synnemata; exudate and soluble pigment are absent; conidia are pale olive brown or olive brown en masse; reverse is brown to grayish brown. Colonies are restricted on DG18, entire; mycelium is white; conidia, if present, are olive brown; reverse is variable, pale, brown, or pale with a dark brown center. Ehrlich reaction is negative.
Sclerotia and ascomata are absent. Conidiophores are biverticillate and appressed, with one subterminal branch in older structures and occasionally a branch further down the stipe. Stipes are coarsely roughened, often 50 to 150 by 2.0 to 3.0 μm. Branches are 20 to 30 μm. Metulae are in terminal whorls of 2 to 4, sometimes slightly vesiculate, roughened, and 11 to 17 by 2.0 to 2.5 μm. Phialides are roughened, consisting of a narrow cylindrical base, tapering more or less abruptly to a narrowed conidium-bearing tube, 4 to 10, and 10 to 14 by 1.5 to 2.5 μm. Conidia are smooth walled, variable in shape and size, ellipsoidal or cylindrical, and 2.0 to 3.5 by 1.7 to 2.5 μm; the mean length/width ratio is between 1.3:1 and 1.7:1.
(v) Extrolites.
There are several uncharacterized extrolites from different chromophore families.
(vi) Distribution and ecology.
Distribution is in Austria, France, Germany, Spain, Sweden, and the United States. The species was isolated from respiratory secretions from a CF patient and a patient with GvHD and from a German shepherd dog, wood chips of Picea abies and Pinus sylvestris, air, and seed of Piper nigrum.
DISCUSSION
Retrospective analysis.
The occurrence of R. argillacea in nine different CF patients in France was studied previously by Giraud et al. (10), and isolates obtained from successive clinical samples from the same patient were identified by using ITS sequencing. In this study, a large set of these isolates was subjected to a multigene sequence typing analysis. Our data indicate that the isolates obtained from the same patient at different time points were identical (Fig. 1), more accurately confirming the suggestion that species belonging to the R. argillacea complex chronically colonize the airways of CF patients (10). Reidentification of these isolates shows that R. argillacea was the causal agent in three of nine cases, R. aegroticola was the causal agent in four cases, and R. piperina was the causal agent in two cases. Similar results were obtained in a multigene sequencing analysis of R. aegroticola strains from Dutch CF patients: the same R. aegroticola genotype persisted in the same patient for more than a year (Dutch CF Consortium, unpublished data). This is in agreement with data on Aspergillus fumigatus, where a single genotype predominated when chronic colonization of the CF lung was established (17–20). Giraud et al. (10) previously reported that the colonization of the airways by R. argillacea always succeeded infections by various bacteria and that all but one of the patients colonized by R. argillacea were also chronically colonized by Staphylococcus aureus. However, there is no clear correlation between the presence of R. argillacea sensu lato in CF lungs and pulmonary deterioration, although this species could play an as-yet-unrecognized role. The fact that this fungus may cause invasive infections in dogs as well as in patients with CGD or GvHD suggests that one may be cautious with patients colonized by this fungus if lung transplantation is undertaken. Moreover, R. argillacea sensu lato is not a commensal of the airways, and even in the absence of clinical signs, this mold (like other molds encountered in the CF airways, including Aspergillus fumigatus) might contribute, through the release of polysaccharides or the production of secreted proteins, to the inflammatory reaction which progressively leads to the deterioration of lung function. Another role of R. argillacea in pulmonary deterioration could be that this species forms a biofilm consortium with bacteria. Gaining insight in the microbial community in the CF lung in the future might lead to the discovery of polymicrobial effects (21). Also, the CFTR (cystic fibrosis transmembrane conductance regulator) genotype affects the microbial community in the CF lung, and R. argillacea sensu lato seems to be associated with the F508del mutation, with all cases reported until now having been diagnosed in patients homozygous (64.7% of the patients) or heterozygous (35.3%) for this mutation (9, 10). In the near future, analysis of sputum samples by culture-independent detection methods (e.g., high-throughput sequencing) will give more insight into the microbiome of the CF lung and consequently in the understanding of this disease (17).
Previous studies by Barton et al. (9) and Houbraken et al. (11) used mainly the same R. argillacea strains. Strains NCPF 7594 (CBS 124445) and NCPF 7596 (CBS 124446) were isolated from blood cultures and peritoneal dialysis fluid from a patient with peritonitis (United Kingdom) and are reidentified here as R. eburnea (Fig. 1). An isolate originating from a blood culture (CBS 124447) (pseudo-outbreak in United Kingdom) is also reidentified as R. eburnea, and a strain collected from the sputum of a CF patient (NCPF 2801 [DTO 049-D4]) proved to be R. aegroticola. Grant et al. (5) previously isolated R. argillacea from systemic mycosis in a German shepherd dog (thoracic vertebrae and urine). Our multigene analysis shows that these isolates are identical, and both isolates are reidentified as R. piperina. Interestingly, the FRR culture collection (CSIRO Food Research, North Ryde, New South Wales, Australia; FRR 4238) also harbors an R. argillacea sensu lato strain originating from a bone biopsy specimen of a German shepherd dog (isolated in 1992); however, no additional data could be found about this case.
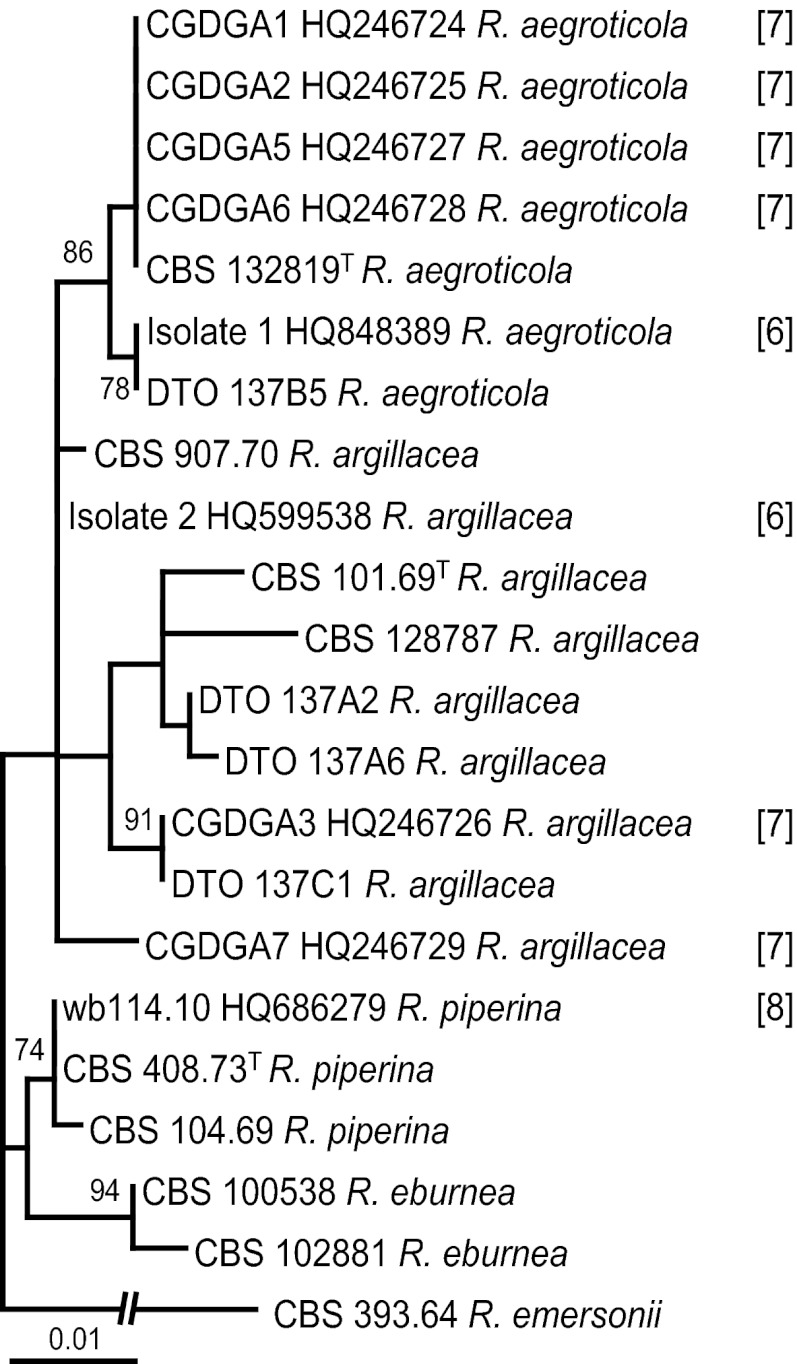
Machouart et al. (7) and De Ravin et al. (6) isolated R. argillacea strains from two and seven different CGD patients, respectively. These strains were not available for examination, and therefore, the ITS sequences deposited in GenBank were used for identification purposes. A comparison of the two sequences reported in the study by Machouart et al. (7) shows that isolate 1 (GenBank accession number HQ848389) belongs to R. aegroticola and that isolate 2 (GenBank accession number HQ599538) is closely related to R. argillacea (Fig. 4). Applying the newly proposed taxonomy to the seven cases described previously by De Ravin et al. (6), we show that four strains (CGDGA1, CGDGA2, CGDGA5, and CGDGA6 [cases 1, 2, 5, and 6, respectively]) could be assigned to R. aegroticola. Similar to the data shown in Fig. 1, large sequence variation was also observed for the R. argillacea strains included in Fig. 4. Strain CGDGA3 (case 3) is closely related to CBS 907.70, a strain identified as R. argillacea in the taxonomical part of this study. The position of strain CGDGA7 (case 7) is uncertain and could represent a new species. More data are needed to determine its relationship with other members of this complex. No sequences were deposited in GenBank for case 4. Valentin et al. (8) reported the detection of G. argillacea in a patient with graft-versus-host disease (GvHD), and an evaluation of the ITS and β-tubulin sequences deposited in GenBank (accession numbers HQ686279 and HQ686280) showed that this strain belongs to R. piperina.
Fig 4.
Best-scoring maximum likelihood tree based on ITS sequences, including type strains, unique R. argillacea sensu lato sequences generated in this study, and sequences deposited in GenBank which were described in previous studies by Machouart et al. (7), De Ravin et al. (6), and Valentin et al. (8).
Distribution and identification.
Species belonging to the R. argillacea complex have a worldwide distribution, are thermophilic, and are associated with hot environments. In recent years, R. argillacea was frequently reported in humans and animals and can be considered an emerging pathogen (5–11). However, a search of collection databases showed that strains belonging to this species complex were isolated from humans and animals earlier; e.g., a strain identified as G. argillacea was isolated from a bone biopsy specimen of a German shepherd dog in 1992 (FRR 4238), and strain IHEM 16291 (DTO 137A7) was isolated from respiratory secretions of a CF patient in 1999. Although there is a bias in our study toward strains originating from respiratory secretions of CF patients from Europe, the occurrence of R. argillacea sensu lato in bronchial wash specimens is more widespread. Four strains originating from bronchial secretions are present in the collection of the University of Alberta (UAMH) and were isolated from patients of different areas of Canada (UAMH 8639 in British Columbia, UAMH 10232 in Ontario, UAMH 9854 in Quebec, and UAMH 7717 in Vancouver), indicating a wider distribution than in Europe. The limited reports of detection of this species in clinical samples might also be due to its resemblance to Paecilomyces variotii, and misidentifications have been reported (6, 11). In the last decade, new insights have determined that certain well-known species appear to be species complexes (22–24). In line with those taxonomic studies, we show that Rasamsonia argillacea is a species complex consisting of at least four species. Correct identification by routine laboratories based solely on phenotypic characteristics is therefore becoming increasingly difficult. Nowadays, molecular-based techniques, especially DNA sequencing, are frequently used for identification. Recently, the ITS region was accepted as the prime fungal barcode (25). In contrast to other related genera, such as Aspergillus and Penicillium (26–28), all Rasamsonia species can be identified with ITS sequences. It has been suggested that clinical laboratories refer to a complex rather than a species in their report when no molecular-based identification is undertaken (29, 30). Results reported in this manner should be interpreted as an indication of the section/group of species to which a strain belongs.
Species identification and susceptibility data.
The results obtained were in agreement with data reported previously. Caspofungin was the most active agent in vitro against R. argillacea sensu lato and the related species R. cylindrospora and R. brevistipitata. With the exception of R. brevistipitata, all strains had high MICs for voriconazole (5–7, 9, 11). The in vitro antifungal susceptibility profiles were similar among most of the investigated strains of the R. argillacea species complex. The main exception is the susceptibility profile of R. argillacea strains against itraconazole. Three of the six strains had MICs above 16 mg/liter. This was also observed for two isolates (case 3 and IHEM 22640) in previous studies by De Ravin et al. (6) and Giraud et al. (10). Similar to our results, these strains also belong to the newly defined R. argillacea complex. Figure 1 shows a high level of sequence variation within R. argillacea, and the variation in the susceptibility of itraconazole could also be due to the presence of cryptic species in the R. argillacea complex.
ACKNOWLEDGMENTS
J.F.M. acknowledges the Dutch Cystic Fibrosis Fungal Bank for support and access to unpublished data. S.G. thanks Loïc Favennec and the Pasteur Institute, and J.H. is grateful to Uwe Braun for suggestions on the new species names.
Footnotes
Published ahead of print 17 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02147-12.
REFERENCES
- 1. Ogawa H, Yoshimura A, Sugiyama J. 1997. Polyphyletic origins of species of the anamorphic genus Geosmithia and the relationships of the cleistothecial genera: evidence from 18S, 5S and 28S rDNA sequence analyses. Mycologia 89:756–771 [Google Scholar]
- 2. Ogawa H, Sugiyama J. 2000. Evolutionary relationships of the cleistothecial genera with Penicillium, Geosmithia, Merimbla and Sarophorum anamorphs as inferred from 18S rDNA sequence divergence, p 149–161 In Samson RA, Pitt JI. (ed), Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Plenum Press, New York, NY [Google Scholar]
- 3. Houbraken J, Spierenburg H, Frisvad JC. 2012. Rasamsonia, a new genus comprising thermotolerant and thermophilic Talaromyces and Geosmithia species. Antonie Van Leeuwenhoek 101:403–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Su Y-Y, Cai L. 25 May 2012. Rasamsonia composticola, a new thermophilic species isolated from compost in Yunnan, China. Mycol. Prog. doi:10.1007/s11557-012-0827-9 [Google Scholar]
- 5. Grant DC, Sutton DA, Sandberg CA, Tyler RD, Jr, Thompson EH, Romanelli AM, Wickes BL. 2009. Disseminated Geosmithia argillacea infection in a German shepherd dog. Med. Mycol. 47:221–226 [DOI] [PubMed] [Google Scholar]
- 6. De Ravin SS, Challipalli M, Anderson V, Shea YR, Marciano B, Hilligoss D, Marquesen M, Decastro R, Liu YC, Sutton DA, Wickes BL, Kammeyer PL, Sigler L, Sullivan K, Kang EM, Malech HL, Holland SM, Zelazny AM. 2011. Geosmithia argillacea: an emerging cause of invasive mycosis in human chronic granulomatous disease. Clin. Infect. Dis. 52:e136–e143 doi:10.1093/cid/ciq250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Machouart M, Garcia-Hermoso D, Rivier A, Hassouni N, Catherinot E, Salmon A, Debourgogne A, Coignard H, Lecuit M, Bougnoux M-E, Blanche S, Lortholary O. 2011. Emergence of disseminated infections due to Geosmithia argillacea in patients with chronic granulomatous disease receiving long-term azole antifungal prophylaxis. J. Clin. Microbiol. 49:1681–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Valentin T, Neumeister P, Pichler M, Rohn A, Koidl C, Haas D, Heiling B, Asslaber M, Zollner-Schwetz I, Hoenigl M, Salzer HJ, Krause R, Buzina W. 2012. Disseminated Geosmithia argillacea infection in a patient with gastrointestinal GvHD. Bone Marrow Transplant. 47:734–736 [DOI] [PubMed] [Google Scholar]
- 9. Barton RC, Borman AM, Johnson EM, Houbraken J, Hobson RP, Denton M, Conway SP, Brownlee KG, Peckham D, Lee TW. 2010. Isolation of the fungus Geosmithia argillacea in sputum of people with cystic fibrosis. J. Clin. Microbiol. 48:2615–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giraud S, Pihet M, Razafimandimby B, Carrère J, Degand N, Mely L, Favennec L, Dannaoui E, Bouchara JP, Calenda A. 2010. Geosmithia argillacea: an emerging pathogen in cystic fibrosis patients? J. Clin. Microbiol. 48:2381–2386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Houbraken J, Verweij PE, Rijs AJ, Borman AM, Samson RA. 2010. Identification of Paecilomyces variotii in clinical samples and settings. J. Clin. Microbiol. 48:2754–2761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Samson RA, Houbraken J, Thrane U, Frisvad JC, Andersen B. 2010. Food and indoor fungi, CBS laboratory manual series 2. CBS-Fungal Biodiversity Centre, Utrecht, the Netherlands [Google Scholar]
- 13. Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput, Nucleic Acids Res. 32:1792–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst. Biol. 75:758–771 [DOI] [PubMed] [Google Scholar]
- 16. Clinical and Laboratory Standards Institute 2008. Reference method for broth dilution antifungals susceptibility testing of conidium-forming filamentous fungi: approved standard, 2nd ed M38-A2. CLSI, Wayne, PA [Google Scholar]
- 17. Delhaes L, Monchy S, Fréalle E, Hubans C, Salleron J, Leroy S, Prevotat A, Wallet F, Wallaert B, Dei-Cas E, Sime-Ngando T, Chabé M, Viscogliosi E. 2012. The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS One 7:e36313 doi:10.1371/journal.pone.0036313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Neuvéglise C, Sarfati J, Debeaupuis JP, Vu Thien H, Just J, Tournier G, Latgé JP. 1997. Longitudinal study of Aspergillus fumigatus strains isolated from cystic fibrosis patients. Eur. J. Clin. Microbiol. Infect. Dis. 16:747–750 [DOI] [PubMed] [Google Scholar]
- 19. de Valk HA, Klaassen CH, Yntema JB, Hebestreit A, Seidler M, Haase G, Müller FM, Meis JF. 2009. Molecular typing and colonization patterns of Aspergillus fumigatus in patients with cystic fibrosis. J. Cyst. Fibros. 8:110–114 [DOI] [PubMed] [Google Scholar]
- 20. Vanhee LM, Symoens F, Bouchara JP, Nelis HJ, Coenye T. 2008. High resolution genotyping of Aspergillus fumigatus isolates recovered from chronically colonised patients with cystic fibrosis. Eur. J. Clin. Microbiol. Infect. Dis. 27:1005–1007 [DOI] [PubMed] [Google Scholar]
- 21. Rybtke MT, Jensen PØ, Høiby N, Givskov M, Tolker-Nielsen T, Bjarnsholt T. 2011. The implication of Pseudomonas aeruginosa biofilms in infections. Inflamm. Allergy Drug Targets 10:141–157 [DOI] [PubMed] [Google Scholar]
- 22. Rivera KG, Seifert KA. 2011. A taxonomic and phylogenetic revision of the Penicillium sclerotiorum complex. Stud. Mycol. 70:139–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Samson RA, Varga J, Meijer M, Frisvad JC. 2011. New taxa in Aspergillus section Usti. Stud. Mycol. 69:81–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Soares C, Rodrigues P, Peterson SW, Lima N, Venâncio A. 2012. Three new species of Aspergillus section Flavi isolated from almonds and maize in Portugal. Mycologia 104:682–697 [DOI] [PubMed] [Google Scholar]
- 25. Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, Chen W, Fungal Barcoding Consortium 2012. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc. Natl. Acad. Sci. U. S. A. 109:6241–6246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Houbraken J, Frisvad JC, Samson RA. 2011. Taxonomy of Penicillium section Citrina. Stud. Mycol. 70:53–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Skouboe P, Frisvad JC, Lauritsen D, Boysen M, Taylor JW, Rossen L. 1999. Nucleotide sequences from the ITS region of Penicillium species. Mycol. Res. 103:873–881 [Google Scholar]
- 28. Varga J, Frisvad JC, Kocsubé S, Brankovics B, Tóth B, Szigeti G, Samson RA. 2011. New and revisited species in Aspergillus section Nigri. Stud. Mycol. 69:1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Balajee SA, Houbraken J, Verweij PE, Hong SB, Yaghuchi T, Varga J, Samson RA. 2007. Aspergillus species identification in the clinical setting. Stud. Mycol. 59:39–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Howard SJ, Harrison E, Bowyer P, Varga J, Denning DW. 2011. Cryptic species and azole resistance in the Aspergillus niger complex. Antimicrob. Agents Chemother. 55:4802–4809 [DOI] [PMC free article] [PubMed] [Google Scholar]