Abstract
Coagulase-negative staphylococci have been identified as major causes of late-onset neonatal bacteremia in neonatal intensive care units. Sixty isolates of Staphylococcus capitis obtained from blood cultures of neonates between 2000 and 2005 were examined in this study. Biochemical analysis confirmed that 52 of these isolates belonged to the subsp. urealyticus, and the remaining 8 belonged to the subsp. capitis. Isolates of the predominant subsp. urealyticus clones were characterized by their resistance to penicillin, erythromycin, and oxacillin and their biofilm formation ability, whereas subsp. capitis isolates were generally antibiotic susceptible and biofilm negative. Pulsed-field gel electrophoresis (PFGE) after SacII digestion separated the 60 isolates into five major clusters. Sequence analysis showed that, in S. capitis, the ica operon plus the negative regulator icaR was 4,160 bp in length. PCRs demonstrated the presence of the ica operon in all isolates. Further analysis of five isolates (two biofilm-positive subsp. urealyticus, one biofilm-negative subsp. urealyticus, and two biofilm-negative subsp. capitis) revealed that the ica operons were identical in all of the biofilm-positive subsp. urealyticus strains; however, the biofilm-negative isolates showed variations. The distinctive phenotypic and genotypic characteristics revealed by this study may affect the epidemiology of the two subspecies of S. capitis in the clinical setting. These results may provide a better understanding of the contribution of these two species to bloodstream infections in neonates.
INTRODUCTION
Coagulase-negative staphylococci (CoNS) have emerged as major causes of nosocomial infections and of nosocomial bacteremia in particular. These microorganisms usually infect premature neonates and immunocompromised patients, particularly those hospitalized for chemotherapy and managed with indwelling devices such as central venous catheters (1, 2).
The ability to form biofilm on the surfaces of inserted devices is the most important virulence mechanism of CoNS. Research on biofilm formation in Staphylococcus epidermidis started relatively early and has served as a model for other staphylococci, including Staphylococcus aureus and other CoNS species. In contrast to S. epidermidis, S. capitis displays the biofilm-forming capacity under particular experimental conditions, for example, in medium with high osmolarity. Quantitative biofilm production is linearly induced by sodium chloride (3). Polysaccharide intercellular adhesin (PIA) is the best-studied factor involved in S. epidermidis biofilm formation. It consists of glycan of β-1,6-linked 2-acetamido-2-deoxy-d-glucopyranosyl subunits, and its synthesis has been shown to be essential for S. epidermidis virulence. The ica operon encodes for proteins that synthesize PIA and includes the icaA, icaD, icaB, and icaC genes, in that order, downstream from the icaA promoter that controls expression of the entire operon. In addition, a regulatory gene called icaR is located upstream and is transcribed in the opposite direction (4). Other surface molecules involved in the accumulation phase of biofilm formation include accumulation-associated protein (Aap) (5), extracellular matrix-binding protein (Embp) (6), and extracellular DNA (7). Aap is a 220-kDa LPXTG protein (8). Embp was recently found to be a multifunctional cell surface protein that mediates attachment to host extracellular matrix, biofilm accumulation, and escape from phagocytosis (9).
Staphylococcus capitis has been implicated in biofilm-related infections such as endocarditis (10), urinary tract infection (11), and catheter-related bacteremia (12). Humans are likely to be the main sources or vehicles of transmission of S. capitis (13). Several reports suggest that S. capitis is an emerging opportunistic pathogen in newborn babies being cared for in newborn intensive care units (NICUs) (14–16). This species has been endemic in the NICU of the Royal Women's Hospital in Melbourne, Australia, for several years, and we have accumulated a collection of isolates from neonates believed to have bloodstream infections with this species.
S. capitis can be further divided into two subspecies: subsp. urealyticus and subsp. capitis. Subspecies urealyticus can be distinguished from subsp. capitis by its urease activity, ability to produce acid from maltose in anaerobic conditions, fatty acid profile, larger colony size, and DNA sequence differentiation (17).
Since no previous study has examined the prevalence, phenotypic characteristics, and molecular epidemiology of the two subspecies of S. capitis as opportunistic pathogens in NICUs, our aim was to characterize our collection of isolates, with respect to subspecies, antimicrobial susceptibility, structure of the ica operon, and expression of the biofilm phenotype under conditions of stress. We hypothesize that the two subspecies differ in prevalence and in their capacity to become opportunistic pathogens in NICUs.
MATERIALS AND METHODS
Bacterial isolates.
Sixty clinical S. capitis isolates from neonates at the NICU, Royal Women's Hospital, Melbourne, Australia, were collected between 2000 and 2005. These isolates were considered to be clinically significant based on the isolation of the same organism from more than one blood culture collected within a 14-day period and/or the presence of clinical or laboratory findings suggestive of neonatal sepsis (18). They had been identified by the ID 32 Staph system (bioMérieux, Marcy l'Etoile, France). All isolates were stored in nutrient broth (Oxoid, Australia) with 15% glycerol at −80°C and were recovered for the present study on tryptone soy agar (Oxoid) incubated at 37°C for 24 to 48 h. Urease activity and maltose fermentation tests were performed to discriminate subsp. urealyticus and capitis (17).
Antibiotic susceptibility testing and screening for mecA gene.
Six antibiotics—penicillin, erythromycin, clindamycin, teicoplanin, vancomycin, and oxacillin—were chosen for the present study based on their use in the hospital. Antibiotic susceptibility was determined by MIC and inducible clindamycin resistance according to Clinical and Laboratory Standards Institute guidelines (19).
The mecA gene was detected by PCR, using the forward primer MECAP4 and the reverse primer MECAP7 (11), obtained from Sigma-Genosys, Sydney, Australia. Nucleotide sequence determination was performed by Microcom Sequencing Facilities, Monash University, Melbourne, Australia.
Detection of biofilm production.
Quantitative determination of biofilm production was performed using a microtiter plate assay. Each plate contained S. epidermidis RP62a and SP2 as positive and negative biofilm-producing controls, respectively (20). Four wells were inoculated per isolate in a given experiment, and all isolates were tested independently on three occasions.
The ability to produce extracellular polysaccharide (slime) (15) was assessed and interpreted using Congo red agar (CRA), according to the method of Freeman et al. (21) and Arciola et al. (22).
The ica operon was amplified according to the Expand long-template PCR system protocol (Roche Applied Science). PCR products were analyzed by 1% agarose (Bioline, Australia) gel electrophoresis and sequenced by Microcom Sequencing Facilities, Monash University, Australia.
PFGE analysis.
PFGE was performed according to the method of Murchan et al. (23) with minor modifications. The SacII enzyme was used instead of SmaI since preliminary studies showed that it provided better discrimination. PFGE gel images were stored electronically as JPG files and analyzed visually with GelCompar II (version 6.0; Applied Maths), using the Dice coefficient, represented by the unweighted pair group method using arithmetic averages with 1% optimization and 1.5% tolerance setting. Cutoff values of 75 and 80% were applied to assess the similarity. A dendrogram was drawn to show the relatedness of the clones.
Hydrophobicity analysis.
Hydrophobicity analysis was carried out using the web-predictor TopPred II (24). All of the possible topologies that include the certain transmembrane segments and either include or exclude each of the candidate segments were automatically generated.
Statistical analysis.
Antibiotic susceptibility experiments were performed at least twice. A binary logistic regression analysis was performed to assess whether the biofilm formation and antibiotic resistance was associated independently with the two subspecies and how the biofilm formation was related to the antibiotic profile. Statistical analyses were performed using GraphPad Prism 5.0 (25). This program computes P values using the Fisher exact test contingency table and summarizes the data by computing the odds ratio, along with the 95% confidence intervals.
Nucleotide submission.
The sequenced complete ica operon of clinical biofilm-positive S. capitis isolate 6 has been deposited in GenBank under accession number JF930147.
RESULTS
Molecular epidemiology of clinical S. capitis isolates over a 6-year period.
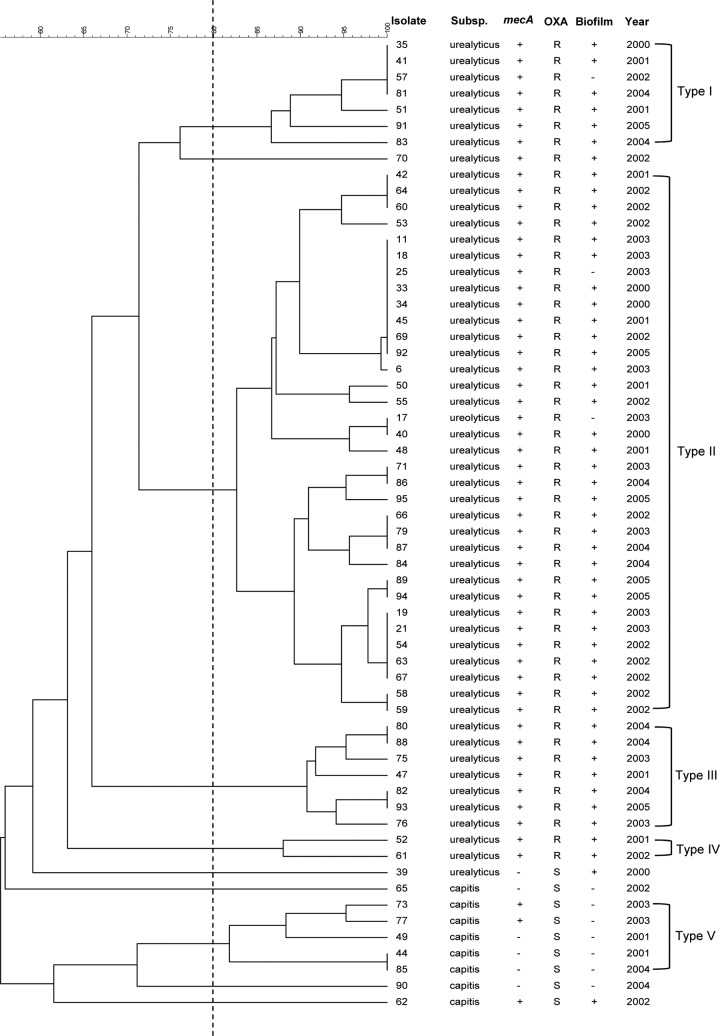
The annual number of episodes of sepsis at the NICU varied over the 6 years of the study, peaking in 2002 and 2003 (Fig. 1). Analysis of PFGE gels by GelCompar II yielded four major clusters of at least five isolates with ≥80% similarity, comprising 53 (88.3%) of all isolates. Another minor cluster comprised two isolates (i.e., isolates 52 and 61). Five isolates (i.e., isolates 39, 62, 65, 70, and 90) that were randomly distributed over the years could not be typed. There was no difference in the allocation of clusters between the 75 and 80% cutoff levels. The only exception was one isolate clustered in the PFGE type I according to the 75% cutoff and nonclustered according to the 80% cutoff level.
Fig 1.
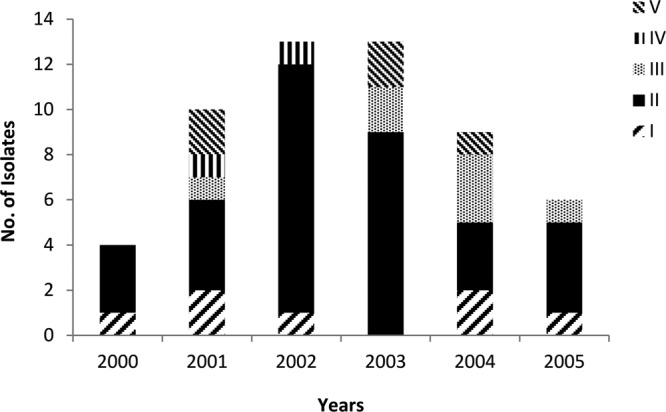
Contributions of PFGE clusters to total infections from 2000 to 2005.
Cluster II, the largest cluster, comprising 34 isolates of S. capitis subsp. urealyticus, was associated with more than half of all cases in the NICU and was apparently capable of persisting for a prolonged period. Isolates belonging to this cluster were distributed throughout the 6-year period. The highest occurrence was in 2002 (11 isolates, 84.6% of isolates in this year). Clusters I, III, and IV (S. capitis subsp. urealyticus) were less prevalent but appeared in most years throughout the period of the study. Isolates classified as cluster V (S. capitis subsp. capitis) appeared sporadically in low numbers throughout the study period (Fig. 1).
Quantitative and qualitative biofilm formation and correlation with icaADBC prevalence and sequences.
Fifty isolates were determined to be biofilm positive by the microtiter plate assay in TSB supplemented with 4% NaCl, and the remaining 10 isolates were biofilm negative. There was complete agreement between the results of the microtiter plate assay and the production of extracellular polysaccharide indicated by colonial appearance on CRA plates.
Sequence analysis showed that the ica operon plus the negative regulator icaR is 4,160 bp in length, and PCR analysis demonstrated that it was present in all S. capitis isolates. The five genes (icaR, icaA, icaD, icaB, and icaC) were closely related to those of S. caprae, S. epidermidis, and S. aureus, and the corresponding polypeptides exhibited 65 to 94% amino acid identity. Further sequence analysis was performed on the ica operon in five isolates: two biofilm-positive subsp. urealyticus isolates (isolates 6 and 70), one biofilm-negative subsp. urealyticus isolate (isolate 17), and two biofilm-negative subsp. capitis (isolates 44 and 65). These isolates were selected by identifying different restriction patterns of the ica operon in accordance with PCR-restriction fragment length polymorphism digestion with TaqI (data not shown). Sequencing showed that DNA sequences of the two biofilm-positive isolates were 100% identical, but the three biofilm-negative isolates showed sequence variations. Isolate 44 (subsp. capitis) had a deletion mutation in the −10 promoter element compared to that of biofilm-positive isolates. This mutation might prevent the transcription of the ica genes, resulting in the inability to express PIA. In isolate 65 (subsp. capitis), a stop codon occurred in the icaB gene; this nonsense mutation resulted in a predicted truncated protein, missing 218 C-terminal amino acids. An in-frame three amino acid deletion in the icaA gene of isolate 17 (subsp. urealyticus) was identified. Protein structural prediction of the IcaA from the deduced sequences of isolates 6 and 17 was performed with TopPred II. It revealed that the IcaA protein sequence of strain 6 exhibited five predicted transmembrane segments, whereas four candidate membrane-spanning segments were shown in strain 17. In the region from amino acids 178 to 198, the hydrophobicity of isolate 17 was below the threshold value compared to that of isolate 6. This structural change might lead to a functionally defective protein and result in abolishment of biofilm production in this isolate.
Antibiotic susceptibility profile and relationship to biofilm production.
Biofilm-positive isolates were, in general, more resistant than biofilm-negative isolates. The great majority of isolates (n = 52) were resistant to penicillin and oxacillin (87 and 85%, respectively). Carriage of mecA gene was consistently high and was almost always associated with biofilm positivity. Forty-three isolates (72%) showed resistance to erythromycin. Four isolates were resistant or intermediate in resistance to clindamycin (6.7%), with only one of this showing inducible resistance. Six isolates (10%) were resistant to teicoplanin (Table 1). Based on MICs, no isolates of S. capitis displayed reduced susceptibility to vancomycin; however, this method may not detect small populations of resistant cells, which can only be detected by population analysis profiling. Our previous studies show that such populations do exist in S. capitis (26).
Table 1.
MIC antibiotic susceptibility patterns and mecA gene carriage and their correlations between biofilm formation phenotype in clinical S. capitis
| Patient group | No. (%) of resistant isolates by yr | No. (%) of resistant isolates by biofilm phenotypea | Pb | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | Biofilm+ (n = 50) | Biofilm− (n = 10) | ||
| Treated with: | |||||||||
| Penicillin | 5 (100) | 9 (90) | 14 (87.5) | 10 (76.9) | 9 (90) | 5 (83.3) | 48 (96) | 4 (40) | 0.0001 |
| Erythromycin | 4 (80) | 8 (80) | 8 (50) | 9 (69.2) | 8 (80) | 6 (100) | 41 (82) | 2 (20) | 0.0003 |
| Clindamycinc | 0 | 1 (10) | 2 (12.5) | 0 | 0 | 1 (16.7) | 4 (8) | 0 | 1.0000 |
| Teicoplanin | 0 | 3 (30) | 2 (12.5) | 1 (7.7) | 0 | 0 | 5 (10) | 1 (10) | 1.0000 |
| Vancomycin | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1.0000 |
| Oxacillin | 4 (80) | 8 (80) | 14 (87.5) | 11 (84.6) | 8 (80) | 6 (100) | 48 (96) | 3 (30) | <0.0001 |
| mecA gene carrier | 4 (80) | 8 (80) | 15 (93.8) | 13 (100) | 8 (80) | 6 (100) | 49 (98) | 5 (50) | 0.0003 |
n, number of isolates.
A binary logistic regression analysis was performed using the Fisher exact test from a contingency table. P < 0.05 is considered a significant difference.
Includes inducible resistance.
Genotype of subspecies and their relatedness to antibiotic resistance and biofilm production.
Fifty-two isolates identified as subsp. urealyticus were mainly distributed in three major PFGE clusters (I, II and III) and one minor cluster (IV) (Fig. 2). Two nonclustered isolates were also grouped into this subspecies. Members of subsp. urealyticus were generally resistant to oxacillin (98%) and biofilm positive (94%). One exception was a nonclustered isolate, which were oxacillin susceptible according to the MIC, although it carried mecA (isolate 39).
Fig 2.
PFGE dendrogram of the clinical S. capitis isolates. The scale bar at the top of the dendrogram indicates the similarity. The dotted line indicates the cutoff value of 80% that was applied to separate the clusters.
As expected, S. capitis subsp. capitis isolates were clustered separately from the isolates belonging to S. capitis subsp. urealyticus. The eight isolates either appeared in PFGE cluster V or remained unclustered (Fig. 2). These isolates were susceptible to all antibiotics according to the MIC data, although mecA was identified in three of them. Only one (isolate 62) produced biofilm (Fig. 2), although all eight contained the entire icaADBC operon. There were no major insertions or deletions in the ica operon in these biofilm-negative isolates of S. capitis subsp. capitis (data not shown); however, this does not exclude the possibility of point mutations resulting in nonfunctional proteins.
DISCUSSION
Significance of antibiotic resistance profile.
Consistent with previous studies on CoNS from NICUs (27), we found a high level of oxacillin resistance 51 (85%) among the isolates in the present study. Oxacillin resistance and mecA carriage reached 100% among isolates belonging to the three major PFGE clusters (I, II, and III) and a minor cluster IV (Table 2). All such isolates were S. capitis subsp. urealyticus. Although there was generally good agreement between the prevalence of mecA and expression of oxacillin resistance, there were some exceptions. Four isolates that carried mecA failed to express oxacillin resistance (Fig. 2). This could be explained by low levels of expression of mecA due to the presence of small subpopulations that were not detected by phenotypic methods. Alternatively, the mecA gene may be defective in these isolates (28). We did not detect isolates expressing non-mecA-mediated resistance to oxacillin. High levels of resistance to erythromycin and clindamycin in S. epidermidis were reported by many studies (21, 26). In the present study, 42 isolates (71.7%) were resistant to erythromycin, and only four (6.7%) were resistant or showed inducible resistance to clindamycin.
Table 2.
Correlations of antibiotic susceptibility profiles and mecA gene carriage with PFGE clusters in S. capitis clinical isolates
| Patient group | No. (%) of resistant isolates in PFGE clusters | No. (%) of nonclustered resistant isolates | ||||
|---|---|---|---|---|---|---|
| I | II | III | IV | V | ||
| Treated with: | ||||||
| Penicillin | 6 (85.7) | 34 (100) | 7 (100) | 2 (100) | 0 | 2 (40) |
| Erythromycin | 7 (100) | 28 (82.4) | 6 (85.7) | 2 (100) | 0 | 0 |
| Clindamycin | 0 | 1 (2.9) | 0 | 2 (100) | 0 | 1 (20) |
| Teicoplanin | 2 (28.6) | 3 (8.8) | 1 (14.3) | 0 | 0 | 0 |
| Vancomycin | 0 | 0 | 0 | 0 | 0 | 0 |
| Oxacillin | 7 (100) | 34 (100) | 7 (100) | 2 (100) | 0 | 1 (20) |
| mecA gene carrier | 7 (100) | 34 (100) | 7 (100) | 2 (100) | 2 (40) | 2 (40) |
Biofilm formation of clinical S. capitis isolates and their prevalence in the NICU.
The predominant S. capitis clones in the NICU setting were biofilm-producing S. capitis subsp. urealyticus. Three isolates of S. capitis subsp. urealyticus failed to produce biofilm and yet were classified into the major PFGE clusters and presented similar antibiotic resistance profiles to the other members in the major clusters. The other isolates displaying a biofilm-negative phenotype belonged to subsp. capitis, were susceptible to all antibiotics, and were members of cluster V. These isolates were few in number and appeared only sporadically over the study period (Fig. 2). Since both subspecies reside on the human skin, the considerably higher prevalence of S. capitis subsp. urealyticus suggests that this subspecies has greater potential as an opportunistic pathogen or greater transmissibility than S. capitis subsp. capitis.
Consistent with previous studies (29), biofilm formation of the S. capitis isolates showed a strong association with the antibiotic resistance profile. It is likely that antibiotic use led to the selection of PFGE types that contained resistant strains that also expressed biofilm. As a consequence, both the ability to express biofilm under specific conditions encountered in the NICU and resistance to multiple antibiotics could provide S. capitis subsp. urealyticus with a selective advantage. Biofilms form a barrier around bacteria, protecting them from antibiotics and phagocytes and thus making the treatment of infections very difficult (4, 30). However, given the likely value of biofilm formation in providing a positive selective advantage, the question remains as to why biofilm-negative antibiotic-susceptible S. capitis subsp. capitis isolates remained in the NICU, albeit at low prevalence. One possibility is that the ica operon is activated under conditions that are not the same as for S. capitis subsp. urealyticus encountered in the hospital environment and thus not under the in vitro conditions examined here. Alternatively, protein-mediated biofilm could be activated within the clinical setting. These hypotheses are supported by our previous conclusions that most S. epidermidis can be induced to produce biofilm in response to environmental stimuli encountered in the clinical setting, including products used in neonatal units (31).
ica operon in biofilm-negative S. capitis isolates and its origin.
Sequence analysis showed variable mutations in biofilm-negative isolates. Whether these mutations were critical factors causing the biofilm-negative phenotype in these isolates remains a question to be answered. The variations in the ica operons of biofilm-negative S. capitis subsp. capitis (isolates 44 and 65) might simply be natural variation in this group of isolates. However, this does not exclude the probability of point mutations resulting in nonfunctional proteins.
The genetic origin of ica genes in staphylococci is not known. It is also uncertain how biofilm-forming isolates are established and disseminated within the hospital environment. Previous studies (18, 32) based on genome-wide comparisons of S. aureus genomes showed that mobile DNA is exchanged readily in S. aureus populations. It was suggested that horizontal gene transfer between staphylococci and other low-GC Gram-positive bacteria is common and contributes to resistance and virulence development. Occurrence of the ica operon in S. capitis strains of different genetic backgrounds suggests mobility and horizontal transfer of the cluster of biofilm-mediating genes among these strains. The close contact of bacteria within a biofilm may facilitate horizontal exchange of genetic information, including antimicrobial resistance genes and virulence determinants. However, a recent study (33) revealed that the enhanced virulence in epidemic community-associated methicillin-resistant S. aureus, strain USA300, is attributable to differential expression of core genome-encoded virulence determinants rather than the acquisition of additional virulence genes via mobile genetic elements. We are currently investigating the differential ica gene expression in biofilm-negative and biofilm-positive S. capitis isolates.
Relatedness of biofilm production and antibiotic resistance to two subspecies.
One striking observation in the present study is that biofilm formation and the presence of the mecA gene in these clinical S. capitis isolates were mainly displayed only in subsp. urealyticus (Fig. 2). A global comparison of the genomes of diverse clinical strains of these two subspecies with known endemic potential would contribute our understanding S. capitis survival and infections in hospitals.
In conclusion, our findings suggest that the endemic S. capitis clones confirmed to be S. capitis subsp. urealyticus are more important causes of bloodstream infections in very-low-birth-weight infants than is subsp. capitis. It would therefore be beneficial to subspeciate S. capitis isolates, especially those isolated from neonates. The results also emphasize the importance of examining the composition and expression of the ica operon to the pathogenic potential of S. capitis.
ACKNOWLEDGMENTS
We thank Vennessa Fleming and Nerida Thurbon for excellent technical assistance.
This study was supported by an Australian Postgraduate Award to B.C.
Footnotes
Published ahead of print 10 October 2012
REFERENCES
- 1. Chaves F, Garciá-Alvarez M, Sanz F, Alba C, Otero JR. 2005. Nosocomial spread of a Staphylococcus hominis subsp. novobiosepticus. J. Clin. Microbiol. 43:4877–4879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pessoa-Silva CL, Miyasaki CH, de Almeida MF, Kopelman BI, Raggio RL, Wey SB. 2001. Neonatal late-onset bloodstream infection: attributable mortality, excess of length of stay, and risk factors. Eur. J. Epidemiol. 17:715–720 [DOI] [PubMed] [Google Scholar]
- 3. Møretrø T, Hermansen L, Holck AL, Sidhu MS, Rudi K, Langsrud S. 2003. Biofilm formation and the presence of the intercellular adhesion locus ica among staphylococci from food and food processing environments. Appl. Environ. Microbiol. 69:5648–5655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Begun J, Gaiani JM, Rohde H, Mack D, Calderwood SB, Ausubel FM, Sifri CD. 2007. Staphylococcal biofilm exopolysaccharide protects against Caenorhabditis elegans immune defenses. PLoS Pathog. 3:e57 doi:10.1371/journal.ppat.0030057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, Knobloch JKM, Heilmann C, Herrmann M, Mack D. 2005. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol. Microbiol. 55:1883–1895 [DOI] [PubMed] [Google Scholar]
- 6. Latasa C, Solano C, Penadés JR, Lasa I. 2006. Biofilm-associated proteins. Immunology 329:849–857 [DOI] [PubMed] [Google Scholar]
- 7. Lou Q, Zhu T, Hu J, Ben H, Yang J, Yu F, Liu J, Wu Y, Fischer A, Francois P, Schrenzel J, Qu D. 2011. Role of the SaeRS two-component regulatory system in Staphylococcus epidermidis autolysis and biofilm formation. BMC Microbiol. 11:146 doi:10.1186/1471-2180-11-146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vandecasteele SJ, Peetermans WE, Merckx RR, Rijnders BJA, Van Eldere J. 2003. Reliability of the ica, aap, and atlE genes in the discrimination between invasive, colonizing and contaminant Staphylococcus epidermidis isolates in the diagnosis of catheter-related infections. Clin. Microbiol. Infect. 9:114–119 [DOI] [PubMed] [Google Scholar]
- 9. Christner M, Franke GC, Schommer NN, Wendt U, Wegert K, Pehle P, Kroll G, Schulze C, Buck F, Mack D, Aepfelbacher M, Rohde H. 2010. The giant extracellular matrix-binding protein of Staphylococcus epidermidis mediates biofilm accumulation and attachment to fibronectin. Mol. Microbiol. 75:187–207 [DOI] [PubMed] [Google Scholar]
- 10. Nalmas S, Bishburg E, Meurillio J, Khoobiar S, Cohen M. 2008. Staphylococcus capitis prosthetic valve endocarditis: report of two rare cases and review of literature. Heart Lung 37:380–384 [DOI] [PubMed] [Google Scholar]
- 11. Oren I, Merzbach D. 1990. Clinical and epidemiological significance of species identification of coagulase-negative staphylococci in a microbiological laboratory. Isr. J. Med. Sci. 26:125–128 [PubMed] [Google Scholar]
- 12. Tristan A, Lina G, Etienne J, Vandenesch F. 2000. Biology and pathogenicity of staphylococci other than Staphylococcus aureus and Staphylococcus epidermidis, p 572–586 In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI. (ed), Gram-positive pathogens. ASM Press, Washington, DC [Google Scholar]
- 13. Hira V, Sluijter M, Goessens WHF, Ott A, de Groot R, Hermans PWM, Kornelisse RF. 2010. Coagulase-negative staphylococcal skin carriage among neonatal intensive care unit personnel: from population to infection. J. Clin. Microbiol. 48:3876–3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. de Silva GDI, Justice A, Wilkinson AR, Buttery J, Herbert M, Day NPJ, Peacock SJ. 2001. Genetic population structure of coagulase-negative staphylococci associated with carriage and disease in preterm infants. Clin. Infect. Dis. 33:1520–1528 [DOI] [PubMed] [Google Scholar]
- 15. Rasigade J-P, Raulin O, Picaud J-C, Tellini C, Bes M, Grando J, Ben Saïd M, Claris O, Etienne J, Tigaud S, Laurent F. 2012. Methicillin-resistant Staphylococcus capitis with reduced vancomycin susceptibility causes late-onset sepsis in intensive care neonates. PLoS One 7:e31548 doi:10.1371/journal.pone.0031548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Der Zwet WC, Debets-Ossenkopp Y-J, Reinders E, Kapi M, Savelkoul PHM, Van Elburg RM, Hiramatsu K, Vandenbroucke-Grauls CMJE. 2002. Nosocomial spread of a Staphylococcus capitis strain with heteroresistance to vancomycin in a neonatal intensive care unit. J. Clin. Microbiol. 40:2520–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bannerman TL, Kloos WE. 1991. Staphylococcus capitis subsp. urealyticus subsp. nov. from human skin. Int. J. Syst. Bacteriol. 41:144–147 [DOI] [PubMed] [Google Scholar]
- 18. Holden MTG, Lindsay JA, Corton C, Quail MA, Cockfield JD, Pathak S, Batra R, Parkhill J, Bentley SD, Edgeworth JD. 2010. Genome sequence of a recently emerged, highly transmissible, multi-antibiotic-and antiseptic-resistant variant of methicillin-resistant Staphylococcus aureus, sequence type 239 (TW). J. Bacteriol. 192:888–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Clinical and Laboratory Standards Institute 2011. Performance standards for antimicrobial disk and dilution susceptibility testing; 21st informational supplement M100-S21. CLSI, Wayne, PA [Google Scholar]
- 20. Christensen GD, Simpson WA, Younger JJ. 1985. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 22:996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Freeman DJ, Falkiner FR, Keane CT. 1989. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 42:872–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arciola CR, Campoccia D, Gamberini S, Cervellati M, Donati E, Montanaro L. 2002. Detection of slime production by means of an optimised Congo red agar plate test based on a colorimetric scale in Staphylococcus epidermidis. Clinical isolates genotyped for ica locus. Biomaterials 23:4233–4239 [DOI] [PubMed] [Google Scholar]
- 23. Murchan S, Kaufmann ME, Deplano A, de Ryck R, Struelens M, Zinn CE, Fussing V, Salmenlinna S, Vuopio-Varkila J, El Solh N, Cuny C, Witte W, Tassios PT, Legakis N, van Leeuwen W, van Belkum A, Vindel A, Laconcha I, Garaizar J, Haeggman S, Olsson-Liljequist B, Ransjo U, Coombes G, Cookson B. 2003. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. J. Clin. Microbiol. 41:1574–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Von Heijne G. 1992. Membrane protein structure prediction, hydrophobicity analysis and the positive-inside rule. J. Mol. Biol. 225:487–494 [DOI] [PubMed] [Google Scholar]
- 25. Motulsky HJ. 1999. Analyzing data with GraphPad Prism. GraphPad Software, Inc., San Diego, CA [Google Scholar]
- 26. D'Mello D, Daley AJ, Rahman MS, Qu Y, Garland S, Pearce C, Deighton MA. 2008. Vancomycin heteroresistance in bloodstream isolates of Staphylococcus capitis. J. Clin. Microbiol. 46:3124–3126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Akinkunmi EO, Lamikanra A. 2010. Species distribution and antibiotic resistance in coagulase-negative staphylococci colonizing the gastrointestinal tract of children in Ile-Ife, Nigeria. Trop. J. Pharmacol. Res. 9:35–43 [Google Scholar]
- 28. Al-Talib H, Yean CY, Al-khateeb A, Singh KK, Hasan H, Al-Jashamy K, Ravichandran M. 2010. Comparative evaluation of five culture media with triplex PCR assay for detection of methicillin-resistant Staphylococcus aureus. Curr. Microbiol. 61:1–6 [DOI] [PubMed] [Google Scholar]
- 29. Ziebuhr W, Hennig S, Eckart M, Kränzler H, Batzilla C, Kozitskaya S. 2006. Nosocomial infections by Staphylococcus epidermidis: how a commensal bacterium turns into a pathogen. Int. J. Antimicrob. Agents 28:14–20 [DOI] [PubMed] [Google Scholar]
- 30. Boynukara B, Gulhan T, Gulhan T, Gurturk K, Alisarli M, Ogun E. 2007. Evolution of slime production by coagulase-negative staphylococci and enterotoxigenic characteristics of Staphylococcus aureus strains isolated from various human clinical specimens. J. Med. Microbiol. 56:1296–1300 [DOI] [PubMed] [Google Scholar]
- 31. Bradford R, Abdul Manan R, Daley A, Pearce C, Ramalingam A, D'Mello D, Mueller Y, Uahwatanasakul W, Qu Y, Grando D, Garland S, Deighton M. 2006. Coagulase-negative staphylococci in very-low-birth-weight infants: inability of genetic markers to distinguish invasive strains from blood culture contaminants. Eur. J. Clin. Microbiol. 25:283–290 [DOI] [PubMed] [Google Scholar]
- 32. Holden MTG. 2004. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc. Natl. Acad. Sci. U. S. A. 101:9786–9791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li M, Diep BA, Villaruz AE, Braughton KR, Jiang XF, DeLeo FR, Chambers HF, Lu Y, Otto M. 2009. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 106:5883–5888 [DOI] [PMC free article] [PubMed] [Google Scholar]