Abstract
Since 1995, a methicillin-resistant Staphylococcus aureus (MRSA) clone has spread in southern Germany. The strain was assigned to the Rhine-Hesse pulsed-field gel electrophoresis (PFGE) type by the staphylococcal reference center and was highly similar to epidemic clones known to belong to clonal complex 5 (CC5; USA100) based on multilocus sequence typing (MLST). Here we analyzed a defined collection of strains assigned to the Rhine-Hesse/USA100 PFGE type. Using sequence-based typing methods (MLST, spa), the isolates were divided into two distinct clusters, ST5 and its single-locus variant ST225. These two lineages are not distinguishable by PFGE or phage typing. Most of the ST5 isolates were derived from patients and volunteers from the Tübingen area in southwest Germany, whereas the ST225 isolates were mostly from other locations in Germany. The locally restricted ST5 isolates were shown to contain different SSCmec islands and exhibited different antibiotic resistance profiles. In contrast, the ST225 isolates form a highly homogenous group and are emerging all over Germany. The two lineages are clearly distinguishable by their phage content and spa type: ST5 strains from Tübingen are characterized by a Sa7int phage that carries the virulence gene sak, which codes for staphylokinase, and ST225 isolates are characterized by a Sa1int phage. In conclusion, based on sequence typing and phage content, CC5 strains can be subdivided into two distinct lineages with different epidemicities.
INTRODUCTION
The occurrence and spread of methicillin-resistant Staphylococcus aureus (MRSA) have become a major challenge in many parts of the world. The initially used phage typing method already indicated the global spread of certain MRSA lineages (1). Later, pulsed-field gel electrophoresis (PFGE) analysis and multilocus sequence typing (MLST) revealed that the species can be divided into a number of distinct clonal complexes (CC) (2–8). CC5 isolates such as USA100, the Pediatric clone (USA), EMRSA-3 (Great Britain), and the Tokyo clone (Japan) have worldwide prevalence but harbor several different SCCmec elements, indicating polyclonal genesis (9, 10). Based on sequence analyses of American ST5 and European ST225 isolates, which both belong to CC5, it was postulated that the ST225 isolates are derived from an U.S. ancestor that was introduced into Europe around 2001. Six years earlier, in 1995, MRSA isolates had emerged in southern Germany that were designated the “Rhine-Hesse” clone by the German reference center based on PFGE typing (11). Similar isolates were later described in studies from different parts of Germany (12–15).
There are now powerful sequence-based typing methods available to define CCs based on the core genome. However, the role of mobile elements, especially phages, in the microevolution of bacterial pathogens cannot be detected by these methods. Recently, there has been increasing evidence that phages (e.g., those carrying pvl or sasX) may drive virulence and epidemicity (16, 17). Here we assessed the diversification of CC5 isolates using a defined collection of strains from different regions in Germany. Molecular typing methods were used to discriminate the isolates into two distinct clusters, each of which is characterized by high prevalence of a lineage-specific phage.
MATERIALS AND METHODS
Bacterial strains.
Based on PFGE typing, the following Rhine-Hesse isolates were selected from the strain collections for further analysis. (i) A total of 49 isolates (41 MRSA and 8 methicillin-susceptible S. aureus [MSSA] strains) were selected from the PFGE database of the Institute for Medical Microbiology and Hygiene in Tübingen containing 251 Rhine-Hesse S. aureus isolates. The strains were chosen to represent isolates with minor differences in the banding pattern. They originated from different previous studies and from in- and outpatients from two medical centers located in southwestern and southeastern Germany. (ii) A total of 27 Rhine-Hesse MRSA isolates from the former German reference laboratory of staphylococci were also chosen due to differences in banding pattern. They originated mainly from North Rhine-Westphalia. (iii) Three well-characterized reference strains (N315, Pediatric clone, and New York clone) with similar PFGE patterns were provided by NARSA (i.e., the Network on Antimicrobial Resistance in Staphylococcus aureus).
Strain typing.
spa typing (12, 14), MLST (4), and SCCmec typing (18) were performed according to previously published references. PFGE and Southern hybridization were performed as described previously (19) using probes listed in Table S1 in the supplemental material. The gels were evaluated by use of WinCam3 software (Cybertech, Berlin, Germany). A similarity index was determined for strains by use of the Dice coefficient. Clustering correlation coefficients were calculated by the unweighted-pair group method. Strains showing only minor differences in the banding pattern (>87% similarity as assessed by the Dice coefficient) were assigned to the same genome type (GT). GTs were evaluated blindly with the aid of the computer program, and all types were verified by visual examination of the original gel. In cases of doubt, the strains were reanalyzed in a second PFGE run that placed the isolates in question next to each other. Phage typing was performed with the international set of typing phages, including MRSA phages and additional regional phages (D11, -16, -92, -187, and -192), according to standard rules. To determine relationships between isolates based on phage typing results, the obtained lysis patterns were converted into numbers (not sensitive = 0, low = 1, sensitive = 2, high = 3). Clusters were detected by hierarchical clustering analysis using the Ward algorithm.
Antimicrobial susceptibility.
Susceptibility was tested either by agar diffusion according to established guidelines (Clinical and Laboratory Standards Institute) or by broth microdilution using the Vitek2 system (bioMérieux, Nürtingen, Germany). Inducible clindamycin resistance was determined employing a disk diffusion D-zone test.
Prophage content.
S. aureus phages were identified by a multiplex PCR strategy targeting the integrase gene sequence as described previously (20). The presence of phage Φ6390 and the phage-carried virulence factor sak was detected using the primers listed in Table S1 in the supplemental material.
RESULTS AND DISCUSSION
Emergence and prevalence of the Rhine-Hesse MRSA clone in Germany.
Between 1991 and 1996, an infection control program was established in the neonatal ward and intensive care unit in Tübingen. All S. aureus isolates (n = 1,908) were sent to the former national reference laboratory of staphylococci for analysis. The frequency of MRSA isolates was below 5% until 1995, when a sudden increase was observed in the surgical department. Among these isolates, a new PFGE pattern was detected that was clearly distinct from the so-called southern German epidemic strain (ST228) that was most prevalent at that time (11, 21). The new PFGE type was later designated the “Rhine-Hesse” clone by the Robert Koch Institute. Between 1999 and 2006, an increase in the total number of isolates of this MRSA type was observed among the specimens from North Rhine-Westphalia sent to the Bonn laboratory for typing (Fig. 1).
Fig 1.
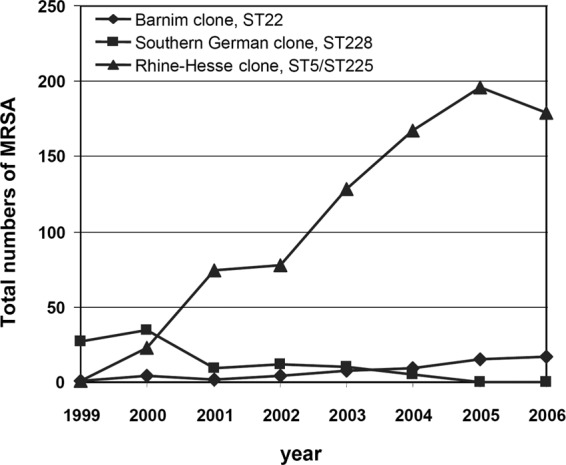
Total numbers of Rhine-Hesse PFGE types collected from 1999 to 2006 among the isolates sent to the former reference laboratory of staphylococci from Bonn hospitals. A large increase in the number of Rhine-Hesse MRSA isolates is evident.
In 2006, the analysis of all MRSA strains from the Institute for Medical Microbiology and Hygiene in Tübingen revealed that 59.4% (95/160) of the isolates still exhibited a PFGE pattern similar to that of the Rhine-Hesse type. For comparison, 203 colonizing strains of methicillin-susceptible S. aureus (MSSA) from healthy volunteers from the Tübingen region were collected between 2000 and 2007. Nine percent of these isolates were assigned to the Rhine-Hesse type by PFGE.
spa typing of Rhine-Hesse strains and correlation with MLST.
Although all Rhine-Hesse strains could clearly be distinguished from other strains by PFGE, a high variability within this cluster was observed. To clarify the great variability within this S. aureus lineage, the isolates were typed using spa typing and MLST. A subset of 79 Rhine-Hesse isolates were selected to represent isolates with minor differences in their banding patterns: 49 isolates (41 MRSA and 8 MSSA strains) originating from in- and outpatients from two medical centers located in southwest and southeast Germany, 27 Rhine-Hesse isolates selected from the former reference laboratory of staphylococci, and 3 reference strains (N315, Pediatric clone, and New York clone).
The isolates exhibited 8 different spa types; spa types t002 (45 isolates) and t003 (27 isolates) were observed most frequently. The others were observed only twice (t045) or once (t105, t264, t535, t1305, t1486). Types clustered into spa-CC 003/045 (t003, t045, t264, and t1486) and spa-CC 105 (t002, t105, t1305) using based-upon-repeat-pattern (BURP) analysis. One spa type (t535) could not be clustered due to there being fewer than 5 repeats. MLST was performed on isolates of spa types t002 and t003 (3 isolates each) and on all single spa types. All isolates could be assigned to either ST5 or its single-locus variant ST225. Only for the t045 isolates did the BURP clustering not correlate with the MLST designation. For the following analysis, we grouped all 79 isolates as belonging to either ST5 (t002, t1305, t105) or ST225 (t003, t264, t1486). Thus, sequence-based methods can clearly separate the isolates into two distinct lineages, both of which have been recently described to contain prevalent, globally spreading MRSA isolates (9).
Regional distribution of ST5 and ST225 isolates.
Interestingly, 94% (31/33) of the isolates from the Tübingen area belonged to the ST5 complex. In contrast, 80% (28/35) of the isolates from elsewhere in Germany were ST225. Thus, the predominant ST5 isolates seem to have persisted in this small area of the Tübingen region in southwest Germany over several decades without nationwide dissemination. The high prevalence of ST5/t002 isolates in the southwestern part of Germany was recently shown in another independent study (22).
PFGE analysis and phage typing for the discrimination of CC5 isolates.
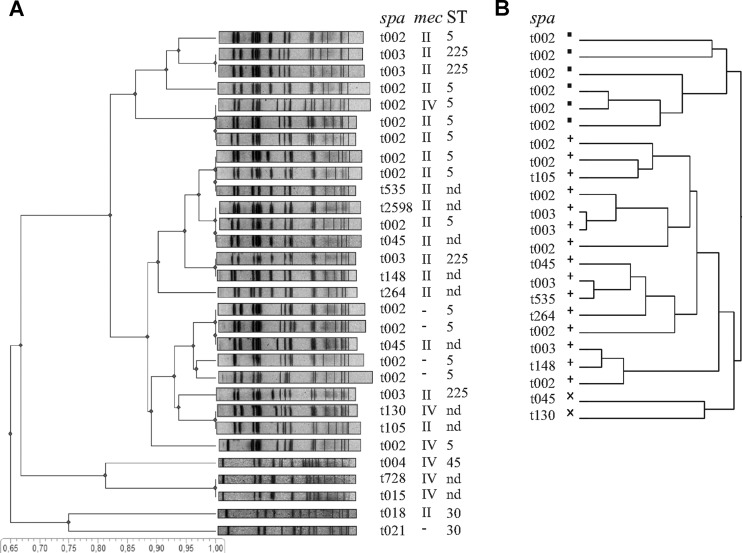
To investigate whether the clusters obtained by spa and MLST typing could be reproduced by other typing techniques, dendrograms based on PFGE and phage typing were generated. The PFGE analysis showed clear clustering of the 79 strains with a similarity index of approximately 87% but demonstrated also the great variability within this S. aureus cluster. The analysis of a subset of 25 isolates is exemplified in Fig. 2A. There is no defined clustering that reflects the spa clonal complexes or MLST types. In contrast, strains of other clonal lineages defined by spa typing could be clearly separated from the Rhine-Hesse cluster. Previously, PFGE results were shown to correlate well with the results from molecular typing methods (spa and MLST) (23, 24). However, our results indicate that PFGE is not suitable to distinguish the ST5 and ST225 isolates, in line with previous observations (25).
Fig 2.
Clustering by PFGE and phage typing. (A) Unweighted-pair group method using average linkages (UPGMA) dendrogram illustrating the similarity (based on the Dice coefficient) of the PFGE profiles of 25 representative Rhine-Hesse isolates and 5 reference isolates. The similarity scale is shown at the bottom. spa clonal complexes (spa), mec types (mec), and MLST types (ST) seem to be randomly distributed in the Rhine-Hesse cluster. (B) Cluster dendrogram of the phage typing results of 25 representative Rhine-Hesse isolates. Data were analyzed by hierarchical clustering computing distances using the Ward method.
Next we analyzed whether the two lineages are distinguishable by phage typing. Phage typing has a discriminatory power that is, at least for the Rhine-Hesse PFGE type, comparable to that of PFGE typing. The first Tübingen isolates that showed the Rhine-Hesse PFGE type belonged to lysogroup III (21). In the meantime, subtle shifts have led to a strong diversification of phage patterns, and the strains that belong to the Rhine-Hesse PFGE type can be assigned to at least 20 related subtypes. We performed a cluster analysis of all strains included in the study, based on phage susceptibility determined using 38 different typing phages. The analysis revealed three distinct clusters without correlation to spa and MLST types (Fig. 2B). Thus, the two spa and MLST lineages (ST5, ST225) could not be recognized by either PFGE or phage typing.
SCCmec typing and the antibiotic resistance pattern of CC5 isolates.
All of the ST225 isolates contained a SCCmec type II cassette. Interestingly, ST5 isolates were significantly (P < 0.001) more diverse: 69% of the isolates belonging to this clonal complex harbored SCCmec type II, 12% harbored SCCmec type IV, and one ST5 isolate could not be assigned to a definite SCCmec type, although it was positive for the mecA gene.
Antibiotic susceptibility testing was correlated to sequence and SCCmec types (see Table S2 in the supplemental material). All ST225 isolates (n = 30) showed resistance to oxacillin, erythromycin, clindamycin, and ciprofloxacin. In three of these isolates, clindamycin resistance was inducible. One isolate demonstrated additional resistance to gentamicin and one to tetracycline. ST5 exhibited resistance profiles that were more diverse: among SCCmec type II isolates, six different resistance patterns were observed. Most of the isolates were resistant to oxacillin, erythromycin, clindamycin, and ciprofloxacin. The other isolates demonstrated various antibiotic resistance patterns, including resistance to gentamicin, rifampin, or fusidic acid. Two isolates lacked phenotypic erythromycin and clindamycin resistance. ST5 SCCmec type IV isolates exhibited four different antibiotic resistance patterns.
Presumably, ST225 was disseminated throughout Germany during the last decade (10), whereas in the Tübingen region, ST5 isolates have remained predominant. Despite their limited distribution, these isolates are more prone to variation regarding antibiotic resistance. The occurrence of ST5 clones with different SSCmec islands indicates that these clones have arisen independently from ST5 MSSA strains. Interestingly, all of the MSSA strains from the region included in this study belong to ST5 and may have served as the ancestors of this locally distributed MRSA strain. This is in line with previous results showing that ST5 strains are heterogeneous with respect to resistance patterns and SSCmec types (9).
Phage content of CC5 isolates.
The molecular basis for the discriminatory power of phage typing is not well understood. Variation in the phage receptor is not likely to account for the strain discrimination, because it was recently shown that most S. aureus phages are recognized by the conserved wall teichoic acids (26). Phage immunity factors (27), leading to the mutual inhibition of phages of the same or other types, may determine the susceptibility to the different phages used for typing. Thus, the differences in phage typing as well as the differences in the banding pattern observed by PFGE might be primarily due to mobilization of phages and thus might reflect short-term changes within the accessory genome that are not reflected by sequence-based methods such as MLST and spa typing. Additionally, certain prophages and/or prophage-carried virulence factors may account for the high epidemicity of these lineages.
To detect prophages, a multiplex PCR assay was applied (20), which revealed that each of the tested phages could be found in CC5 isolates (Table 1), ranging in prevalence from 2.5% (2/79) for Sa4int phages to 92.4% (73/79) for Sa3int phages. An analysis of the distribution of the phages detected in ST5 and ST225 isolates showed a significant (P < 0.0001) association of Sa1int phages with ST225 isolates and of Sa7int phages with ST5 isolates.
Table 1.
Prophage content in the two Rhine-Hesse clusters
| Phage group | % prevalence (no. of isolates) |
Pa | |
|---|---|---|---|
| ST5 (n = 49) | ST225 (n = 30) | ||
| Sa1 | 12 (6) | 57 (17) | <0.0001 |
| Sa2 | 20 (10) | 23 (7) | 1 |
| Sa3 | 90 (44) | 97 (29) | 1 |
| Sa4 | 2 (1) | 3 (1) | 1 |
| Sa5 | 5 (2) | 7 (2) | 1 |
| Sa6 | 2 (1) | 23 (7) | 0.028 |
| Sa7 | 55 (27) | 10 (3) | <0.0001 |
Fisher's exact test with Bonferroni adjustment.
Sa1int phages are found in approximately 10% of S. aureus isolates (20), and in some cases they carry the accessory virulence gene eta, which codes for exfoliative toxin A. Recently, Nübel et al. (10) found a strong association between a certain phage and ST225. Based on the published genome sequence, we could confirm that this phage belongs to the Sa1int phage group, as assigned here.
Sak-encoding Sa7 phages are associated with ST5 SCCmec type II isolates.
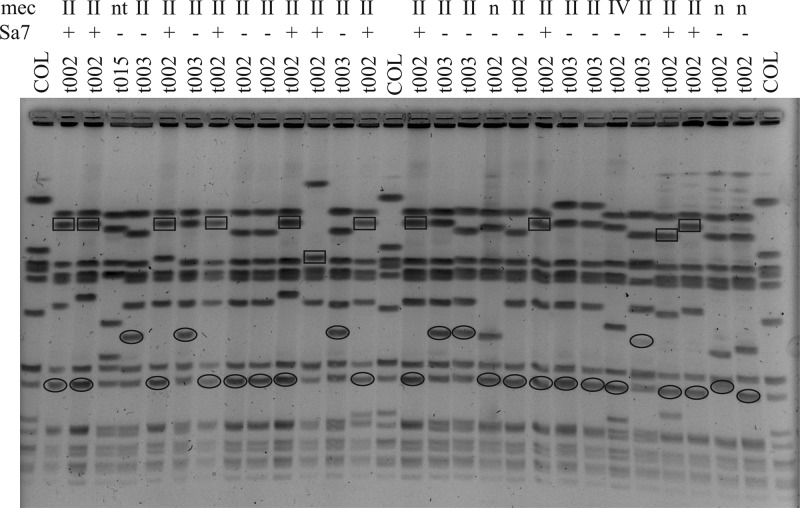
Sa7int phages as detected in the ST5 isolates have not been thoroughly characterized. They were shown to integrate into an attachment site located in an intergenic region upstream of isdB (28). Some of these Sa7int phages carry the virulence determinant staphylokinase (sak), which is usually located on a Sa3int phage. Therefore, we analyzed how many of the Sa7int phages harbor sak using Southern hybridization experiments (Fig. 3). The results revealed that all but one of the Sa7int phages from ST5 SCCmec type II isolates carry sak. In some cases, these isolates harbored an additional sak copy on a Sa3int phage. In contrast, the ST225 and ST5 SCCmec type IV isolates carried sak only on Sa3int phages. The three Sa7int phages detected in ST225 isolates all were sak negative. Thus, the sak-containing Sa7 phage is strongly associated with Tübingen ST5 SCCmec type II isolates.
Fig 3.
Localization of sak on Sa7int and Sa4int phages. SmaI-digested DNAs from randomly chosen isolates were separated by PFGE and hybridized with probes specific for sak and Sa3 and Sa7. Sak could be localized to Sa3 phages (ovals) or Sa7 phages (squares). The corresponding MLST and mec types are indicated above the lanes. COL, SmaI-digested S. aureus strain COL.
To our knowledge, sak-carrying Sa7int phages have not been described in other S. aureus strain collections; thus, this phage may have originated in the Tübingen region through recombination of different phages. Whether this occurred in an ancestral MSSA or MRSA strain is not yet clear. Nevertheless, the now-prevalent ST5 SCCmec type II isolates harboring the Sa7 phage have persisted as nosocomial pathogens over several decades but, so far, have not spread nationwide.
Conclusion.
We report the patterns of variation and dissemination of two related CC5-MRSA clones, ST5 and its single-locus variant ST225, which are not distinguishable by PFGE or phage typing. Interestingly, ST5 strains are locally restricted, are of a high phenotypic diversity, and are characterized by Sa7int phages. In contrast, ST225 isolates are spreading throughout Germany and are highly uniform with regard to SSCmec composition, antibiotic resistance, and the high prevalence of Sa1int phages. The reason why the related lineages ST5 and ST225 differ with respect to local spreading remains to be elucidated. In general, the reason for the high success of CC5 isolates as hospital-associated lineages is unclear, especially because data from whole-genome sequencing did not reveal any obvious mutations within the core genome that might be associated with this trait (9). One may speculate that the accessory elements might drive the spreading of these strains, because the most obvious difference is the phage content. The assumption that mobile genetic elements promote the spreading of bacterial clones was recently emphasized by the emergence of highly virulent MRSA strains carrying a phage harboring a new adherence factor (16).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Wo578/6).
We thank Stefan Borgmann for providing MRSA strains from southeast Germany and Marion Oedenkoven and Monika Pinkwart for expert technical assistance. MRSA reference strains were obtained through the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) program supported under NIAID/NIH contract no. HHSN272200700055C.
Footnotes
Published ahead of print 7 November 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01967-12.
REFERENCES
- 1.Crisóstomo MI, Westh H, Tomasz A, Chung M, Oliveira DC, de Lencastre H. 2001. The evolution of methicillin resistance in Staphylococcus aureus: similarity of genetic backgrounds in historically early methicillin-susceptible and -resistant isolates and contemporary epidemic clones. Proc. Natl. Acad. Sci. U. S. A. 98:9865–9870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Deleo FR, Otto M, Kreiswirth BN, Chambers HF. 2010. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 375:1557–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deurenberg RH, Vink C, Kalenic S, Friedrich AW, Bruggeman CA, Stobberingh EE. 2007. The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 13:222–235 [DOI] [PubMed] [Google Scholar]
- 4.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. 2002. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc. Natl. Acad. Sci. U. S. A. 99:7687–7692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindsay JA, Holden MT. 2004. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 12:378–385 [DOI] [PubMed] [Google Scholar]
- 7.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan HL, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willems RJ, Hanage WP, Bessen DE, Feil EJ. 2011. Population biology of Gram-positive pathogens: high-risk clones for dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35:872–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nübel U, Roumagnac P, Feldkamp M, Song JH, Ko KS, Huang YC, Coombs G, Ip M, Westh H, Skov R, Struelens MJ, Goering RV, Strommenger B, Weller A, Witte W, Achtman M. 2008. Frequent emergence and limited geographic dispersal of methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U. S. A. 105:14130–14135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nübel U, Dordel J, Kurt K, Strommenger B, Westh H, Shukla SK, Zemlickova H, Leblois R, Wirth T, Jombart T, Balloux F, Witte W. 2010. A timescale for evolution, population expansion, and spatial spread of an emerging clone of methicillin-resistant Staphylococcus aureus. PLoS Pathog. 6:e1000855 doi:10.1371/journal.ppat.1000855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Witte WB, C, Heuck D. 2000. MRSA situation in Germany. Hyg. Med. 9:347–354 [Google Scholar]
- 12.Harmsen D, Claus H, Witte W, Rothganger J, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 41:5442–5448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Köck R, Mellmann A, Schaumburg F, Friedrich AW, Kipp F, Becker K. 2011. The epidemiology of methicillin-resistant Staphylococcus aureus (MRSA) in Germany. Dtsch. Arztebl. Int. 108:761–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellmann A, Friedrich AW, Rosenkotter N, Rothganger J, Karch H, Reintjes R, Harmsen D. 2006. Automated DNA sequence-based early warning system for the detection of methicillin-resistant Staphylococcus aureus outbreaks. PLoS Med. 3:e33 doi:10.1371/journal.pmed.0030033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersdorf S, Oberdorfer K, Wendt C. 2006. Longitudinal study of the molecular epidemiology of methicillin-resistant Staphylococcus aureus at a university hospital. J. Clin. Microbiol. 44:4297–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li M, Du X, Villaruz AE, Diep BA, Wang D, Song Y, Tian Y, Hu J, Yu F, Lu Y, Otto M. 2012. MRSA epidemic linked to a quickly spreading colonization and virulence determinant. Nat. Med. 18:816–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy ME, Etienne J. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oliveira DC, de Lencastre H. 2002. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 46:2155–2161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goerke C, Matias y Papenberg S, Dasbach S, Dietz K, Ziebach R, Kahl BC, Wolz C. 2004. Increased frequency of genomic alterations in Staphylococcus aureus during chronic infection is in part due to phage mobilization. J. Infect. Dis. 189:724–734 [DOI] [PubMed] [Google Scholar]
- 20.Goerke C, Pantucek R, Holtfreter S, Schulte B, Zink M, Grumann D, Broker BM, Doskar J, Wolz C. 2009. Diversity of prophages in dominant Staphylococcus aureus clonal lineages. J. Bacteriol. 191:3462–3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dommke A, Lenz W, Bierbaum G, Werner H. 2000. Typing of Staphylococcus aureus strains isolated from prematurely born infants and patients of intensive care units. Hyg. Med. 25:129–134 [Google Scholar]
- 22.Schaumburg F, Kock R, Mellmann A, Richter L, Hasenberg F, Kriegeskorte A, Friedrich AW, Gatermann S, Peters G, von Eiff C, Becker K. 2012. Population dynamics among methicillin-resistant Staphylococcus aureus isolates in Germany during a 6-year period. J. Clin. Microbiol. 50:3186–3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cookson BD, Robinson DA, Monk AB, Murchan S, Deplano A, de Ryck R, Struelens MJ, Scheel C, Fussing V, Salmenlinna S, Vuopio-Varkila J, Cuny C, Witte W, Tassios PT, Legakis NJ, van Leeuwen W, van Belkum A, Vindel A, Garaizar J, Haeggman S, Olsson-Liljequist B, Ransjo U, Muller-Premru M, Hryniewicz W, Rossney A, O'Connell B, Short BD, Thomas J, O'Hanlon S, Enright MC. 2007. Evaluation of molecular typing methods in characterizing a European collection of epidemic methicillin-resistant Staphylococcus aureus strains: the HARMONY collection. J. Clin. Microbiol. 45:1830–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grundmann H, Hori S, Enright MC, Webster C, Tami A, Feil EJ, Pitt T. 2002. Determining the genetic structure of the natural population of Staphylococcus aureus: a comparison of multilocus sequence typing with pulsed-field gel electrophoresis, randomly amplified polymorphic DNA analysis, and phage typing. J. Clin. Microbiol. 40:4544–4546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strommenger B, Kettlitz C, Weniger T, Harmsen D, Friedrich AW, Witte W. 2006. Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J. Clin. Microbiol. 44:2533–2540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia G, Corrigan RM, Winstel V, Goerke C, Grundling A, Peschel A. 2011. Wall teichoic acid-dependent adsorption of staphylococcal siphovirus and myovirus. J. Bacteriol. 193:4006–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 8:317–327 [DOI] [PubMed] [Google Scholar]
- 28.Goerke C, Wirtz C, Fluckiger U, Wolz C. 2006. Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol. Microbiol. 61:1673–1685 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.