Abstract
A transrectal prostate biopsy is the most common procedure used to establish the diagnosis of prostate cancer. Prior to biopsy, patients are commonly given ciprofloxacin for prophylaxis. However, a complication of the procedure is infection with ciprofloxacin-resistant organisms, in particular resistant Escherichia coli. In order to identify patients carrying ciprofloxacin-resistant E. coli, so as to tailor their antibiotic prophylaxis, rectal swabs are screened using selective broth and/or solid medium. In our evaluation, we compared broth enrichment and direct plating techniques by using brain heart infusion broth and MacConkey agar containing 1 μg/ml or 10 μg/ml of ciprofloxacin. Of the 100 patients included in the study, 20 were colonized with ciprofloxacin-resistant organisms, 19 of which were E. coli. There was no significant difference (P > 0.1) between the culture methods or the ciprofloxacin concentrations in the medium when identifying patients with ciprofloxacin-resistant E. coli; however, broth enrichment using 1 μg/ml ciprofloxacin was the most sensitive at 100%, but it was the least specific. Direct plating of rectal swabs onto MacConkey agar containing 10 μg/ml of ciprofloxacin was 100% specific and missed only 1 positive specimen, with a sensitivity of 94.7%; this method was the most cost-effective. Therefore, direct plating of rectal swabs onto selective medium proved to be a sensitive and cost-effective approach in identifying patients colonized with ciprofloxacin-resistant E. coli.
INTRODUCTION
Transrectal ultrasound-guided prostate biopsy (TRUSP) is one of the most common methods to provide a tissue diagnosis of prostate cancer, with over 1 million such procedures performed in the United States each year (1). Urinary tract infection, prostatitis, epididymo-orchitis, and sepsis have been traditionally cited as the most common post-TRUSP infectious complications (2). Fluoroquinolones, in particular ciprofloxacin, have been the most widely used antibiotics for prophylaxis prior to the biopsy procedure, as they have been shown to significantly decrease the infection rate (3, 4). However, an increasing trend in infectious complications, largely due to fluoroquinolone-resistant organisms, in particular Escherichia coli, has been reported (2, 5, 6).
The mechanism thought to cause the increasing infection rate is the carriage of ciprofloxacin-resistant organisms in the rectal vault, which at the time of the biopsy are introduced by the biopsy needle transversing the contaminated rectal wall, thereby introducing them directly into the prostate and occasionally the bladder (7). This has prompted investigations into the prevalence of ciprofloxacin-resistant organisms in the rectal flora at the time of the biopsy, which has been estimated to be between 10 and 22% (7–9).
Two recent studies investigated the use of rectal cultures to provide a targeted approach to prophylaxis (10, 11). Both studies showed a 0% infection rate after implementation of rectal cultures to guide prophylactic antibiotic therapy and also a cost reduction related to the prevention of hospitalization due to infectious complications. Unfortunately, neither of these studies was a true randomized study nor sufficiently powered to detect a substantial difference. In addition, previous studies investigating ciprofloxacin resistance in fecal flora used different methods of collection, isolation techniques, and timing relative to the prostate biopsy (7–12).
We have now expanded on a previous study in which we reported using an enrichment broth containing 10 μg/ml of ciprofloxacin to detect ciprofloxacin-resistant E. coli from rectal swabs (13). In the present report, we compared a broth enhancement culture to direct plating for the laboratory detection of ciprofloxacin-resistant E. coli in rectal swabs from patients prior to antibiotic prophylaxis preceeding a TRUSP. In addition, we also compared these two approaches using two different concentrations of ciprofloxacin, 1 μg/ml and 10 μg/ml, in the selective culture medium.
MATERIALS AND METHODS
Specimens.
After Institutional Review Board approval, patients at the Long Beach Veterans Medical Center (Long Beach, CA) who were scheduled for a TRUSP and were not presently on antibiotics were enrolled, from 9/12/2011 to 4/20/2012. Three rectal swabs (Copan Diagnostics, Murrieta, CA) were obtained in the office. The swabs were transported to the laboratory on the day of collection and refrigerated until processed.
Media and cultures.
All media containing ciprofloxacin were obtained from Hardy Diagnostics (Santa Maria, CA), and all other media were obtained from BBL (Becton, Dickinson, Sparks, MD).
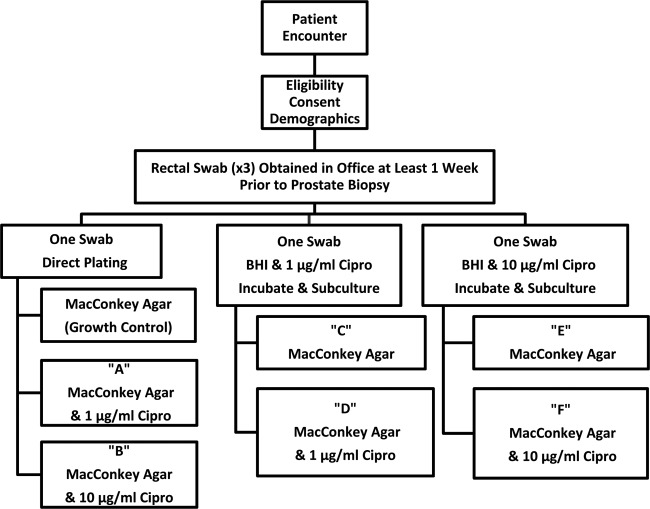
The overall plating scheme for the three different swabs is shown in Fig. 1. Within 24 h of receipt in the laboratory, one rectal swab was emulsified in 0.5 ml of sterile saline and ∼0.05 ml was inoculated directly onto MacConkey agar, which served as a specimen growth control, MacConkey agar containing 1 μg/ml of ciprofloxacin, and MacConkey agar containing 10 μg/ml of ciprofloxacin. The other two swabs were used to inoculate either 5 ml of brain heart infusion (BHI) broth containing 1 μg/ml of ciprofloxacin or BHI broth containing 10 μg/ml of ciprofloxacin. Broth cultures were incubated overnight at 35°C and then subcultured onto MacConkey agar without and with the corresponding broth-ciprofloxacin concentration. Plates were incubated overnight at 35°C in ambient air. Organisms that grew directly from ciprofloxacin-containing agar or a broth subculture were identified using the Vitek 2 GN-ID card (bioMérieux, Durham, NC). The MICs for nalidixic acid and ciprofloxacin were established using an Etest (bioMérieux), and the Clinical and Laboratory Standards Institute guidelines were used to perform and interpret the MIC results (14).
Fig 1.
Schematic of the different culture approaches used in this evaluation.
Statistics.
Predictive values were calculated, and a Fisher exact test was used to compare the different methods to one another. A consensus gold standard was used to define a true positive as being a specimen from which a ciprofloxacin-resistant E. coli isolate was detected from any of the three swabs collected, regardless of the medium from which it was detected.
RESULTS
A total of 106 men were enrolled into the study. The average age of the population was 65 years, and average body mass index was 27. Of the patients, 71% were presenting for their first-time prostate biopsy. Other patient characteristics are displayed in Table 1. Six men were excluded due to an insufficient sample, as determined by absence of growth when their rectal swab was plated directly onto MacConkey agar without ciprofloxacin. These men did not differ in characteristics from the study population.
Table 1.
Patient characteristics
| Patient characteristic | N | Median value (range) |
|---|---|---|
| Age (yrs) | 100 | 65 (48–84) |
| Body mass index (kg/m2) | 100 | 28.7 (20.5–50.7) |
| Prostate specific antigen (ng/ml) | 100 | 6.0 (0.4–59.1) |
| No. of days prior to biopsy that culture was obtained | 83 | 14 (4–119) |
| Charlson comorbidity index | 100 | 2 (0–8) |
| First-time prostate biopsy | 71 | |
| No. of previous biopsies | ||
| 1 | 19 | |
| 2 | 6 | |
| >2 | 4 | |
| No. with diabetes | 29 | |
| Race or ethnicitya | ||
| Caucasian | 72 | |
| Black | 7 | |
| Hispanic | 15 | |
| Asian | 5 | |
| Hawaiian | 2 |
One subject had two race/ethnicity identities.
The overall results of the culture methods are summarized in Table 2. There were 20 individuals from whom a ciprofloxacin-resistant organism was recovered. All ciprofloxacin-resistant isolates were E. coli, except for one Pseudomonas putida isolate. Therefore, for purposes of data analysis, there were 19 true positives, i.e., individuals from which ciprofloxacin-resistant E. coli was isolated. All 19 isolates had a MIC to ciprofloxacin of ≥32 μg/ml and a MIC to nalidixic acid of ≥256 μg/ml.
Table 2.
Overall results with the different culture methods
| Test parameter | Direct plating on MacConkey agar with indicated ciprofloxacin concn |
1-μg/ml ciprofloxacin broth subcultured to: |
10-μg/ml ciprofloxacin broth subcultured to: |
|||
|---|---|---|---|---|---|---|
| 1 μg/ml (method A) | 10 μg/ml (method B) | 0 μg/ml (method C) | 1 μg/ml (method D) | 0 μg/ml (method E) | 10 μg/ml (method F) | |
| True positivea | 17 | 18 | 19 | 19 | 18 | 18 |
| False positive | 2 | 0 | 13b | 4c | 1 | 0 |
| False negative | 2 | 1 | 0 | 0 | 1 | 1 |
| True negative | 79 | 81 | 68 | 77 | 80 | 81 |
| Sensitivity (%) | 89.5 | 94.7 | 100 | 100 | 94.7 | 94.7 |
| Specificity (%) | 97.5 | 100 | 84.0 | 95.1 | 98.8 | 100 |
| Positive predictive value (%) | 85.0 | 100 | 59.4 | 82.6 | 94.7 | 100 |
| Negative predictive value (%) | 97.6 | 98.8 | 100 | 100 | 98.8 | 98.8 |
| Cost of media and supplies ($)d,e | 6.35 | 4.62 | 7.63 | 10.11 | 5.54 | 7.36 |
| Labor (min)e | 5.0 | 4.9 | 6.6 | 6.3 | 6.0 | 5.9 |
P > 0.1 by Fischer's exact test for each method compared to one another regarding ability to detect true-positive specimens.
P = 0.0016 for method C compared to methods B and F.
P = 0.03 for method D compared to methods B and F.
The cost of media included selective and nonselective agar and broth, Vitek GN-ID cards, and the ciprofloxacin Etest.
Labor was calculated in minutes and included processing of specimens, computer entry, culture evaluation, susceptibility testing, identification, and computer reporting. The total cost for 100 specimens for each method was calculated, including the added cost required for the true-positive and false-positive specimens, and this total was divided by 100. Shown are the average minutes required for each specimen.
There were no significant differences between any of the methods (P > 0.1) in the detection of specimens containing ciprofloxacin-resistant E. coli. All 19 true positives were recovered from the broth containing 1 μg/ml of ciprofloxacin that was plated onto MacConkey agar with or without 1 μg/ml of ciprofloxacin. In contrast, the method where a rectal swab was directly inoculated onto MacConkey agar containing 1 μg/ml of ciprofloxacin was the least sensitive, detecting 89.5% (17/19) of the positive specimens.
The largest difference among the methods was seen with the 14 specimens that yielded a false positive, which was defined as an organism that was recovered from the ciprofloxacin-containing media that was susceptible to ciprofloxacin, i.e., a ciprofloxacin MIC of ≤1 μg/ml (14). In these instances, the BHI broth containing 1 μg/ml ciprofloxacin when subcultured to MacConkey without ciprofloxacin (method C [Table 2]) had the largest number of false-positive specimens, 13. This was significant compared to specimens directly plated onto medium containing the higher concentration of ciprofloxacin, 10 μg/ml, whether the rectal swab was plated directly (method B [Table 2]) or plated after overnight growth in BHI broth containing the higher concentration of ciprofloxacin (method F [Table 2]).
Of the 14 specimens that resulted in false positives, a single organism was recovered from 12 specimens, while two organisms were isolated from each of two specimens. Organisms isolated were 8 isolates of E. coli, 3 of Klebsiella, 2 of Pseudomonas, 1 of Citrobacter, 1 of Morganella, and 1 of Stenotrophomonas. Susceptibilities to nalidixic acid and ciprofloxacin were determined on 10 of these isolates. Here, 60% (6/10) were resistant to nalidixic acid but susceptible to ciprofloxacin, while 4 were susceptible to both antibiotics. All the false positives recovered from direct plating onto MacConkey agar containing 1 μg/ml of ciprofloxacin were resistant to nalidixic acid but susceptible to ciprofloxacin. The one false-positive isolate recovered from the 10-μg/ml ciprofloxacin broth enhancement culture subcultured to MacConkey agar grew a Klebsiella spp. resistant to nalidixic acid but susceptible to ciprofloxacin. Therefore, the majority of the false positives were attributed to organisms isolated from media containing the lower concentration of ciprofloxacin, 1 μg/ml, and many of these isolates, while susceptible to ciprofloxacin, were resistant to nalidixic acid.
The cost of the different culture approaches varied, as can be seen in Table 2. Direct plating onto MacConkey agar containing 10 μg/ml ciprofloxacin, method B, was the least expensive in terms of both media and supplies as well as labor, as measured by time. In comparison, subculturing from BHI broth containing 1 μg/ml ciprofloxacin was the most expensive, i.e., methods C and D. This was due to the additional cost of the ciprofloxacin-containing broth and the additional cost for the labor and supplies required for susceptibility and identification of the false positives. In general, methods using media containing 1 μg/ml of ciprofloxacin were more expensive than the identical approach using media with 10 μg/ml of ciprofloxacin. This is illustrated in Table 2, in which method A is compared to B, method C to E, and method D to F.
DISCUSSION
Due to the increasing awareness of infectious complications after a prostate biopsy and the strong association of these infections with ciprofloxacin-resistant E. coli, rectal cultures to guide targeted antibiotic prophylaxis have been suggested as an approach to reduce post-biopsy infections (1, 2, 7, 9–11). In a series of patients undergoing TRUSP, Steensels et al. (9) reported a direct correlation between the increasing isolation of ciprofloxacin-resistant E. coli from urine cultures of men undergoing TRUSP from 2003 to 2009 and the incidence of sepsis after TRUSP biopsy. This same study group reported that in a prospective study, of 236 patients undergoing TRUSP biopsy there were 7 who had infectious complications, and all were caused by ciprofloxacin-resistant E. coli (9). Feliciano et al. (5) noted that samples from 89% of those patients presenting with post-TRUSP infectious complications contained E. coli, of which 80% were ciprofloxacin resistant. Therefore, it is clear that the majority of complications from TRUSP are due to ciprofloxacin-resistant E. coli (8).
Multiple groups have performed studies on ciprofloxacin-resistant E. coli colonization prior to TRUSP by using different culture approaches, including broth enhancement, direct plating, media employed that were nonselective or selective, and use of different concentrations of ciprofloxacin incorporated into the media (8, 11–13). In order to determine which approach is the most sensitive and specific yet cost-effective, we evaluated both broth enrichment and direct plating, using MacConkey agar and two concentrations of ciprofloxacin. We found that broth enrichment using 1 μg/ml ciprofloxacin was the most sensitive; however, results under these conditions were not statistically different from those with direct plating onto ciprofloxacin-containing agar and were the most expensive. In addition, broth enhancement using 1 μg/ml of ciprofloxacin doubled the cost of the culture and resulted in the workup of significantly more false-positive organisms, also adding to the overall cost.
False-positive rates would incur a substantial cost and therefore decrease the cost savings of a targeted prophylaxis approach compared to the prevention per TRUSP infectious complication. The financial aspect of targeted antibiotic prophylaxis using rectal culture screening must have a favorable cost-benefit ratio in order for the test to be widely accepted. Taylor et al. (11) provided a cost analysis in a recent study in which 457 men were enrolled. In that study, 112 men were in the targeted antibiotic prophylaxis group. A rectal swab was obtained to screen for ciprofloxacin-resistant organisms. When a ciprofloxacin-resistant organism was recovered from the specimen, prophylaxis was adjusted prior to TRUSP biopsy. This group was compared to 345 controls who were not screened for ciprofloxacin-resistant organisms and who were all given ciprofloxacin prior to biopsy. There were no infectious complications in the targeted antibiotic prophylaxis group, compared to 9/345 (2.6%) of men in the control group who had an infectious complication. Those authors described a cost savings of $4,499, based on comparison of targeted prophylaxis to the costs of managing post-TRUSP infectious complications. Here, rectal swabs were directly plated onto MacConkey agar containing 1 μg/ml ciprofloxacin. This culture approach in our study was the third least expensive, with direct plating using 10 μg/ml ciprofloxacin being the least expensive. The difference in these two direct methods was due to the false positives when we used the lower ciprofloxacin concentration, which had to be further tested for ciprofloxacin susceptibility.
While a limitation of our findings is the relatively small sample size, our study revealed that overall there was not a significant difference in culture approaches. A valid concern when using selective agar or broth containing 10 μg/ml of ciprofloxacin is that specimens containing organisms with a ciprofloxacin MIC >1 μg/ml and <10 μg/ml may be missed. However, all specimens identified as a true positive contained a ciprofloxacin-resistant E. coli with a high MIC to ciprofloxacin, ≥32 μg/ml. Therefore, based on this limited sample, resistant isolates with a ciprofloxacin MIC of <10 μg/ml were not encountered. In conclusion, the least expensive and specific approach for detecting patients colonized with ciprofloxacin-resistant E. coli was direct plating using MacConkey agar containing 10 μg/ml ciprofloxacin, although this method did fail to identify one specimen containing ciprofloxacin-resistant E. coli.
ACKNOWLEDGMENTS
We thank Hardy Diagnostics for providing the ciprofloxacin-containing media used for this study, as well as Amy Nakama and Kaye Evans for their technical support.
Footnotes
Published ahead of print 14 November 2012
REFERENCES
- 1. Loeb S, Carter HB, Berndt SI, Ricker W, Schaeffer EM. 2011. Complications after prostate biopsy: data from SEER-Medicare. J. Urol. 186:1830–1834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nam RK, Saskin R, Lee Y, Liu Y, Law C, Klotz LH, Loblaw DA, Trachtenberg J, Stanimirovic A, Simor AE, Seth A, Urbach DR, Narod SA. 2010. Increasing hospital admission rates for urological complications after transrectal ultrasound guided prostate biopsy. J. Urol. 183:963–968 [DOI] [PubMed] [Google Scholar]
- 3. Aron M, Rajeev TP, Gupta NP. 2000. Antibiotic prophylaxis for transrectal needle biopsy of the prostate: a randomized controlled study. BJU Int. 85:682–685 [DOI] [PubMed] [Google Scholar]
- 4. Wolf JS, Jr, Bennett CJ, Dmochowski RR, Hollenbeck BK, Pearle MS, Schaeffer AJ. 2008. Best practice policy statement on urologic surgery antimicrobial prophylaxis. J. Urol. 179:1379–1390 [DOI] [PubMed] [Google Scholar]
- 5. Feliciano J, Teper E, Ferrandino M, Macchia RJ, Blank W, Grunberger I, Colon I. 2008. The incidence of fluoroquinolone resistant infections after prostate biopsy: are fluoroquinolones still effective prophylaxis? J. Urol. 179:952–955 [DOI] [PubMed] [Google Scholar]
- 6. Loeb S, van den Heuvel S, Zhu X, Bangma CH, Schroder FH, Roobol MJ. 2012. Infectious complications and hospital admissions after prostate biopsy in a European randomized trial. Eur. Urol. 61:1110–1114 [DOI] [PubMed] [Google Scholar]
- 7. Liss MA, Chang A, Santos R, Nakama-Peeples A, Peterson EM, Osann K, Billimek J, Szabo RJ, Dash A. 2011. Prevalence and significance of fluoroquinolone resistant Escherichia coli in patients undergoing transrectal ultrasound guided prostate needle biopsy. J. Urol. 185:1283–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Batura D, Rao GG, Nielsen PB. 2010. Prevalence of antimicrobial resistance in intestinal flora of patients undergoing prostatic biopsy: implications for prophylaxis and treatment of infections after biopsy. BJU Int. 106:1017–1020 [DOI] [PubMed] [Google Scholar]
- 9. Steensels D, Slabbaert K, De Wever L, Vermeersch P, Van Poppel H, Verhaegen J. 2012. Fluoroquinolone-resistant E. coli in intestinal flora of patients undergoing transrectal ultrasound-guided prostate biopsy: should we reassess our practices for antibiotic prophylaxis? Clin. Microbiol. Infect. 18:575–581 [DOI] [PubMed] [Google Scholar]
- 10. Duplessis CA, Bavaro M, Simons MP, Marguet C, Santomauro M, Auge B, Collard DA, Fierer J, Lesperance J. 2012. Rectal cultures before transrectal ultrasound-guided prostate biopsy reduce post-prostatic biopsy infection rates. Urology 79:556–563 [DOI] [PubMed] [Google Scholar]
- 11. Taylor AK, Zembower TR, Nadler RB, Scheetz MH, Cashy JP, Bowen D, Murphy AB, Dielubanza E, Schaeffer AJ. 2012. Targeted antimicrobial prophylaxis using rectal swab cultures in men undergoing transrectal ultrasound guided prostate biopsy is associated with reduced incidence of postoperative infectious complications and cost of care. J. Urol. 187:1275–1279 [DOI] [PubMed] [Google Scholar]
- 12. Minamida S, Satoh T, Tabata K, Kimura M, Tsumura H, Kurosaka S, Matsumoto K, Fujita T, Iwamura M, Baba S. 2011. Prevalence of fluoroquinolone-resistant Escherichia coli before and incidence of acute bacterial prostatitis after prostate biopsy. Urology 78:1235–1239 [DOI] [PubMed] [Google Scholar]
- 13. Liss MA, Peeples AN, Peterson EM. 2011. Detection of fluoroquinolone-resistant organisms from rectal swabs by use of selective media prior to a transrectal prostate biopsy. J. Clin. Microbiol. 49:1116–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clinical and Laboratory Standards Institute 2010. Performance standards for antimicrobial susceptibility testing. Twentieth informational supplement M100-S20. CLSI, Wayne, PA [Google Scholar]