Abstract
The profiles of 61 Candida tropicalis isolates from 43 patients (28 adults and 15 children) diagnosed with candidemia at two teaching hospitals in São Paulo, Brazil, were characterized by multilocus sequence typing (MLST). For the 14 patients who had bloodstream infections, 32 isolates were serially collected from their blood and/or catheters. Thirty-nine diploid sequence types (DSTs) were differentiated. According to the C. tropicalis MLST database (http://pubmlst.org/ctropicalis/), 36 DSTs and 23 genotypes identified from the 61 isolates had not previously been described. This report represents the first study to characterize sequential isolates of C. tropicalis from candidemia cases in South America. Microvariation in a single gene was found in the sequential isolates from 7 patients. The main polymorphisms occurred in the alleles of the XYR1 gene, specifically at nucleotide positions 215, 242, and 344. Macrovariation in six gene fragments was detected in the isolates from 3 patients. eBURST analysis added two new groups to this study (groups 6 and 18). Additionally, susceptibility tests indicate that 3 isolates were resistant to fluconazole. No correlation was found between the DSTs and susceptibility to fluconazole and/or selective antifungal pressure. Two patients were sequentially infected with resistant and susceptible strains. MLST is an important tool for studying the genetic diversity of multiple/sequential isolates of patients with candidemia, allowing the comparison of our data with those from other regions of the world, as well as allowing an analysis of the genetic relationship among several clones in sequential isolates from the same or different candidemia patient sites (blood or catheter).
INTRODUCTION
Infections caused by Candida spp. have been characterized as important causes of nosocomial infections and are associated with high morbidity and mortality. Additionally, these infections have been related to higher hospitalization costs because of longer hospitalization times and intensive care needs (1–8).
Candida tropicalis is one of the most frequently isolated species in invasive candidiasis, ranging from 4% to 24% of all candidemia cases, depending on the geographical area and other predisposing conditions, such as cancer, neutropenia, diabetes in adults and the elderly, and lung disease in pediatric patients (9–21).
In the ARTEMIS DISK Global Antifungal Surveillance Study, which covered the period from 1997 to 2007, Pfaller et al. (22) studied fluconazole in vitro susceptibility test results for 256,882 isolates of Candida spp. collected from 142 sites in 41 countries. Increased rates of isolation of C. tropicalis (5.4% to 8.0%) were noted. Overall, 90.2% of Candida isolates tested were susceptible to fluconazole. In addition, 91% of C. tropicalis isolates were fluconazole susceptible and 4.1% were resistant. Fluconazole resistance varies according to geographic region. The highest rates of resistance were observed in isolates from the Asia-Pacific region (6.5%). Europe presented 2.9% resistance and North America 4.4%. The lowest rates of resistance were observed from the Africa/Middle East region and Latin America, with both presenting 2.6% resistance.
Fluconazole resistance in C. tropicalis isolates has been described in Brazil, although it is a rare event. Colombo et al. (23) reported that from 2003 to 2004, in Brazilian hospitals, there were 712 cases of candidemia, with an incidence of 2.49 cases per 1,000 admissions. Candida albicans was the most common species (40%), followed by C. tropicalis (20.9%) and C. parapsilosis (20.5%). Fluconazole resistance occurred in 6 (0.8%) isolates (C. krusei, C. glabrata, and C. albicans). In addition, 27 cases (4%) were classified as having dose-dependent susceptibility to fluconazole (C. glabrata, C. krusei, C. tropicalis, and C. albicans).
The largest Brazilian study of fungemia by C. tropicalis was published by Nucci and Colombo (18), with a total of 924 episodes of candidemia in 906 patients from 12 Brazilian institutions. C. albicans was the most frequent (384 cases, 41.5%), followed by C. tropicalis (188 cases, 20%) and C. parapsilosis (187 cases, 20%). The proportion of candidemia caused by C. tropicalis ranged from 15.7% to 25.8% among the 12 institutions. Antifungal resistance was a rare event and was restricted to one isolate of C. albicans and one of C. tropicalis.
In a multicenter study in São Paulo (Brazil), Da Matta et al. (24) analyzed 1,000 isolates of candidemia from 1995 to 2003. In the first period, from 1995 to 1999, C. tropicalis was the second most frequent species (74/299). In the second period, from 2000 to 2003, C. tropicalis was the third most frequent (169/701), whereas C. albicans and C. parapsilosis were the two most frequent. All C. tropicalis isolates were fluconazole susceptible.
According to Costa et al. (25), at the Clinics Hospital of the University of São Paulo Medical School (FMUSP) in São Paulo (Brazil), 86 fungemia patients, treated between 1994 and 1996, were described as having C. albicans (50%), C. parapsilosis (17%), and C. tropicalis (12%). The fungemia mortality rate was 70%. At the same hospital in 2006, Motta et al. (28) found an incidence of 1.87 candidemia cases/1,000 admissions. The main species isolated were C. albicans (52.2%), C. parapsilosis (22.1%), C. tropicalis (14.8%), and C. glabrata (6.6%). The C. tropicalis isolates were 100% susceptible to fluconazole.
Several molecular techniques with high discriminatory power and reproducibility have been described, such as multilocus sequence typing (MLST), which is based on the analysis of single-nucleotide polymorphisms (SNPs) in housekeeping gene fragments of up to 500 bp (as detected by molecular sequencing) (29–31). Fragments of the C. tropicalis genes ICL1, MDR1, SAPT2, SAPT4, XYR1, and ZWF1a have already been analyzed (32, 33). Previous reports have indicated that MLST can provide additional information on the mechanisms of genetic variability, such as the persistence, replacement, and microevolution of strains in the hosts (30, 34, 35).
MLST has been used to establish the relationship between the DSTs with susceptibility profile to fluconazole and the trailing phenomenon (27, 32, 36). In Brazil, fluconazole is an effective systemic antifungal drug against most isolates of C. tropicalis (24). No studies in this country have reported on the MLST of C. tropicalis and its correlation with antifungal susceptibility tests.
The objective of this study was to use MLST to analyze the presence of genetic variation among sequential isolates from blood and catheter samples from the same patient and to characterize the fluconazole susceptibility pattern in patients with bloodstream infections caused by C. tropicalis at two teaching hospitals in São Paulo, Brazil, between 1998 and 2003.
MATERIALS AND METHODS
Case definition and C. tropicalis isolates.
A total of 61 C. tropicalis isolates from 43 patients diagnosed with bloodstream infection and who were admitted to two hospitals (Clinics Hospital of the University of São Paulo Medical School [HCFMUSP] and the Clinics Hospital of the University of Campinas [UNICAMP]) and isolated from 1998 to 2003 were studied. In 14 patients with bloodstream infection, 32 isolates were serially collected from their blood and/or catheters. The cultures were initially identified by the mycology section at the HCFMUSP central laboratory using automated methods (Vitek, bioMérieux, Nürtingen, Germany) and carbon assimilation using an API20C strip (bioMérieux SA, Paris, France). The UNICAMP central laboratory used a Vitek system and a chromogenic method (CHROMagar Candida Microbiology, Paris, France). All of the isolates were reidentified using the ID 32C method (BioMerieux SA, Paris, France) for standardization. The yeasts were maintained on Sabouraud agar (Oxoid, Basingstoke, United Kingdom) and BHI (BHI-Difco) glycerol at −80°C until use. The standard strains of C. krusei (ATCC 6258) and C. parapsilosis (ATCC 22019) were used.
DNA extraction.
DNA extraction was performed according to the method described by Buchman et al. (37). The samples were cultured in liquid Sabouraud medium (Oxoid, Basingstoke, United Kingdom), stirred for 48 h at 30°C and then treated with lyticase (Sigma-Aldrich). This procedure was followed by the addition of lysis buffer consisting of sorbitol, β-mercaptoethanol, and proteinase K (Invitrogen). A 1:1 mixture of phenol and chloroform was used to perform the DNA purification. The DNA precipitation was performed with 3 M sodium acetate and 1 ml of cold ethanol. The DNA was resuspended in 100 ml of TE, measured, and stored at −20°C until use.
MLST typing.
The MLST method and PCR amplification were performed as described by Tavanti et al. (33). Briefly, oligonucleotide primers were used for six gene fragments: ICL1, MDR1, SAPT2, SAPT4, XYR1, and ZWF1a. The PCR results were revealed using 2% agarose gel electrophoresis, which displayed a single band. The DNA was purified with GFX PCR and a gel band purification kit (GE Health Care Life Sciences, United Kingdom).
Forward and reverse sequencing of the fragments was performed with an ABI 3500 automated DNA sequencer (Applied Biosystems) using the original PCR primers and the BigDye Terminator v3.1 cycle sequencing kit (Applied Biosystems) according to the manufacturer's guidelines. The quality of each electropherogram was evaluated using Phred-Phrap software (38, 39), and consensus sequences were obtained using CAP3 software, which is available at http://asparagin.cenargen.embrapa.br/phph/.
The chromatograms were visually checked and scrutinized for heterozygosity. Subsequently, the SNPs in the six previously sequenced gene fragments were concatenated into a single sequence, as described by Tavanti et al. (33). The alignment was performed with the MEGA 5.0 (40) and BioEdit (http://www.mbio.ncsu.edu/bioedit/page2.html) software. The sequences were compared with others from GenBank (http://www.ncbi.nlm.nih.gov/GenBank/tbl2asn2/) and submitted to the Candida tropicalis MLST database (http://pubmlst.org/ctropicalis/), as described by Jolley et al. (41).
The new allelic profiles and new allele combinations (new DSTs) were submitted to the C. tropicalis MLST database. Heterozygosity was defined by the presence of overlapping peaks in the forward and reverse chromatograms. A haplotype analysis was subsequently performed. Microvariation (referred to as “microevolution”) was defined as minor genetic changes and minor variations in genetically determined strain types in surveys of multiple isolates from individual patients (30, 31). “Macrovariation” was defined as C. tropicalis strains with variations/genetic changes in five or six gene fragments.
Phylogenetic analysis.
A phylogenetic analysis was obtained through the following clustering methods: unweighted pair group method with arithmetic averages (UPGMA), neighbor-joining, maximum parsimony, maximum likelihood, and Bayesian inference. Unrooted trees were created using MEGA 5.0 and MrBayes 3.2.1 software (http://mrbayes.sourceforge.net/download.php) and the BEAST software package (http://beast.bio.ed.ac.uk/Programs), which includes the programs BEAST 1.7.2, BEAUti, LogCombiner, Treelog Analyzer, Tracer, and Figtree. The best evolutionary history was inferred using the maximum likelihood method, which is based on the Tamura-Nei model (42). The bootstrap consensus tree inferred from 2,000 replicates is taken to represent the evolutionary history of the analyzed taxa. A discrete gamma distribution was used to model the evolutionary rate differences among the sites (16 categories, +G [discrete gamma distribution], parameter = 0.584). The analysis involved 39 nucleotide sequences. All of the ambiguous positions were removed for each sequence pair. There were 416 positions in the final data set. The evolutionary analyses were conducted in MEGA 5.0. Finally, the 267 sequences described in the C. tropicalis MLST database were inferred using Bayesian inference. The groups were numbered to be consistent with bootstrap values (43) greater than 70%. The discriminatory power was calculated using Simpson's index of diversity (44, 45). The ancestor-descendant relationship among the isolates was studied with the eBURST package, v3.0 (http://eburst.mlst.net/).
Antifungal susceptibility testing.
The stock solutions of antifungal agents, inocula, and plates were prepared according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI) document M27-A2/A3 (46, 47). The results were read visually and using a spectrophotometer (Versa Max tunable microplate reader) with a wavelength of 530 nm. The MICs of fluconazole were defined as the lowest concentration of the drug that was able to inhibit the growth of at least 50% of the organisms. The cutoff points were defined according to the standards established for C. tropicalis by CLSI document M27-A3/S3. The trailing phenomenon was analyzed, and when there was doubt between trailing and fluconazole resistance, the susceptibility tests were repeated using RPMI at a pH of 5.0.
RESULTS
Among the 61 isolates, 48 were isolated from the blood and 13 from catheter tips. The study included 9 patients (no. 1, 2, 3, 4, 6, 8, 9, 13, and 14) whose blood cultures and central venous catheter tip cultures were both positive and 5 patients with more than one positive blood culture. The positivity interval between the cultures ranged from the same day to 12 days, with a median of 6 days. Isolates 17, 19, 20, and 26 came exclusively from catheter tips. The patient admission units included pediatrics (23%), adult and pediatric intensive care (18%), gastrointestinal surgery (14.75%), trauma surgery (8.2%), neurology and neurosurgery (8.2%), emergency (6.6%), gastroenterology clinic (6.6%) and “other.” Investigating the hypothesis of an outbreak was not possible because the isolates were not from the same unit and were not obtained during the same period of time. The data from the 43 patients with sequential isolates are shown in Table 1.
Table 1.
Details of 61 C. tropicalis isolates tested by MLST, listed in order of their DST, showing genotypes for six DNA fragments sequenced
| Patient/location | Age | Isolate | Sample | Date (mo/day/yr) | Ward | Genotype |
eBURST group | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ICL1 | MDR1 | SAPT2 | SAPT4 | XYR1 | ZWF1a | DST | |||||||
| 1/U | 44 yr | 1a | Blood | 3/24/03 | ICU | 1 | 24 | 3 | 7 | 53 | 6 | 239 | 4 |
| 1b | Blood | 3/28/03 | ICU | 1 | 7 | 4 | 11 | 4 | 3 | 234 | 6 | ||
| 1c | CVC | 3/31/03 | ICU | 1 | 24 | 3 | 7 | 24 | 6 | 232 | 4 | ||
| 2/U | 79 yr | 2a | CVC | 7/25/02 | Gastrosurgery | 1 | 7 | 4 | 11 | 50 | 3 | 236 | 6 |
| 2b | Blood | 7/30/02 | Gastrosurgery | 3 | 58 | 4 | 13 | 76 | 1 | 265 | Singleton | ||
| 3/U | 9 yr | 3a | Blood | 6/25/02 | Pediatric | 1 | 29 | 4 | 1 | 51 | 1 | 249 | Singleton |
| 3b | CVC | 6/25/02 | Pediatric | 28 | 84 | 28 | 7 | 86 | 28 | 258 | Singleton | ||
| 4/U | 9 mo | 4a | CVC | 4/21/03 | Pediatric ICU | 3 | 4 | 4 | 17 | 77 | 4 | 264 | Singleton |
| 4b | Blood | 4/21/03 | Pediatric ICU | 3 | 4 | 4 | 17 | 77 | 4 | 264 | Singleton | ||
| 5/U | 71 yr | 5a | Blood | 7/13/02 | Trauma surgery | 1 | 24 | 3 | 7 | 53 | 6 | 239 | 4 |
| 5b | Blood | 7/24/02 | Emergency | 1 | 24 | 3 | 7 | 48 | 6 | 124 | 4 | ||
| 6/U | 2 yr | 6a | Blood | 6/17/02 | Pediatric | 1 | 24 | 3 | 7 | 53 | 6 | 239 | 4 |
| 6b | CVC | 6/17/02 | Pediatric | 1 | 24 | 3 | 7 | 24 | 6 | 232 | 4 | ||
| 6c | Blood | 6/17/02 | Pediatric | 1 | 87 | 3 | 7 | 89 | 6 | 261 | Singleton | ||
| 7/U | 2 yr | 7a | Blood | 9/2/02 | Pediatric | 29 | 48 | 29 | 48 | 87 | 30 | 267 | Singleton |
| 7b | Blood | 9/8/02 | Pediatric | 1 | 7 | 4 | 6 | 52 | 4 | 238 | 1 | ||
| 8/U | 65 yr | 8a | Blood | 9/8/01 | Gastroclinic | 1 | 7 | 1 | 6 | 52 | 4 | 237 | 1 |
| 8b | CVC | 9/10/01 | Gastroclinic | 1 | 7 | 1 | 6 | 84 | 4 | 243 | 1 | ||
| 9/U | 36 yr | 9a | CVC | 1/15/00 | Neurosurgery | 10 | 58 | 4 | 8 | 31 | 3 | 246 | 18 |
| 9b | Blood | 1/15/00 | Neurosurgery | 10 | 58 | 4 | 8 | 31 | 3 | 246 | 18 | ||
| 9c | Blood | 1/17/00 | Neurology | 10 | 58 | 4 | 8 | 3 | 3 | 247 | 18 | ||
| 10/U | 44 yr | 10a | Blood | 12/15/02 | Gastrosurgery | 1 | 7 | 4 | 11 | 19 | 3 | 241 | 6 |
| 10b | Blood | 12/22/02 | Gastroclinic | 1 | 7 | 4 | 11 | 51 | 3 | 235 | 6 | ||
| 11/U | 9 yr | 11a | Blood | 9/12/01 | Pediatric ICU | 3 | 7 | 4 | 6 | 52 | 4 | 203 | 1 |
| 11b | Blood | 9/24/01 | Pediatric | 1 | 24 | 3 | 7 | 24 | 6 | 232 | 4 | ||
| 12/U | 58 yr | 12a | Blood | 10/28/02 | Internal medicine | 1 | 7 | 4 | 6 | 52 | 4 | 238 | 1 |
| 12b | Blood | 10/31/02 | Gastrosurgery | 1 | 7 | 4 | 6 | 84 | 4 | 248 | 1 | ||
| 12c | Blood | 10/31/02 | Gastrosurgery | 1 | 1 | 18 | 1 | 31 | 1 | 256 | 3 | ||
| 13/U | 64 yr | 13a | Blood | 10/14/02 | Gastroclinic | 3 | 7 | 4 | 6 | 84 | 4 | 245 | 1 |
| 13b | CVC | 10/17/02 | Gastrosurgery | 3 | 7 | 1 | 6 | 52 | 4 | 134 | 1 | ||
| 14/U | 3 yr | 14a | Blood | 1/3/01 | Trauma surgery | 1 | 24 | 3 | 7 | 24 | 6 | 232 | 4 |
| 14b | CVC | 1/3/01 | Trauma surgery | 1 | 87 | 3 | 7 | 89 | 6 | 261 | Singleton | ||
| 15/U | 69 yr | 15 | Blood | 1/11/01 | ICU | 1 | 24 | 3 | 7 | 90 | 6 | 262 | 4 |
| 16/U | 11 mo | 16 | Blood | 2/13/01 | Pediatric | 1 | 1 | 18 | 1 | 31 | 1 | 256 | 3 |
| 17/U | 70 yr | 17 | CVC | 2/4/02 | Pneumology | 1 | 7 | 4 | 6 | 67 | 6 | 244 | Singleton |
| 18/U | 25 yr | 18 | Blood | 2/5/02 | Oncology | 30 | 85 | 30 | 7 | 88 | 29 | 259 | Singleton |
| 19/U | 72 yr | 19 | CVC | 2/19/02 | Hemodialysis | 1 | 49 | 18 | 1 | 31 | 1 | 266 | 3 |
| 20/U | 1 yr | 20 | CVC | 5/19/02 | Gastrosurgery | 1 | 24 | 3 | 7 | 48 | 6 | 124 | 4 |
| 21/U | 9 yr | 21 | Blood | 6/25/02 | Pediatric | 3 | 39 | 3 | 7 | 90 | 7 | 268 | Singleton |
| 22/U | 27 yr | 22 | Blood | 8/5/02 | Neurology | 1 | 24 | 3 | 7 | 47 | 6 | 240 | 4 |
| 23/U | 65 yr | 23 | Blood | 9/25/02 | Emergency | 1 | 87 | 3 | 7 | 89 | 6 | 261 | Singleton |
| 24/U | 17 yr | 24 | Blood | 12/14/02 | Trauma surgery | 3 | 1 | 4 | 11 | 19 | 1 | 255 | Singleton |
| 25/U | 62 yr | 25 | Blood | 3/4/03 | Trauma surgery | 1 | 7 | 4 | 6 | 52 | 4 | 238 | 1 |
| 26/U | 72 yr | 26 | CVC | 3/28/03 | Gastrosurgery | 1 | 7 | 4 | 6 | 52 | 4 | 238 | 1 |
| 27/H | 65 yr | 27 | Blood | 3/30/01 | Emergency | 1 | 7 | 4 | 11 | 76 | 3 | 251 | 6 |
| 28/H | 5 yr | 28 | Blood | 2/1/01 | Pediatric | 3 | 86 | 12 | 11 | 73 | 3 | 260 | Singleton |
| 29/H | 32 yr | 29 | Blood | 7/4/99 | ICU | 1 | 17 | 3 | 7 | 1 | 3 | 242 | Singleton |
| 30/H | 66 yr | 30 | Blood | 3/6/01 | Emergency | 1 | 7 | 4 | 11 | 76 | 3 | 251 | 6 |
| 31/H | 70 yr | 31 | Blood | 1/18/00 | Vascular surgery | 3 | 4 | 4 | 7 | 77 | 7 | 254 | Singleton |
| 32/H | 40 yr | 32 | Blood | 4/16/01 | Neurology | 1 | 7 | 4 | 11 | 76 | 7 | 263 | 6 |
| 33/H | 16 yr | 33 | Blood | 1/29/99 | Hematology | 1 | 24 | 3 | 7 | 24 | 6 | 232 | 4 |
| 34/H | 15 yr | 34 | Blood | 4/7/98 | Pediatric | 1 | 83 | 1 | 11 | 2 | 3 | 253 | Singleton |
| 35/H | 40 yr | 35 | Blood | 12/11/99 | Burns unit | 1 | 24 | 3 | 7 | 24 | 7 | 233 | 4 |
| 36/H | 2 yr | 36 | Blood | 9/14/00 | Pediatric | 1 | 7 | 4 | 11 | 76 | 3 | 251 | 6 |
| 37/H | 18 yr | 37 | Blood | 9/24/01 | Pediatric | 1 | 24 | 3 | 7 | 47 | 6 | 240 | 4 |
| 38/H | 23 yr | 38 | Blood | 3/12/98 | Hematology | 1 | 82 | 3 | 7 | 24 | 6 | 250 | 4 |
| 39/H | 28 yr | 39 | Blood | 4/3/01 | Gastrosurgery | 1 | 7 | 4 | 11 | 76 | 3 | 251 | 6 |
| 40/H | 70 yr | 40 | Blood | 3/17/00 | Burns unit | 1 | 66 | 3 | 7 | 9 | 7 | 252 | Singleton |
| 41/H | 75 yr | 41 | Blood | 7/29/98 | Nephrology | 1 | 7 | 4 | 11 | 76 | 3 | 251 | 6 |
| 42/H | 30 yr | 42 | Blood | 10/20/98 | Emergency | 1 | 24 | 3 | 7 | 24 | 6 | 232 | 4 |
| 43/H | 15 yr | 43 | Blood | 4/18/01 | Gynecology | 1 | 24 | 3 | 7 | 24 | 6 | 232 | 4 |
H, HCFMUSP; U, UNICAMP; CVC, central venous catheter; DST, diploid sequence type. FCZ, fluconazole. The alleles/genotypes and a new representative from each of the surrounding 23 alleles are shown in bold. Underlining indicates resistant DSTs; italic type indicates previously described DSTs.
Of the 14 patients with sequential isolates, three had positive blood cultures despite the use of antifungals (patients 1, 2 and 11). Patients 1, 7, and 11 maintained the catheter despite the candidemia. Patients 7 and 11 died. The treatment was started prior to the blood culture only for patients 1 and 4, based on the diagnosis of candidemia. In our study, no antifungal prophylaxis had been prescribed during the 6 months prior to the candidemia.
In the sequencing analysis, the sizes of the DNA fragments that were obtained for each of the six genes studied ranged from 370 bp to 525 bp. A total of 543 cases of heterozygosity were detected: Y = C+T (59.5%), R = A+G (22.3%), W = A+T (13.6%), M = A+C (1.5%), K = G+T (2.2%) and S = G+C (0.9%). In the six analyzed gene fragments, 154 polymorphic sites were identified (20 in ICL1, 27 in MDR1, 39 in SAPT2, 34 in SAPT4, 19 in XYR1, and 15 in ZWF1a).
The C. tropicalis MLST database identified 23 new alleles, specifically in ICL1 (3 new), MDR1 (6 new), SAPT2 (3 new), SAPT4 (1 new), XYR1 (7 new), and ZWF1a (3 new). The most frequently detected genotypes were the following: genotype 1 in 45 isolates (73.8%) in ICL1; genotype 7 in 21 isolates (34.43%) and genotype 24 in 17 isolates (27.87%) in MDR1; genotype 4 in 27 isolates (44.26%) and genotype 3 in 22 isolates (30.07%) in SAPT2; genotype 7 in 26 isolates (42.62%) and genotype 11 in 13 isolates (21.3%) in SAPT4; and genotype 6 in 20 isolates (32.79%), genotype 3 in 15 isolates (23%) and genotype 4 in 12 isolates (19.67%) in ZWF1a. The XYR1 gene presented a great variety of genotypes (a total of 24).
Regarding the DSTs, 39 were found in the 61 isolates and were submitted to the C. tropicalis MLST database on April 2, 2012 (Table 1). Of the 39 DSTs, types 124, 134, and 203 were previously known. DST 232 was the most frequently observed (isolates 1c, 6b, 11b, and 14a [UNICAMP] and isolates 33, 42, and 43 [HCFMUSP]). Interestingly, five of the seven isolates with DST 232 were obtained from children. DST 251 was the second most frequently observed (isolates 27, 30, 36, 39, and 41 [HCFMUSP]).
Minor genetic changes characterized as variations in a single genotype (microvariation) were more common in the XYR1 gene fragment: 1a versus 1c, 5a versus 5b, 6a versus 6b, 8a versus 8b, 9a = 9b versus 9c, 10a versus 10b, 12a versus 12b (Table 2). Differences in a single nucleotide occurred among isolates 8a versus 8b, 9a = 9b versus 9c, 10a versus 10b, and 12a versus 12b. As an example, isolate 8a (genotype 52) was different from isolate 8b (allele 84) because of a loss of heterozygosity at position 215. Multiple variations in the same gene occurred between isolates 1a and 1c (3 nucleotide differences), 5a and 5b (5 nucleotide differences), and 6a and 6b (3 nucleotide differences). The main polymorphisms in the XYR1 gene occurred at positions 215, 242, and 344 (Table 2). Genetic changes in two gene fragments were found between isolates 6b and 6c in MDR1 (a single variation) and XYR1 (3 nucleotide differences), between 13a and 13b in SAPT2 (4 nucleotide differences) and XYR1 (a single variation), and between 14a and 14b in MDR1 (a single variation) and XYR1 (3 nucleotide differences).
Table 2.
Similarity of 32 selected isolates of C. tropicalisa
| Patient | Isolates compared | Genotyping difference | Nucleotide difference(s) | Similarity (%) | eBURST result |
|---|---|---|---|---|---|
| 1 | 1a and 1c | XYR1: 53–24 | 215, C-Y; 242, C-Y; 344, T-Y | 99.19 | 4 |
| 1a/1c and 1b | 5 alleles | 5 alleles | 4 versus 6 | ||
| 2 | 2a and 2b | 5 alleles | 5 alleles | 6 versus singleton | |
| 3 | 3a and 3b | 6 alleles | 6 alleles | Different singleton | |
| 4 | 4a and 4b | Identical | Identical | 100 | Identical singleton |
| 5 | 5a and 5b | XYR1: 53–48 | 11, Y-T; 14, Y-T; 215, C-Y; 242, C-Y; 344, T-Y | 98.65 | 4 |
| 6 | 6a and 6b | XYR1: 53–24 | 215, C-Y; 242, C-Y; 344, T-Y | 99.19 | 4 |
| 6a = 6b and 6c | MDR1: 24–87 | 7, T-Y | 99.76 | ||
| 6a and 6c | XYR1: 53–89 | 14, Y-C; 344, T-Y | 99.46 | 4 versus singleton | |
| 6b and 6c | XYR1: 24–89 | 14, Y-C; 215, Y-C; 242, Y-C | 99.19 | ||
| 7 | 7a and 7b | 6 alleles | 6 alleles | Singleton versus 1 | |
| 8 | 8a and 8b | XYR1: 52–84 | 215, Y-C | 99.73 | 1 |
| 9 | 9a = 9b and 9c | XYR1: 31–3 | 242, C-Y | 99.73 | 18 |
| 10 | 10a and 10b | XYR1: 19–51 | 344, Y-T | 99.73 | 6 |
| 11 | 11a and 11b | 6 alleles | 6 alleles | 1 versus 4 | |
| 12 | 12a/12b and 12c | 5 alleles | 5 alleles | 1 versus 3 | |
| 12a and 12b | XYR1: 52–84 | 215, Y-C | 99.73 | 1 | |
| 13 | 13a and 13b | SAPT2: 4–1 | 55, W-T; 262, G-R; 310, R-A; 421, C-Y; 215, C-Y | 99.06 | 1 |
| XYR1: 84–52 | 99.73 | ||||
| 14 | 14a and 14b | MDR1: 24–87 | 7, T-Y | 99.76 | 4 versus singleton |
| XYR1: 24–89 | 14, Y-C; 215, Y-C; 242, Y-C | 99.19 |
Isolates were obtained from 14 patients with fungemia, according to identical patterns generated by sequencing the fragments of six genes (ICL1, MDR1, SAPT2, SAPT4, XYR1, and ZWF1a) and determining the differences in the genotypes. All gene fragments were submitted to the official MLST website (http://pubmlst.org/ctropicalis/). Isolates from patients 1 to 14 are from UNICAMP. In the “Genotyping difference” column, italicized entries represent the different respective genotypes of the compared isolates (e.g., “XYR1: 53-24” in the first row indicates that 1a has XYR1 genotype 53 and 1c has XYR1 genotype 24).
Greater differences (macrovariation) occurred in isolates from 6 patients (no. 1, 2, 3, 7, 11, and 12), particularly those from patients 3, 7, and 11, which displayed differences in all six gene fragments.
All isolates had sufficient growth for MIC determination using the CLSI method. In the majority of the cases (95%), the CLSI method categorized an isolate as susceptible (MIC ≤ 4 μg/ml). A MIC greater than or equal to 32 μg/ml was observed in at least one isolate from patients 5 (5b, DST 124), 12 (12c, DST 256), and 25 (DST 238) (Table 3). Patient 5 had a good response to treatment with fluconazole, despite a MIC of 64 μg/ml.
Table 3.
Relationship among DST, alleles, and fluconazole susceptibility testing
| DST or allele | Isolate(s) |
|
|---|---|---|
| S | R | |
| DSTs | ||
| 124 | 20 | 5b |
| 256 | 16 | 12c |
| 238 | 7b, 12a, 26 | 25 |
| Alleles | ||
| ICL1 | ||
| 3 | 2b, 11a, P13, 21, 24, 28, 31 | |
| 1 | P1, 2a, 3a, 5a, P6, 7b, P8, P10, 11b, 12a, 12b, P14, 15–17, 19, 20, 22, 23, 26, 27, 29, 30, 32–43 | 5b, 12c, 25 |
| MDR1 | ||
| 1 | 16, 24 | 12c |
| 7 | 1b, 2a, 7b, P8, P10, 11a, 12a, 12b, P13, 17, 26, 27, 30, 32, 36, 39, 41 | 25 |
| 24 | 1a, 1c, 5a, 6a, 6b, 11b, P14, 15, 20, 22, 33, 35, 37, 42, 43 | 5b |
| SAPT2 | ||
| 1 | P8, 13b, 34 | |
| 3 | 1a, 1c, 5a, P6, 11b, P14, 15, 20–23, 29, 33, 35, 37, 38, 40, 42, 43 | 5b |
| 4 | 1b, P2, 3a, P4, 7b, P9, P10, 11a, 12a, 12b, 13a, 17, 24, 26, 27, 30–32, 36, 39, 41 | 25 |
| 18 | 16, 19 | 12c |
| SAPT4 | ||
| 1 | 3a, 16, 19 | 12c |
| 6 | 7b, P8, 11a, 12a, 12b, P13, 17, 26 | 25 |
| 7 | 1a, 1c, 3b, 5a, P6, 11b, P14, 15, 18, 20–23, 29, 31, 33, 35, 37, 38, 40, 42, 43 | 5b |
| XYR1 | ||
| 31 | 9a, 9b, 16, 19 | 12c |
| 48 | 20 | 5b |
| 52 | 7b, 8a, 11a, 12a, 13b, 26 | 25 |
| ZWF1a | ||
| 1 | 2b, 3a, 16, 19, 24 | 12c |
| 4 | P4, 7b, P8, 11a, 12a, 12b, P13, 26 | 25 |
| 6 | 1a, 1c, 5a, P6, 11b, P14, 15, 17, 20, 22, 23, 33, 37, 38, 42, 43 | 5b |
DST, diploid sequence type; P, total isolates from the same patient. The DSTs are those of isolates 5b, 12c, and 25, which had MICs of >32 μg/ml for fluconazole (resistant [R]) or a MIC of <16 μg/ml (susceptible [S]).
Of the total of 14 patients with sequential isolates, one patient received no treatment and 11 received antifungal agents. Amphotericin B (given to eight patients) was the most common. Fluconazole was given to two patients, and a combination of fluconazole and amphotericin B were used in one patient. It is important to note that the antifungal drugs were prescribed at doses recommended by existing protocols of each hospital at that time.
Additionally, two patients (5 and 12) were simultaneously infected by resistant and susceptible isolates. For patient 5, isolate 5b (DST 124) was considered resistant to fluconazole, and 5a (DST 239) was considered susceptible to fluconazole. For patient 12, isolate 12c (DST 256) was resistant to fluconazole, and 12a (DST 238) and 12b (DST 248) were considered fluconazole susceptible. DSTs 124, 238, and 256 were not exclusive to the resistant isolates. As shown in Table 3, isolates 20 (DST 124), 16 (DST 256), 7b, 12, and 26 (DST 238) were all susceptible to fluconazole.
Phylogenetic analysis.
The genetic distance between the strains of C. tropicalis included in this study was illustrated by the construction of an unrooted dendrogram. The clustering methods chosen for the phylogenetic analysis were maximum likelihood and Bayesian inference. The result of the phylogenetic analysis of the six gene fragments is displayed in the concatenated haplotypes shown in Fig. 1. The discriminatory power was 97.4%.
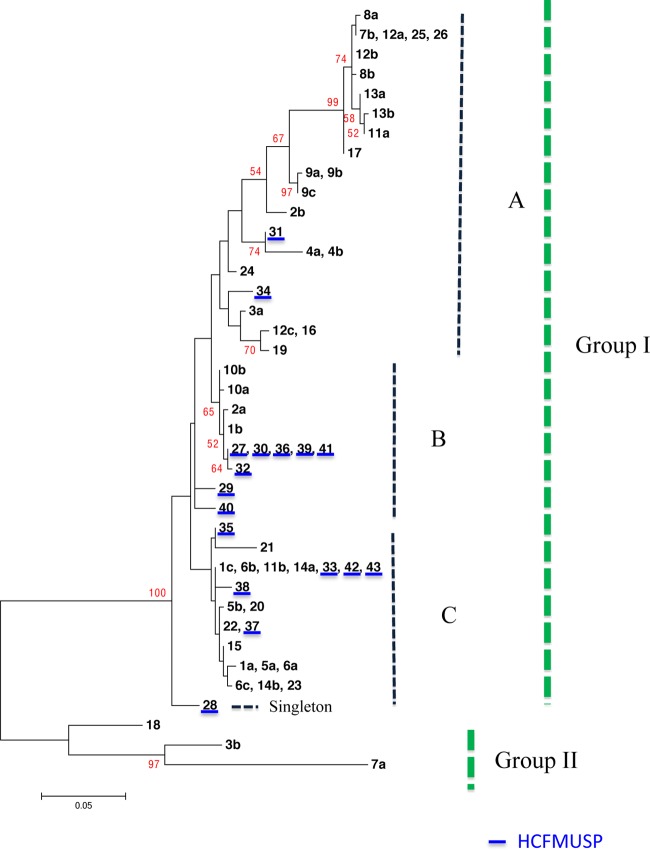
Fig 1.
Maximum-likelihood dendrogram generated from the MLST of six gene fragments for the 61 isolates of C. tropicalis from HCFMUSP and UNICAMP. The dendrogram shows two groups, I and II, and a singleton. Group I was divided into three subgroups (A, B, and C). The HCFMUSP isolates are underlined in blue. Group II comprised isolates 7a, 3b, and 18.
The 61 sequences of the six gene fragments in the present study were classified into two major groups (I and II) and one singleton group, supported by a 99% bootstrap value. Group I was divided into 3 subgroups, with 57 isolates: subgroup A, with 24 isolates (39.3%), 22 from UNICAMP and 2 (isolates 31 and 34) from HCFMUSP; subgroup B, with 12 isolates (19.7%), 8 from HCFMUSP and 4 from UNICAMP; and subgroup C, with 21 isolates (34.4%), 6 from HCFMUSP and 15 from UNICAMP. The singleton group consists of isolate 28. Group II consists of isolates 3b, 7a, and 18 (Fig. 1). The fluconazole-resistant isolates remained distant in the dendrogram. Isolates 12c and 25 were clustered in subgroup A and separated by a 99% bootstrap value. The isolates from patient 5 were placed in subgroup C. The DSTs (234, 236, 235, 241, and 251) associated with genotypes (ICL1, 1; MDR1, 7; SAPT2, 4; SAPT4, 11; ZWF1a, 3; and XYR1, variable) were grouped in subgroup B on the dendrogram. We tried to correlate the results with the original hospital, clinic, year, age, and susceptibility to antifungal drugs unsuccessfully.
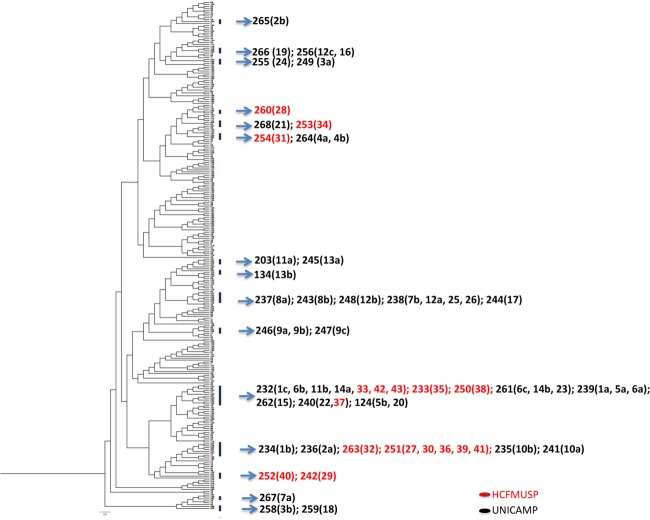
The isolates from Brazil, when analyzed by the Bayesian inference clustering method, were grouped into a variety of possibly existent clades. Figure 2 shows the arrangement of the isolates with the addition of the analysis of all 267 DSTs obtained from the C. tropicalis MLST database. The distribution of the DSTs from the study is shown in the tree. Isolates 3b, 7a, and 18 are clustered and distant from the other isolates.
Fig 2.
Bayesian inference dendrogram generated from the MLST polymorphic sites, concatenated and converted in haplotype form for 267 DSTs of C. tropicalis. The analysis includes the 61 isolates of C. tropicalis from UNICAMP and HCFMUSP that were submitted on 2 April 2012. The HCFMUSP isolates are in red.
eBURST analysis.
Among the 267 DSTs from the C. tropicalis MLST database, eBURST software found 28 groups and 131 solitary DSTs, known as singletons. Of the 39 DSTs observed in the study, 24 DSTs were grouped into five groups (groups 1, 3, 4, 6, and 18), and 15 DSTs were classified as singletons. Group 1 comprised 27 DSTs, 7 from this study. Isolate 13b (DST 134) assumed the central position of this group. Group 3 included 10 DSTs, 2 from this study. Group 4 comprised 9 DSTs, 7 from this study. Groups 6 and 18 are new groups added from this study comprising only DSTs identified only in the present work. Among the singletons, isolates 3b, 7a, and 18 remained far from the other DSTs according to the ancestry and phylogenetic analyses.
DISCUSSION
This study is the first MLST analysis to characterize the sequential isolates of C. tropicalis candidemia in South America and accounted for 36 new DSTs and 23 alleles that were incorporated into the official database (2 April 2012; http://pubmlst.org/ctropicalis/), allowing the comparison of our data with that from other countries. We compared both the phenotypic (through in vitro antifungal susceptibility testing) and genotypic patterns of the samples found in two large cities and analyzed the DNA polymorphisms—microvariations and macrovariations—in patients with candidemia who had sequential isolates.
Until July 2012, the C. tropicalis MLST database had a total of 401 isolates worldwide and 267 DSTs compared with 2062 DSTs for C. albicans (http://calbicans.mlst.net/). The rate of the description of new alleles in our study was lower than the number of DSTs found, as observed by Odds and Jacobsen (31) in C. albicans, suggesting that there might be a tendency toward the conservation of these alleles and an increase in the combination of them. Our data reinforce their same theory for C. albicans.
The limitations of this study are represented by the interpretation of heterozygosity in diploid microorganisms (30, 31, 49) and the evaluation of only DNA fragments and not the genome as a whole (33). In both participating hospitals, great genotypic diversity was detected among the isolates. MLST was able to detect microvariation and macrovariation in the nucleotide polymorphisms of the gene fragments studied in the isolates of 13 of the 14 patients who had sequential isolates. Microvariation in a single gene fragment occurred more frequently in the sequential isolates from seven patients. These minor genetic changes occurred at 1 to 5 nucleotide positions. Macrovariation occurred in the isolates from 6 patients, 1, 2, 3, 7, 11, and 12, who presented differences in at least five sequenced gene fragments. Genetic changes in two gene fragments were found in 3 patients. Our results were consistent with those of Jacobsen et al. (49), who found 2 of 18 paired isolates of C. tropicalis with multiple losses of heterozygosity in a single gene and 2 pairs with differences in multiple genes. Of the 5 other isolates from the same source, 2 of 4 genes presented homozygous and heterozygous differences, and the fifth isolate showed a complete loss of heterozygosity in five of the six sequenced genes. During a prospective surveillance study in adult ICU patients, in specimens from different anatomical sites of the same patient, Chen et al. (32) found several cases of heterozygosity changes in single and multiple genes among 11 patients who had C. tropicalis isolates.
Da Matta et al. (26) analyzed 21 C. albicans isolates from 8 patients diagnosed with persistent or recurrent candidemia in a multicenter surveillance study. Except for one patient with 3 different DSTs, all of the isolates from the remaining 7 patients showed the same DST, the same ABC type and the same susceptibility to antifungal agents. In another study, Bougnoux et al. (50) used MLST to demonstrate that 32 C. albicans isolates were responsible for candidemia and 118 isolates for candiduria in an ICU. Of the 9 patients with candidemia and candiduria, only one presented isolates of different DSTs, and these isolates differed at one locus, which suggested that they were evolutionarily related. In our study, the sequential isolates from 13 of 14 patients had different DSTs (which was not a rare event), and sequential isolates from two patients presented different fluconazole susceptibility profiles.
In contrast to our study, Shin et al. (51) demonstrated that among 41 sequential C. glabrata isolates from 15 patients with persistent candidemia, the sequential isolates had the same sequence type by MLST. In addition, they confirmed that acquiring azole resistance during the course of candidemia under the pressure of azole therapy is possible. In a study examining C. glabrata bloodstream and noninvasive isolates, Lott et al. (48) found no significant association between the sequence type and fluconazole resistance, with the MLST possessing low discriminatory power.
According to previous studies, variations in the isolates from the same patient could be explained by cross-contamination during the transport and storage times (33) or by polymorphisms that are related to antifungal-drug use (52). The present study was conducted in one of the laboratories from HCFMUSP, and the isolates have been monitored since 1998 to exclude the possibility of contamination. UNICAMP isolates were maintained in the Mycology Laboratory of the Clinics Hospital of UNICAMP from 2001 to 2003. All of the isolates were directly transferred to the Medical Investigation Laboratory (LIM 48), where they were immediately reidentified and confirmed as C. tropicalis before the beginning of the analyses. Since then, the isolates have been maintained in a unique laboratory and were carefully monitored during this study. DNA extraction was performed. Although contamination within the laboratory may have occurred, it is unlikely. In the present work, MLST was able to elucidate aspects of the differences between the sequential isolates from blood and catheter samples. Among isolates from patients whose blood and catheter cultures were both positive, the isolates from patients 2 and 3 deserve a special mention. In these patients, macrovariation was observed between the isolates from the blood and catheter, suggesting that each isolate apparently had no relationship with the isolate from the other site. In our study, we could not determine whether there was more than one clone in the host in the blood, the catheter, or both.
No relationships between antifungal MICs and defined DSTs and/or genotypes were observed in our study. Fluconazole resistance was found in only 3 of 61 isolates. Although the blood and/or catheter isolates presented different patterns by MLST and different susceptibility profiles in our study, no relationship between the antifungal MICs and defined DSTs and/or genotypes was observed. The relationship between the DSTs and the antifungal resistance profile or trailing phenomenon has been described. Chen et al. (32) found that in C. tropicalis, DST 164 in MLST group II was associated with a high MIC of flucytosine. In other studies, Chou et al. (27) and Li et al. (36) found an association between DST 140 and a reduced susceptibility to fluconazole. Chou et al. (27) also reported an association between the DSTs and the C. tropicalis trailing phenomenon. Additionally, Li et al. (36) described several alleles associated with low MICs of fluconazole (alleles 3 of ICL1, 9 of MDR1, 1 of SAPT2, 3, 6, and 10 of SAPT4, 48 of XYR1, and 7 of ZWF1a). In our study, we found no isolate with DST 140, and there was an association of allele 48 (XYR1) and allele 6 (SAPT4) with fluconazole-susceptible and fluconazole-resistant isolates, respectively.
In conclusion, MLST is an important tool for studying the genetic diversity of multiple/sequential isolates of patients with candidemia, particularly with respect to the polymorphisms in gene fragments. This technique allowed us to compare our data with data from other regions of the world and added new groups to the MLST database. It also allowed us to compare the genetic profiles of isolates from different sites, such as blood and catheters, and sequential isolates from patients with candidemia. Fluconazole resistance was found in only 3 of 61 isolates, making any correlative analysis of DST with drug resistance impossible. In addition, the phylogenetic and eBURST analyses also suggested that there was no correlation between the DSTs and the susceptibility to fluconazole, and the selective pressure of antifungal drugs most likely exerted no influence. Prospective studies will be necessary to assess the relationship between genetic variability and the pathogenicity of C. tropicalis isolates.
ACKNOWLEDGMENTS
This research was financially supported by the Fundação de Apoio à Pesquisa do Estado de São Paulo (FAPESP No. 06/61438-0) and the Fundação Faculdade de Medicina (FFM).
We thank Angelica Zaninelli Schreiber of Clinical Pathology Department, Faculty of Medical Sciences UNICAMP for providing the strains. We thank Gisele Duboc at Microbiology Laboratory, Division of Central Laboratory, Clínics Hospital, Medical School, University of São Paulo, São Paulo, Brazil. We thank Alexei Bender Haydu for his technical support in some PCRs. We thank Marcia de Souza Carvalho Melhem and Elisabeth Heins Vaccari for their contribution in antifungal susceptibility test. We thank Sergio Russo Matioli of Department of Genetics and Evolutionary Biology, Institute of Biosciences, University of São Paulo, for his contribution in phylogenetic analysis. We also express our sincere appreciation to Frank C. Odds of C. tropicalis MLST Database for carefully reading the sequences.
We declare no conflicts of interest in the presentation of these findings.
Footnotes
Published ahead of print 14 November 2012
REFERENCES
- 1. Almirante B, Rodríguez D, Park BJ, Cuenca-Estrella M, Planes AM, Almela M, Mensa J, Sanchez F, Ayats Gimenez M, Saballs P, Fridkin SK, Morgan J, Rodriguez-Tudela Warnock DW, Pahissa A, Barcelona Candidemia Project Study Group 2005. Epidemiology and predictors of mortality in cases of Candida bloodstream infection: results from population-based surveillance, Barcelona, Spain, from 2002 to 2003. J. Clin. Microbiol. 43:1829–1835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fraser VJ, Jones M, Dunkel J, Storfer S, Medoff G, Dunagan WC. 1992. Candidemia in a tertiary care hospital: epidemiology, risk factors, and predictors of mortality. Clin. Infect. Dis. 15:414–421 [DOI] [PubMed] [Google Scholar]
- 3. Gudlaugsson O, Gillespie S, Lee K, Vande BJ, Hu J, Messer S, Herwaldt L, Pfaller M, Diekema D. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin. Infect. Dis. 37:1172–1177 [DOI] [PubMed] [Google Scholar]
- 4. Morgan J, Meltzer MI, Plikaytis BD, Sofair AN, Huie-White S, Wilcox S, Harrison LH, Seaberg EC, Hajjeh RA, Teutsch SM. 2005. Excess mortality, hospital stay, and cost due to candidemia: a case-control study using data from population-based candidemia surveillance. Infect. Control Hosp. Epidemiol. 26:540–547 (Erratum, 26: 675) [DOI] [PubMed] [Google Scholar]
- 5. Pfaller MA, Jones RN, Messer SA, Edmond MB, Wenzel RP. 1998. National surveillance of Nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE program. Diagn. Microbiol. Infect. Dis. 30:121–129 [DOI] [PubMed] [Google Scholar]
- 6. Rentz AM, Halpern MT, Bowden R. 1998. The impact of candidemia on length of hospital stay, outcome, and overall cost of illness. Clin. Infect. Dis. 27:781–788 [DOI] [PubMed] [Google Scholar]
- 7. Sheng WH, Wang JT, Lu DC, Chie WC, Chen YC, Chang SC. 2005. Comparative impact of hospital-acquired infections on medical costs, length of hospital stay and outcome between community hospitals and medical centres. J. Hosp. Infect. 59:205–214 [DOI] [PubMed] [Google Scholar]
- 8. Voss A, Kluytmans JAJW, Koeleman JGM, Spanjaard L, Vandenbroucke-Grauls CMJE, Verbrugh HA, Vos MC, Weersink AYL, Hoogkamp-Korstanje JAA, Meis JFGM. 1996. Occurrence of yeast bloodstream infections between 1987 and 1995 in five Dutch university hospitals. Eur. J. Clin. Microbiol. Infect. Dis. 15:909–912 [DOI] [PubMed] [Google Scholar]
- 9. Abi-Said D, Anaissie E, Uzun O, Raad I, Pinzcowski H, Vartivaria S. 1997. The epidemiology of hematogenous candidiasis caused by different Candida species. Clin. Infect. Dis. 24:1122–1128 [DOI] [PubMed] [Google Scholar]
- 10. Bedini A, Venturelli C, Mussini C, Guaraldi G, Codeluppi M, Borghi V, Rumpianesi F, Barchiesi F, Esposito R. 2006. Epidemiology of candidaemia and antifungal susceptibility patterns in an Italian tertiary-care hospital. Clin. Microbiol. Infect. 12:75–80 [DOI] [PubMed] [Google Scholar]
- 11. Chakrabarti A, Chatterjee SS, Rao KL, Zameer MM, Shivaprakash MR, Singhi S, Singh R, Varma SC. 2009. Recent experience with fungaemia: change in species distribution and azole resistance. Scand. J. Infect. Dis. 41:275–284 [DOI] [PubMed] [Google Scholar]
- 12. Falagas ME, Roussos N, Vardakas KZ. 2010. Relative frequency of albicans and the various non-albicans Candida spp among candidemia isolates from inpatients in various parts of the world: a systematic review. Int. J. Infect. Dis. 14:954–966 [DOI] [PubMed] [Google Scholar]
- 13. Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, Marr KA, Pfaller MA, Chang CH, Webster KM. 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin. Infect. Dis. 48:1695–1703 [DOI] [PubMed] [Google Scholar]
- 14. Kontoyiannis DP, Vaziri I, Hanna HA, Boktour M, Thornby J, Hachem R, Bodey GP, Raad II. 2001. Risk factors for Candida tropicalis fungemia in patients with cancer. Clin. Infect. Dis. 33:1676–1681 [DOI] [PubMed] [Google Scholar]
- 15. Leroy O, Gangneux JP, Montravers P, Mira JP, Gouin F, Sollet JP, Carlet J, Reynes J, Rosenheim M, Regnier B, Lortholary O, AmarC and Study Group 2009. Epidemiology, management, and risk factors for death of invasive Candida infections in critical care: a multicenter, prospective, observational study in France (2005–2006). Crit. Care Med. 37:1612–1618 [DOI] [PubMed] [Google Scholar]
- 16. Myoken Y, Kyo T, Fujihara M, Sugata T, Mikami Y. 2004. Clinical significance of breakthrough fungemia caused by azole-resistant Candida tropicalis in patients with hematologic malignancies. Haematologica 89:378–380 [PubMed] [Google Scholar]
- 17. Muñoz P, Giannella M, Fanciulli C, Guinea J, Valerio M, Rojas L, Rodríguez-Créixems M, Bouza E. 2011. Candida tropicalis fungaemia: incidence, risk factors and mortality in a general hospital. Clin. Microbiol. Infect. 17:1538–1545 [DOI] [PubMed] [Google Scholar]
- 18. Nucci M, Colombo AL. 2007. Candidemia due to Candida tropicalis: clinical, epidemiologic, and microbiologic characteristics of 188 episodes occurring in tertiary care hospitals. Diagn. Microbiol. Infect. Dis. 58:77–82 [DOI] [PubMed] [Google Scholar]
- 19. Sipsas NV, Lewis RE, Tarrand J, Hachem R, Rolston KV, Raad II, Kontoyiannis DP. 2009. Candidemia in patients with hematologic malignancies in the era of new antifungal agents (2001–2007): stable incidence but changing epidemiology of a still frequently lethal infection. Cancer 115:4745–4752 [DOI] [PubMed] [Google Scholar]
- 20. Vigouroux S, Morin O, Moreau P, Jean-Luc Harousseau Milpied N. 2006. Candidemia in patients with hematologic malignancies: analysis of 7 years' experience in a single center. Haematologica 91:717–718 [PubMed] [Google Scholar]
- 21. Viscoli C, Girmenia C, Marinus A, Collette L, Martino P, Vandercam B, Doyen C, Lebeau B, Spence D, Krcmery V, De Pauw B, Meunier F. 1999. Candidemia in cancer patients: a prospective, multicenter surveillance study by the Invasive Fungal Infection Group (IFIG) of the European Organization for Research and Treatment of Cancer (EORTC). Clin. Infect. Dis. 28:1071–1079 [DOI] [PubMed] [Google Scholar]
- 22. Pfaller MA, Diekema DJ, Gibbs DL, Newell VA, Ellis D, Tullio V, Rodloff A, Fu W, Ling TA, the Global Antifungal Surveillance Group 2010. Results from the ARTEMIS DISK Global Antifungal Surveillance Study, 1997 to 2007: a 10.5-year analysis of susceptibilities of Candida species to fluconazole and voriconazole as determined by CLSI standardized disk diffusion. J. Clin. Microbiol. 48:1366–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Colombo AL, Nucci M, Park BJ, Nouér SA, Arthington-Skaggs B, DA Matta DA, Warnock D, Morgan J, Brazilian Network Candidemia Study 2006. Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J. Clin. Microbiol. 44:2816–2823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. DA Matta DA, de Almeida LP, Machado AM, Azevedo AC, Kusano EJ, Travassos NF, Salomão R, Colombo AL. 2007. Antifungal susceptibility of 1000 Candida bloodstream isolates to 5 antifungal drugs: results of a multicenter study conducted in São Paulo, Brazil, 1995–2003. Diagn. Microbiol. Infect. Dis. 57:399–404 [DOI] [PubMed] [Google Scholar]
- 25. Costa SF, Marinho I, Araújo EA, Manrique AE, Medeiros EA, Levin AS. 2000. Nosocomial fungaemia: a 2-year prospective study. J. Hosp. Infect. 45:69–72 [DOI] [PubMed] [Google Scholar]
- 26. DA Matta DA, Melo AS, Guimarães T, Frade JP, Lott TJ, Colombo AL. 2010. 2010. Multilocus sequence typing of sequential Candida albicans isolates from patients with persistent or recurrent fungemia. Med. Mycol. 48:757–762 [DOI] [PubMed] [Google Scholar]
- 27. Chou HH, Lo HJ, Chen KW, Liao MH, Li SY. 2007a. Multilocus sequence typing of Candida tropicalis shows clonal cluster enriched in isolates with resistance or trailing growth of fluconazole. Diagn. Microbiol. Infect. Dis. 58:427–433 [DOI] [PubMed] [Google Scholar]
- 28. Motta AL, Almeida GM, Almeida Júnior Burattini JNMN, Rossi F. 2010. Candidemia epidemiology and susceptibility profile in the largest Brazilian teaching hospital complex. Braz. J. Infect. Dis. 14:441–448 [PubMed] [Google Scholar]
- 29. Bougnoux ME, Morand S, d'Enfert C. 2002. Usefulness of multilocus sequence typing for characterization of clinical isolates of Candida albicans. J. Clin. Microbiol. 40:1290–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Odds FC, Davidson AD, Jacobsen MD, Tavanti A, Whyte JA, Kibbler CC, Ellis DH, Maiden MC, Shaw DJ, Gow NA. 2006. Candida albicans strain maintenance, replacement, and microvariation demonstrated by multilocus sequence typing. J. Clin. Microbiol. 44:3647–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Odds FC, Jacobsen MD. 2008. Multilocus sequence typing of pathogenic Candida species. Eukaryot. Cell 7:1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen KW, Chen YC, Lin YH, Chou HH, Li SY. 2009. The molecular epidemiology of serial Candida tropicalis isolates from ICU patients as revealed by multilocus sequence typing and pulsed-field gel electrophoresis. Infect. Genet. Evol. 9:912–920 [DOI] [PubMed] [Google Scholar]
- 33. Tavanti A, Davidson AD, Johnson EM, Maiden MC, Shaw DJ, Gow NA, Odds FC. 2005. Multilocus sequence typing for differentiation of strains of Candida tropicalis. J. Clin. Microbiol. 43:5593–5600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen KW, Chen YC, Lo HJ, Odds FC, Wang TH, Lin CY, Li SY. 2006. Multilocus sequence typing for analyses of clonality of Candida albicans strains in Taiwan. J. Clin. Microbiol. 44:2172–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lin CY, Chen YC, Lo HJ, Chen KW, Li SY. 2007. Assessment of Candida glabrata strain relatedness by pulsed-field gel electrophoresis and multilocus sequence typing. J. Clin. Microbiol. 45:2452–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li SY, Yang YL, Lin YH, Ko HC, Wang AH, Chen KW, Wang CW, Chi H, Lo HJ, Hospitals TSARY 2009. Two closely related fluconazole-resistant Candida tropicalis clones circulating in Taiwan from 1999 to 2006. Microb. Drug Resist. 15:205–210 [DOI] [PubMed] [Google Scholar]
- 37. Buchman TG, Rossier M, Merz WG, Charache P. 1990. Detection of surgical pathogens by in vitro DNA amplification of Candida albicans by in vitro amplification of fungus-specific gene. Surgery 108:338–347 [PubMed] [Google Scholar]
- 38. Ewing B, Green P. 1998b. Base calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8:186–194 [PubMed] [Google Scholar]
- 39. Ewing B, Hillier L, Wendl MC, Green P. 1998a. Base calling of automated sequencer traces using Phred. I. Accuracy assessment. Genome Res. 8:175–185 [DOI] [PubMed] [Google Scholar]
- 40. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28:2731–2739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jolley KA, Chan MS, Maiden MC. 2004. mlstdbNet—distributed multi-locus sequence typing (MLST) databases. BMC Bioinformatics 5:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tamura K, Nei M. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512–526 [DOI] [PubMed] [Google Scholar]
- 43. Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791 [DOI] [PubMed] [Google Scholar]
- 44. Hunter PR, Fraser CA. 1989. Application of a numerical index of discriminatory power to a comparison of four physiochemical typing methods for Candida albicans. J. Clin. Microbiol. 27:2156–2160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hunter PR. 1990. Reproducibility and indices of discriminatory power of microbial typing methods. J. Clin. Microbiol. 28:1903–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clinical Laboratory Standards Institute 2008. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard, 3rd ed CLSI document M27-A3. CLSI, Wayne, PA [Google Scholar]
- 47. National Committee for Clinical Laboratory Standards 2002. Reference method for broth dilution antifungal susceptibility testing of yeasts: approved standard— 2nd ed NCCLS document M27-A2. National Committee for Clinical Laboratory Standards, Wayne, PA [Google Scholar]
- 48. Lott TJ, Frade JP, Lyon GM, Iqbal N, Lockhart SR. 2012. Bloodstream and non-invasive isolates of Candida glabrata have similar population structures and fluconazole susceptibilities. Med. Mycol. 50:136–142 [DOI] [PubMed] [Google Scholar]
- 49. Jacobsen MD, Rattray AM, Gow NA, Odds FC, Shaw DJ. 2008. Mitochondrial haplotypes and recombination in Candida albicans. Med. Mycol. 46:647–654 [DOI] [PubMed] [Google Scholar]
- 50. Bougnoux ME, Kac G, Aegerter P, d'Enfert C, Fagon JY, CandiRea Study Group. 2008. Candidemia and candiduria in critically ill patients admitted to intensive care units in France: incidence, molecular diversity, management and outcome. Intensive Care Med. 34:292–299 [DOI] [PubMed] [Google Scholar]
- 51. Shin JH, Chae MJ, Song JW, Jung SI, Cho D, Kee SJ, Kim SH, Shin MG, Suh SP, Ryang DW. 2007. Changes in karyotype and azole susceptibility of sequential bloodstream isolates from patients with Candida glabrata candidemia. J. Clin. Microbiol. 45:2385–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Odds FC, Hanson MF, Davidson AD, Jacobsen MD, Wright P, Whyte JA, Gow NA, Jones BL. 2007. One year prospective survey of Candida bloodstream infections in Scotland. J. Med. Microbiol. 56:1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]