Abstract
Pericentrin is a large coiled-coil protein in mammalian centrosomes that serves as a multifunctional scaffold for anchoring numerous proteins. Recent studies have linked numerous human disorders with mutated or elevated levels of pericentrin, suggesting unrecognized contributions of pericentrin-related proteins to the development of these disorders. In this study, we characterized AnPcpA, a putative homolog of pericentrin-related protein in the model filamentous fungus Aspergillus nidulans, and found that it is essential for conidial germination and hyphal development. Compared to the hyphal apex localization pattern of calmodulin (CaM), which has been identified as an interactive partner of the pericentrin homolog, GFP-AnPcpA fluorescence dots are associated mainly with nuclei, while the accumulation of CaM at the hyphal apex depends on the function of AnPcpA. In addition, the depletion of AnPcpA by an inducible alcA promoter repression results in severe growth defects and abnormal nuclear segregation. Most interestingly, in mature hyphal cells, knockdown of pericentrin was able to significantly induce changes in cell shape and cytoskeletal remodeling; it resulted in some enlarged compartments with condensed nuclei and anucleate small compartments as well. Moreover, defects in AnPcpA significantly disrupted the microtubule organization and nucleation, suggesting that AnPcpA may affect nucleus positioning by influencing microtubule organization.
INTRODUCTION
Pericentrin is an integral component of the centrosome that serves as a multifunctional scaffold for anchoring numerous proteins and protein complexes (12, 37). Its orthologs kendrin (Homo sapiens), pericentrin (Mus musculus), Spc110 (Saccharomyces cerevisiae), Pcp1p (Schizosaccharomyces pombe), and centrosomin (Drosophila melanogaster) all contain the predicted coiled-coil secondary structure and the COOH-terminal calmodulin-binding region (6, 8, 9, 26, 41). In the budding yeast Saccharomyces cerevisiae, Spc110p facilitates mitotic spindle formation from the fungal centrosome, or spindle pole body (SPB) (8). Similarly, the fission yeast pericentrin-like protein Pcp1 regulates multiple functions of the SPB by recruiting two critical factors: the γ-tubulin complex and polo kinase (10). Moreover, previous studies have linked spindle formation to mutated or elevated levels of pericentrin, as the mutation of Pcp1 results in defective monopolar spindle microtubules (10), while overexpression of Pcp1 causes multiple mitotic spindle fragments and chromosome missegregation (8). Clearly, proper expression of centrosomal pericentrin-related protein plays a pivotal role in mitotic spindle formation during cell division. In addition, recent studies have linked numerous mammalian disorders to the function of pericentrin (5, 18, 37). For example, mice receiving transplants of pericentrin-depleted islets exhibit abnormal fasting hypoglycemia and the inability to regulate blood glucose during a glucose challenge (19). Moreover, mutations in pericentrin have also been related to human microcephalic osteodysplastic primordial dwarfism II (MOPDII), a rare genetic disease characterized by severe growth retardation and early onset of type 2 diabetes (36, 37). Such findings suggest unknown functions for these pericentrin-related proteins in mediating the cellular process of these disorders (19). In addition, previous research has highlighted the ability of these scaffolding proteins to adapt and regulate diverse cellular processes (5). The disruption of pericentrin function has recently been suggested to contribute to human disease through multiple mechanisms. To date, pericentrin function has been linked to four human disorders: primordial dwarfism, human cancer, mental disorders, and ciliopathies (5, 36, 37). Thus, studies that examine pericentrin's function at cellular, molecular, and organismal levels can help to generate models through which the etiology of these disorders may be better understood. Previous studies on disruption of the pericentrin gene in mice by insertional mutagenesis using gene trap technology that leads to embryonic lethality suggest that the Pcp1 homolog in mammalian cells is essential for animal survival (5, 6). Thus, research on the function of the Pcp1 homolog has been conducted using RNA interference (RNAi)-mediated knockdown in cultured mammalian cells instead of knockout of pericentrin (54). However, when cellular and molecular changes are being explored in order to explain the connection between pericentrin and cellular disorders, approaching the localization of Pcp1 globally using green fluorescent protein (GFP) tagging techniques in live cells represents a challenge. Thus, using simple eukaryotic microorganisms as a model can facilitate our understanding of many cellular processes that are common in mammalian cells. Unlike yeasts, the filamentous fungus Aspergillus nidulans contains the mycelium of multinucleate cells as well as attached daughter cells after cell division; therefore, A. nidulans can endure defects in cytokinesis, enabling clear identification of the regulation features that characterize cell division (7, 38, 53). In this study, the functional characterization of a pericentrin-related protein homolog in A. nidulans was examined using recombination strains at the targeted pcpA gene with either an inducible alcA promoter or null deletion. Our data collectively provide direct evidence that AnPcpA not only plays an important role in nucleus positioning by affecting microtubule organization and nucleation but also has a unique and complex function in cell polarity.
MATERIALS AND METHODS
Strains, media, culture conditions, and transformation.
A list of A. nidulans strains used in this study is provided in Table 1. The media YAG, YUU, YUUK, MMGR, MMGPR, and MMGTPR are described elsewhere (20, 46). Growth conditions, crosses, and inducing conditions for alcA(p)-driven expression were as previously described (45). Transformation was performed as described previously (27, 35). Expression of tagged genes under the control of the alcA promoter was regulated by different carbon sources: repressed on glucose, derepressed on glycerol, and induced on threonine.
Table 1.
A. nidulans strains in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| GR5 | pyrG89; pyroA4; wA3; veA1 | FGSC |
| TN02A7 | pyrG89; pyroA4, nkuA::argB2; riboB2, veA1 | 31 |
| CPA01 | pyrG89; pyroA4, nkuA::argB2; ΔAnpcpA::pyrG;riboB, veA1 | This study |
| CPA02 | pyrG89; pyroA4, nkuA::argB2; alcA(p)::GFP-AnpcpA::pyr4; riboB2, veA1 | This study |
| CSA02-2 | pyrG89; pabaA1; yA2; nkuA::argB2; cam::RFP::pyrG; veA1 | This study |
| CPA03 | nkuA::argB2; alcA(p)::GFP-AnpcpA::pyr4; cam::RFP::pyrG; veA1 | This study |
| SJW100 | wA3; ΔargB::trpCΔB; pyroA4;alcA(p)::GFP::tubA; gpd(p)::stuA(NLS)::DsRedT4; veA1 | 43, 44 |
| CPA04 | wA3; ΔargB::trpCΔB; pyroA4; ΔAnpcpA::pyroA; alcA(p)::GFP::tubA; gpd(p)::stuA(NLS)::DsRedT4; veA1 | This study |
| WJA01 | pyroA4, nkuA::argB2; riboB2, veA1 | 4 |
| CSA01 | pyrG89; pyroA4, nkuA::argB2; alcA(p)::GFP-cam::pyr-4; riboB2, veA1 | 4 |
| CSA02 | pyrG89; pyroA4, nkuA::argB2; cam::RFP::pyrg; riboB2, veA1 | 4 |
| SCA02 | pyroA4, nkuA::argB2; CaM::RFP::pyrG; riboB2, alcA(p)::GFP::tubA, veA1 | This study |
Construction of the AnpcpA deletion strain.
A strain containing AnpcpA-null mutation was created by double joint PCR (52). The Aspergillus fumigatus pyrG gene in plasmid pXDRFP4 was used as a selectable nutritional marker for fungal transformation. Linearized DNA fragment 1, which included a sequence of about 996 bp that corresponded to the regions immediately upstream of the AnpcpA start codon, was amplified with the primers Diag-del-AnpcpA-5′ and AnpcpA-pre-3′ (see Table S1 in the supplemental material). Linearized DNA fragment 2, including a sequence of about 992 bp that corresponded to the regions immediately downstream of the AnpcpA stop codon, was amplified with primers AnpcpA-post-5′ and AnpcpA-post-3′ (see Table S1). Lastly, purified linearized DNA fragments 1 and 2 plus pyrG were mixed and used in a fusion PCR with primers Full5′ and Full3′. The final fusion PCR product was purified and used to transform into A. nidulans strains TN02A7. A diagnostic PCR assay was performed to identify the deletion of the AnpcpA gene with the primers Diag-del-AnpcpA-5′ and Diag-AfpyrG-3′. A similar strategy was used to construct the AnpcpA deletion strain in the SJW100 background. The A. nidulans pyroA gene in plasmid pQa was used as a selectable nutritional marker for fungal transformation. Linearized DNA fragment 1 was amplified with the primers Diag-del-AnpcpA-5′ and P3-SJW100-de (see Table S1). Linearized DNA fragment 2 was amplified with primers P4-SJW100-de and AnpcpA-post-3′ (see Table S1). Lastly, purified linearized DNA fragments 1 and 2 plus pyroA were mixed and used in a fusion PCR with the primers Full5′ and Full3′. The final fusion PCR product was purified and used for transformation into A. nidulans strains SJW100. A diagnostic PCR assay was performed to detect the deletion of the AnpcpA gene using primers Diag-del-AnpcpA-5′ and Diag-pyroA-3′.
Tagging of AnPcpA with GFP.
To generate an alcA(p)-GFP-AnpcpA fusion construct, a 1,051-bp fragment of AnpcpA was amplified from GR5 genomic DNA with primer AnpcpA-5′ (NotI site included) and primer AnpcpA-3′ (XbaI site included) (see Table S1 in the supplemental material). The 1,051-bp amplified DNA fragment was cloned into pLB01 (22, 24), yielding pLB01-AnpcpA5′. This plasmid was transformed into TN02A7. Homologous recombination of this plasmid into the AnpcpA locus should result in N-terminal GFP fusion of the entire AnpcpA gene under the control of the alcA promoter. The transformant, which was able to form a normal colony under inducing conditions but could not grow under repressing conditions, was subjected to PCR analysis using a forward primer (Diag-GFP-5′) designed to recognize the gfp sequence and a reverse primer (Diag-AnpcpA-3′) designed to recognize the 3′ AnpcpA sequence.
Microscopy and imaging processing.
Several sterile glass coverslips were placed on the bottom of petri dishes, and gently overlaid with 1 ml liquid medium amended with 0.07 ml spore suspension (5 × 107/ml). Strains were grown on the coverslips at related temperature prior to observation under microscope. The GFP-AnPcpA signal was observed in live cells by placing coverslips on a glass slide. DNA and chitin were stained using 4′,6-diamidino-2-phenylindole (DAPI) and calcofluor white (CFW) (Sigma-Aldrich, St. Louis, MO), respectively, after the cells had been fixed with 4% paraformaldehyde (Polysciences, Warrington, PA) (15). For immunofluorescent detection of actin, the growing hyphae were fixed in phosphate-buffered saline (PBS) (pH 7.4) with 4% paraformaldehyde at room temperature for 30 min. After three washes in PBS, cells were permeabilized with 0.5% Triton X-100 in PBS for 15 min, washed three times with PBS, and blocked in blocking buffer (1% bovine serum albumin in PBS) at room temperature for 30 min. Cells were then stained with mouse anti-actin monoclonal antibody (1:200; MP Biomedicals) as the primary antibody, washed three times with PBS, and then incubated with Alexa Fluor 546-labeled rabbit anti-mouse IgG (1:400; Invitrogen) as the secondary antibody. Differential interference contrast (DIC) images of the cells were collected with a Zeiss Axio Imager A1 microscope (Zeiss, Jena, Germany). These images were then collected and analyzed with a Sensicam QE cooled digital camera system (Cooke Corporation, Germany) with MetaMorph/MetaFluor combination software package (Universal Imaging, West Chester, PA), and the results were assembled in Adobe Photoshop 7.0 (Adobe, San Jose, CA).
RESULTS
AnPcpA is a putative pericentrin-related protein homolog in A. nidulans.
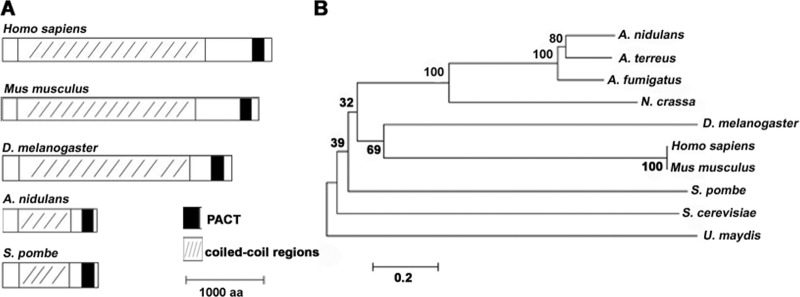
To search the homolog of pericentrin-related protein in A. nidulans, the pericentrin homologs Pcp1 in S. pombe and kendrin in H. sapiens were used to Blast search the A. nidulans protein database of the National Center for Biotechnology Information (NCBI) and the Central Aspergillus Data Repository (CADRE). A putative coiled-coil protein was consistently obtained (GenBank accession no. AN3062.4 in NCBI and ANIA_03062 in CADRE) and was named AnPcpA; this is the same as An110p, identified by Flory et al. (8). AnPcpA shares 30% amino acid sequence identity with Pcp1 in S. pombe and 28% with kendrin in H. sapiens. Furthermore, the AnpcpA gene is 3,915 nucleotides long and contains three introns and four exons. It translates into a predicted protein of 1,207 amino acids, including a coiled-coil domain and a conserved PACT (pericentrin–AKAP-450 domain of centrosomal targeting) protein (Fig. 1) (11). To understand the molecular characterization of pericentrin homologs in filamentous fungi compared to single-cell yeasts and other eukaryotic organisms, we carried out conserved-domain Blast searches for these homologs. As shown in Fig. 1, all selected homologs included a coiled-coil domain in the central portion of the protein and a PACT domain located near the C terminus. In addition, alignment analyses indicated that the percent identity for whole sequences of fungi and animals is not high but that the region of the PACT domain in homologs is relatively conserved.
Fig 1.
Phylogenetic analysis of AnPcpA homologs in selected organisms. (A) Comparison of amino acid sequence characterization among AnPcpA homologs in selected species. The black box indicates an area that belongs to a conserved PACT domain, as determined by searching conserved domains in NCBI. (B) Phylogenetic tree representing the evolutionary relationships between AnPcpA homologs. The tree was constructed with MEGA5 software by the neighbor-joining method from amino acid sequences of pericentrin homologs: Aspergillus nidulans, AN3062.4; Aspergillus terreus, Q0CXP2; Aspergillus fumigatus, Q4WXF9; Neurospora crassa, XP_959539; Drosophila melanogaster, Q400N2; Homo sapiens, AAD10838; Mus musculus, NP_006022.3; Schizosaccharomyces pombe, Q92351.1; Saccharomyces cerevisiae, P32380; and Ustilago maydis, Q4PCA7). Sequences were obtained from NCBI and CADRE.
AnpcpA is an essential gene.
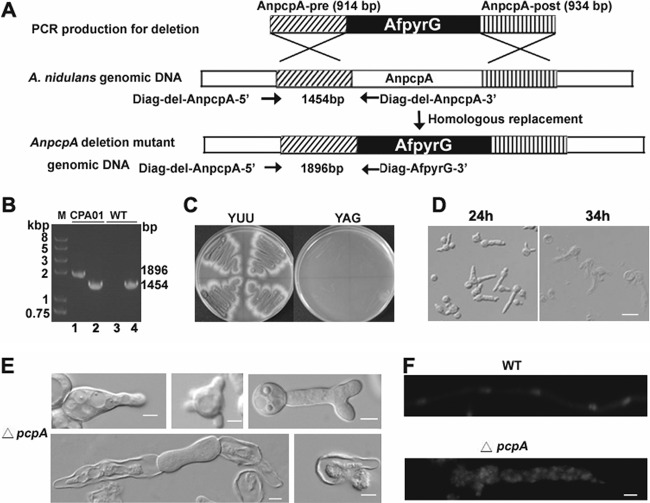
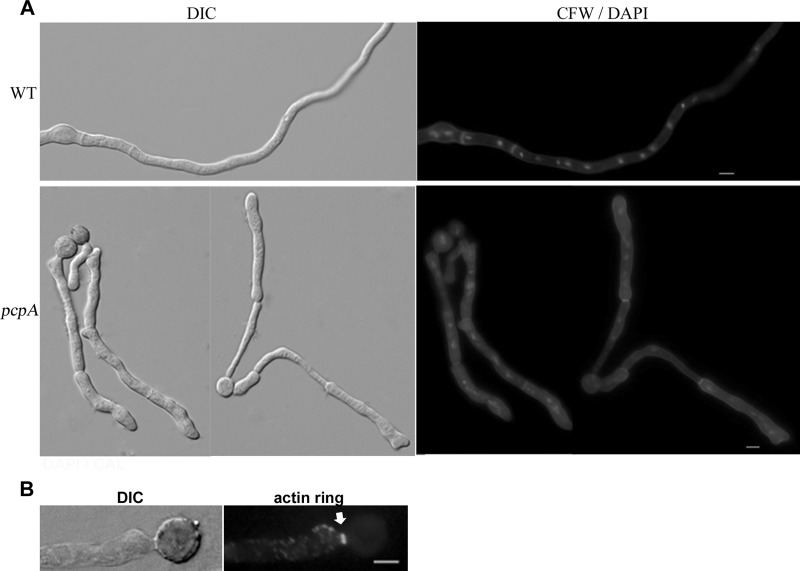
To further determine the function of the pericentrin-related protein PcpA in A. nidulans, we created the AnpcpA deletion strain CPA01, in which the pcpA open reading frame was replaced with the nutritional marker AfpyrG (Fig. 2A). PCR analysis showed that the AfpyrG nutritional marker had been integrated into the genome at the original AnpcpA locus, but the AnpcpA gene could still be detected, indicating the possibility that heterokaryons existed (Fig. 2B). To better observe the resultant phenotype, we inoculated spores from a transformant onto selective medium (YAG); they were unable to germinate normally or form detectable colonies. However, when inoculated onto nonselective YUU medium, the spores germinated and formed colonies in the same time period as in YAG (Fig. 2C). Moreover, microscopic examination indicated that germlings from transformants cultured on the selective YAG medium had severe defects in establishing and/or maintaining polarity, accompanied by abnormal septum shape and random nucleus distribution (Fig. 2E and F). With increasing culture time, most mutants exhibited a lethal phenotype in which protoplasm erupted from the cell (Fig. 2D). According to the standard protocol used to detect the essential gene as previously described (34), these results clearly indicate that the transformant was a heterokaryon and that AnpcpA is an essential gene in A. nidulans.
Fig 2.
Identification and analysis of the essential A. nidulans gene AnpcpA using the heterokaryon rescue technique. (A) Diagram showing the deletion strategy for AnpcpA. (B) PCR analysis showing the integration of the AfpyrG nutritional marker into the genome at the original AnpcpA locus in the CPA01 strain while AnpcpA can still be detected, suggesting that transformants were heterokaryons. For lanes 2 and 4, the primers Diag-del-AnpcpA-5′ and Diag-del-AnpcpA-3′ were used to determine whether AnpcpA still exists in the genome, and the expected size is 1,454 bp. For lanes 1 and 3, primers Diag-del-AnpcpA-5′ and Diag-AfpyrG-3′ were used to determine whether there was a homologous recombination to replace AnpcpA with the nutritional marker gene pyrG in the genome, and the expected size is 1,896 bp. In lanes 3 and 4, genomic DNA of TN02A7 was used as a PCR template; for lanes 1 and 2, genomic DNA of transformants was used as a PCR template. (C) Replica transformant spores from heterokaryons on YAG and YUU. (D) Conidial spores from heterokaryon transformants cultured in liquid YAG for 24 h, showing two kinds of genotypes in the progeny: ungerminated spores were pcpA wild type in the pyrG mutation background, and the other germlings had the pcpA deletion. Bar, 10 μm. (E) Phenotypes of abnormal germlings in progeny of AnpcpA deletion transformants. (F) Germlings from progeny of AnpcpA deletion transformants were stained with DAPI, showing the abnormal distribution of nuclei. Bar, 5 μm.
GFP-AnPcpA fluorescence dots are associated with nuclei.
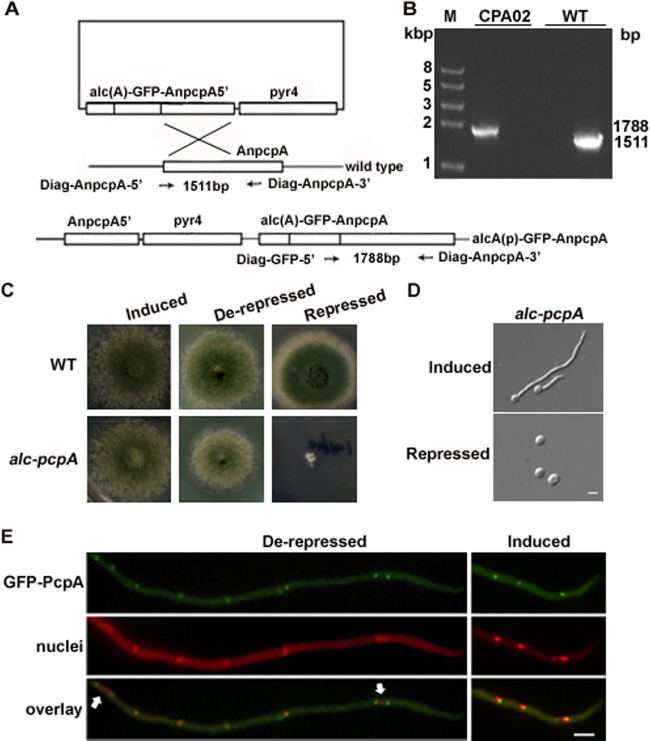
Because AnpcpA is essential, the deletion strain CPA01 could not be used for further study. Instead, we constructed a conditional strain in which the AnpcpA gene was under the control of the inducible/repressible alcA promoter [alcA(p)] as regulated by the carbon source: repressed on glucose, derepressed on glycerol, and induced on threonine. As shown in Fig. 3A, the AnpcpA gene was fused to the original locus under the control of alcA(p), resulting in a strain which we refer to as CPA02. When CPA02 was inoculated into inducing or derepressing medium, it showed slightly slow colony growth compared to the wild type at the beginning, but with increasing culture time, the CPA02 colonies were mostly similar in size to the wild type. This indicates that the AnPcpA with GFP fused to its N terminus was functional. In comparison, under the repressing conditions, CPA02 formed almost no detectable colonies when cultured for 24 h at 37°C. However, when culturing was extended to 48 h, CPA02 displayed very small, densely packed colonies (Fig. 3C). Thus, the global deletion strain CPA01 showed a more severe lethal phenotype than did the conditional knockdown strain CPA02.
Fig 3.
Phenotypic characterization and the localization of GFP-AnPcpA in the alcA(p)::GFP-AnpcpA strain CPA02. (A) Outline of the strategy for creating the functional strain CPA02 by highly efficient homologous gene integration. (B) PCR analysis showing the integration of pLB01-AnpcpA5′ into the genome at the original AnpcpA locus in CPA02. (C) Colony phenotype comparison of CPA02 cultured on the solid inducing medium MMGTPR, the derepressing medium MMGPR, and the repressing medium YAG for 48 h at 37°C. (D) Germling phenotype comparison of CPA02 cultured in MMGPR and YAG for 9 h at 37°C. (E) Localization pattern of GFP-AnPcpA in CPA02 in different media. The nuclei were stained with DAPI. Arrows indicate GFP-AnPcpA, which was found occasionally on opposite ends of the elongated nucleus. Bars, 5 μm.
Using live-cell imaging of CPA02, we examined the subcellular localization of AnPcpA throughout the cell cycle. As shown in Fig. 3E, GFP-AnPcpA fluorescence was distributed along the mature hypha as spot accumulations, which seem to have been associated with the nuclei, most likely at the SPBs. During the interphase, GFP-AnPcpA was localized as a dot associated with a nucleus; however, in mitosis with an elongated nucleus, two of the GFP-AnPcpA fluorescence dots moved to opposite ends of the nucleus (Fig. 3E). To further analyze the localization of AnPcpA under the control of its own promoter, we constructed both GFP and red fluorescent protein (RFP) fusions at the C terminus of AnPcpA under the control of its promoter (see Fig. S3 in the supplemental material). Unfortunately, for an unknown reason, we could not find any GFP or RFP signal. Thus, we used CPA02 in which AnPcpA had a GFP fusion at the N terminus to detect the localization of AnPcpA in subsequent experiments. In addition, to test whether the expression of AnPcpA under different culture conditions could cause the mislocalization of GFP-PcpA in CPA02, we assessed the localization of GFP-PcpA. It showed that the GFP-PcpA localization pattern under derepressing conditions on glycerol was similar to patterns obtained under inducing conditions on threonine (Fig. 3E). This suggests that the overexpression of alcA promoter could not cause mislocalization of GFP-PcpA in CPA02.
A defect in AnPcpA disturbs hyphal polarity and localization of calmodulin.
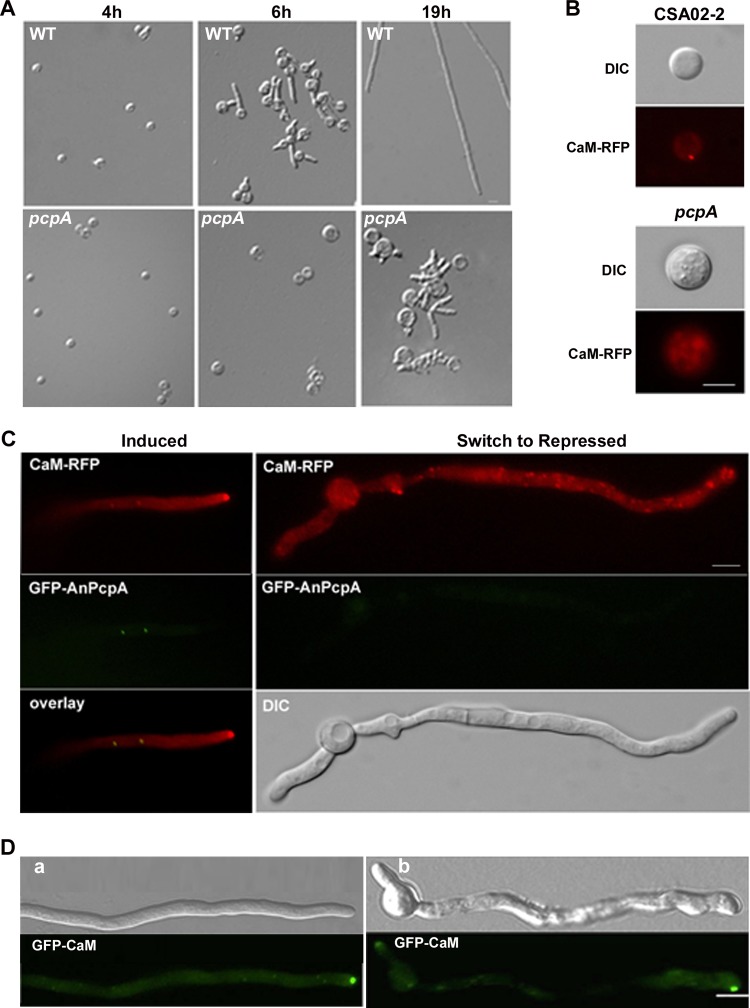
To further verify the above finding (i.e., that the deletion of AnPcpA caused the polarity defect), the spores of the conditional strain CPA02 were cultured on YAG medium, where AnPcpA expression was turned off (Fig. 4A). Consequently, depletion of AnPcpA resulted in a prolonged spore isotropic growth period, so that the wild-type strain started to germinate when the cells were cultured for 6 h at 37°C, while CPA02 was unable to germinate at all and instead showed an abnormally swollen conidiospore. After incubation for about 19 h on YAG, CPA02 displayed a hyperbranching phenotype with multiple polarity axes in germlings. These data suggest that AnPcpA may help establish and maintain polar growth. Because genomic information analysis indicated that a calmodulin (CaM) binding domain exists at the C terminus of AnPcpA, we wondered whether the polarity problems caused by the depletion of AnPcpA could be associated with the function of CaM. Our previous research demonstrated that CaM plays an important role in maintaining hyphal polarity, as indicated by the dense accumulation of CaM at the hyphal apex (4). To test whether CaM localization during polar growth of hyphae is influenced by AnPcpA depletion, we crossed CPA02 with the RFP-CaM-expressing strain CSA02-2 to produce strain CPA03, which included CaM-RFP under the control of the CaM native promoter and the fusion of GFP-AnPcpA under the control of the alcA promoter. Notably, when the expression of GFP-AnPcpA was inhibited in the repressing medium, as shown in Fig. 4B and C (right), the localization pattern of CaM-RFP changed dramatically, resulting in diffused and random CaM localization in germlings and mature hyphal cells. In contrast, when the expression of AnPcpA was induced, a high concentration of CaM accumulated at the tip of the germinated spore (Fig. 4B, top) and the hyphal apex (Fig. 4C, left), as expected. This indicates that the accumulation of CaM at the hyphal apex depends on proper functioning of AnPcpA. In comparison, when the inducible alcA(p)::GFP-CaM strain CSA01 was treated with 10 μg/ml nocodazole, a reagent that interferes with the polymerization of microtubules, germlings also showed diffused fluorescence distribution throughout the hyphal cell along with noncentral GFP-CaM localization at the tip (Fig. 4D). This suggests that AnPcpA affected the localization of CaM and may be related to the function of microtubules.
Fig 4.
Defect of AnPcpA disturbs the hyphal polarity and the localization pattern of calmodulin. (A) Phenotypic comparison between the alcA(p)::GFP-AnpcpA strain CPA02 and the wild-type strain WJA01 at 37°C under repressed culture conditions for 4, 6, and 19 h, respectively. (B and C) Comparison of CaM-RFP localization patterns between CPA03 [alcA(p)::GFP-AnpcpA; CaM-RFP] and CSA02-2 (CaM-RFP) cultured on inducing and repressing media. (B) CPA03 and CSA02-2 cultured on YAG at 37°C for 18 h and 5 h, respectively. (C) (Left) Localization of CaM-RFP with the induced expression of AnPcpA on MMGPR in CPA03. (Right) After being shifted from MMGPR to YAG for 4 h, with decreasing GFP-AnPcpA expression (middle), CPA03 cells showed a diffused and random localization pattern of CaM-RFP through the hyphal cell. (D) Normal localization of CaM at the apex depends on the function of microtubule organization. (a) Normal localization pattern of GFP-CaM in a growing hyphal cell, showing hyphal tip accumulation and related DIC in the CSA01 strain cultured in MMGPR medium for 12 h. (b) Noncentral GFP-CaM localization at the tip along with diffuse fluorescence throughout the hyphal cell in CSA01 cultured in MMGPR medium for 11 h and then switched to MMGPR medium amended with 10 μg/ml nocodazole for 1 h. Bars, 5 μm.
Down-regulation of AnPcpA results in the formation of abnormal septa.
Previous studies verified that mitotic signals are able to direct the assembly of the septal band (28, 49); furthermore, the pcpA deletion strain CPA01 has been shown to exhibit a clearly abnormal septum shape. To confirm whether the depletion of AnPcpA affects the developmental process of septation, we switched the cultured hyphal cells in CPA02 from the inducing MMGPR medium to the repressing YAG medium, in which the expression of AnPcpA was turned off. After the cells had been switched to the YAG medium for 4 h, septa in mature hyphal cells could be detected by DIC microscopy. However, the shape of the hyphal cells had been mostly changed in such a way that the shank of the hyphal cell had been twisted to become an uneven hyphal tube (Fig. 5A, bottom). Especially at the septum site, the septum seems to be much narrower than that in other regions, resulting in an abnormally contracted septum that was significantly different from that seen in the presence of AnPcpA. This suggests that the maintenance of septum in mature cells requires a role performed by AnPcpA. Normally, septum formation requires the assembly of a septal band, which is accomplished by the contraction of a contractile actin ring along with the deposition of chitin at the septum (14). We wondered whether this abnormal septum phenotype was related to the defect in the formation of the actin ring or the deposition of chitin. As shown in Fig. 5A, septa of hyphal cells CPA02 were able to be stained with calcofluor white during the depletion of AnPcpA, indicating chitin deposition on the septum. Meanwhile, immunofluorescence analysis indicated that actin rings could be stained at some of the abnormal septum sites when induced mycelium was switched to repressing medium for 4 h. This suggested that the depletion of AnPcpA did not affect actin ring formation (Fig. 5B). These data suggest that AnPcpA is involved in the maintenance of septum shape but is not required for actin ring formation and chitin deposition at the septum site during septation.
Fig 5.
AnPcpA is involved in the maintenance of septum shape but not required for actin ring formation and chitin disposition. (A) The mycelium phenotypes of wild-type WJA01 and CPA02 stained with DAPI and CFW. (Top) Conidial spores of WJA01 were cultured at 37°C for 10 h; (bottom) hyphal phenotypes in CPA02 cultured in the repressing medium YAG for 4 h after being cultured in the inducing medium MMGPR for 10 h. (B) Actin ring visualized by indirect immunofluorescence at sites of septum formation (arrow) in CPA02 after a shift from the inducing medium MMGPR to the repressing medium YAG for 4 h. Bars, 5 μm.
AnPcpA is required for nuclear positioning.
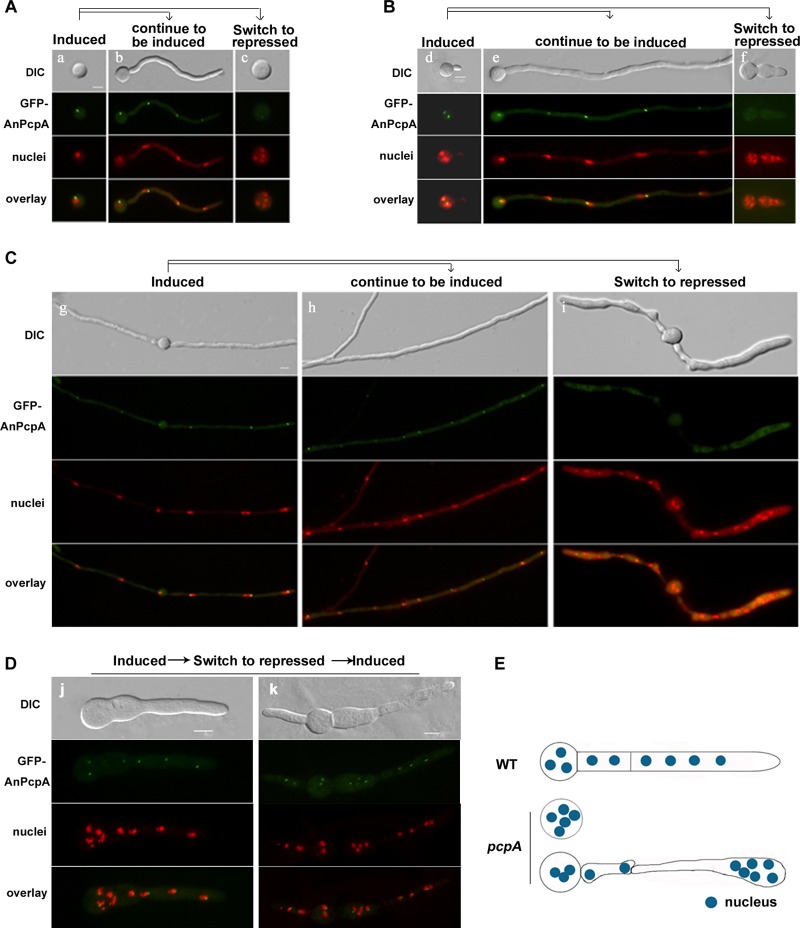
Based on the finding that germlings in the AnpcpA deletion mutant have a highly abnormal distribution of nuclei, we asked how AnPcpA affects the positioning of nuclei. To answer this question, the spores of the conditional strain CPA02 were cultured on the inducing medium MMGPR for 5 h and were then switched to the repressing medium YAG for 4 h. At this time point, the fluorescence signal of GFP-PcpA appeared to be very weak and diffused throughout the cytoplasm (Fig. 6), which indicated that the expression of AnPcpA had been successfully turned off. Notably, due to the decrease of GFP-PcpA, the conidia of the CPA02 strain showed an abnormally extended isotropic swelling pattern accompanied by an increased number of nuclei in the spore in YAG (Fig. 6A). Thus, inhibited expression of AnPcpA might be capable of stopping the polarized growth but not able to block mitosis; this results in localization of multiple nuclei in the spore. Perhaps most interestingly, shifting developed mycelia or germlings from inducing MMGPR medium to repressing YAG medium resulted in hyphal remodeling ((Fig. 6A, B, and C; also, see Movie S1 in the supplemental material) and reorganized nuclear distribution (Fig. 6A, B, and C) with the decrease of GFP-AnPcpA. Consequently, depletion of AnPcpA resulted in random nuclear distribution accompanied by uneven hyphal shape. Interestingly, the size of the mycelium seemed to depend on the accumulation of nuclei so that the expanded region of mycelium included numerous nuclei, whereas the anucleate region shrank (Fig. 6C and E). These uneven shapes of hyphal cells were accompanied by random nuclear distribution with depletion of AnPcpA. This result indirectly implies that the defect of AnPcpA results in nuclear abnormal positioning, possibly by affecting nuclear anchoring. In contrast, after being switched from YAG to MMGPR, the cells displayed the recovery of the GFP-AnPcpA signal as well as polarized growth (Fig. 6D), suggesting that under repressive conditions, CPA02 might not undergo loss of viability at that time point.
Fig 6.
Depletion of AnPcpA resulted in an abnormally extended isotropic swelling pattern of the spore and a reorganized nuclear distribution with hyphal remodeling. Conditional alcA(p)::GFP-AnpcpA strain CPA02 was used in all experiments at 37°C for the indicated cultured times. Nuclei were stained with DAPI, and the cell shape was observed by DIC. (A) Conidial spores were inoculated in the inducing medium MMGPR (a) for 5 h, and then replicate plates were switched to the repressing medium YAG (c) for 4 h or left in MMGPR (b) for 4 h. (c) With the decrease in GFP-PcpA fluorescence signal, the spore showed an abnormally extended isotropic swelling pattern with an increased number of nuclei. (B) Conidial spores were inoculated in MMGPR (d) for 6 h, and then replicate germlings were switched to the repressing medium YAG (f) for 4 h or left in MMGPR (e) for 4 h. (f) Inhibition of AnPcpA expression affected the normal polarized growth but was unable to block mitosis. (C) Replicate mycelia grown in inducing medium were switched to repressing medium YAG (i) for 4 h or left in MMGPR (h) for 4 h. (i) Decrease of GFP-AnPcpA expression caused a reorganized nuclear distribution and hyphal remodeling. (D) Conidia of CPA02 were cultured in MMGPR for 6 h, and then the germlings were switched to YAG for 4 h, and lastly, mycelia were transferred from YAG to MMGPR for 2 h. Cells displayed recovery of the GFP-AnPcpA signal as well as polarized hyphal growth. (E) Model of the hyphal shape caused by the inhibition of AnPcpA expression showing a reorganized nuclear distribution in mature mycelia. Bars, 5 μm.
Deletion of AnpcpA causes a defect in microtubule nucleation.
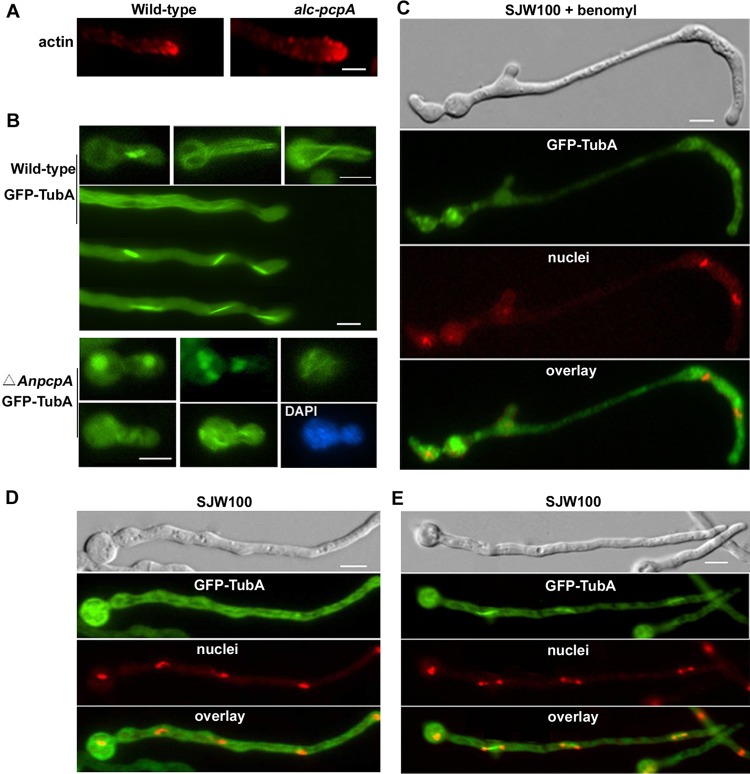
To further determine whether the abnormal nucleus distribution caused by depletion of AnPcpA was due to the defect in the cytoskeleton, we examined actin localization with an immunostaining experiment. Unexpectedly, no detectable difference in the actin distribution pattern in the growing hyphal tips between mutant and wild-type strains was observed (Fig. 7A). Because fungal nuclear migration during interphase depends on the presence of intact microtubules (25, 29, 33), these results suggest that AnPcpA may act as a cortical anchor for interphase microtubules, thereby stabilizing the position of the nucleus. To further analyze the relationship between AnPcpA and microtubule organization, the AnpcpA gene was deleted in the background of the SJW100 strain, which had a visual GFP-TubA fusion in live cells. PCR analysis showed the integration of the AnpyroA4 nutritional marker into the genome at the original AnpcpA site in the background of the GFP-TubA fusion strain (see Fig. S1 in the supplemental material). As indicated by the results in Fig. 2, AnpcpA is an essential gene. Thus, we obtained the heterokaryons only in transformants in which both the AnpcpA deletion strain with AnpyroA4 integration and the wild type without AnpyroA4 insertion coexisted. As predicted, two different types of transformants were observed. Based on the phenotypes that the AnpcpA deletion CPA01 strain had displayed, we selected germlings that had phenotypes similar to those of CPA01 in terms of transformants to check spindle formation visually by GFP-TubA fusion. In the control strain SJW100, as shown in Fig. 7B, GFP-TubA displayed the GFP fluorescence signal with a clear parallel bundle of interphase microtubules in most cases. Moreover, by using time-lapse images, we can clearly see the process of mitotic spindle development from a short spindle to a full-length spindle. In comparison, when AnpcpA was deleted, all selected mutants displayed abnormal irregular accumulation of GFP-TubA. We could not find any clear spindle structure, even though some of them possibly belonged to monopolar or multipolar spindle microtubules, suggesting remarkable defects in microtubule organization. After staining with DAPI, there were abnormally shaped nuclei localized in the basal cells or germ tubes of mutants (Fig. 7B).
Fig 7.
Deletion of AnPcpA causes defects in microtubule organization. (A) Actin patches were visualized by indirect immunofluorescence microscopy in hyphal cells of wild-type strain WJA01 and conditional strain CPA02. (B) (Top) In control strain SJW100, GFP-TubA displayed a clear parallel bundle of interphase microtubules and mitotic spindles. (Bottom) Germlings from a strain with a deletion of AnpcpA in the SJW100 background showed very severe defects in microtubule organization and irregular GFP-TubA accumulation. Nuclei were stained with DAPI. (C) Conidial spores from SJW100 labeled with GFP-TubA were inoculated in MMGPR for 9 h and then switched to MMGPR amended with 5 μg/ml benomyl for 3 h. The nuclei were stained with DAPI. (D and E) As the control for cells shown in panel C, conidial spores from SJW100 were inoculated in MMGPR without benomyl for 12 h, the cytoplasmic microtubule was labeled with GFP-TubA, and the nuclei were stained with DAPI during interphase (D) and mitosis (E). Bars, 5 μm.
To analyze how the microtubule organization was affected by AnPcpA expression, we constructed a TubA-RFP fusion at the C terminus of TubA in the alc(p)-GFP-AnPcpA background (see Fig. S4 in the supplemental material). This RFP-pyrG cassette has been successfully used in construction of RFP-tagged CaM strain CSA02. Unfortunately, however, after checking all homologous TubA-RFP integration transformants, we could not find any RFP signal. Previous research has demonstrated that TubA tagged with GFP at the N terminus is functional (13, 17). Possibly, the RFP signal fused at the C terminus of TubA under the control of the native promoter was blocked by this fusion protein, or the native promoter of tubA is not strong enough to induce RFP to be seen at the C terminus of TubA. To further test whether this abnormal phenotype was linked to microtubule organization, the germlings of SJW100 were treated with 5 μg/ml benomyl (a reagent that inhibits microtubule polymerization and dynamics). Consequently, with the increase of defects in microtubule organization labeled by GFP-TubA, the selected transformants showed a phenotype with abnormal hyphal shape similar to that caused by the defect of AnPcpA (Fig. 7C). Under the same culture conditions, GFP-TubA displayed a clear parallel bundle of microtubules and some spindle structures in SJW100 without benomyl treatment (Fig. 7D and E). These data suggest that AnPcpA may be involved in directing the assembly of microtubule organization.
DISCUSSION
Recent studies have linked a growing list of human disorders to mutated or elevated levels of pericentrin (2, 5, 6, 21). Notably, pericentrin mutations have been shown to be present in all MOPDII patients examined, which confirmed the genetic homogeneity of this disorder (37, 48). Although the disruption of pericentrin function could contribute to human disease through multiple mechanisms, a unifying theme among the diverse deficiencies associated with pericentrin-associated disorders is the loss of microtubule integrity (5, 54). To further explore the connection between pericentrin and cellular function, this study used the model filamentous fungus A. nidulans and verified the global localization and the functional characterization of a Pcp1 homolog by GFP tagging techniques in live cells and by the null deletion technique. Our data collectively provide direct evidence that AnPcpA, a putative pericentrin-related protein homolog in A. nidulans, is essential and not only plays important roles in nuclear positioning by affecting microtubule organization and nucleation but also has a function in polar growth.
Relationship between AnPcpA and CaM.
From domain analysis results for the molecular characterization of pericentrin homologs in selected eukaryotic organisms (Fig. 1), it is clear that all pericentrin homologs include a coiled-coil region present in the central area of the protein and a PACT domain located near the C terminus. Previous studies determined that the pericentrin homologs Spc110p in S. cerevisiae, Pcp1p in S. pombe, and kendrin in H. sapiens contain a calmodulin-binding site at the COOH terminus (8, 9, 41). In S. cerevisiae, Spc110p directs CaM via its CaM-binding site to the central plaque of the SPB to mediate the process of mitosis (39). Mutations that block the binding of CaM to Spc110p have been reported to cause multiple mitotic defects (40). In A. nidulans, as shown with live-cell images in Fig. 4C, both GFP-AnPcpA and CaM-RFP showed clear colocalization as spot accumulations along the mature hypha at the location associated with nuclei. Our previous work verified that, during mitosis, CaM was localized to the mitotic spindle poles (see Fig. S2 in the supplemental material); thus, it is probable that GFP-AnPcpA and CaM-RFP were colocalized at the SPBs during mitosis. Notably, when AnPcpA was downregulated by switching the conditional strain CPA02 to the repressing YAG medium, the germlings showed severe defects, with abnormally shaped septa and a random distribution of the diffused nuclei, which is a phenotype similar to that caused by the defect of microtubules (Fig. 7C). In addition, we found that AnPcpA depletion resulted in a severe defect in polar growth, which was accompanied by the hyperbranching phenotype with multiple polarity axes (Fig. 4A). This is a phenotype similar to that caused by a defect in CaM (4), which suggests that AnPcpA may have a complex function with CaM and may be involved in polar growth. Unexpectedly, there was no detectable accumulation of GFP-AnPcpA at the hyphal apex, which indicated that AnPcpA may not be directly required for polar growth. In this respect, CaM is completely different from AnPcpA (4). Our results imply that the depletion of AnPcpA causes this cell polarity defect, possibly by affecting the trafficking of CaM, which is a key regulator of polar growth. Figure 4C shows that, as predicted, CaM accumulation at the hyphal apex can be dramatically blocked by the depletion of AnPcpA, resulting in a random distribution of the CaM-RFP signal along the whole hyphal cell. Because the deletion of AnPcpA caused a defect in microtubule organization (Fig. 7B), resulting in the elimination of CaM accumulation at the hyphal apex (Fig. 4D), the abnormal accumulation of CaM in hyphal cells may have been a result of the defect in microtubule organization.
Depletion of AnPcpA resulted in abnormal microtubule organization and nuclear distribution.
A large body of evidence supports a role for pericentrin in microtubule nucleation and spindle organization (30). Dysfunction of pericentrin results in mitotic spindle defects, chromosome missegregation, and mitotic failure, which could lead to cell cycle arrest and/or cell death (37). However, as shown in Fig. 2E and Fig. 6, deletion or depletion of AnPcpA caused an abnormally extended isotropic swelling pattern; this pattern was accompanied by an increased number of nuclei present within the spore and hyphal cell. It can be inferred that deletion of AnpcpA or inhibition of AnPcpA expression might be unable to block mitosis; possibly when strains were switched from inducing medium to repressing medium, AnPcpA could not be used up immediately, and the residual activity of the alcA promoter under repressing condition allowed mitosis to occur. Consequently, depletion or deletion of AnpcpA caused the irregular spindle formation, resulting in abnormal segregation of DNA and the defect in the mitotic-specific reorganizations. Perhaps most interestingly, when mature hyphal cells were switched from the inducing to the repressing medium, the shape of the hyphal cells as well as the nuclear distribution inside the cells was remarkably changed or reorganized, and these changes were accompanied by decreased AnPcpA expression. As a result, more nuclei accumulated in the abnormally expanded region of the mycelium. In contrast, in most of the constricted regions, no nuclei were detected under conditions that repressed AnPcpA expression (Fig. 6C). This finding clearly indicates that AnPcpA is necessary for nuclear normal positioning.
Previous studies have shown that proper nuclear distribution first requires the active migration of nuclei, followed by the anchoring of nuclei to the cytoplasm microtubule and then their attachment to the plasma membrane (44, 50, 51). During this process, microtubule organization, actin cytoskeleton, and motor proteins play important roles; thus, blocking any of their components results in abnormal nuclear trafficking and nuclear distribution (3, 32, 50, 51). In S. cerevisiae, Spc110 operates as a spacer protein in SPBs, and its amino and carboxy termini are anchored at the inner and central plaques of the SPB, respectively (16, 39, 42). The N terminus of Spc110 has an affinity for its protein complex, for recruiting the yeast γ-tubulin complex at the inner plaque, and for initiating the nucleation of nuclear microtubules (22, 54); another component of SPB that is located at the outer plaque is important for docking the γ-tubulin complex to initiate the nucleation of cytoplasmic microtubules (1, 23, 47). Similarly, in S. pombe, it has been shown that the pericentrin protein homolog Pcp1p plays the essential role of connecting microtubules to the SPB and that Pcp1p links the polo kinase and γ-tubulin entry to spindle formation in both SPB assembly and microtubule nucleation (10). Our results (Fig. 7) show that deletion of AnpcpA caused the severe defect in cytoplasmic microtubule organization with abnormal and irregular GFP-TubA accumulation. This suggests that the abnormal nuclear distribution caused by the depletion of AnPcpA may be the result of the defect of microtubule organization. Thus, AnPcpA not only is required for nucleation of cytoplasmic microtubules but also may play a role in anchoring interphase microtubules to cortical membranes for stabilizing the position of the nucleus. Future studies on how AnPcpA affects microtubule organization should clarify the details of AnPcpA's function in nucleus positioning.
Supplementary Material
ACKNOWLEDGMENTS
This work was financially supported by grants from the National Natural Science Foundation of China (NSFC31070031), the Natural Science Foundation of the Jiangsu Higher Education Institutions of China (grant no. 11KJA180005), and the Priority Academic Program Development (PAPD) of Jiangsu Higher Education Institutions and the Research to L.L.
A. nidulans strain TN02A7 was a gift from B. R. Oakley (Ohio State University, Columbus, OH); A. nidulans strain SJW100 was a gift from R. Fischer (University of Karlsruhe, Applied Microbiology, Karlsruhe, Germany); Plasmid pLB01 was a gift from B. Liu (University of California, Davis); plasmid pXDRFP4 and pFNO3 were from FGSC (http://www.fgsc.net); and plasmid pQa was a gift from H. M. Park (Chungnam National University, South Korea).
Footnotes
Published ahead of print 19 October 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Adams IR, Kilmartin JV. 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145: 809–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anitha A, et al. 2008. Gene and expression analyses reveal enhanced expression of pericentrin 2 (PCNT2) in bipolar disorder. Biol. Psychiatry 63: 678–685 [DOI] [PubMed] [Google Scholar]
- 3. Cadot B, et al. 2012. Nuclear movement during myotube formation is microtubule and dynein dependent and is regulated by Cdc42, Par6 and Par3. EMBO Rep. 13: 741–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen S, et al. 2010. Localization and function of calmodulin in live-cells of Aspergillus nidulans. Fungal Genet. Biol. 47: 268–278 [DOI] [PubMed] [Google Scholar]
- 5. Delaval B, Doxsey SJ. 2010. Pericentrin in cellular function and disease. J. Cell Biol. 188: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Endoh-Yamagami S, et al. 2010. A mutation in the pericentrin gene causes abnormal interneuron migration to the olfactory bulb in mice. Dev. Biol. 340: 41–53 [DOI] [PubMed] [Google Scholar]
- 7. Fischer R. 1999. Nuclear movement in filamentous fungi. FEMS Microbiol. Rev. 23: 39–68 [DOI] [PubMed] [Google Scholar]
- 8. Flory MR, Morphew M, Joseph JD, Means AR, Davis TN. 2002. Pcp1p, an Spc110p-related calmodulin target at the centrosome of the fission yeast Schizosaccharomyces pombe. Cell Growth Differ. 13: 47–58 [PubMed] [Google Scholar]
- 9. Flory MR, Moser MJ, Monnat RJ, Jr, Davis TN. 2000. Identification of a human centrosomal calmodulin-binding protein that shares homology with pericentrin. Proc. Natl. Acad. Sci. U. S. A. 97: 5919–5923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fong CS, Sato M, Toda T. 2010. Fission yeast Pcp1 links polo kinase-mediated mitotic entry to gamma-tubulin-dependent spindle formation. EMBO J. 29: 120–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gillingham AK, Munro S. 2000. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 1: 524–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Griffith E, et al. 2008. Mutations in pericentrin cause Seckel syndrome with defective ATR-dependent DNA damage signaling. Nat. Genet. 40: 232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han G, et al. 2001. The Aspergillus cytoplasmic dynein heavy chain and NUDF localize to microtubule ends and affect microtubule dynamics. Curr. Biology. 11: 719–724 [DOI] [PubMed] [Google Scholar]
- 14. Harris SD. 2001. Septum formation in Aspergillus nidulans. Curr. Opin. Microbiol. 4: 736–739 [DOI] [PubMed] [Google Scholar]
- 15. Harris SD, Morrell JL, Hamer JE. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136: 517–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Helfant AH. 2002. Composition of the spindle pole body of Saccharomyces cerevisiae and the proteins involved in its duplication. Curr. Genet. 40: 291–310 [DOI] [PubMed] [Google Scholar]
- 17. Horio T, Oakley BR. 2005. The role of microtubules in rapid hyphal tip growth of Aspergillus nidulans. Mol. Biol. Cell 16: 918–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang-Doran I, et al. 2011. Genetic defects in human pericentrin are associated with severe insulin resistance and diabetes. Diabetes 60: 925–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jurczyk A, et al. 2010. A novel role for the centrosomal protein, pericentrin, in regulation of insulin secretory vesicle docking in mouse pancreatic beta-cells. PLoS One 5: e11812 doi:10.1371/journal.pone.0011812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kafer E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19: 33–131 [DOI] [PubMed] [Google Scholar]
- 21. Klingseisen A, Jackson AP. 2011. Mechanisms and pathways of growth failure in primordial dwarfism. Genes Dev. 25: 2011–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knop M, Pereira G, Geissler S, Grein K, Schiebel E. 1997. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 16: 1550–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Knop M, Schiebel E. 1998. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 17: 3952–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu B, Morris NR. 2000. A spindle pole body-associated protein, SNAD, affects septation and conidiation in Aspergillus nidulans. Mol. Gen. Genet. 263: 375–387 [DOI] [PubMed] [Google Scholar]
- 25. Malone CJ, Fixsen WD, Horvitz HR, Han M. 1999. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development 126: 3171–3181 [DOI] [PubMed] [Google Scholar]
- 26. Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW. 2004. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165: 673–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. May GS. 1989. The highly divergent beta-tubulins of Aspergillus nidulans are functionally interchangeable. J. Cell Biol. 109: 2267–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Momany M, Hamer JE. 1997. Relationship of actin, microtubules, and crosswall synthesis during septation in Aspergillus nidulans. Cell Motil. Cytoskeleton 38: 373–384 [DOI] [PubMed] [Google Scholar]
- 29. Morris NR, Xiang X, Beckwith SM. 1995. Nuclear migration advances in fungi. Trends Cell Biol. 5: 278–282 [DOI] [PubMed] [Google Scholar]
- 30. Muhlhans J, Brandstatter JH, Giessl A. 2011. The centrosomal protein pericentrin identified at the basal body complex of the connecting cilium in mouse photoreceptors. PLoS One 6: e26496 doi:10.1371/journal.pone.0026496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nayak T, et al. 2006. A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172: 1557–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oakley BR. 2004. Tubulins in Aspergillus nidulans. Fungal Genet. Biol. 41: 420–427 [DOI] [PubMed] [Google Scholar]
- 33. Oakley BR, Morris NR. 1980. Nuclear movement is β-tubulin-dependent in Aspergillus nidulans. Cell 19: 255–262 [DOI] [PubMed] [Google Scholar]
- 34. Osmani AH, Oakley BR, Osmani SA. 2006. Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat. Protoc. 1: 2517–2526 [DOI] [PubMed] [Google Scholar]
- 35. Osmani SA, Pu RT, Morris NR. 1988. Mitotic induction and maintenance by overexpression of a G2-specific gene that encodes a potential protein kinase. Cell 53: 237–244 [DOI] [PubMed] [Google Scholar]
- 36. Piane M, et al. 2009. Majewski osteodysplastic primordial dwarfism type II (MOPD II) syndrome previously diagnosed as Seckel syndrome: report of a novel mutation of the PCNT gene. Am. J. Med. Genet. A 149A: 2452–2456 [DOI] [PubMed] [Google Scholar]
- 37. Rauch A, et al. 2008. Mutations in the pericentrin (PCNT) gene cause primordial dwarfism. Science 319: 816–819 [DOI] [PubMed] [Google Scholar]
- 38. Seiler S, Justa-Schuch D. 2010. Conserved components, but distinct mechanisms for the placement and assembly of the cell division machinery in unicellular and filamentous ascomycetes. Mol. Microbiol. 78: 1058–1076 [DOI] [PubMed] [Google Scholar]
- 39. Spang A, Grein K, Schiebel E. 1996. The spacer protein Spc110p targets calmodulin to the central plaque of the yeast spindle pole body. J. Cell Sci. 109: 2229–2237 [DOI] [PubMed] [Google Scholar]
- 40. Stirling DA, Rayner TF, Prescott AR, Stark MJ. 1996. Mutations which block the binding of calmodulin to Spc110p cause multiple mitotic defects. J. Cell Sci. 109: 1297–1310 [DOI] [PubMed] [Google Scholar]
- 41. Stirling DA, Stark MJ. 2000. Mutations in SPC110, encoding the yeast spindle pole body calmodulin-binding protein, cause defects in cell integrity as well as spindle formation. Biochim. Biophys. Acta 1499: 85–100 [DOI] [PubMed] [Google Scholar]
- 42. Sundberg HA, Goetsch L, Byers B, Davis TN. 1996. Role of calmodulin and Spc110p interaction in the proper assembly of spindle pole body components. J. Cell Biol. 133: 111–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Toews MW, et al. 2004. Establishment of mRFP1 as a fluorescent marker in Aspergillus nidulans and construction of expression vectors for high-throughput protein tagging using recombination in vitro (GATEWAY). Curr. Genet. 45: 383–389 [DOI] [PubMed] [Google Scholar]
- 44. Veith D, Scherr N, Efimov VP, Fischer R. 2005. Role of the spindle-pole-body protein ApsB and the cortex protein ApsA in microtubule organization and nuclear migration in Aspergillus nidulans. J. Cell Sci. 118: 3705–3716 [DOI] [PubMed] [Google Scholar]
- 45. Wang G, Lu L, Zhang CY, Singapuri A, Yuan S. 2006. Calmodulin concentrates at the apex of growing hyphae and localizes to the Spitzenkorper in Aspergillus nidulans. Protoplasma 228: 159–166 [DOI] [PubMed] [Google Scholar]
- 46. Wang J, et al. 2009. The important role of actinin-like protein (AcnA) in cytokinesis and apical dominance of hyphal cells in Aspergillus nidulans. Microbiology 155: 2714–2725 [DOI] [PubMed] [Google Scholar]
- 47. Wigge PA, et al. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Willems M, et al. 2010. Molecular analysis of pericentrin gene (PCNT) in a series of 24 Seckel/microcephalic osteodysplastic primordial dwarfism type II (MOPD II) families. J. Med. Genet. 47: 797–802 [DOI] [PubMed] [Google Scholar]
- 49. Wolkow TD, Harris SD, Hamer JE. 1996. Cytokinesis in Aspergillus nidulans is controlled by cell size, nuclear positioning and mitosis. J. Cell Sci. 109: 2179–2188 [DOI] [PubMed] [Google Scholar]
- 50. Xiang X. 2012. Nuclear positioning: dynein needed for microtubule shrinkage-coupled movement. Curr. Biol. 22: R496–R499 [DOI] [PubMed] [Google Scholar]
- 51. Xiang X, Fischer R. 2004. Nuclear migration and positioning in filamentous fungi. Fungal Genet. Biol. 41: 411–419 [DOI] [PubMed] [Google Scholar]
- 52. Yu JH, et al. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41: 973–981 [DOI] [PubMed] [Google Scholar]
- 53. Zekert N, Veith D, Fischer R. 2010. Interaction of the Aspergillus nidulans microtubule-organizing center (MTOC) component ApsB with gamma-tubulin and evidence for a role of a subclass of peroxisomes in the formation of septal MTOCs. Eukaryot. Cell 9: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zimmerman WC, Sillibourne J, Rosa J, Doxsey SJ. 2004. Mitosis-specific anchoring of gamma tubulin complexes by pericentrin controls spindle organization and mitotic entry. Mol. Biol. Cell 15: 3642–3657 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.