Abstract
Microsporidia are unicellular fungi that are obligate endoparasites. Although nematodes are one of the most abundant and diverse animal groups, the only confirmed report of microsporidian infection was that of the “nematode killer” (Nematocida parisii). N. parisii was isolated from a wild Caenorhabditis sp. and causes an acute and lethal intestinal infection in a lab strain of Caenorhabditis elegans. We set out to characterize a microsporidian infection in a wild nematode to determine whether the infection pattern of N. parisii in the lab is typical of microsporidian infections in nematodes. We describe a novel microsporidian species named Sporanauta perivermis (marine spore of roundworms) and characterize its infection in its natural host, the free-living marine nematode Odontophora rectangula. S. perivermis is not closely related to N. parisii and differs strikingly in all aspects of infection. Examination by transmission electron microscopy (TEM) revealed that the infection was localized in the hypodermal and muscle tissues only and did not involve the intestines. Fluorescent in situ hybridization (FISH) confirmed infection in the muscle and hypodermis, and surprisingly, it also revealed that the parasite infects O. rectangula eggs, suggesting a vertical mode of transmission. Our observations highlight the importance of studying parasites in their natural hosts and indicate that not all nematode-infecting microsporidia are “nematode killers”; instead, microsporidiosis can be more versatile and chronic in the wild.
INTRODUCTION
Microsporidia are obligate intracellular parasites whose hosts include diverse animals and one protist group (20, 26). Microsporidia are also unicellular fungi with over 1,300 recognized species (7, 14, 20). Their life cycles vary considerably for different species, but they all have at least one extracellular (spore) stage and one intracellular (merogonial/sporogonial) stage (12). Microsporidian spores are distinguished by the presence of an internal tube known as the polar filament (34). Fixed at the apex of the spore by the anchoring disk, the polar filament extends to the posterior of the spore where it is usually found coiled around one or two nuclei. The number of coils and their angle of inclination can often distinguish species. Microsporidian spores are also characterized by having a thick, protective chitinous wall around the cell membrane that allows them to survive outside their hosts. The spore wall thins near the anchoring disk; this is thought to facilitate the exit of the polar filament through the anchoring disk. During infection, the polar filament everts and, upon contacting a host's cell membrane, transfers the contents of the spore (sporoplasm) into the host cell. The sporoplasm differentiates into the proliferative (meront) stage that goes on to differentiate into spores (sporogony). Mature spores may then infect adjacent host cells or be released into the environment.
Microsporidian infections (microsporidiosis) are highly variable and cause a wide range of symptoms depending on the host and the microsporidium involved. Infections are usually acquired by ingestion of spores (horizontal transmission), but vertical transmission (from mother to offspring) has been observed (mainly in insects) (1). Microsporidian infections may be acute or chronic, affect one or multiple tissue types, cause mild or lethal symptoms, and even change the host's sex (18, 26).
Nematodes, commonly known as roundworms, are ubiquitous in the environment. In marine sediments, nematodes account for more than 90% of individual animals and over 50% of the total biomass (16), yet reports of microsporidian infections in this animal group are limited and unclear. Indeed, most reports of microsporidian infections in nematodes, including marine nematodes, failed to clearly demonstrate morphologically (presence of polar filament) and genetically that the infecting agent was a microsporidian (17, 21, 28, 29). The only exception is the microsporidian infection described 4 years ago in a lab strain (N2) of the soil nematode Caenorhabditis elegans (33). In this strain, ingestion of microsporidian spores (Nematocida parisii) causes an acute and extremely virulent (100% lethal) infection that affects only the intestinal tissues. N. parisii was originally isolated from a wild Caenorhabditis sp. and transferred to the lab strain; effects of the parasite on the native host are unclear. Given that nematodes are arguably the most abundant and diverse animal phylum (15), the dearth of knowledge regarding their microsporidian parasites is surprising. Here we present the first morphological and molecular description of a microsporidian infection in a marine nematode. We describe Sporanauta perivermis, a novel microsporidian species infecting the free-living marine nematode, Odontophora rectangula (family Axonolaimida). This is, to our knowledge, the first confirmed case of an interaction between a fungal group and nematodes in marine environments (3, 4). Moreover, the characteristics of S. perivermis infection challenge our current understanding of how microsporidia and nematodes interact in the wild by providing a completely different infection pattern from that observed in the lab with N. parisii. Unlike infection with N. parisii, infection with S. perivermis is mild and not severe, chronic and not acute, and it affects the hypodermal and muscles tissues and not the intestines. S. perivermis even differs in its mode of transmission, which appears to be vertical (infection of the eggs) and not exclusively horizontal (ingestion of spores) as it is in N. parisii. Indeed, by comparison, S. perivermis infection represents the opposing end of the spectrum of microsporidian infection variability, suggesting that host-parasite relationships between nematodes and microsporidia are considerably more complex and diverse than previously considered.
MATERIALS AND METHODS
Marine nematode sampling.
Marine nematodes were collected in July and August 2010 and 2011 from Boundary Bay beach (49.013N, 123.036W), Tsawwassen (British Columbia, Canada). Surface sand and sediment samples (1 to 10 cm deep) were collected at low tide using sterile 50-ml falcon tubes (filled 3/4 with sand and 1/4 with seawater). All samples were kept on ice until they were stored permanently at 4°C in the dark and with loosened caps. For each tube, 1 to 2 g of sand was transferred to a sterile petri dish and flooded with cold (4°C) sterile seawater. Live nematodes were observed using a dissecting microscope and transferred individually to a sterile petri dish containing cold sterile seawater. Marine nematodes were then used for DNA extraction, fluorescent in situ hybridization (FISH), and transmission electron microscopy (TEM).
Microsporidian and nematode rDNA extraction and sequencing.
Microsporidian and marine nematode rDNA were initially extracted from 0.5 to 1 g of sand using the Mobio UltraClean soil DNA isolation kit (Bio/Can Scientific). Beads (included in this kit) and a beadbeater (Mini-Beadbeater-1; BioSpec Products Inc.) were used to disrupt the tissues during the lysis step. This step was modified from the manufacturer's protocol by adding a 15-min incubation period at 70°C with a 20-s beadbeating (2,500 rpm) treatment every 5 min. Microsporidian small-subunit (SSU) ribosomal DNA (rDNA) sequences were amplified using the microsporidian-specific forward primer 18ss (35) and a novel microsporidian-specific primer Micro33R (5′-TAGAGACCGTTGTAGTTCCG-3′) (this study). This microsporidian primer set amplifies fragments of 550 to 650 bp long from the 5′ end of the microsporidian SSU rDNA. PCR amplicons were separated on agarose gels (2%), purified with the UltraClean 15 Mobio DNA purification kit (Bio/Can Scientific), and cloned using the TOPO TA cloning kit with pCR 2.1 vector (Invitrogen). At least three independent clones were sequenced for each DNA extraction. The same procedures were performed on individual live nematodes to determine the host and prevalence of microsporidian infections. Marine nematodes were barcoded using 18S rDNA marine nematode-specific primers (M18F 5′-AGRGGTGAAATYCGTGGAC-3′ and M18R 5′-TCTCGCTCGTTATCGGAAT-3′) (2) and an S. perivermis-specific primer set (5′-CGAGATGTGCAGTATGTCTGGG-3′) designed from previously amplified sequences. These primers were also used in combination with 530 and 1492 microsporidian primers (35) to generate a full-length (1,385 bp) S. perivermis SSU rDNA sequence (GenBank accession no. JN195782).
Phylogenetic analysis.
The small-subunit rDNA sequence of S. perivermis was examined phylogenetically by comparing it with that of 43 other microsporidian species representing the five major microsporidian groups (36). We also included an environmental microsporidian sequence (accession no. FJ756061.1) identified from the nucleotide BLAST analysis. The fungal species Basidiobolus ranarum and Conidiobolus coronatus were included as outgroups (36). All sequences were obtained from GenBank.
All sequences were viewed and aligned using the molecular sequence analysis multiplatform SeaView version 4. The alignment was adjusted automatically (trmAL;Phylemon) and by eye. The final alignment was used in three phylogenetic analyses: Bayesian inference, maximum likelihood (ML), and approximate likelihood ratio test based on Shimodaira-Hasegawa-like procedure (aLRT-SH).
To perform Bayesian inference, we used the program MrBayes version 3.2. One million generations were run using the general time reversible (GTR) model with two separate runs of four chains each. Sampling was performed every 1,000 trees of which the initial 25% (approximately 25 thousand trees) were discarded before the generation of the best supported consensus tree. To perform ML and aLRT-SH tests, we used Phyml version 3.0.
Transmission electron microscopy.
Live O. rectangula nematodes were transferred into a sterile polyethylene embedding capsule (size 3; BEEM Inc.) containing 30 μl of cold (4°C) sterile seawater. Nematodes were centrifuged at 3,000 × g for 30 s, and cuticles were nicked with a razor blade. Fixation proceeded immediately by capping the capsules with a piece of Whatman filter paper soaked in a fixative solution containing 1% (wt/vol) osmium tetraoxide in 0.2 M sodium cacodylate buffer (pH 7.2) and left on ice for 30 min. Fixative solution was then added directly to each sample and kept on ice for 2 h. This was followed by three 5-min washes with 0.2 M sodium cacodylate buffer (pH 7.2). The samples were dehydrated through an ethanol series (25%, 50%, 75%, 90%, 95%, and 100%) of 20 min at each step. Ethanol was substituted with an ethanol and acetone mixture (1:1) twice (20 min each) and then repeated twice with pure acetone. Acetone was removed, and a mixture (1:1) of acetone and Spurr's resin was added and left at room temperature for 24 h. Spurr's resin was changed twice (24 h each) before polymerizing overnight (12 h) at 70°C. Sectioning of each resin was performed using a Leica EM UC6 ultramicrotome (Leica Microsystems) and then stained twice with 2% (wt/vol) uranyl acetate and lead citrate (Reynolds 1963). Each ultrathin section was examined on a Hitachi H7600 TEM.
Fluorescent in situ hybridization.
Staining of S. perivermis was modified from methods described previously (33). A specific S. perivermis probe was purified by high-performance liquid chromatography (HPLC) and synthesized with Quasar 570 (Cy3) at the 5′ end (Biosearch Technologies, Inc.). Live O. rectangula nematodes were placed in a 1.5-ml Eppendorf tube containing 30 μl of sterile seawater and aggregated into a pellet by centrifugation (3,000 × g for 30 s). Excess seawater was removed, and cuticles were nicked with a razor blade before a 2-h fixation with 4% paraformaldehyde. The samples were then washed twice with 250 μl of phosphate-buffered saline (PBS) plus 0.1% Tween 20 and hybridized at 46°C for 12 h with 60 μl of hybridization buffer (900 mM NaCl, 20 mM Tris [pH 7.5], 0.01% SDS, and 5 ng/μl probe). Following this, samples were washed with 500 μl of wash buffer (900 mM NaCl, 20 mM Tris [pH 7.5], 0.01% SDS, 5 mM EDTA) and placed at 48°C for 1 h. Samples were then stained and mounted with 4′,6′-diamidino-2-phenylindole (DAPI) Vectashield mounting medium (Vector Laboratories) and examined using an Olympus FV10i confocal microscope (Olympus Inc.). Marine nematodes collected from the same sand samples as O. rectangula were collected and stained alongside as negative controls.
DNA sequence.
The full-length sequence of the SSU rDNA gene is deposited as GenBank accession no. JN195782.
RESULTS
The free-living marine nematode Odontophora rectangula is infected with a microsporidian parasite.
Live marine nematodes were collected at low tide from Boundary Bay beach (Tsawwassen, British Columbia, Canada). DNA was extracted from the nematodes, and microsporidian infection was assessed by PCR amplifying the microsporidian small-subunit (SSU) rRNA gene. An identical 668-bp fragment was amplified, and this sequence was most similar (88% similarity by BLAST) to an undescribed microsporidian sp. (accession no. FJ756061.1), suggesting the presence of a novel and single microsporidian infection in local nematodes.
Given that the initial isolations were done with a pooled, mixed marine nematode sample, we performed DNA extractions on individual marine nematodes to determine the host. Using a primer set specific for a fragment of the SSU sequence described above, individual nematodes were screened for both their infection status with the novel microsporidian and with a set of nematode barcoding primers. In all cases, microsporidian infections were observed only in a single nematode species, predicted to be Odontophora rectangula (Axonolaimida). Overall, the majority of O. rectangula nematodes (84%; n = 27) were infected, and the proportion of infected individuals in each sex was almost the same (83% males and 84% females). Morphologically, these nematodes match the predicted barcoding identification; O. rectangula nematodes are distinguished by their body size and tail shape and for having a distinctive set of large teeth (25, 27).
Phylogenetic position of the novel microsporidian, Sporanauta perivermis.
To establish the phylogenetic placement of the novel microsporidian SSU rDNA sequence, we compared it with 43 microsporidian species that span the known diversity of the group. Overall, recovered trees reflected our current understanding of microsporidian relationships, with the five major microsporidian clades strongly supported by all methods (Fig. 1) (36). Surprisingly, the new species did not show an affiliation with the only other known nematode-infecting microsporidian (N. parisii). Instead, all trees strongly support placing the microsporidian infecting O. rectangula within clade IV (Fig. 1). We have named this species Sporanauta perivermis (for marine spores of roundworms).
Fig 1.
Phylogenetic placement of the novel microsporidium, S. perivermis. The tree shown was obtained by Bayesian inference using microsporidian SSU rDNA sequences. The same tree topology was obtained by maximum likelihood. Support values from Bayesian inference, maximum likelihood bootstrapping, and the approximate likelihood ratio test Shimodaira-Hasegawa-like procedure (BI/MLB/aLRT-SH) are indicated in the figure. Support values below 50% are not shown. Other nematode-infecting microsporidia are denoted by an asterisk. Bar at the bottom indicates estimated number of changes per site.
The subclade containing S. perivermis includes four other microsporidian lineages: (i) the bark louse-infecting microsporidian Mockfordia xanthocaecila (32), (ii) the daphnia-infecting microsporidia Ordospora colligata (30), (iii) an undescribed microsporidian species (top BLAST hit), and (iv) the genus Encephalitozoon (five species) that infect a variety of animal hosts (Fig. 1). The position of S. perivermis relative to the rest of its subclade could not be fully resolved, even when restricting the analysis to members of this group and outgroup representatives within clade IV (data not shown).
Microscopic characterization of S. perivermis.
O. rectangula nematodes were examined using standard light microscopy techniques for any behavioral changes (i.e., less active) or any morphological abnormalities such as enlargement of specific tissues that could be signs of microsporidian infection. However, no external signs of infection were evident. Therefore, to examine the parasite's effect on the nematode and confirm morphologically that S. perivermis is a microsporidian parasite, we examined O. rectangula nematodes by transmission electron microscopy (TEM). All nematode digestive tissues (pharynx, intestine, and lumen) appeared normal. However, dense oval structures were located within the hypodermal and muscle tissues (Fig. 2A and B). These structures were morphologically consistent with microsporidian spores by their size (2.0 to 2.5 μm by 1.1 to 1.5 μm) and ovoid shape. Conclusive evidence that these structures are indeed microsporidian spores includes the presence of a polar filament (3 or 4 coils), an anchoring disk surrounded by polaroplast membranes, and a thin electron-dense exospore over a thick electron-transparent endospore surrounding the cell membrane (Fig. 2C and D).
Fig 2.
Ultrastructure of S. perivermis spores by TEM. (A) Cross section of an infected O. rectangula nematode showing the localization of S. perivermis spores (S) in the hypodermal tissues (H). Note the absence of spores in the lumen (L) and intestinal (In) tissues. Localization of the spores in the muscle tissues (M) is also shown. Bar = 0.5 μm. (B) S. perivermis spores surrounded by muscle fibers (M). Bar = 2 μm. (C) S. perivermis spore showing the four coils of its polar filament (Pf), the exospore (Ex), the endospore (En), the polaroplast membranes (Pm), and a vesicle (V) enclosing the spore. Bar = 0.5 μm. (D) S. perivermis spores showing four coils of the polar filament (Pf), the anchoring disk (Ad), and the polaroplast membranes (Pm). Bar = 0.5 μm.
Spores were not observed to be in direct contact with the host cytoplasm. Instead, each microsporidian spore appeared to be enclosed within a vesicle that was often observed to be connected to adjacent vesicles (Fig. 2). This resulted in a large “cluster” of interconnected vesicles containing S. perivermis spores. Proliferative microsporidian life cycle stages (i.e., meronts) were not observed in the tissues examined.
Fluorescent in situ hybridization reveals S. perivermis infection of O. rectangula eggs.
To further assess the extent of S. perivermis infection in O. rectangula, we performed fluorescent in situ hybridization (FISH) using a fluorescent probe targeting the S. perivermis SSU rDNA sequence. Surprisingly, FISH indicated infection differences in male and female nematodes. In males, FISH confirmed our previous TEM observations in which S. perivermis infection was localized to the hypodermal and muscular tissues, surrounding the intestine but never within it (Fig. 3). Within these tissues, spores were observed in clusters (few to hundreds of spores) which appeared to be distributed along the length of the body but were never observed in the head. In addition, spore “clusters” appeared enclosed within large vesicles. We interpret these as vesicles of microsporidian origin due to the hybridization of the microsporidian tag. S. perivermis spores were oval and consistent in size (∼2 μm by 1 μm) with those observed by TEM.
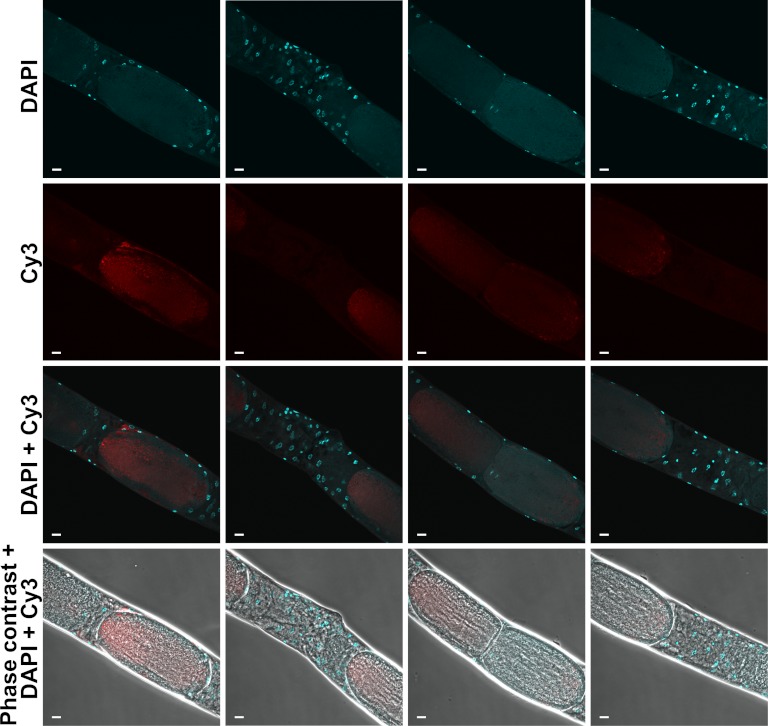
Fig 3.
Localization of S. perivermis infecting the free-living marine nematode O. rectangula by fluorescent in situ hybridization (FISH). The columns show cross sections of nematode tissues at sequential depths, moving from interior to exterior (left to right) to show S. perivermis surrounding the intestine and localized to hypodermal and muscle tissues (long white arrows). Note the absence of S. perivermis in the intestine, whose position is clearly marked by the concentric rings of DAPI-stained nematode nuclei in the leftmost column. Each row shows a different light filter showing the nuclear stain DAPI (shown in blue), the S. perivermis fluorescent probe (Cy3 [shown in red]), a combination of the two dyes, and the view of the image under phase contrast. Bars = 10 μm.
In females, spores were also observed in clusters within the hypodermal and muscle tissues; however, the spores appeared to be markedly less abundant. Surprisingly, FISH revealed that in female nematodes the microsporidian infection was localized to O. rectangula eggs (Fig. 4). In all infected females examined, the eggs showed a very strong signal for S. perivermis, but the parasite did not appear to be in the spore stage, as spores were not observed within the eggs (Fig. 4). Aside from the eggs, there appeared to be no other infection of female reproductive tissues.
Fig 4.
S. perivermis infection in O. rectangula eggs by FISH. Each column shows a sequence of images taken on the same plane to show the localization of S. perivermis inside eggs. Each row shows a different light filter: the nuclear stain DAPI (shown in blue), the S. perivermis fluorescent probe (Cy3 [shown in red]), a combination of the two dyes, and the view of the image under phase contrast. Bars = 10 μm.
A small number of juveniles were observed to be infected. Generally, the infection pattern was the same as in adults, but in some cases, the reproductive tissues were labeled with the FISH probe. The sex of most nematodes, including O. rectangula, cannot be resolved in juveniles, so it is unclear how this labeling pattern correlates with the gender differences observed in adults.
TAXONOMY
Sporanauta n. gen. Ardila-Garcia & Fast 2012.
This is a novel microsporidian lineage that belongs to microsporidian clade IV (36) based on SSU rDNA phylogenetic analyses. The closest relatives to Sporanauta are the microsporidian genera Mockfordia, Encephalitozoon, and Ordospora. The type species is Sporanauta perivermis Ardila-Garcia & Fast 2012. The Latin “sporo” (spore) and the Latin “nauta” (mariner) refer to microsporidian spores to the habitat (marine).
Sporanauta perivermis n. sp. Ardila-Garcia & Fast 2012.
The spores are ovoid and measured 2.0 to 2.5 μm by 1.1 to 1.5 μm. The polar filament has 3 or 4 coils. Spores are found in vesicles that prevent them from being in direct contact with the host cells. Vesicles may contain a few spores to hundreds of spores. Infection is localized to the hypodermal and muscle tissues in adults, mature eggs in adult females, and the reproductive tissues of juveniles. Infection does not affect the intestinal tissues. Infection is chronic and appears to cause minor symptoms (if any) on the host. Transmission appears to be vertical (transovary). The type host is the free-living marine nematode Odontophora rectangula (family Axonolaimidae). The Latin “peri” (round) and the Latin “vermis” refer to roundworms, the common name for nematodes.
DISCUSSION
Fungi and nematodes are highly abundant and diverse in marine ecosystems (19, 22), but the nature of their interactions remains largely unknown (3, 4). The present study is, to our knowledge, the first to provide morphological and genetic evidence to show that fungi (Microsporidia) can parasitize marine nematodes. We describe the infection of a novel microsporidian species, S. perivermis, in its wild host, the free-living marine nematode O. rectangula. The characteristics of S. perivermis infection challenge the current view of the nature of microsporidian infections in nematodes. The only other described microsporidian infection in nematodes involves inoculation of N. parisii from wild nematodes to a C. elegans lab strain (8, 13, 33). Here the exposure to a microsporidian parasite was lethal, but it raises questions as to whether this particular parasite or any other nematode-infecting microsporidian is as damaging and virulent in its natural host. Our observations on S. perivermis infection in its wild host indicate that microsporidian infections in the wild can vary considerably in almost every aspect compared to that observed with N. parisii.
S. perivermis represents a novel microsporidian lineage that grouped with eight other species in a subclade of microsporidian clade IV. Its closest relatives were isolated from a diversity of hosts, including mammals (9–11, 37), lizards (6), crustaceans (30), and insects (23, 32). Despite their differences in hosts, all members of this subclade are morphologically similar in that they are considerably small (spore size, 2 to 4 μm) by microsporidian standards (spore size range, 2 to 40 μm). In addition, their spores are always found within vesicles that prevent them from being in direct contact with the host cytoplasm. Other members of the S. perivermis subclade, O. colligata and the Encephalitozoon group, also develop in isolation from their hosts' tissues. However, their separation is achieved by means of a membrane-bound parasitophorous vacuole (host origin) instead of a vesicle (24, 31). The host-parasite interface varies considerably for other members of clade IV, as some (i.e., Endoreticulatus species) develop in parasitophorous vacuoles, while others (i.e., Nosema) develop in direct contact with the host tissues (5). Even though N. parisii spores were also observed inside vesicles (33), N. parisii and the undescribed Nematocida sp. strain 1247 did not group in clade IV with S. perivermis. Instead, Nematocida falls within microsporidian clade II with their closest relative being the beetle-infecting microsporidian Ovavesicula popillae (a position consistent with reference 33).
S. perivermis and N. parisii also have stark differences in how they interact with their respective hosts. S. perivermis causes minor lesions (if any) in the hypodermal and muscle tissues and does not infect the intestinal tissues of O. rectangula nematodes. The parasites do not appear to affect O. rectangula's motility, behavior, or life expectancy. In fact, the infection was highly prevalent (84%), and it was not possible to distinguish uninfected from infected nematodes with standard light microscopy. Conversely, N. parisii infection of the C. elegans N2 lab strain was reported to invade the intestine and be highly destructive, sickening N2 nematodes severely, reducing their motility considerably, and causing rapid death (13, 33). Each case represents opposing ends on the full spectrum of the host-parasite relationships between animals and microsporidia (9). The features of S. perivermis are typical of chronic microsporidian infections, which are usually asymptomatic and cause minor effects on the host. On the other hand, N. parissii infection is typical of unbalanced, opportunistic, acute infections which are usually characterized by uncontrolled microsporidian propagation, severe symptoms, and high host lethality (9). The striking differences between the two may be explained by the fact that our observations were performed in the natural host of S. perivermis. Our observations on S. perivermis suggest that nematodes are capable of dealing with microsporidian infections and that N. parisii is perhaps a “nematode killer” of Caenorhabditis (particularly N2) but that its lethality should not be generalized as a feature of all nematode-infecting microsporidia. Indeed, one would expect microsporidian-nematode relationships to encompass the full spectrum of infection (9) once additional species are described and their interactions with their hosts are understood.
Transmission of microsporidian parasites in their host is generally horizontal (ingestion of spores) as was reported for N. parisii, while microsporidian vertical transmission in this animal group remains unknown. Vertical transmission of microsporidia is common in insects, in which it is usually transovarial and involves the transfer of the parasite within the female's eggs (1). In the present study, we provided evidence that microsporidian parasites may be transmitted in this manner in nematodes. S. perivermis infects the eggs of adult O. rectangula females and the reproductive tissues of S. perivermis juveniles, suggesting that S. perivermis is transmitted from mother to offspring via the eggs.
Our observations of the infection of S. perivermis in the free-living marine nematode O. rectangula provide the first description of a fungal parasite of nematodes in marine environments. They provide the first indications that the interactions between microsporidia and nematodes in the wild can be highly intricate and involve versatile adaptations such as microsporidian vertical transmission. Our understanding of microsporidia in nematodes is no longer limited to “nematode killers” but has broadened to include interactions more indicative of coexistence.
ACKNOWLEDGMENTS
We thank N. Raghuram for assistance with sampling, N. Yabuki and K. Wakeman for assistance with imaging, and F. Burki for assistance with the phylogenetic analysis.
This work was supported by an NSERC Discovery Grant to N. M. Fast.
Footnotes
Published ahead of print 19 October 2012
REFERENCES
- 1. Becnel JJ, Andreadis TG. 1999. Microsporidia in insects, p 447–501. In Wittner M, Weiss LM. (ed), The microsporidia and microsporidiosis. ASM Press, Washington, DC [Google Scholar]
- 2. Bhadury P, Austen MC. 2010. Barcoding marine nematodes: an improved set of nematode 18S rRNA primers to overcome eukaryotic co-interference. Hydrobiologia 641:245–251 [Google Scholar]
- 3. Bhadury P, et al. 2011. Molecular diversity of fungal phylotypes co-amplified alongside nematodes from coastal and deep-sea marine environments. Plos One 6:e26445 doi:10.1371/journal.pone.0026445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bhadury P, Bridge PD, Austen MC, Bilton DT, Smerdon GR. 2009. Detection of fungal 18S rRNA sequences in conjunction with marine nematode 18S rRNA amplicons. Aquat. Biol. 5:149–155 [Google Scholar]
- 5. Cali A, Takvorian PM. 1999. Developmental morphology and life cycles of the microsporidia, p 85–128. In Wittner M, Weiss LM. (ed), The microsporidia and microsporidiosis. ASM Press, Washington, DC [Google Scholar]
- 6. Canning EU. 1981. Encephalitozoon lacertae n. sp., a microsporidian parasite of the lizard Podarcis muralis, p 57–64. In Parasitological Topics. Society of Protozoologists Special Publication 1. Allen Press, Lawrence, KS [Google Scholar]
- 7. Capella-Gutierrez S, Marcet-Houben M, Gabaldon T. 2012. Phylogenomics supports microsporidia as the earliest diverging clade of sequenced fungi. BMC Biol. 10:47 doi:10.1186/1741-7007-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cuomo CA, et al. 18 July 2012. Microsporidian genome analysis reveals evolutionary strategies for obligate intracellular growth. Genome Res. doi:10.1101/gr.142802.112. [DOI] [PMC free article] [PubMed]
- 9. Didier ES, Bessinger GT. 1999. Host-parasite relationships in microsporidiosis: animal models in immunology, p 225. In Wittner M, Weiss LM. (ed), The microsporidia and microsporidiosis. ASM Press, Washington, DC [Google Scholar]
- 10. Didier ES, et al. 1991. Isolation and characterization of a new human microsporidian, Encephalitozoon hellem (n. sp.), from 3 AIDS patients with keratoconjunctivitis. J. Infect. Dis. 163:617–621 [DOI] [PubMed] [Google Scholar]
- 11. Didier ES, et al. 1994. Experimental microsporidiosis in immunocompetent and immunodeficient mice and monkeys. Folia Parasitol. 41:1–11 [PubMed] [Google Scholar]
- 12. Dunn AM, Smith JE. 2001. Microsporidian life cycles and diversity: the relationship between virulence and transmission. Microb. Infect. 3:381–388 [DOI] [PubMed] [Google Scholar]
- 13. Estes KA, Szumowski SC, Troemel ER. 2011. Non-lytic, actin-based exit of intracellular parasites from C. elegans intestinal cells. PLoS Pathog. 7:e1002227 doi:10.1371/journal.ppat.1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fischer WM, Palmer JD. 2005. Evidence from small-subunit ribosomal RNA sequences for a fungal origin of Microsporidia. Mol. Phylogenet. Evol. 36:606–622 [DOI] [PubMed] [Google Scholar]
- 15. Fonseca VG, et al. 2010. Second-generation environmental sequencing unmasks marine metazoan biodiversity. Nat. Commun. 1:98 doi:10.1038/ncomms1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giere O. 2009. Meiobenthology: the microscopic motile fauna of aquatic sediments. Springer-Verlag, Berlin, Germany [Google Scholar]
- 17. Hopper BE, Meyers SP, Cefalu R. 1970. Microsporidian infection of a marine nematode Metoncholaimus scissus. J. Invertebr. Pathol. 16:371–377 [DOI] [PubMed] [Google Scholar]
- 18. Hurst LD. 1991. The incidences and evolution of cytoplasmic male killers. Proc. R. Soc. Lond. Ser. B Biol. Sci. 244:91–99 [Google Scholar]
- 19. Hyde KD, et al. 1998. Role of fungi in marine ecosystems. Biodivers. Conserv. 7:1147–1161 [Google Scholar]
- 20. Keeling P. 2009. Five questions about Microsporidia. PLoS Pathog. 5:e1000489 doi:10.1371/journal.ppat.1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kudo R, Hetherington DC. 1922. Notes on a microsporidian parasite of a nematode. J. Parasitol. 8:129–132 [Google Scholar]
- 22. Lambshead PJD, Boucher G. 2003. Marine nematode deep-sea biodiversity - hyperdiverse or hype? J. Biogeogr. 30:475–485 [Google Scholar]
- 23. Lange CE, Johny S, Baker MD, Whitman DW, Solter LF. 2009. A new Encephalitozoon species (Microsporidia) isolated from the lubber grasshopper, Romalea microptera (Beauvois) (Orthoptera: Romaleidae). J. Parasitol. 95:976–986 [DOI] [PubMed] [Google Scholar]
- 24. Larsson JIR, Ebert D, Vavra J. 1997. Ultrastructural study and description of Ordospora colligata gen. et sp. nov. (Microspora, Ordosporidae fam. nov.), a new microsporidian parasite of Daphnia magna (Crustacea, Cladocera). Eur. J. Protistol. 33:432–443 [Google Scholar]
- 25. Lorenzen S. 1971. Die Nematodenfauna im Verklappundsgebiet fur Industrieabwasser nordwestlich von Helgoland: I. Araeolaimida und Monhysterida. Zool. Anz., Leipzig. 187:223–248 [Google Scholar]
- 26. Murray W, Weiss LM. 1999. The microsporidia and microsporidiosis. ASM Press, Washington, DC [Google Scholar]
- 27. Platt HM, Warwick RM. 1988. Part II: British chromadorids, p 58–59. In Brill EJ, Backuys W. (ed), Free living marine nematodes. The Linnean Society of London and The Estuarine and Brackish-Water Science Association, Avon, Great Britain [Google Scholar]
- 28. Poinar GO. 1988. A microsporidian parasite of Neoplectana glaseri (Steinernematidae: Rhabditida). Rev. Nematol. 11:359–361 [Google Scholar]
- 29. Poinar GO, Hess R. 1986. Microsporidium rhabdophilum sp. n. (Microsporida: Pansporoblastina), a parasite of the nematode, Rhabditis myriophila (Rhabditina: Rhabditidae). Rev. Nematol. 9:369–375 [Google Scholar]
- 30. Refardt D, et al. 2002. Small subunit ribosomal DNA phylogeny of microsporidia that infect Daphnia (Crustacea: Cladocera). Parasitology 124:381–389 [DOI] [PubMed] [Google Scholar]
- 31. Schottelius J, et al. 2000. Presentation by scanning electron microscopy of the life cycle of microsporidia of the genus Encephalitozoon. Microb. Infect. 2:1401–1406 [DOI] [PubMed] [Google Scholar]
- 32. Sokolova YY, Sokolov IM, Carlton CE. 2010. New microsporidia parasitizing bark lice (Insecta: Psocoptera). J. Invertebr. Pathol. 104:186–194 [DOI] [PubMed] [Google Scholar]
- 33. Troemel ER, Felix MA, Whiteman NK, Barriere A, Ausubel FM. 2008. Microsporidia are natural intracellular parasites of the nematode Caenorhabditis elegans. PLoS Biol. 6:2736–2752 doi:10.1371/journal.pbio.0060309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vavra J, Larsson JIR. 1999. Structure of the Microsporidia, p 7–84. In Wittner M, Weiss LM. (ed), The microsporidia and microsporidiosis. ASM Press, Washington, DC [Google Scholar]
- 35. Vossbrinck CR, Baker MD, Didier ES, Debrunner-Vossbrinck BA, Shadduck JA. 1993. Ribosomal DNA sequences of Encephalitozoon hellem and Encephalitozoon cuniculi: species identification and phylogenetic construction. J. Eukaryot. Microbiol. 40:354–362 [DOI] [PubMed] [Google Scholar]
- 36. Vossbrinck CR, Debrunner-Vossbrinck BA. 2005. Molecular phylogeny of the Microsporidia: ecological, ultrastructural and taxonomic considerations. Folia Parasitol. 52:131–142 [DOI] [PubMed] [Google Scholar]
- 37. Weber R, Bryan RT, Schwartz DA, Owen RL. 1994. Human microsporidial infections. Clin. Microbiol. Rev. 7:426–461 [DOI] [PMC free article] [PubMed] [Google Scholar]