Abstract
In Trichomonas vaginalis, a novel nuclear localization signal spanning the folded R2R3 DNA-binding domain of a Myb2 protein was previously identified. To study whether a similar signal is used for nuclear translocation by other Myb proteins, nuclear translocation of Myb3 was examined in this report. When overexpressed, hemagglutinin-tagged Myb3 was localized to nuclei of transfected cells, with a cellular distribution similar to that of endogenous Myb3. Fusion to a bacterial tetracycline repressor, R2R3, of Myb3 that spans amino acids (aa) 48 to 156 was insufficient for nuclear translocation of the fusion protein, unless its C terminus was extended to aa 167. The conserved isoleucine in helix 2 of R2R3, which is important for Myb2's structural integrity in maintaining DNA-binding activity and nuclear translocation, was also vital for the former activity of Myb3, but less crucial for the latter. Sequential nuclear influx and efflux of Myb3, which require further extension of the nuclear localization signal to aa 180, were immediately induced after iron repletion. Sequence elements that regulate nuclear translocation with cytoplasmic retention, nuclear influx, and nuclear efflux were identified within the C-terminal tail. These results suggest that the R2R3 DNA-binding domain also serves as a common module for the nuclear translocation of both Myb2 and Myb3, but there are intrinsic differences between the two nuclear localization signals.
INTRODUCTION
With an estimated ∼180 million cases annually of Trichomonas vaginalis infection worldwide (42), the protozoan parasite imposes a heavy burden on public health, especially when trichomoniasis is recognized as one of the major risk factors in promoting transmission of the human immunodeficiency virus (HIV) and papillomaviruses (13). Manifestations of the disease are usually mild to asymptomatic, and it is easy to cure, but increasing reports on drug-resistant clinical isolates are alarming (13, 34). The organism displays intriguing biological features distinct from those of model organisms and is reputed to have branched out early from eukaryotic lineages (40). The parasite exploits rather simplified general transcription machinery likely to express more than 26,000 protein genes in its genome (9), which is surprisingly complicated for a unicellular organism that survives only in human hosts. The study of unique features not seen in the human host may shed light on the prevention of the disease and alternative treatments to curb it.
A number of parasitic virulence factors, especially those inducible under iron repletion (1, 2, 5, 16, 25), were identified decades ago. Since the parasite persistently colonizes the human urogenital tract without a dormant cyst stage, its cytoadherence to vaginal epithelial cells is crucial for its survival. To date, multiple parasitic factors that contribute to cytoadherence, including some carbohydrate moieties and several reputed adhesion proteins on plasma membranes, have been identified (1–6, 16, 26). Serial studies on inducible transcription of the adhesion protein gene, ap65-1, revealed that its transcription is critically coregulated by three Myb proteins, Myb1, Myb2, and Myb3, through competitive entries of multiple Myb recognition elements in the distal promoter region in a space- and time-dependent manner (18, 28–30, 38), suggesting that these Myb proteins are differentially regulated in response to various stimuli from host environments. Myb proteins are ubiquitous in eukaryotes that share a conserved DNA-binding domain (DBD) composed of one to three repeat sequences termed R1, R2R3, and R1R2R3. In vertebrates, there are three Myb proteins in the R1R2R3 subfamily, exemplified by human c-Myb, B-Myb, and A-Myb (32). Recently, the structures of the DBDs in Myb1, Myb2, and Myb3 were each resolved (21, 24, 41). Like human c-Myb (27), the R2R3 domains of Myb1 and Myb2 display optimal DNA-binding activities and are comprised of six α-helices arranged in two bundles (21, 24), while the optimal DNA-binding activity of Myb3 requires an additional unique hairpin structure in the flexible tail C-terminal to R2R3 (41) (see Fig. S4 in the supplemental material).
Timely shuttling of transcription factors between the cytoplasm and nucleus is a critical step in gene-specific transcription (8, 20, 23, 39). In general, a nuclear localization signal (NLS) embedded in the sequence of a cargo protein is necessary for binding to a specific nuclear transporter of the importin family prior to its nuclear importation via a Ran-mediated pathway (36). Nuclear proteins without such a signal often interact with another cargo protein containing an NLS. On the other hand, protein nuclear export also requires a nuclear export signal (NES) on the cargo protein (14, 35). The classical NES, with a consensus hydrophobic sequence of L-X(2–3)-L-X(2–3)-L-X-L (where L can be Leu, Lys, Val, Ile, Phe, or Met and X denotes any amino acid) (7), binds the exportin CRM1 for transport out of nuclei, also via a Ran-mediated pathway (14, 35). CRM1-mediated nuclear protein export can be inhibited by the fungal metabolite leptomycin B (LMB) (22). The classical NLS (cNLS) comprises a loose consensus polybasic sequence, K(K/R)X(K/R) (23), of the simian virus 40 T antigen that allows the nuclear import of nonnuclear proteins in many heterologous systems but does not function in T. vaginalis (11). Instead, a novel NLS comprising an ordered structure embedded in the R2R3 domain of Myb2 was defined in the parasite (11) (see Fig. S4 in the supplemental material), but its nuclear transporter remains unknown. In contrast, human c-Myb and B-Myb use cNLS-mediated nuclear import machinery (12, 19, 37), suggesting that T. vaginalis may have evolved a distinct mechanism for the nuclear importation of numerous Myb proteins to properly regulate transcription.
To test whether other Myb proteins in the parasite share a module for nuclear importation, Myb3's NLS was studied here. Myb3 was found to harbor an NLS spanning the entire DBD and further extending to a short, flexible C-terminal tail. Intriguingly, iron was demonstrated to induce the sequential nuclear influx of Myb3, but not Myb2, and the activity requires further extension of the C-terminal tail, which comprises multiple sequence elements to regulate nuclear translocation at distinct steps. As proof of this concept, different Myb proteins may share a highly organized module, which is essential but might not be sufficient in all cases, for constitutive nuclear import, and additional sequence elements may be required for inducible nuclear import.
MATERIALS AND METHODS
Cultures.
A T. vaginalis T1 isolate and derived transgenic cell lines were maintained as previously described (38). Iron repletion or depletion was achieved, respectively, with the addition of 250 μM ferrous ammonium sulfate or 50 μM 2,2′-dipyridyl, an iron chelator, to normal growth medium.
DNA transfection and selection of stable transfectants.
Plasmids were transfected into T. vaginalis for paromomycin selection of stable transfectants, and cloned cell lines were established as previously described (28).
Oligonucleotides.
The sequences of the oligonucleotides used in the present study are listed in Table S1 in the supplemental material, unless reported elsewhere.
Plasmid construction.
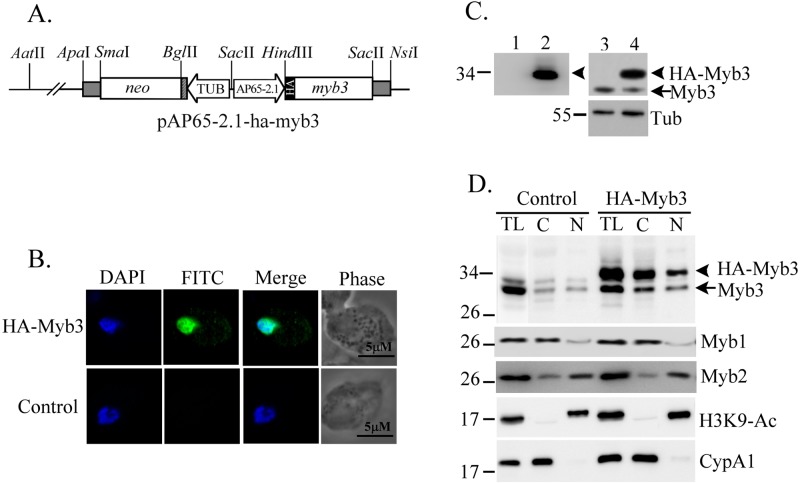
A T. vaginalis stable cell line transfected with the expression plasmid pAP65-1-m(MRE-1)-ha-myb3/TUBneo expressed hemagglutinin (HA)-Myb3 to a level detected in only a small fraction of cells by an immunofluorescence assay (IFA) using a 100× dilution of a mouse anti-HA antibody (29). To generate a plasmid with a higher HA-tagged Myb3 (HA-Myb3) expression level than that conferred by pAPm(MRE-1)-ha-myb3, the sequence encompassing the ap65-2.1 promoter was amplified from pAP65-2.1-ha-myb2/TUBneo (30) by a PCR using the primer pair pAP65-2.1-sac2-5′ and ap65-2.1-hind3-3′ (see Table S1 in the supplemental material). The amplified DNA was restricted by SacII/HindIII and subcloned into a vector backbone from SacII/NsiI-digested pAPm(MRE-1)-ha-myb3 (18), along with the HindIII/NsiI fragment from pAPm(MRE-1)-ha-myb3, to generate pAP65-2.1-ha-myb3 (Fig. 1A).
Fig 1.
Overexpression of HA-Myb3 in T. vaginalis. (A) In pAP65-2.1-ha-myb3, an ap65-2.1 promoter drives expression of the HA-tagged myb3 gene and the β-tubulin (TUB) promoter drives the neo gene. Restriction sites for constructing the plasmid are indicated. Cells for the experiments were grown in normal medium overnight. (B) Cells overexpressing HA-Myb3 (top) or the control (bottom) were examined by an immunofluorescence assay using the anti-HA antibody and FITC-conjugated mouse IgG. The nucleus was stained with DAPI, and the cell morphology was recorded by phase-contrast microscopy. (C) Cell lysates from the control (lanes 1 and 3) and cells overexpressing HA-Myb3 (lanes 2 and 4) were examined by Western blotting using anti-HA (lanes 1 and 2), anti-Myb3 (lanes 3 and 4, top), and anti-α-tubulin (lanes 3 and 4, bottom) antibodies. (D) Cell lysates (TL) from the control and cells overexpressing HA-Myb3 were fractionated into cytosolic (C) and nuclear (N) fractions for Western blotting using anti-Myb3, anti-Myb1, anti-Myb2, anti-H3K9-Ac, and anti-CyPA1 antibodies. In panels C and D, molecular weight markers (left) and the detected proteins (right) are indicated.
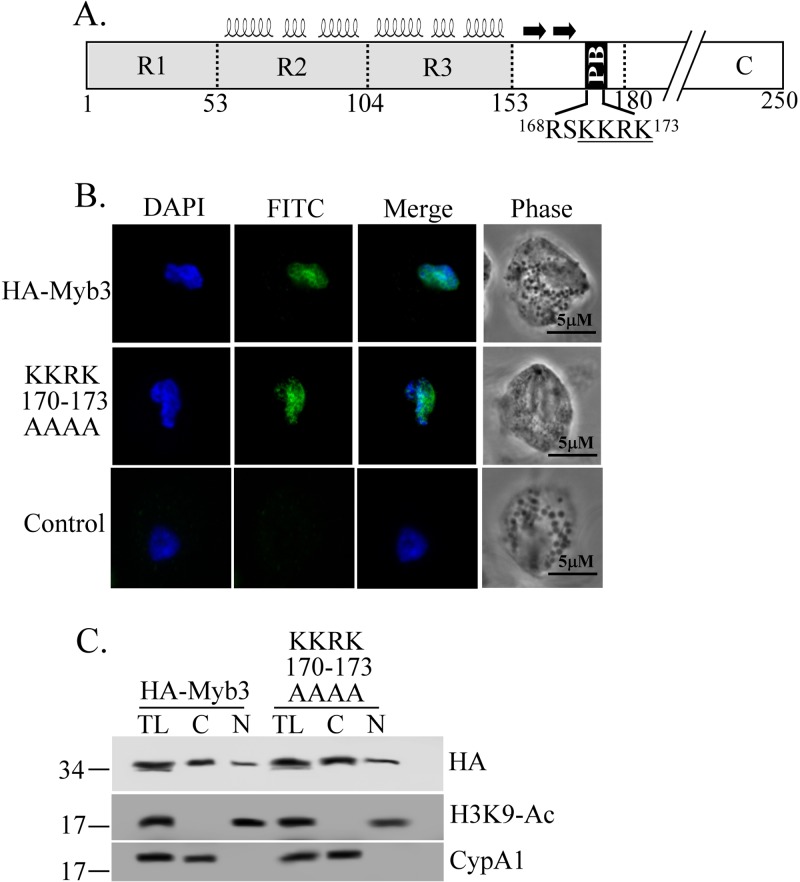
Site-directed mutagenesis was performed to mutate specific amino acid residues in HA-Myb3. To do this, a 5′ DNA fragment was amplified from pAP65-2.1-ha-myb3 using the primer pair ap65-2.1-sac2-5′ (35) and myb3(XnX′)-3′ (X is the amino acid to be mutated, n is the numeric location of the residue, and X′ is the mutated counterpart, for example, I79A or I79P, as shown in Table S1 in the supplemental material), whereas a 3′ fragment was amplified using the primer pair myb3(XnX′)-5′ and SP6. The PCR products were gel purified, mixed, denatured, and annealed for the second-round PCR using the primer pair ap65-2.1-sac2-5′ and SP6. The DNA thus amplified was restricted by HinDIII/NsiI and cloned into a HinDIII/NsiI-restricted pAP65-2.1-ha-myb3 backbone. A similar approach was exploited to generate clustered mutations in the polybasic 170KKRK173 sequence (Fig. 2) and the sequence spanning positions 157 to 165 of HA-Myb3 of polyalanines (see Fig. 4).
Fig 2.
The polybasic sequence and Myb3's nuclear localization. (A) Relative locations of Myb3′ R1R2R3 structural domains (helices), a β-hairpin (two arrows), and a polybasic (PB) sequence are depicted. The PB sequence was mutated to produce a mutant protein, KKRK170–173AAAA. Cells overexpressing HA-Myb3, KKRK170–173AAAA, or the control were grown overnight in normal medium. (B) Cells were examined by an immunofluorescence assay using a mouse anti-HA antibody and FITC-conjugated mouse IgG. The nucleus was stained with DAPI, and the cell morphology was recorded by phase-contrast microscopy. (C) Cell lysates (TL) were fractionated into cytosolic (C) and nuclear (N) fractions for Western blotting using anti-HA (top), anti-H3K9-Ac (middle), and anti-CypA1 (bottom) antibodies. Molecular weight markers (left) and the proteins detected (right) are indicated.
Fig 4.
Roles of the NLS C-terminal tail in the nuclear translocation of Myb3. The sequence spanning aa 157 to 165 of HA-Myb3 was mutated to produce NSN157–159AAA, HKE160–162AAA, or ILL163–165AAA. Cells were grown overnight in normal medium. (A) Cells were examined by an immunofluorescence assay using a mouse anti-HA antibody and FITC-conjugated mouse IgG. The nuclei were stained with DAPI, and the cell morphology was recorded by phase-contrast microscopy. (B) Cell lysates (TL) were fractionated into cytosolic (C) and nuclear (N) fractions for Western blotting using anti-HA (top), anti-H3K9-Ac (middle), and anti-CyPA1 (bottom) antibodies. Molecular weight markers (left) and the detected proteins (right) are indicated.
To map the NLS, a 5′ DNA fragment spanning amino acids (aa) 1 to 180 of Myb3 was amplified from pAP65-2.1-ha-myb3 using the primer pair ap65-2.1-sac2-5′ and myb3(180)tetR-3′. A 3′ DNA fragment encoding a bacterial tetracycline repressor (TetR) was amplified from p48-143-tetR, which encodes a Myb2-tetR fusion protein (11), using the primer pair myb3(180)tetR-5′ and tetR-sac2-3′. The 5′ and 3′ DNA fragments thus generated were mixed, denatured, and annealed for a second-round PCR using the primer pair ap65-2.1-sac2-5′ and tetR-sac2-3′. The PCR product was digested with SacII/HindIII, and the resulting DNA was ligated together with two DNA pieces restricted from pAP65-2.1-ha-myb3 by AatII/HinDIII and AatII/SacII to generate pMyb3(1∼180)-tetR (see the restriction map in Fig. S1 in the supplemental material). A 5′ fragment and a 3′ fragment were each amplified from pMyb3(1∼180)-tetR using the primer pairs ap65-2.1-sac2-5′ and myb3(48)-3′, and myb3(48)-5′ and tetR-sac2-3′, respectively. The 5′ and 3′ fragments were mixed, denatured, and annealed for PCR using the primer pair ap65-2.1-sac2-5′ and tetR-sac2-3′. The product was digested by SacII/HindIII, and the resulting DNA was ligated, together with two DNA pieces, respectively, restricted from pAP65-2.1-ha-myb3 by AatII/HinDIII and AatII/SacII to generate pMyb3(48∼180)-tetR. A similar approach was exploited to create the deletion mutants of pMyb3(1∼180)-tetR, as shown in Fig. 3A, each using a specific primer pair listed in Table S1 in the supplemental material.
Fig 3.
Deletion mapping to define Myb3's NLS. (A) The relative locations of the R1, R2, and R3 domains and a polybasic (PB) sequence in Myb3 are indicated by numbers. Regions spanning various fragments of Myb3 were each fused with an N-terminal HA tag and a C-terminal TetR to produce fusion proteins, the names of which are shown on the left to identify the fusion protein, while the nuclear or cytoplasmic localization of a protein was examined in panel B (indicated by N or C, respectively). The cells were grown in normal medium overnight. (B) Cells overexpressing various proteins or the control, as indicated on the left, were examined by an immunofluorescence assay using a mouse anti-HA antibody and FITC-conjugated mouse IgG. Nuclei were stained with DAPI, and the cell morphology was recorded by phase-contrast microscopy. The relative intensity of the FITC signal in the nucleus (N) versus the entire cell (N+C) for each protein is shown as a histogram at the bottom. The error bars indicate standard deviations. (C) Cell lysates (TL) were fractionated into cytosolic (C) and nuclear (N) fractions for Western blotting using anti-HA (top), anti-H3K9-Ac (middle), and anti-CyPA1 (bottom) antibodies. Signal intensities in samples from nuclear fractions were quantified and are shown as a histogram at the bottom. Molecular weight markers (left) and the detected proteins (right) are indicated. Fusion proteins are indicated by arrowheads and HA-Myb3 by an arrow.
To produce the recombinant protein, the sequence spanning aa 48 to 180 of Myb3 was amplified from pAP65-2.1-ha-myb3 by a PCR using the primer pair sac1-myb3(48)-5′ and myb3(180)-bgl2-3′ and was cloned into pGEM-T. The insert restricted by SacI/BglII was cloned into SacI/BamHI-restricted pET6H (11) to produce pET6H/Myb3(48∼180). Site-directed mutagenesis using a two-step PCR as described above and primer pairs listed in Table S1 in the supplemental material was performed to mutate the I79 isoleucine to a proline or alanine to produce pET6H/Myb3-I79P(48∼180) or pET6H/Myb3-I79A(48∼180), respectively.
Production of the recombinant protein.
pET6H/Myb3(48∼180), pET6H/I79A(48∼180), or pET6H/I79P was transformed into an Escherichia coli strain, BL21-codonplus(DE3)-RIL (Stratagene). A His-tagged recombinant protein, rMyb3(48–180), rMyb3 I79A(48–180), or rMyb3 I79P(48–180), was produced and purified by a nickel column, as described by the supplier (Novagen).
Immunofluorescence assay.
T. vaginalis was fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 30 min and permeabilized in 0.2% Triton X-100 in PBS for 15 min at room temperature prior to the IFA using a mouse anti-HA monoclonal antibody (400×; HA-7; Sigma) and fluorescein isothiocyanate (FITC)-conjugated mouse immunoglobulin G (IgG), as previously described (29). The nuclear DNA was stained with DAPI (4′,6-diamidino-2-phenylindole). The fluorescence signals from FITC and DAPI recorded by a confocal microscope were quantified with Metamorph software, as described by the supplier (Molecular Devices). The intensity of the FITC signal from each cell was normalized using DAPI signals. In Fig. 3B, the intensity of the fluorescence signal in the entire area of a cell (referred to as N + C) or in its nucleus (referred to as N) was measured, and the proportion of the nuclear signal versus the entire cellular signal was calculated. The values are averages from measurements of 300 randomly selected cells from 5 microscopic fields.
Western blotting.
Cytoplasmic and nuclear fractions of the T. vaginalis lysate were prepared using a cellular fractionation kit, NE-PER, as described by the supplier (Thermo Scientific). In the Western blot assay, protein samples equivalent to lysate from 105 to 106 cells were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in 12% gels and transferred to polyvinylidene difluoride (PVDF) membranes with a semidry electroblotter using Immobilon-P (Millipore). Sequential immunoreactions were performed, and an enhanced chemiluminescence (ECL) system was used for signal detection as instructed by the supplier (Thermo Scientific). Reaction conditions for the antibodies from commercial sources, including the mouse monoclonal anti-α-tubulin antibody (10,000×; DM1A; Sigma), rabbit anti-acetyl-histone H3K9 (H3K9-Ac) (3,000×; Upstate), and rat monoclonal anti-HA antibody (2,000×; 3F10; Roche), were as described by the suppliers. Myb1, Myb2, Myb3, and CyPA1 were each detected using mouse anti-Myb1 (1,000×), rabbit anti-Myb2 (3,000×), rabbit anti-Myb3 (3,000×), and mouse anti-CyPA1 (5,000×), respectively. The signal intensity of each protein on a blot was measured and quantified as previously described (18).
EMSA.
Probe labeling and an electrophoretic mobility shift assay (EMSA) for testing the DNA-binding activities of proteins were performed as previously described (38).
CD and fluorescence spectroscopy.
The secondary structure and ternary folding of proteins were measured by circular dichroism (CD) and fluorescence spectroscopy, respectively, as previously described (11).
RESULTS
To study the nuclear translocation of Myb3, a stable cell line overexpressing HA-tagged Myb3 was established from T. vaginalis transfected with an expression plasmid, pAP65-2.1-ha-myb3 (Fig. 1A), which drives protein expression to a higher level than a previous version for better detection (18). HA-Myb3 was mainly localized in nuclei, with much weaker punctate signals also observed in the cytoplasm of all transfected cells examined, but not the control, according to an IFA using a mouse anti-HA antibody (Fig. 1B). HA-Myb3 (at ∼33-kDa) was detected in samples from transfected cells, but not the control, by Western blotting using a rat anti-HA antibody (Fig. 1C, left). The same protein in a duplicate blot was also detected by an anti-Myb3 antibody, which also detected endogenous Myb3 (referred to as Myb3 below) in samples from both control and transfected cells (Fig. 1C, right). The cell lysates were then fractionated for Western blotting using the anti-Myb3 antibody (Fig. 1D). Consistent with previous findings (18), Myb3 and HA-Myb3 were each detected at higher levels in the cytosolic than in the nuclear fractions. A similar distribution of Myb1 was observed. In contrast, Myb2 was detected at a much higher level in the nuclear than in the cytosolic fractions. In these blots, cyclophilin A1, a cytosolic marker, and histone H3 acetylated at lysine 9, a nuclear marker, were detected only in the cytosolic and nuclear fractions, respectively. The purity of the cellular fractions, as shown in subsequent experiments, was examined in the same way and is not discussed further.
Identification of the NLS.
The structural organization of Myb3 is comprised of a highly ordered R1R2R3 domain, followed by a small hairpin and a polybasic sequence in a direction C-terminal to the structured domain (Fig. 2A) (41). Clustered mutations were first introduced into the polybasic sequence to study its role in Myb3's nuclear translocation. KKRK170–173AAAA was localized to the nucleus like HA-Myb3 (Fig. 2B). When examined by Western blotting (Fig. 2C), HA-Myb3 and the mutant protein were expressed at similar levels, but both were detected at higher levels in the cytosolic than in the nuclear fractions to similar extents, suggesting that 170KKRK173 does not regulate nuclear translocation of Myb3 when cells are cultured in normal medium.
To explore whether Myb3 exploits structural components similar to those of Myb2 for nuclear translocation (11), a series of Myb3 deletion mutants in fusion with the C-terminal TetR were produced (Fig. 3A), and their subcellular localizations were examined. 48–180-TetR, 48–173-TetR, and 48–167-TetR were largely localized to nuclei (Fig. 3B), while 56–180-TetR and 48–156-TetR were evenly localized to the cytoplasm. In contrast, 48–160-TetR was equally localized to nuclei and the cytoplasm in a subset of cells but exclusively to the cytoplasm in another subset of cells. The fusion proteins were each expressed at a level higher than that of HA-Myb3, as revealed by cellular fractionation and Western blotting (Fig. 3C). Unlike HA-Myb3, 48–180-TetR, 48–173-TetR, and 48–167-TetR were detected in both the cytosolic and nuclear fractions to similar extents, while 56–180-TetR, 48–156-TetR, and 48–160-TetR were detected at much higher levels in the cytosolic than in the nuclear fractions. These results suggest that the Myb3′ NLS for efficient nuclear translocation resides within aa 48 to 167, which encompass the entire helical structure of R2R3, followed by a short β-hairpin and a flexible random coil (Fig. 2A; see Fig. S4 in the supplemental material) (41).
Clustered mutations were then introduced into the sequence spanning aa 157 to 165 of HA-Myb3. NSN157–159AAA, HKE160–162AAA, and ILL163–165AAA were each localized to nuclei at levels similar to that of HA-Myb3 when cells were cultured in normal medium (Fig. 4A). The expression level and subcellular distribution of each of these mutant proteins as shown by Western blotting were similar to those of HA-Myb3 (Fig. 4B). These results suggest that, in addition to the R2R3 structural domain, a more flexible C-terminal tail (41) is essential for the nuclear translocation of Myb3 in cells under normal growth conditions.
Structural integrity of the Myb3 NLS.
To test whether the structural integrity of the NLS is essential for the nuclear translocation of Myb3, as is that of Myb2 (11), the conserved isoleucine I79 in HA-Myb3 was mutated to proline or alanine. As shown by IFA, I79A was distinctly localized to nuclei, with rather faint staining in the cytoplasm, similar to that of HA-Myb3 (Fig. 5A), while I79P was localized to nuclei to a lesser extent than HA-Myb3, but it was also detected in the cytoplasm as punctate signals at a level substantially higher than that of HA-Myb3. The expression level of the mutant protein, as shown by Western blotting, was similar to that of HA-Myb3 (Fig. 5B). I79A and I79P were also each detected at higher levels in the cytosolic than in the nuclear fractions, like HA-Myb3, but the concentration of I79A in the nuclear fraction was slightly higher, while that of I79P was lower, than that of HA-Myb3.
Fig 5.
Effects of a conserved isoleucine, I79, on nuclear importation, DNA-binding activity, and the structural integrity of Myb3. I79 in HA-Myb3 was mutated to alanine (I79A) or proline (I79P). Cells were grown overnight in normal medium. (A) Cells were examined by an immunofluorescence assay using a mouse anti-HA antibody and FITC-conjugated mouse IgG. Nuclei were stained with DAPI, and the cell morphology was recorded by phase-contrast microscopy. (B) Cell lysates (TL) were fractionated into cytosolic (C) and nuclear (N) fractions for Western blotting using anti-HA (top), anti-H3K9-Ac (middle), and anti-CyPA1 (bottom) antibodies. Molecular weight markers (left) and the detected proteins (right) are indicated. (C) I79 in the recombinant protein rMyb3(48–180) (lane 2) was mutated to alanine or proline to produce rI79A(48–180) (lane 3) or rI79P(48–180) (lane 4), respectively. An electrophoretic mobility shift assay was performed with coincubation of the recombinant proteins with a γ-32P-labeled MRE-1 probe (lane 1). (D) The secondary structures of the recombinant proteins rMyb3(48–180), rI79A(48–180), and rI79P(48–180) were monitored by far-UV CD spectra. (E) The ternary folding of rMyb3(48–180), rI79A(48–180), and rI79P(48–180) was examined by fluorescence spectroscopy. All fluorescence emission spectra were measured at 300 to 400 nm and 25°C. Protein samples were dissolved in PBS buffer (pH 7.4) with 1 mM dithiothreitol (DTT). rMyb3(48–180) was also denatured with 6 M guanidine hydrochloride.
To further study the physical and biochemical properties of the Myb3′ NLS, the recombinant protein rMyb3(48–180) and the related rI79P(48–180) and rI79A(48–180) were produced and purified to near homogeneity, as examined by SDS-PAGE (data not shown). When examined by an EMSA in reaction with the 32P-labeled DNA probe MRE-1 (38), rI79A(48–180) exhibited DNA-binding activity lower than that of rMyb3(48–180), whereas that of rI79P was lost (Fig. 5C). Since I79 is unlikely to be involved in protein-DNA interactions (21, 24, 27), these observations suggest that abolition of the DNA-binding activity caused by I79 mutation may have been due to a perturbation of Myb3's structure.
The secondary structure of each Myb3 recombinant protein was examined by circular dichroism spectroscopy (Fig. 5D). In contrast to the effect of the I74 mutation in Myb2 (11), the far-UV spectra, with absorbance minima at ∼222 and 208 nm, from samples of rMyb3(48–180), rI79A(48–180), and rI79P(48–180), were approximate, indicating that the mutation of I79 to either proline or alanine exerted only a slight effect on the α-helical content of the recombinant Myb3 protein.
When examined by fluorescence spectroscopy (Fig. 5E), the emission maximum of rMyb3(48–180) was detected at 335.9 nm, suggesting that all five tryptophan residues in the protein were probably located in a hydrophobic environment. The emission maximum for rI79A(48–180) was at 339.6 nm, a value slightly larger than that of rMyb3(48–180), implying that the ternary structure of Myb3 had become looser with the mutation of I79 to alanine. In contrast, the emission maximum of rI79P(48–180) had shifted to 347.9 nm, a value that was almost midway between those of rMyb3(48–180) and the denatured rMyb3(48–180), implying that some of the tryptophan residues in this mutant protein were exposed to the solvent.
Iron-inducible nuclear import of Myb3.
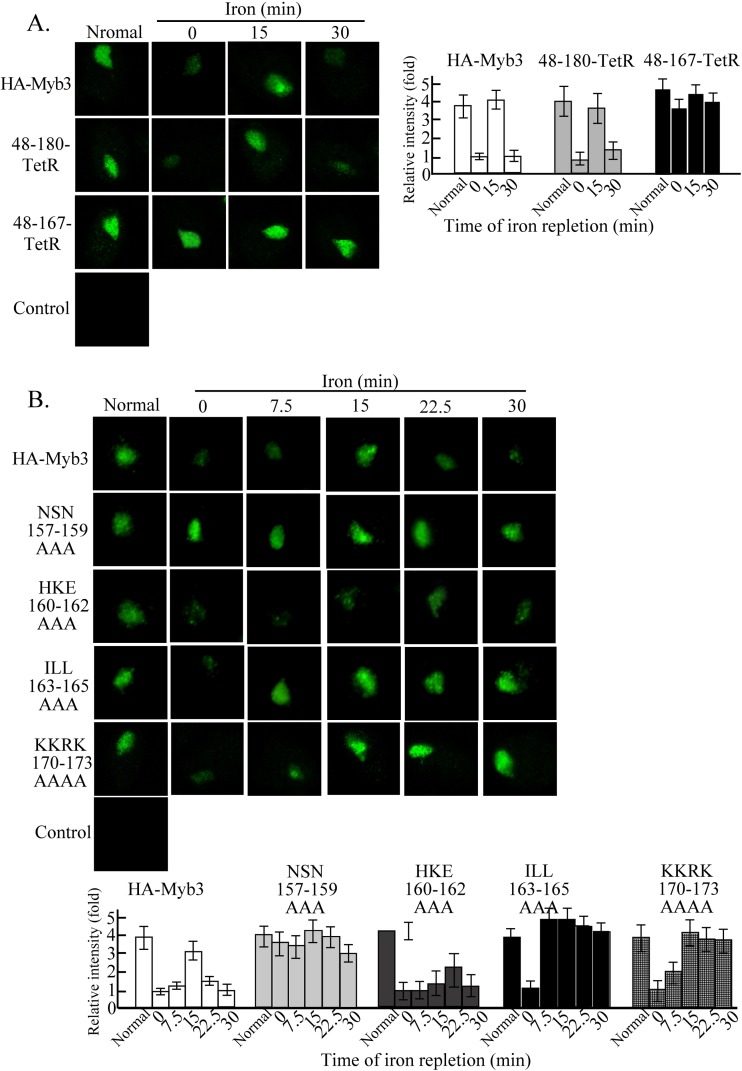
The Myb3 level in the nuclear fraction was lower in cells cultivated under iron depletion than repletion (18). Similarly, a much lower level of HA-Myb3 was seen in nuclei of transfected cells depleted of iron than in those cultivated in normal medium (see Fig. 7). Upon iron repletion, the intensity of the nuclear signal increased contiguously to a much greater level within an ∼15-min period (Fig. 6A), a phenomenon referred to as iron-inducible nuclear import. The signal intensity had substantially decreased at 22.5 min and was barely detectable at 30 to 60 min after iron repletion. To test whether this phenomenon is unique to Myb3, cells overexpressing 4HA-Myb2 (11) were concurrently examined, and the level of 4HA-Myb2 in nuclei remained unchanged. When iron-depleted cells were pretreated with LMB, a nuclear export inhibitor (Fig. 6B), the signal intensity of HA-Myb3 in nuclei also reached an optimal level at 15 min after iron repletion but changed little thereafter, suggesting that Myb3 that had been imported into nuclei upon iron repletion was rapidly exported. To test whether iron also affected the nuclear translocation of endogenous Myb3, cell lysates from nontransfected cells were fractionated for Western blotting. In these experiments, Myb1 and Myb3 were enriched in the cytosolic fraction, with only minor amounts in the nuclear fraction, whereas Myb2 was slightly more enriched in the nuclear than in the cytosolic fraction in cells depleted of iron (Fig. 6C). Upon iron repletion for 15 min, the level of nuclear Myb3 increased, with a concomitant decrease in cytosolic Myb3. By 30 min, Myb3's subcellular distribution had returned to the original level. In contrast, the subcellular distributions of Myb1 and Myb2 changed little upon iron repletion. Consistent with the transgenic studies (Fig. 6A and B), these observations suggest that iron specifically induces the nuclear influx of Myb3. Similar results were observed in transfected cells overexpressing HA-Myb3 (see Fig. S2 in the supplemental material).
Fig 7.
Sequence elements in the C-terminal tail of Myb3's NLS for iron-inducible nuclear translocation. Cells overexpressing HA-Myb3 and its derived proteins (48–180-TetR or 48–167-TetR [A]; NSN157–159AAA, HKE160–162AAA, or ILL163–165AAA [B]; KKRK170–173AAAA [C]) were grown in normal or iron-depleted medium overnight, and the latter were also supplied with iron and sampled at the indicated time intervals for an immunofluorescence assay using a mouse anti-HA antibody and FITC-conjugated mouse IgG. Images of DAPI-stained nuclei and cell morphology are shown in Fig. S3 in the supplemental material. Each bar in the histograms of the micrographs shows the average signal intensity from the measurements of 300 cells in five microscopic fields.
Fig 6.
Iron-inducible nuclear translocation of Myb3. Cells were cultivated overnight in growth medium depleted of iron. (A) Cells overexpressing HA-Myb3 and 4HA-Myb2 were replete with iron and were fixed at the indicated time intervals. (B) Cells overexpressing HA-Myb3 were pretreated with methanol or LMB for 30 min prior to iron repletion and fixed at the indicated time intervals. In panels A and B, an immunofluorescence assay was performed using a mouse anti-HA antibody and FITC-conjugated mouse IgG. Images of DAPI-stained nuclei and cell morphology are shown in Fig. S2 in the supplemental material. Signal intensities were quantified and are shown as a histogram at the bottom in panel A and on the right in panel B. (C) Cell lysates (TL) from nontransfected cells were fractionated into cytosolic (C) and nuclear (N) fractions for Western blotting using anti-Myb3, anti-Myb1, anti-Myb2, anti-H3K9-Ac, and anti-CypA1 antibodies. Molecular weight markers (left) and the detected proteins (right) are indicated.
Regulatory elements for iron-inducible nuclear translocation.
To study whether Myb3's NLS (aa 48 to 167), as defined in Fig. 3, is sufficient for iron-inducible nuclear translocation, subcellular localizations of 48–180-TetR and 48–167-TetR in cells under inducible conditions were examined (Fig. 7A). Like HA-Myb3, 48–180-TetR was localized to nuclei with a signal intensity much lower in cells with iron depletion than in those cultured in normal growth medium. Inducible nuclear translocation was observed for 48–180-TetR at a level slightly lower than that of HA-Myb3. In contrast, 48–167-TetR was persistently localized to nuclei at a level substantially higher than that of HA-Myb3, but its expression level in nuclei varied little with changes in the iron concentration, suggesting that the flexible region C terminus of the R2R3 domain may contain sequence elements regulating the nuclear accumulation of Myb3.
We then examined by IFA the subcellular localizations of mutant proteins derived from the NLS tail region upon iron repletion (Fig. 7B). NSN157–159AAA was localized to nuclei, but its signal intensity changed little in cells either depleted of iron or cultured in normal medium to a level similar to that of HA-Myb3 in cells from normal growth medium. The signal intensity varied only slightly within a 30-min period when cells depleted of iron were supplied with iron, suggesting that 157NSN159 regulates cytoplasmic retention of HA-Myb3 when iron is limited. Under iron-depleted conditions, HKE160–162AAA was detected as speckles adjacent to the nucleus. Upon iron repletion, the nuclear influx of the mutant protein was observed at 22.5 min, a time when the nuclear efflux of wild-type protein occurred, indicating that 160HKE162 may regulate translocation of Myb3 through nuclear pores. The ILL163–165AAA mutant protein was also detected in nuclei, with an intensity much lower under iron depletion than in normal medium. Its nuclear influx reached the maximal level at 7.5 min after iron repletion, with a signal intensity much greater than that of HA-Myb3. The signal intensity varied slightly over the time course studied, implying that 163ILL165 likely plays dual roles in retarding nuclear import and facilitating nuclear export of Myb3. Intriguingly, KKRK170–173AAAA accumulated in the nuclei of cells upon iron repletion without an apparent efflux within a 30-min period, suggesting that the polybasic sequence is important for the nuclear export of Myb3.
DISCUSSION
Myb proteins are conceivably the major group of transcription regulators in T. vaginalis, with more than 400 members of the myb gene family in its genome (9). For Myb1, Myb2, and Myb3, each conserved DBD preserves structural components similar to those of human c-Myb (21, 24, 27), but each also possesses subtle differences in binding to DNA elements and selecting promoters with changes in extrinsic and intrinsic factors (18). Moreover, the structural components conferring DNA-binding activity on Myb2 also mediate its nuclear translocation in a manner uncoupled from its ability to bind DNA (11), implying that Myb proteins in the parasite may exploit a common module to mediate nuclear translocation and DNA binding. This notion was tested in Myb3, which belongs to the R1R2R3 subfamily and the DBD of which contain a unique small β-hairpin C-terminal to the bundled helical structure (see Fig. S4 in the supplemental material) (41). In our previous study (18), subcellular localization of HA-Myb3 in transfected T. vaginalis was detectable in only a small fraction of cells, while the tagged protein was not detected in cells depleted of iron (data not shown). To circumvent this problem, a new HA-Myb3 expression system was developed here, using a stronger promoter to drive protein expression (Fig. 1A). The expression level of this tagged protein was substantially higher than that of Myb3 without affecting the growth of the parasite (data not shown). Unequivocal detection of the overexpressed protein in nearly all transfected cells was achieved, even in cells depleted of iron (Fig. 6). Such an expression system allowed us to dissect the course of Myb3′ iron-inducible nuclear translocation (Fig. 6). Similar to Myb3, HA-Myb3 was persistently detected at a substantially higher level in the cytosolic than in the nuclear fractions, whereas Myb2 was much more enriched in nuclear fractions (Fig. 1D), suggesting that nuclear translocations of Myb2 and Myb3 are differentially regulated.
Although the overall expression of Myb3 and its nuclear level were both upregulated by iron, it was always much more abundant in the cytoplasm than in nuclei (18). It is not uncommon for a transcription factor to be sequestered in the cytoplasm and to be imported into the nucleus upon stimulation (10, 20). Transcription factors can be retained in the cytoplasm in unstimulated cells by binding to inhibitors, as exemplified by the binding of NF-κB to IκB (10, 15). When cells are exposed to a variety of extracellular stimuli, IκB is modified and degraded to release NF-κB, which is in turn transported to the nucleus (17). In contrast, transcription factors in the SMAD and STAT families exist as monomers that remain latent in the cytoplasm. When cells are stimulated, these proteins are phosphorylated to form dimers and are transported to nuclei (31, 33). These transcription factors are recycled back to the cytoplasm when the effects of the stimuli subside. Other than persistent localization in the nucleus, Myb3's nuclear translocation is iron inducible, but how it is retained in the cytoplasm and transported to the nucleus remains elusive.
The length and structural flexibility of Myb3's NLS differ from those of Myb2's NLS. The NLS of Myb2 is embedded within R2R3 and is devoid of a flexible C-terminal tail and half of helix 6 (11) (see Fig. S4 in the supplemental material), while that of Myb3 stretches beyond the highly ordered structure to include a more flexible tail (Fig. 3). A conserved isoleucine in helix 2 essential for maintaining the structural integrity of DNA binding of both Myb proteins is less crucial for the nuclear translocation of Myb3 than for that of Myb2 (Fig. 5). In both cases, nuclear translocation conferred by each NLS is independent of its DNA-binding activity. Thus, the intact folded structure of the R2R3 domain in Myb2 or Myb3 may be more crucial for DNA binding than for nuclear translocation, and the folded structure of Myb3's NLS is probably more flexible than that of Myb2. The R2R3 domains may therefore serve as a common module for both nuclear translocation and DNA binding of Myb2 and Myb3, yet they each may possess intrinsic differences to fulfill their roles in transcriptional regulation. While the C-terminal tail of Myb3's NLS is essential to confer nuclear translocation of a nonnuclear protein, its sequence context imposed little effect on the nuclear accumulation of Myb3 when cells were cultivated in normal medium (Fig. 4). Whether this flexible tail provides some surface areas on the NLS for Myb3 to bind a transporter or other protein partners remains speculative.
With sudden iron overloading, Myb3 can undergo transient nuclear translocation not seen with Myb2 (Fig. 6). Since this phenomenon was observed as early as 3 min after iron repletion (unpublished observations), iron most likely triggers signal transduction leading to the nuclear translocation of Myb3. In this regard, the phenomenon can be a useful model to study iron-triggered signal transduction in T. vaginalis. As suggested by site-directed mutagenesis (Fig. 7), this cellular response can be dissected into three distinct steps: first, the release of restrained Myb3 into the cytoplasm, followed by accelerated nuclear import and ending with accelerated nuclear efflux (10). The C-terminal tail of the NLS that mediates normal nuclear translocation was insufficient to sense iron depletion and did not respond to a sudden iron overload, unless its length was further extended by ∼13 aa without an implicated structure. It is not clear whether the longer flexible tail contributes to the structural stability of the NLS, but it does contain multiple sequence elements for regulating iron-inducible nuclear translocation at distinct steps. Among these sequence elements, 157NSN159 is most crucial for sensing iron depletion and cytoplasmic retention and 160HKE162 is important for nuclear influx, while 163ILL165 and 170KKRK173 are necessary for nuclear efflux (Fig. 6). 170KKRK173 was previously shown to enhances Myb3's DNA-binding activity (41); thus, retention of KKRK170–173AAAA in the nucleus 30 min after iron repletion is unlikely to be due to its binding to DNA. Since Myb3's nuclear efflux was inhibited by LMB, the nuclear export of Myb3 is possibly mediated via a CRM1-like exportin (14, 35). It remains to be tested whether 163ILL165 and 170KKRK173 are part of a noncanonical classical NES that binds exportin for subsequent nuclear export.
The transient nuclear translocation of Myb3 suggests that its amount in the nucleus must be tightly regulated when the parasite faces a sudden iron overload. Given that cytotoxicity may be caused by excessive iron, it is tempting to speculate that the rapid nuclear influx of Myb3 allows the parasite to modulate the expression of genes involved in iron homeostasis and detoxification. Coincidentally, the Myb3 recognition sequence, TAACGA (18), is present in the distal promoter regions of some of the genes that might encode proteins for iron homeostasis or detoxification of oxidative stresses, as well as proteins containing iron-sulfur clusters (see Table S2 in the supplemental material). Whether their transcription is regulated by Myb3 to cope with iron overloading remains to be studied.
In summary, our studies suggest that different Myb proteins in T. vaginalis may share a module for regulating nuclear translocation and DNA binding, but each likely possesses intrinsic differences for nuclear import.
Supplementary Material
ACKNOWLEDGMENTS
We thank Daniel P. Chamberlin for English editing of the manuscript.
This work was supported by grants from the National Science Council, Taiwan (NSC92-2314-B-001-009 and NSC93-2314-B-001-003), and IBMS, Academia Sinica.
Footnotes
Published ahead of print 5 October 2012
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1. Alderete JF. 1999. Iron modulates phenotypic variation and phosphorylation of P270 in double-stranded RNA virus-infected Trichomonas vaginalis. Infect. Immun. 67:4298–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alderete JF, Provenzano D, Lehker HW. 1995. Iron mediates Trichomonas vaginalis resistance to complement lysis. Microb. Pathog. 19:93–103 [DOI] [PubMed] [Google Scholar]
- 3. Alderete JF, et al. 1995. Cloning and molecular characterization of two genes encoding adhesion proteins involved in Trichomonas vaginalis cytoadherence. Mol. Microbiol. 17:69–83 [DOI] [PubMed] [Google Scholar]
- 4. Alderete JF, Milsap KW, Lehker MW, Benchimol M. 2001. Enzymes on microbial pathogens and Trichomonas vaginalis: molecular mimicry and functional diversity. Cell Microbiol. 3:359–370 [DOI] [PubMed] [Google Scholar]
- 5. Alderete JF, Nguyen J, Mundodi V, Lehker HW. 2004. Heme-iron increases level of AP65-mediated adherence by Trichomonas vaginalis. Microb. Pathog. 36:263–271 [DOI] [PubMed] [Google Scholar]
- 6. Bastida-Corcuera FD, Okumura CY, Colocoussi A, Johnson PJ. 2005. Trichomonas vaginalis lipophosphoglycan mutants have reduced adherence and cytotoxicity to human ectocervical cells. Eukaryot. Cell 4:1951–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR. 1996. Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol. Cell. Biol. 16:4207–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Boulikas T. 1994. Putative nuclear localization signals (NLS) in protein transcription factors. J. Cell Biochem. 55:32–58 [DOI] [PubMed] [Google Scholar]
- 9. Carlton JM, et al. 2007. Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science 315:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cartwright P, Helin K. 2000. Nucleocytoplasmic shuttling of transcription factors. Cell Mol. Life. Sci. 57:1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chu CH, et al. 2011. A highly organized structure mediating nuclear localization of a Myb2 transcription factor in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell 10:1607–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dang CV, Lee WM. 1989. Nuclear and nucleolar targeting sequences of c-erb-A, c-myb, N-myc, p53, HSP70, and HIV-tat proteins. J. Biol. Chem. 264:18019–18023 [PubMed] [Google Scholar]
- 13. Fichorova RN. 2009. Impact of T. vaginalis infection on innate immune responses and reproductive outcome. J. Reprod. Immunol. 83:185–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fornerod M, Ohno M, Yoshida M, Mattaj IW. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051–1060 [DOI] [PubMed] [Google Scholar]
- 15. Ganchi PA, Sun SC, Greene WC, Ballard DW. 1992. IκB/MAD-3 masks the nuclear localization signal of NF-κB p65 and requires the transactivation domain to inhibit NF-κB p65 DNA binding. Mol. Cell. Biol. 3:1339–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Garcia AF, et al. 2003. Iron and contact with cells induce expression of adhesions on surface of Trichomonas vaginalis. Mol. Microbiol. 47:1207–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henkel T, et al. 1993. Rapid proteolysis of IκB-α is necessary for activation of transcription factor NFκB. Nature 365:182–185 [DOI] [PubMed] [Google Scholar]
- 18. Hsu HM, Ong SJ, Lee MC, Tai JH. 2009. Transcriptional regulation of an iron-inducible gene by differential and alternate promoter entries of multiple Myb proteins in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell 8:362–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Humbert-Lan G, Pieler T. 1999. Regulation of DNA-binding activity and nuclear transport of B-Myb in Xenopus oocyte. J. Biol. Chem. 274:10293–10300 [DOI] [PubMed] [Google Scholar]
- 20. Jans DA. 1995. The regulaion of protein transport to the nucleus by phosphorylation. Biochem. J. 311:705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiang I, et al. 2011. Molecular basis of the recognition of the ap65-1 gene transcription promoter elements by a Myb protein from the protozoan parasite Trichomonas vaginalis. Nucleic Acids Res. 39:8992–9008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kudo N, et al. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540–547 [DOI] [PubMed] [Google Scholar]
- 23. Lange A, et al. 2007. Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 282:5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lou YC, et al. 2009. NMR structural analysis of DNA recognition by a novel Myb1 DNA-binding domain in the protozoan parasite Trichomonas vaginalis. Nucleic Acids Res. 37:2381–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moreno-Brito V, et al. 2005. A Trichomonas vaginalis 120 kDa protein with identity to hydrogenosome pyruvate:ferredoxin oxidoreductase is a surface adhesin induced by iron. Cell Microbiol. 7:245–258 [DOI] [PubMed] [Google Scholar]
- 26. Mundodi V, Kucknoor AS, Klumpp DJ, Chang TH, Alderete JF. 2004. Silencing the ap65 gene reduces adherence to vaginal epithelial cells by Trichomonas vaginalis. Mol. Microbiol. 53:1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ogata K, et al. 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell 79:639–648 [DOI] [PubMed] [Google Scholar]
- 28. Ong SJ, Huang SC, Liu HW, Tai JH. 2004. Involvement of multiple DNA elements in iron-inducible transcription of the ap65-1 gene in the protozoan parasite Trichomonas vaginalis. Mol. Microbiol. 52:1721–1730 [DOI] [PubMed] [Google Scholar]
- 29. Ong SJ, Hsu HM, Liu HW, Chu CH, Tai JH. 2006. Multifarious transcriptional regulation of adhesion protein gene ap65-1 by a novel Myb1 protein in the protozoan parasite Trichomonas vaginalis. Eukaryot. Cell 5:391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ong SJ, Hsu HM, Liu HW, Chu CH, Tai JH. 2007. Activation of multifarious transcription of an adhesion protein ap65-1 gene by a novel Myb2 protein in the protozoan parasite Trichomonas vaginalis. J. Biol. Chem. 282:6716–6725 [DOI] [PubMed] [Google Scholar]
- 31. Reich NC, Liu L. 2006. Tracking STAT nuclear traffic. Nat. Rev. Immunol. 6:602–612 [DOI] [PubMed] [Google Scholar]
- 32. Rushton JJ, et al. 2003. Distinct changes in gene expression induced by A-Myb, B-Myb and c-Myb proteins. Oncogene 22:308–313 [DOI] [PubMed] [Google Scholar]
- 33. Schmierer B, Hill CS. 2007. TGFβ-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 8:970–982 [DOI] [PubMed] [Google Scholar]
- 34. Shafir SC, Sorvillo FJ, Smith L. 2009. Current issues and considerations regarding trichomoniasis and human immunodeficiency virus in African-Americans. Clin. Microbiol. Rev. 22:37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Stade K, Ford CS, Guthrie C, Weis K. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041–1050 [DOI] [PubMed] [Google Scholar]
- 36. Stoffler D, Schwarz-Herion K, Aebi U, Fahrenkrog B. 2006. Getting across the nuclear pore complex: new insights into the nuclear cytoplasmic import. Can. J. Physiol. Pharmacol. 84:499–507 [DOI] [PubMed] [Google Scholar]
- 37. Takemoto Y, Tashiro S, Handa H, Ishii S. 1994. Multiple nuclear localization signals of the B-Myb gene products. FEBS Lett. 350:55–60 [DOI] [PubMed] [Google Scholar]
- 38. Tsai CD, Liu HW, Tai JH. 2002. Characterization of an iron-responsive promoter in the protozoan pathogen Trichomonas vaginalis. J. Biol. Chem. 277:5153–5162 [DOI] [PubMed] [Google Scholar]
- 39. Vandromme M, Gauthier-Rouvière C, Lamb N, Fernandez A. 1996. Regulation of transcription factor localization: fine-tuning of gene expression. Trends Biochem. Sci. 21:59–64 [PubMed] [Google Scholar]
- 40. Viscogliosi E, Philippe H, Baroin A, Perasso R, Brugerolle G. 1993. Phylogeny of trichomonads based on partial sequences of large subunit rRNA and on cladistic analysis of morphological data. J. Eukaryot. Microbiol. 40:411–421 [DOI] [PubMed] [Google Scholar]
- 41. Wei SY, et al. 2012. Structure of the Trichomonas vaginalis Myb3 DNA-binding domain bound to a promoter sequence reveals a unique C-terminal b-hairpin conformation. Nucleic Acids Res. 40:449–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Weinstock H, Berman S, Cates W., Jr 2000. Sexually transmitted diseases among American youth: incidence and prevalence estimates. Perspect. Sex. Reprod. Health 36:6–10 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.