Abstract
Saccharomyces cerevisiae Snf1 is a member of the conserved Snf1/AMP-activated protein kinase (Snf1/AMPK) family involved in regulating responses to energy limitation, which is detected by mechanisms that include sensing adenine nucleotides. Mitochondrial voltage-dependent anion channel (VDAC) proteins, also known as mitochondrial porins, are conserved in eukaryotes from yeast to humans and play key roles in mediating mitochondrial outer membrane permeability to small metabolites, including ATP, ADP, and AMP. We previously recovered the yeast mitochondrial porin Por1 (yVDAC1) from a two-hybrid screen for Snf1-interacting proteins. Here, we present evidence that Snf1 interacts with Por1 and its homolog Por2 (yVDAC2). Cells lacking Por1 and Por2, but not respiratory-deficient rho0 cells lacking the mitochondrial genome, exhibit reduced Snf1 activation loop phosphorylation in response to glucose limitation. Thus, Por1 and Por2 contribute to the positive control of Snf1 protein kinase. Physical proximity to the VDAC proteins and mitochondrial surface could facilitate Snf1's ability to sense energy limitation.
INTRODUCTION
Mammalian AMP-activated protein kinase (AMPK) is often referred to as the cell's “fuel gauge” (15). According to the standard model, AMPK responds to decreases in cellular energy levels by sensing increased levels of AMP relative to ATP (reviewed in references 13, 14). The AMPK activation mechanism involves AMP binding to the γ subunit of the AMPK complex to the exclusion of ATP, making the catalytic α subunit of AMPK a better substrate for activation loop threonine (Thr172) phosphorylation by upstream kinases while interfering with its dephosphorylation and inactivation. More recent evidence indicates that besides AMP binding, ADP binding also stimulates Thr172 phosphorylation of AMPK and protects it from dephosphorylation (30, 49). Activated AMPK functions to balance the energy “budget” by cutting general energy spending (e.g., by inhibiting cell growth and proliferation) while activating specific energy-generating mechanisms (e.g., glucose uptake). In agreement with these important roles, defects in AMPK signaling have been linked to diabetes, obesity, and cancer, making AMPK a good drug target (reviewed in references 8 and 12).
Decreases in cellular energy levels can be caused by factors ranging from reduced glucose availability to reduced mitochondrial respiration (reviewed in reference 21). Metformin, a drug that is often prescribed to patients with type 2 diabetes, is thought to activate AMPK indirectly, by affecting respiratory complex chain I (2, 6). Metformin also exhibits anti-cancer action, an effect that is mediated largely by AMPK (reviewed in references 5 and 42). These findings indicate that a better understanding of the functional connection between AMPK and mitochondria could provide valuable clues to the molecular etiology, treatment, and prevention of metabolic disorders and cancer.
The Snf1 protein kinase of the yeast Saccharomyces cerevisiae is the homolog of mammalian AMPK and is required for growth on carbon sources that are less preferred than glucose, including nonfermentable carbon sources (reviewed in reference 16). Like AMPK, Snf1 is activated in response to glucose and energy limitation, which is detected by mechanisms that include ADP binding to the γ subunit of the Snf1 complex (Snf4), which similarly protects Snf1 from dephosphorylation of the activation loop threonine (Thr210) (26). Snf1 serves as a good model for studying the general aspects of Snf1/AMPK regulation in eukaryotes, and S. cerevisiae is particularly suitable for studies of the relationship between this kinase and mitochondria because of the well-known ability of this organism to survive with its respiratory function completely disrupted.
We previously recovered the yeast mitochondrial porin Por1 (9, 28) from a two-hybrid screen for Snf1-interacting proteins (46). Mitochondrial porins, also widely known as voltage-dependent anion channel (VDAC) proteins, are conserved in eukaryotes from yeast to humans. These proteins play key roles in mediating mitochondrial outer membrane permeability to small metabolites, notably adenine nucleotides, and are involved in mitochondrial respiration; VDACs also play important roles in apoptosis and represent attractive targets for anti-cancer therapy (reviewed in references 4, 25, and 36).
Here, we present evidence that the yeast mitochondrial porin Por1/yVDAC1 and its homolog Por2/yVDAC2 (referred to herein as Por1 and Por2, respectively) share a role in the positive control of Snf1, revealing a new and counterintuitive layer of regulation. Physical proximity to the VDAC proteins and mitochondrial surface could facilitate Snf1's ability to sense energy limitation.
MATERIALS AND METHODS
Yeast strains and media.
Yeast two-hybrid assays were performed with strain CTY10-5d (MATa gal4 gal80 URA3::lexAop-lacZ his3 leu2 ade2 trp1-901) (R. Sternglanz, SUNY, Stony Brook, NY). Except for CTY10-5d and its por1Δ derivative (see below), all strains were in the W303 background. The snf1Δ::KanMX6 allele has been described (32). To generate the por1Δ::KanMX6 and por2Δ::KanMX6 alleles, the marker sequences were amplified by PCR with primers flanking the corresponding open reading frames. The mutant alleles were introduced into W303-1A (MATa ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1) by transformation; all yeast transformations were performed using standard methods (33). In addition, por1Δ::KanMX6 was introduced into CTY10-5d, and por2Δ::KanMX6 was introduced into W303-1B (MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1). The genotypes of all knockout strains were confirmed by PCR analysis of genomic DNA. Double mutants lacking Por1 and Por2 were constructed by tetrad analysis of diploid cells obtained by crossing por1Δ and por2Δ single mutants of the opposite mating types. The rho0 cells were generated in W303-1A by ethidium bromide treatment (10). The rho0 status of the non-glycerol-utilizing cells thus obtained was verified by the loss of the mitochondrial COX1 and COX3 genes by PCR; the presence of the nuclear COX4 gene was used as a positive control.
Rich medium was yeast extract-peptone (YEP) supplemented with extra tryptophan (40 mg/liter) and adenine (20 mg/liter); synthetic complete (SC) medium lacking appropriate supplements was used to select for plasmids (33). Unless indicated otherwise, the media contained 2% glucose, and cells were grown at 30°C.
Yeast two-hybrid interaction assays.
Plasmids pIT469 (22) and pRJ79 (19) express LexA-Snf1 and VP16-Snf1 from vectors pEG202 (11) and pVP16 (45), respectively. To construct pLexA-Por1 and pGAD-Por1, a PCR fragment encompassing the POR1 open reading frame was inserted at the BamHI site of pEG202 and pACTII (24), respectively. The pLexA-Por1 and pGAD-Por1 plasmids were confirmed for ability to complement the por1Δ mutant derivative of CTY10-5d for the growth defect on glycerol at 37°C (1). Strain CTY10-5d was cotransformed with pairs of plasmids expressing the protein pairs being tested. Transformants were grown to mid-log phase with plasmid selection in SC medium lacking histidine and leucine and containing 2% glucose and then shifted for 3 h to an otherwise identical medium containing 0.05% glucose. β-Galactosidase activity was assayed in permeabilized cells and expressed in Miller units, as described previously (46).
Coimmunoprecipitation assays.
Plasmids pHA-Por1 and pHA-Por2 express N-terminal triple hemagglutinin (HA) epitope-tagged Por1 (HA-Por1) and HA-Por2, respectively, and were constructed by inserting the POR1 and POR2 coding sequences into the BamHI site of vector pWS93 (37). Cells of W303-1A carrying the snf1Δ mutation were transformed with plasmid pSK117 expressing Snf1 (39) or with vector pSK134 (39, 43) and cotransformed with plasmids pHA-Por1, pHA-Por2, or with vector pWS93. Cells were grown to mid-log phase with plasmid selection in SC medium lacking leucine and uracil and containing 2% glucose; where indicated, cells were grown as described above and then shifted for 1 h to an otherwise identical medium containing 0.05% glucose. Protein extracts were prepared, and immunoprecipitations (from 200 μg of protein per reaction) were performed with anti-HA 12CA5 essentially as described previously (39) in a buffer containing 0.1% Triton X-100 and 50 mM NaCl (46). The immunoprecipitates were examined for the presence of Snf1 by immunoblot analysis using anti-polyhistidine antibody H1029 (Sigma-Aldrich), which strongly recognizes Snf1 due to the presence of a natural stretch of 13 consecutive histidines near its N terminus (amino acids 18 to 30) (31). The presence of HA-Por1 and HA-Por2 was analyzed using anti-HA. Signals were detected by enhanced chemiluminescence using ECL Plus (Amersham Biosciences) or HyGlo (Denville Scientific). The presence of Snf1, HA-Por1, and HA-Por2 in the extracts was analyzed by immunoblotting similarly (10 μg protein per lane).
Analysis of SUC2-LEU2-lacZ reporter expression.
To target its genomic integration, reporter plasmid pLS11 (34) was linearized at the NcoI site within the URA3 gene prior to transformation. pLS11 was first introduced into a doubly heterozygous diploid obtained by crossing por1Δ and por2Δ single mutants. Wild-type and mutant haploids carrying pLS11 (Ura+) were then recovered by tetrad analysis. The rho0 derivatives were generated from pLS11-carrying wild-type cells as described above. Cells were grown to mid-log phase in YEP with 2% glucose and then shifted to YEP with 0.05% glucose for 3 h. β-Galactosidase activity was assayed in permeabilized cells and expressed in Miller units.
RESULTS
Snf1 and Por1 interact in the two-hybrid system.
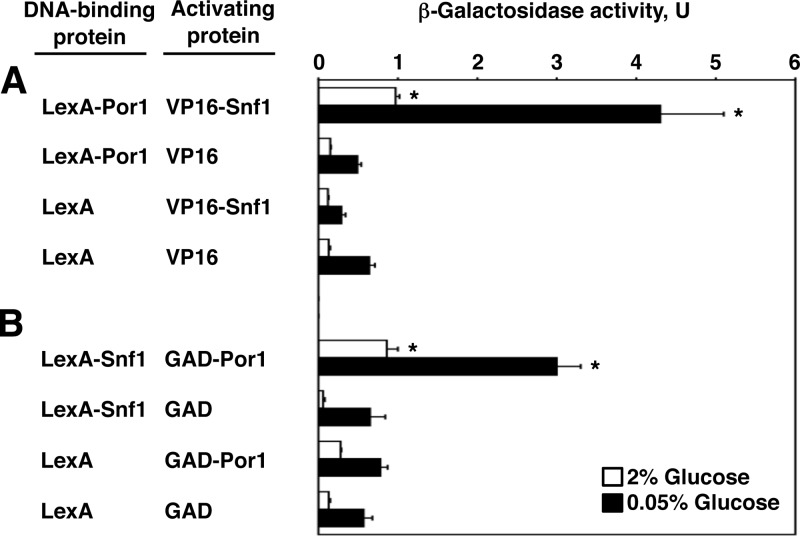
We previously recovered Por1 from a two-hybrid screen for Snf1-interacting proteins (46). To further demonstrate this interaction, fusions to the LexA DNA-binding protein were tested in combination with fusions to the viral VP16 or Gal4 transcription activation domain (GAD) for the ability to activate a reporter with LexA binding sites (lexAop-lacZ) in strain CTY10-5d. LexA-Por1 interacted with VP16-Snf1 (Fig. 1A). In the reciprocal setting, LexA-Snf1 interacted with GAD-Por1 (Fig. 1B). In both cases, the interaction signals exhibited an apparent increase in response to glucose limitation. As discussed further below, we do not necessarily take this to suggest that the affinity of the Snf1-Por1 interaction is stimulated by glucose limitation.
Fig 1.
Snf1 and Por1 interact in the two-hybrid system. Strain CTY10-5d with an integrated lexAop-lacZ reporter was cotransformed with pairs of plasmids expressing the indicated proteins. β-Galactosidase activity was assayed in mid-log cultures grown in selective medium with high (2%) glucose (open bars) and shifted for 3 h to an otherwise identical medium with low (0.05%) glucose (closed bars). Values are averages for at least three transformants. Error bars indicate standard errors. Asterisks indicate statistically significant differences from the corresponding control values (P < 0.03 both in high and low glucose). (A) LexA-Por1 interacts with VP16-Snf1. (B) LexA-Snf1 interacts with GAD-Por1.
In terms of β-galactosidase activity, the observed interaction was weak in comparison to other known interactions of Snf1 (e.g., within the classical Snf1 complex [19]), which was not surprising; in fact, it was more surprising that this interaction could be detected at all. In order for a two-hybrid interaction to be detected, both interacting partners must be present in the nucleus. While this is not a problem for Snf1, which is a cytoplasmic/nuclear protein (44), Por1 is a major mitochondrial outer membrane protein (1). However, porin/VDAC proteins are apparently not absolutely restricted from the nucleus, allowing the yeast two-hybrid system to work and provide valuable preliminary information. For example, some interactions of eukaryotic VDAC proteins that were initially detected in the yeast two-hybrid system were subsequently shown to exist and to be physiologically relevant (3, 7). We therefore decided to explore the Snf1-Por1 interaction further.
Snf1 coimmunoprecipitates with Por1.
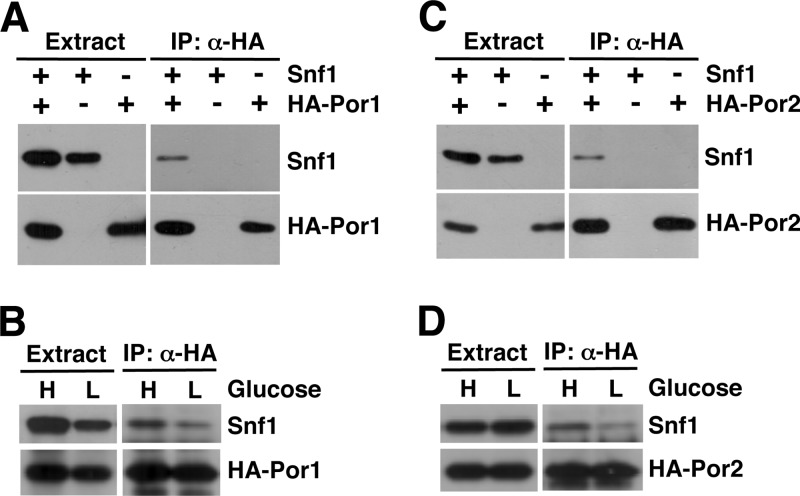
We next conducted coimmunoprecipitation experiments. A snf1Δ strain was cotransformed with plasmids expressing Snf1 and HA-Por1. Cells were grown to mid-log phase in abundant glucose, and protein extracts were prepared and subjected to immunoprecipitation with anti-HA. The immunoprecipitates were then tested for the presence of Snf1 by immunoblot analysis. Snf1 was detected in the immunoprecipitates from cells expressing both HA-Por1 and Snf1 but not in those from cells expressing HA-Por1 but no Snf1 or cells expressing Snf1 but no HA-Por1 (Fig. 2A). Thus, these results provided further evidence that Snf1 interacts with Por1.
Fig 2.
Snf1 coimmunoprecipitates with Por1 and Por2. (A and B) Snf1 coimmunoprecipitates with HA-Por1. (A) Cells carrying the snf1Δ mutation (isogenic to W303-1A) were transformed with a plasmid expressing Snf1 or the appropriate vector, as well as with a plasmid expressing HA-Por1 or the appropriate vector. Cells were grown to mid-log phase with plasmid selection in medium containing 2% glucose, protein extracts were prepared, and HA-Por1 was immunoprecipitated with anti-HA. The presence of Snf1 and HA-Por1 in the extracts and immunoprecipitates (IP: α-HA) was examined by immunoblotting. (B) Cells coexpressing Snf1 and HA-Por1 were grown in 2% glucose as for panel A (H, high glucose) and then shifted for 1 h to an otherwise identical medium with 0.05% glucose (L, low glucose). Coimmunoprecipitation experiments were performed as for panel A. (C and D) Snf1 coimmunoprecipitates with HA-Por2. Experiments for panels C and D were performed like those for panels A and B, respectively, except that HA-Por2 was expressed instead of HA-Por1.
We also compared the ability of Snf1 to coimmunoprecipitate with Por1 under high- and low-glucose conditions. In contrast to the two-hybrid experiments, there was no apparent increase in interaction upon shift from high to low glucose (Fig. 2B). It should be noted that the apparent strength of a two-hybrid interaction reflects not only interaction affinity but also the nuclear content of the interaction partners. The latter factor could at least partly account for the increased two-hybrid signal in low glucose, since there is more Snf1 in the nucleus under carbon stress conditions (44). In addition, the lexAop-lacZ two-hybrid reporter could be more responsive to activation in low glucose, since this reporter is constructed on the basis of the GAL1 promoter, which is glucose regulated (20, 47).
Lack of Por1 and its homolog Por2 affects Snf1 activation.
S. cerevisiae has two VDAC isoforms, Por1 and Por2. Por2 is 49% identical and 70% similar to Por1 and similarly localizes to the mitochondrial outer membrane (1, 50). Unlike Por1, however, Por2 does not appear to possess channel function (1, 23). Unlike the por1Δ mutation, the por2Δ mutation alone does not affect growth on nonfermentable carbon sources but nonetheless enhances por1Δ for this defect (1). This and other evidence strongly suggested the existence of a mechanism whereby Por1 and Por2 share a role in nonfermentable carbon source utilization that is not directly related to channel function (1, 23). Since Snf1 is required for growth on nonfermentable carbon sources (16), and since Snf1 also coimmunoprecipitates with Por2 (Fig. 2C and D), we considered the possibility that Por1 and Por2 share a role in the positive control of Snf1.
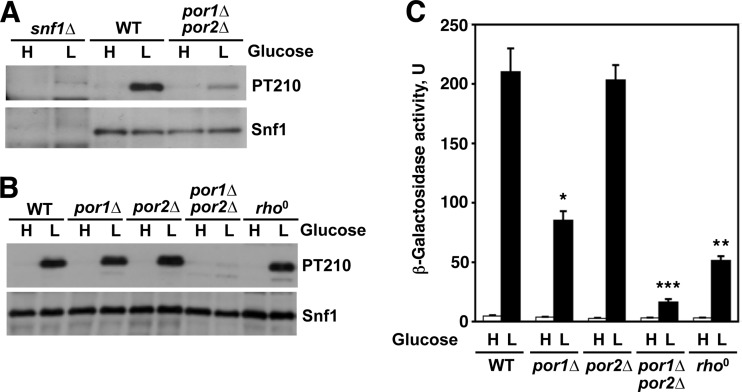
Catalytic activation of Snf1 requires phosphorylation of its conserved activation loop threonine (Thr210, which corresponds to Thr172 of AMPK), which is performed by the upstream kinases Sak1, Tos3, and Elm1 (18, 29, 38). In the presence of abundant glucose, Snf1 is turned off by Thr210 dephosphorylation, which involves the type 1 protein phosphatase Glc7 together with its regulatory protein, Reg1 (27, 40, 41). When wild-type cells are shifted from high to low glucose, the level of activated phospho-Thr210-Snf1 increases dramatically (27). In comparison to the wild type, double-mutant cells lacking Por1 and Por2 exhibited a lower level of phospho-Thr210-Snf1, as hypothesized (Fig. 3A), with a reduction of approximately 5-fold upon normalization to total Snf1 levels. No such Snf1 activation defects were observed in cells lacking Por1 alone or Por2 alone or in respiratory-deficient rho0 cells lacking the mitochondrial genome (Fig. 3B), with the normalized phospho-Thr210-Snf1 levels in these mutants and the wild type differing from each other by no more than 30%.
Fig 3.
Lack of Por1 and Por2 affects Snf1 activation. (A and B) Cells of the indicated genotypes (isogenic to W303-1A) were grown to mid-log phase in YEP containing 2% glucose (H, high glucose) and shifted for 60 min (A) or 90 min (B) to YEP containing 0.05% glucose (L, low glucose). The activation loop Thr210 phosphorylation status of Snf1 was analyzed by immunoblotting as described previously (31): phospho-Thr210-Snf1 (PT210) was detected with anti-phospho-Thr172-AMPK (Cell Signaling Technology), and the total Snf1 protein (Snf1) was detected by reprobing with anti-polyhistidine antibody H1029 (Sigma-Aldrich), which recognizes the natural stretch of 13 histidines present in Snf1. (C) Cells of the indicated genotypes with an integrated SUC2-LEU2-lacZ reporter were grown to mid-log phase in YEP with 2% glucose (H, high glucose) and then shifted to YEP with 0.05% glucose for 3 h (L, low glucose). β-Galactosidase activity was assayed in permeabilized cells and expressed in Miller units. Values are averages for four to six assays. Error bars indicate standard errors. For low-glucose conditions, the value for the por1Δ por2Δ double mutant was significantly different from any other value (triple asterisk, P < 0.001), and the values for por1Δ and rho0 were significantly different from any other value and from each other (single and double asterisks, P < 0.003). Under high glucose conditions, the values were 4.7 units (for the wild type) or lower.
Thus, these results indicated that the combined lack of Por1 and Por2 confers a defect in Snf1 activation loop phosphorylation in low glucose and that this defect is not directly related to compromised respiratory competence.
We also examined the effects of the mutations on the regulation of an integrated SUC2-LEU2-lacZ reporter; expression of this reporter is activated by glucose limitation in a Snf1-dependent manner (34). Under glucose-limiting conditions, the por1Δ por2Δ double mutation caused a 13-fold reporter activation defect relative to the wild type (Fig. 3C). Interestingly, the por1Δ and rho0 mutations also affected activation, albeit to a lesser extent (2.5-fold and 4.1-fold, respectively), raising the possibility that these mutations affect yet another aspect of Snf1 pathway activity that is distinct from Thr210 phosphorylation.
DISCUSSION
Here, we present evidence that the yeast mitochondrial VDAC proteins Por1 and Por2 contribute to the positive control of Snf1 activation-loop phosphorylation and that this role is not directly mediated by the respiratory proficiency of the cell.
The VDAC proteins represent a major component of the interface between the mitochondria and cytoplasm of the eukaryotic cell (4). Proteomic studies in yeast indicate that Por1 and Por2 physically interact with numerous proteins, including components of major nutrient-sensing pathways such as TOR, PKA, casein kinase II, and others (17); the ability of Snf1 to interact with Por1 and Por2 is also consistent with the previous finding that Snf1 associates with the mitochondria (35). If the mitochondrial surface serves as a major substratum for interactions between global regulatory pathways, the Snf1 activation defect observed in the por1Δ por2Δ mutant could represent an indirect consequence of major pathway miscommunication caused by mitochondrial surface perturbation.
However, Por1 and Por2 could also at least partly contribute to Snf1 regulation by a more direct mechanism. Similar to mammalian AMPK, Snf1 activation correlates with increased AMP-to-ATP ratios in vivo; unlike AMPK, however, the yeast Snf1 complex does not respond to regulation by AMP in vitro (48). The identity of the relevant in vivo AMP and/or ATP sensor, if it exists, remains unknown. Following the recent findings that mammalian AMPK is positively regulated by ADP (30, 49), it was shown that ADP protects yeast Snf1 from Thr210 dephosphorylation in vitro (26). Based on analysis of ADP and ATP binding affinities, it was suggested that the yeast Snf1 complex responds to absolute ADP levels rather than to ADP-to-ATP ratios (26). At the same time, according to an earlier report (48), the absolute ADP levels in yeast cells shifted from high to low glucose do not appear to change as much as the AMP-to-ATP or ADP-to-ATP ratios. These considerations suggest a possible functional niche for Por1 and Por2. We envision that an ability to interact with the VDAC proteins and proximity to the mitochondrial surface could put Snf1 in a better position to sense glucose/energy limitation. For example, as glucose levels decrease, the ADP levels could rise more sharply at the mitochondrial surface, making it a good location to monitor. In addition, we cannot exclude the possibility that Por1 and Por2 could also act as “co-sensors” of a stress signal upstream of Snf1. Further experiments will be necessary to address these and other possibilities.
As mentioned above, mitochondria are generally viewed as indirect negative regulators of AMPK. Our findings with yeast Snf1 indicate the existence of a positive regulatory component. Since Snf1/AMPK and VDAC proteins are conserved in evolution, functional relationships similar to the one observed here could exist in other eukaryotes.
ACKNOWLEDGMENTS
We thank D. Krakovich for technical assistance, M. Carlson for reagents, and M. Maziarz for comments on the manuscript.
This work was supported by National Science Foundation grant MCB-0818837 to S.K.
Footnotes
Published ahead of print 26 October 2012
REFERENCES
- 1. Blachly-Dyson E, Song J, Wolfgang WJ, Colombini M, Forte M. 1997. Multicopy suppressors of phenotypes resulting from the absence of yeast VDAC encode a VDAC-like protein. Mol. Cell. Biol. 17:5727–5738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Brunmair B, et al. 2004. Thiazolidinediones, like metformin, inhibit respiratory complex I: a common mechanism contributing to their antidiabetic actions? Diabetes 53:1052–1059 [DOI] [PubMed] [Google Scholar]
- 3. Chen Y, Craigen WJ, Riley DJ. 2009. Nek1 regulates cell death and mitochondrial membrane permeability through phosphorylation of VDAC1. Cell Cycle 8:257–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Colombini M. 2004. VDAC: the channel at the interface between mitochondria and the cytosol. Mol. Cell. Biochem. 256–257:107–115 [DOI] [PubMed] [Google Scholar]
- 5. Dowling RJ, Niraula S, Stambolic V, Goodwin PJ. 2012. Metformin in cancer: translational challenges. J. Mol. Endocrinol. 48:R31–R43 [DOI] [PubMed] [Google Scholar]
- 6. El-Mir MY, et al. 2000. Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J. Biol. Chem. 275:223–228 [DOI] [PubMed] [Google Scholar]
- 7. Feng P, et al. 2007. A novel inhibitory mechanism of mitochondrion-dependent apoptosis by a herpesviral protein. PLoS Pathog. 3:e174 doi:10.1371/journal.ppat.0030174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fogarty S, Hardie DG. 2009. Development of protein kinase activators: AMPK as a target in metabolic disorders and cancer. Biochim. Biophys. Acta 1804:581–591 [DOI] [PubMed] [Google Scholar]
- 9. Forte M, Guy HR, Mannella CA. 1987. Molecular genetics of the VDAC ion channel: structural model and sequence analysis. J. Bioenerg. Biomembr. 19:341–350 [DOI] [PubMed] [Google Scholar]
- 10. Goldring ES, Grossman LI, Krupnick D, Cryer DR, Marmur J. 1970. The petite mutation in yeast. Loss of mitochondrial deoxyribonucleic acid during induction of petites with ethidium bromide. J. Mol. Biol. 52:323–335 [DOI] [PubMed] [Google Scholar]
- 11. Golemis EA, Serebriiskii I, Gjuris J, Brent R. 1997. Current protocols in molecular biology. Wiley, New York, NY [Google Scholar]
- 12. Hardie DG. 2007. AMP-activated protein kinase as a drug target. Annu. Rev. Pharmacol. Toxicol. 47:185–210 [DOI] [PubMed] [Google Scholar]
- 13. Hardie DG. 2007. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat. Rev. Mol. Cell Biol. 8:774–785 [DOI] [PubMed] [Google Scholar]
- 14. Hardie DG. 2007. AMPK and SNF1: snuffing out stress. Cell Metab. 6:339–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hardie DG, Carling D. 1997. The AMP-activated protein kinase—fuel gauge of the mammalian cell? Eur. J. Biochem. 246:259–273 [DOI] [PubMed] [Google Scholar]
- 16. Hedbacker K, Carlson M. 2008. SNF1/AMPK pathways in yeast. Front. Biosci. 13:2408–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ho Y, et al. 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415:180–183 [DOI] [PubMed] [Google Scholar]
- 18. Hong SP, Leiper FC, Woods A, Carling D, Carlson M. 2003. Activation of yeast Snf1 and mammalian AMP-activated protein kinase by upstream kinases. Proc. Natl. Acad. Sci. U. S. A. 100:8839–8843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang R, Carlson M. 1996. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 10:3105–3115 [DOI] [PubMed] [Google Scholar]
- 20. Johnston M. 1987. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol. Rev. 51:458–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kahn BB, Alquier T, Carling D, Hardie DG. 2005. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 1:15–25 [DOI] [PubMed] [Google Scholar]
- 22. Kuchin S, Treich I, Carlson M. 2000. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc. Natl. Acad. Sci. U. S. A. 97:7916–7920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lee AC, Xu X, Blachly-Dyson E, Forte M, Colombini M. 1998. The role of yeast VDAC genes on the permeability of the mitochondrial outer membrane. J. Membr. Biol. 161:173–181 [DOI] [PubMed] [Google Scholar]
- 24. Legrain P, Dokhelar MC, Transy C. 1994. Detection of protein-protein interactions using different vectors in the two-hybrid system. Nucleic Acids Res. 22:3241–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lemasters JJ, Holmuhamedov E. 2006. Voltage-dependent anion channel (VDAC) as mitochondrial governator—thinking outside the box. Biochim. Biophys. Acta 1762:181–190 [DOI] [PubMed] [Google Scholar]
- 26. Mayer FV, et al. 2011. ADP regulates SNF1, the Saccharomyces cerevisiae homolog of AMP-activated protein kinase. Cell Metab. 14:707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCartney RR, Schmidt MC. 2001. Regulation of Snf1 kinase. Activation requires phosphorylation of threonine 210 by an upstream kinase as well as a distinct step mediated by the Snf4 subunit. J. Biol. Chem. 276:36460–36466 [DOI] [PubMed] [Google Scholar]
- 28. Mihara K, Sato R. 1985. Molecular cloning and sequencing of cDNA for yeast porin, an outer mitochondrial membrane protein: a search for targeting signal in the primary structure. EMBO J. 4:769–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nath N, McCartney RR, Schmidt MC. 2003. Yeast Pak1 kinase associates with and activates Snf1. Mol. Cell. Biol. 23:3909–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Oakhill JS, et al. 2011. AMPK is a direct adenylate charge-regulated protein kinase. Science 332:1433–1435 [DOI] [PubMed] [Google Scholar]
- 31. Orlova M, Barrett L, Kuchin S. 2008. Detection of endogenous Snf1 and its activation state: application to Saccharomyces and Candida species. Yeast 25:745–754 [DOI] [PubMed] [Google Scholar]
- 32. Orlova M, Kanter E, Krakovich D, Kuchin S. 2006. Nitrogen availability and TOR regulate the Snf1 protein kinase in Saccharomyces cerevisiae. Eukaryot. Cell 5:1831–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rose MD, Winston F, Hieter P. 1990. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 34. Sarokin L, Carlson M. 1985. Upstream region of the SUC2 gene confers regulated expression to a heterologous gene in Saccharomyces cerevisiae. Mol. Cell. Biol. 5:2521–2526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sickmann A, et al. 2003. The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. U. S. A. 100:13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Simamura E, Shimada H, Hatta T, Hirai K. 2008. Mitochondrial voltage-dependent anion channels (VDACs) as novel pharmacological targets for anti-cancer agents. J. Bioenerg. Biomembr. 40:213–217 [DOI] [PubMed] [Google Scholar]
- 37. Song W, Carlson M. 1998. Srb/Mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 17:5757–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sutherland CM, et al. 2003. Elm1p is one of three upstream kinases for the Saccharomyces cerevisiae SNF1 complex. Curr. Biol. 13:1299–1305 [DOI] [PubMed] [Google Scholar]
- 39. Treitel MA, Kuchin S, Carlson M. 1998. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:6273–6280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tu J, Carlson M. 1994. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:6789–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tu J, Carlson M. 1995. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 14:5939–5946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vakana E, Platanias LC. 2011. AMPK in BCR-ABL expressing leukemias. Regulatory effects and therapeutic implications. Oncotarget 2:1322–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vincent O, et al. 2001. Interaction of the Srb10 kinase with Sip4, a transcriptional activator of gluconeogenic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:5790–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Vincent O, Townley R, Kuchin S, Carlson M. 2001. Subcellular localization of the Snf1 kinase is regulated by specific beta subunits and a novel glucose signaling mechanism. Genes Dev. 15:1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vojtek AB, Hollenberg SM, Cooper JA. 1993. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell 74:205–214 [DOI] [PubMed] [Google Scholar]
- 46. Vyas VK, Kuchin S, Carlson M. 2001. Interaction of the repressors Nrg1 and Nrg2 with the Snf1 protein kinase in Saccharomyces cerevisiae. Genetics 158:563–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. West RW, Jr, Yocum RR, Ptashne M. 1984. Saccharomyces cerevisiae GAL1-GAL10 divergent promoter region: location and function of the upstream activating sequence UASG. Mol. Cell. Biol. 4:2467–2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wilson WA, Hawley SA, Hardie DG. 1996. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 6:1426–1434 [DOI] [PubMed] [Google Scholar]
- 49. Xiao B, et al. 2011. Structure of mammalian AMPK and its regulation by ADP. Nature 472:230–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zahedi RP, et al. 2006. Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins. Mol. Biol. Cell 17:1436–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]