Abstract
The morphogenesis checkpoint in Saccharomyces cerevisiae couples bud formation to the cell division cycle by delaying nuclear division until cells have successfully constructed a bud. The cell cycle delay is due to the mitosis-inhibitory kinase Swe1p, which phosphorylates the cyclin-dependent kinase Cdc28p. In unperturbed cells, Swe1p is degraded via a mechanism thought to involve its tethering to a cortical scaffold of septin proteins at the mother-bud neck. In cells that experience stresses that delay bud formation, Swe1p is stabilized, accumulates, and promotes a G2 delay. The tethering of Swe1p to the neck requires two regulators, called Hsl1p and Hsl7p. Hsl1p interacts with septins, and Hsl7p interacts with Swe1p; tethering occurs when Hsl1p interacts with Hsl7p. Here we created a version of Swe1p that is artificially tethered to the neck by fusion to a septin so that Swe1p no longer requires Hsl1p or Hsl7p for its localization to the neck. We show that the interaction between Hsl1p and Hsl7p, required for normal Swe1p degradation, is no longer needed for septin-Swe1p degradation, supporting the idea that the Hsl1p-Hsl7p interaction serves mainly to tether Swe1p to the neck. However, both Hsl1p and Hsl7p are still required for Swe1p degradation, implying that these proteins play additional roles beyond localizing Swe1p to the neck.
INTRODUCTION
Yeast cells in the environment are exposed to variations in temperature, osmolarity, and nutrient conditions that can delay bud formation. A common response to sudden (presumably stressful) environmental changes is to depolarize the actin cytoskeleton and transiently halt bud growth until the cells adapt to the stress (6, 11). During this interval, there is a concomitant delay of the cell cycle in G2 phase, enforced by the morphogenesis checkpoint, which maintains coordination between the budding and nuclear cycles and prevents the formation of binucleate cells (15). The morphogenesis checkpoint cell cycle delay requires the Wee1-family kinase Swe1p, which inhibits the mitotic Clb/cyclin-dependent kinase (CDK) complex by phosphorylating a conserved tyrosine residue on the CDK, Cdc28p (13, 16, 27). The delay is also thought to involve the inhibition of the counteracting Cdc25-family phosphatase Mih1p, which would otherwise dephosphorylate Cdc28p (12). The best-characterized aspect of the morphogenesis checkpoint concerns the degradation of Swe1p. Swe1p degradation is halted in response to stress (26), promoting a G2 delay, and this has served as a valuable assay to dissect how stresses impact the cell cycle. Here we have tested a prevailing hypothesis for the role of two proteins, Hsl1p and Hsl7p, that are thought to be primary transducers of stress signals to control Swe1p degradation (7, 19, 21, 28).
Swe1p accumulates periodically in the cell cycle, peaking in late S/G2 phase and then falling sharply in late G2/M phase. Swe1p abundance is controlled at the levels of transcription (SWE1 mRNA accumulates in a sharp pulse in late G1 phase and disappears in S phase) and degradation (Swe1p is degraded slowly in G1/S phase and quickly in G2/M phase) (26, 27). The phosphorylation state of Swe1p also changes during the cell cycle, with multiple sites being phosphorylated in G2/M phase by Clb2p/CDK and the polo family kinase Cdc5p (1, 24). Phosphorylation by the CDK primes Swe1p for subsequent phosphorylation by Cdc5p, which is necessary for Swe1p degradation (1). Following its hyperphosphorylation, Swe1p is probably ubiquitylated by the redundant E3 ubiquitin ligases Dma1p and Dma2p, which target it for degradation by the proteasome (23).
Swe1p degradation is also correlated with the subcellular localization of Swe1p. In G1 phase, Swe1p is localized primarily in the nucleus and is stable (17, 26). As the cell builds a bud in S phase, some of the Swe1p accumulates at the bud neck. Neck localization relies on the interaction of Swe1p with Hsl7p (19, 21), which itself localizes to the neck through its interaction with the septin-binding kinase Hsl1p (17, 19, 25). The septins are conserved, filament-forming proteins that assemble a cortical scaffold at the cytoplasmic face of the plasma membrane at the mother-bud neck (29). The genetic perturbation of septin scaffold assembly results in a Swe1p-mediated delay of mitosis, suggesting that septin-mediated Swe1p neck localization is important for Swe1p degradation (3, 17).
Hsl1p and Hsl7p are both required for Swe1p degradation, and mutations that specifically disrupt the Hsl1p-Hsl7p or Hsl7p-Swe1p interactions result in Swe1p delocalization and stabilization (7, 19, 21). These findings suggest that Hsl1p and Hsl7p tether Swe1p to the bud neck, leading to its degradation. Because both Clb2p/CDK (2) and Cdc5p (1) also accumulate at the bud neck, it is attractive to speculate that tethering serves to co-concentrate Swe1p together with its upstream kinases, thereby enhancing the Swe1p phosphorylations that target it for degradation.
The tethering model for Swe1p degradation presents an elegant explanation for how yeast cells couple budding to nuclear division, as the neck exists only after bud emergence, so the degradation of the Swe1p mitotic inhibitor would depend on prior budding. After budding, some stresses can transiently displace Swe1p (and possibly also Hsl7p) from the neck (8, 17), potentially explaining why such stresses can stabilize Swe1p to delay mitosis even after bud emergence. However, actin depolymerization in budded cells does not immediately displace Swe1p from the neck (17), and the basis for Swe1p stabilization in those cells remains unclear.
A prediction of the tethering model is that the need for Hsl1p and Hsl7p in Swe1p degradation could be bypassed if Swe1p were to be tethered directly to the septins at the bud neck. Here we tested this prediction by fusing Swe1p to the septin Cdc3p. The fusion protein was degraded normally in G2/M phase, but although neck localization was no longer dependent on Hsl1p and Hsl7p, we found that both of these proteins were still required for Swe1p degradation, revealing additional roles for Hsl1p and Hsl7p in the degradation pathway.
MATERIALS AND METHODS
Yeast strains and plasmids.
Yeast strains are in the BF264-15DU background and are listed in Table 1. The creation of the following alleles was described previously: SWE1myc (19), SWE1Δ1myc (21), hsl1::URA3 hsl7::URA3 (18), swe1::LEU2 (4), hsl1K110R (28), hsl1Δ8 (10), CDC3-GFP (5), and CDC28E12K (20).
Table 1.
Yeast strains used in this study
| Strain | Genotypea |
|---|---|
| DLY12229 | a bar1 CDC28E12K SWE1myc:HIS2 hsl1::URA3 hsl7::URA3 GFP-SWE1-12×myc:TRP1 |
| DLY12230 | a bar1 CDC28E12K SWE1myc:HIS2 hsl1::URA3 hsl7::URA3 CDC3-GFP-SWE1-12×myc:TRP1 |
| DLY12237 | a bar1 CDC28E12K SWE1myc:HIS2 hsl1::URA3 GFP-SWE1-12×myc:TRP1 |
| DLY12238 | a bar1 CDC28E12K SWE1myc:HIS2 hsl1::URA3 CDC3-GFP-SWE1-12×myc:TRP1 |
| DLY12320 | a bar1 CDC28E12K SWE1myc:HIS2 GFP-SWE1-12×myc:TRP1 |
| DLY12321 | a bar1 CDC28E12K SWE1myc:HIS2 CDC3-GFP-SWE1-12×myc:TRP1 |
| DLY12369 | a bar1 CDC28E12K SWE1myc:HIS2 hsl7::URA3 GFP-SWE1-12×myc:TRP1 |
| DLY12370 | a bar1 CDC28E12K SWE1myc:HIS2 hsl7::URA3 CDC3-GFP-SWE1-12×myc:TRP1 |
| DLY12973 | a bar1 CDC28E12K SWE1myc:HIS2 hsl1K110R CDC3-GFP-SWE1-12×myc:TRP1 |
| DLY13156 | a bar1 CDC28E12K swe1::LEU2 SWE1Δ1myc:URA3 CDC3-GFP-SWE1Δ1-12×myc:TRP1 |
| DLY15224 | a bar1 CDC28E12K SWE1myc:HIS2 hsl1Δ8::URA3 CDC3-GFP-SWE1-12×myc:TRP1 |
| DLY15563 | a bar1 CDC28E12K swe1::LEU2 CDC3-GFP-SWE1-12×myc:TRP1 |
| DLY15597 | a bar1 CDC28E12K SWE1myc:HIS2 hsl1K110R GFP-SWE1-12×myc:TRP1 |
Single colons in genotypes indicate that the gene is integrated at a particular locus without disrupting it, and double colons indicate that the gene is disrupted by the marker.
To create green fluorescent protein (GFP)-SWE1-myc and CDC3-GFP-SWE1-myc, we began with plasmid pDLB2340 (14), which contains the SWE1 promoter, the Swe1p open reading frame (ORF) fused in frame to a 12×myc epitope tag, and SWE1 3′ sequences cloned into the integrating TRP1 vector pRS304. A linker with PstI and SphI sites (underlined) (5′-GCATGCATGTGGGTAAAACTGCAG-3′) was introduced into the NcoI site at the Swe1p start codon in pDLB2340, yielding pDLB3276. PCR products containing either GFP- or CDC3-GFP coding sequences with flanking PstI and SphI sites were then cloned into the corresponding sites in pDLB3276 to make pDLB3290 (GFP-SWE1-12×myc) and pDLB3291 (CDC3-GFP-SWE1-12×myc). The CDC3-GFP PCR template pDLB3137 contains GFP-coding sequences inserted at codon 13 of CDC3, which was shown previously to retain Cdc3p function (5).
CDC3-GFP-SWE1Δ1-12×myc plasmid pDLB3381 was constructed by replacing the EcoRI/BamHI fragment (encoding a Swe1p N-terminal region) in pDLB3291 with the corresponding fragment from pJM1103 (21), which contains the Δ1 mutation.
The plasmids described above were digested at the unique Bsu36I site and transformed into yeast cells to target integration at the TRP1 locus.
Media, growth conditions, and cell cycle synchrony.
Yeast strains were grown in YEPD (1% yeast extract, 2% Bacto peptone, 2% dextrose, 0.012% adenine, and 0.01% uracil) or synthetic complete medium at 30°C. For α-factor arrest-release-rearrest experiments, exponentially growing cells were treated with 100 ng/ml α-factor (Research Genetics, Huntsville, AL) for 3 h, harvested by centrifugation, and released into fresh YEPD, and after 60 min, 100 ng/ml fresh α-factor was added so that cells would arrest in the subsequent G1 phase. Samples for Western blotting and nuclear staining were taken every 15 min.
Fluorescence staining and microscopy.
To visualize nuclear DNA, cells were fixed in 70% ethanol overnight at 4°C. The cells were then harvested by centrifugation, resuspended in 0.2 μg/ml 4′,6′-diamidino-2-phenylindole (DAPI) in phosphate-buffered saline (PBS), centrifuged again, and finally resuspended in mounting medium (90% glycerol, 9.2 mM p-phenylenediamine in PBS [Sigma-Aldrich, St. Louis, MO]). Cells were scored for budding and nuclear division on an Axioskop apparatus with a 100× objective equipped with epifluorescence and differential interference contrast (DIC) optics (Zeiss, Thornwood, NY). For DIC and GFP photography, cells were grown in complete synthetic medium containing dextrose and spotted onto a 2% agarose slab made with synthetic medium. Images were acquired with an Axioimager apparatus with a 100× objective equipped with epifluorescence and differential interference contrast optics (Zeiss) and photographed with an Orca ER monochrome cooled charge-coupled-device camera (Hamamatsu, Bridgewater, NJ). Images were captured by using MetaMorph software (Millipore, Billerica, MA).
Western blotting.
Cell lysis was performed according to the trichloroacetic acid (TCA) method (14). Samples were subjected to SDS-PAGE on a 6% polyacrylamide gel. Proteins were transferred onto a nitrocellulose membrane (Pall, East Hills, NY) and blocked for 1 h with PBS containing 3% nonfat dry milk (Kroger, Cincinnati, OH). Antibodies were diluted in blocking buffer supplemented with 0.1% Tween 20 (Bio-Rad, Hercules, CA), and membranes were probed overnight with a 1:250 dilution of mouse anti-myc antibody 9E10 and a 1:50,000 dilution of rabbit anti-Cdc11p antibody (Santa Cruz Biotechnology, Santa Cruz, CA). Membranes were then washed three times with PBS–0.1% Tween 20 and then probed with a 1:7,500 dilution of goat anti-mouse IRdye800 antibody (Rockland Immunochemicals, Gilbertsville, PA) or a 1:7,500 dilution of goat anti-rabbit AlexaFluor680 antibody (Invitrogen, Carlsbad, CA) for 1 h in blocking buffer containing 0.1% Tween 20 and 0.01% SDS. Blots were then washed as described above and visualized by using an Odyssey scanner (Li-Cor Biosciences, Lincoln, NE). Fluorescence was quantified by using Odyssey software (Li-Cor Biosciences), where the intensities of the myc bands were normalized to the Cdc11 loading controls.
RESULTS
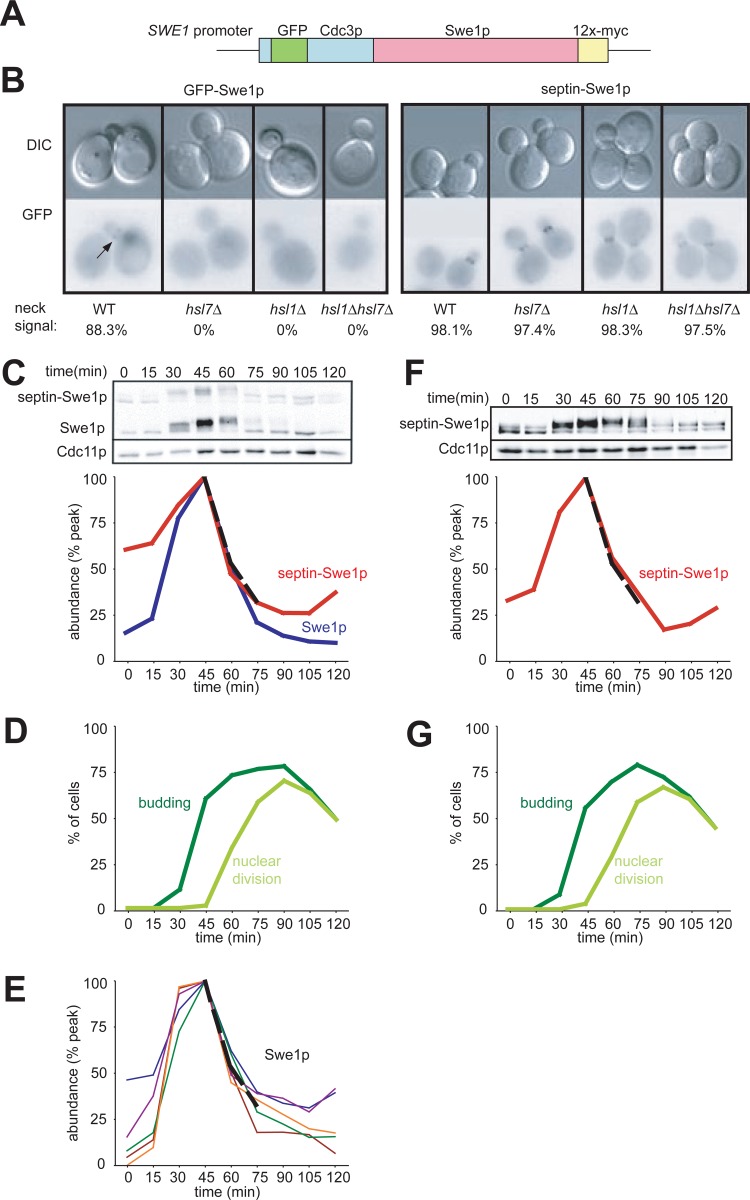
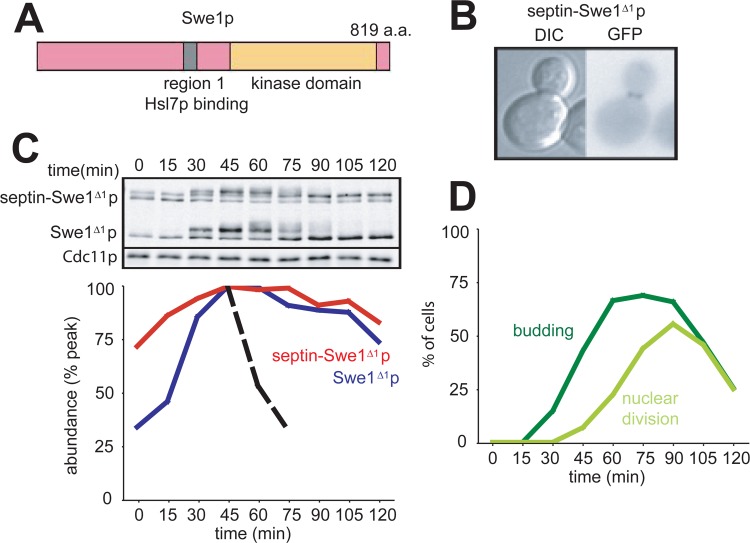
If the role of Hsl1p and Hsl7p is simply to recruit Swe1p to the septin cortex at the neck, then the artificial linkage of Swe1p to a septin should bypass the need for Hsl1p and Hsl7p in Swe1p degradation. To test this hypothesis, we created a fusion protein that joins GFP-Cdc3p (a septin) with Swe1p-myc, under the control of the SWE1 promoter (referred to here as “septin-Swe1p”) (Fig. 1A). We integrated this plasmid at an exogenous locus so that septin-Swe1p did not replace endogenous Swe1p or Cdc3p. As anticipated, septin-Swe1p was localized to the bud neck independent of Hsl1p and Hsl7p (Fig. 1B).
Fig 1.
Localization and degradation of a septin-Swe1p fusion. (A) Schematic of the Cdc3-GFP-Swe1p fusion protein. (B) Localization of Cdc3-GFP-Swe1p to the neck is independent of Hsl1p and Hsl7p. GFP-Swe1p is shown as a control. Cells were grown overnight at 30°C. Representative cells are shown, and over 200 cells of each strain were scored for bud neck signals. Strains containing GFP-Swe1p (with genotypes indicated in parentheses) were DLY12320 (wild type [WT]), DLY12369 (hsl7Δ), DLY12237 (hsl1Δ), and DLY12229 (hsl1Δ hsl7Δ), and strains containing Cdc3-GFP-Swe1p were DLY12321 (wild type), DLY12238 (hsl1Δ), DLY12370 (hsl7Δ), and DLY12230 (hsl1Δ hsl7Δ). (C) Swe1p and septin-Swe1p levels rise and fall in parallel as cells traverse the cell cycle. Cells containing both Swe1p-12×myc and septin-Swe1p-12×myc (DLY12321) were arrested in G1 phase with pheromone and released into fresh medium at 30°C to traverse the cell cycle. Pheromone was reintroduced 60 min after release to rearrest cells at the next G1 phase. Samples taken at 15-min intervals were analyzed by Western blotting with anti-myc antibody (Cdc11 as a loading control) and quantitated (graph). (D) Synchrony parameters (budding and nuclear division) for the experiment depicted in panel C were scored for >200 cells per sample. (E) Swe1p degradation kinetics are reproducible between experiments. Five replicates of the experiment depicted in panel C were quantified, and an average degradation profile for wild-type Swe1p is plotted as a dotted black line. (F) Degradation of septin-Swe1p occurs independent of wild-type Swe1p. A single-cycle synchrony experiment, as described above for panel C, was performed with swe1Δ strain DLY15563. (G) Synchrony parameters (budding and nuclear division) for the experiment depicted in panel F were scored for >200 cells per sample.
To directly compare Swe1p and septin-Swe1p levels, both proteins were myc tagged in the same strain. Cells were synchronized in G1 phase by using mating pheromone, released to traverse the cell cycle, and rearrested in the subsequent G1 phase by the readdition of pheromone. The kinetics of Swe1p accumulation and degradation were highly reproducible by using this protocol (Fig. 1E). The septin-Swe1p fusion accumulated, became hyperphosphorylated, and then disappeared with a timing comparable to that of the wild-type Swe1p-myc control (Fig. 1C), suggesting that the normal Swe1p degradation machinery can target the full septin-Swe1p fusion for degradation. Septin-Swe1p degradation was similar whether or not wild-type Swe1p was present (compare Fig. 1F with Fig. 1C), indicating that its degradation was not due to dimerization with wild-type Swe1p.
If Hsl1p and Hsl7p simply tether Swe1p to the neck for degradation, then septin-Swe1p should be degraded in strains lacking those proteins even though wild-type Swe1p is stable. However, a complication arises in strains containing both wild-type Swe1p and septin-Swe1p, because Swe1p degradation also requires active CDK (26). If wild-type Swe1p is stable, it would inhibit the CDK, and that inhibition could indirectly stabilize septin-Swe1p. To avoid this complication, we introduced an E12K mutation into the CDK Cdc28p. Cdc28pE12K is highly resistant to Swe1p-dependent inhibition, unlike the nonphosphorylatable Cdc28pY19F, which is still susceptible to inhibition by binding to Swe1p (20). We used this CDC28E12K background for all Swe1p stability experiments, and as shown for the controls below, cell cycle progression in this background was unaffected by any of the manipulations of Swe1p, Hsl1p, or Hsl7p.
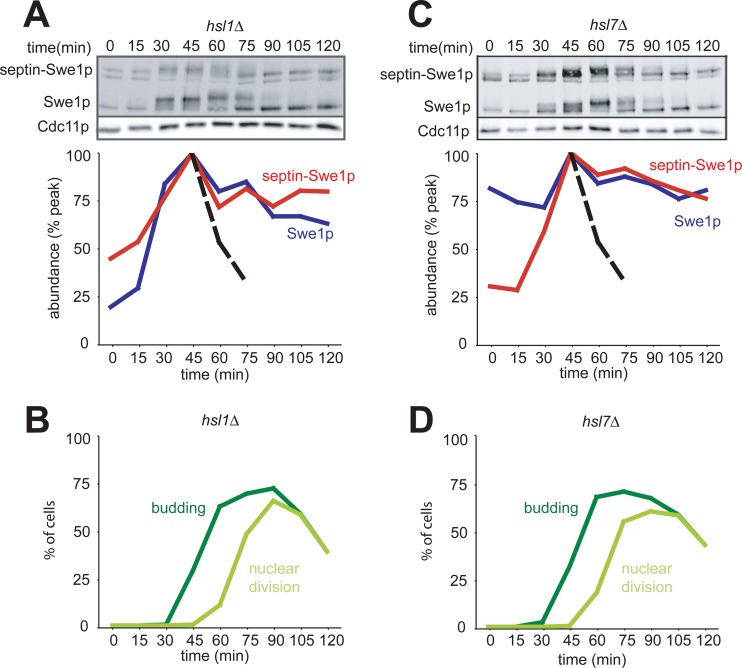
In hsl1Δ or hsl7Δ deletion strains, we found that both Swe1p and septin-Swe1p were stabilized in G2/M phase (Fig. 2). These findings do not support the idea that Hsl1p and Hsl7p act simply by tethering Swe1p to the neck: at minimum, they must play additional roles in promoting Swe1p degradation.
Fig 2.
Hsl1p and Hsl7p are required for degradation of septin-tethered Swe1p in G2/M phase. (A) Swe1p and septin-Swe1p are both stabilized upon the deletion of HSL1. A single-cycle synchrony experiment, as described in the legend of Fig. 1C, was performed with hsl1Δ strain DLY12238. (B) Synchrony parameters (budding and nuclear division) for the experiment depicted in panel A were scored for >200 cells per sample. (C) Swe1p and septin-Swe1p are both stabilized upon the deletion of HSL7. A single-cycle synchrony experiment, as described in the legend of Fig. 1C, was performed with hsl7Δ strain DLY12370. (D) Synchrony parameters (budding and nuclear division) for the experiment depicted in panel C were scored for >200 cells per sample.
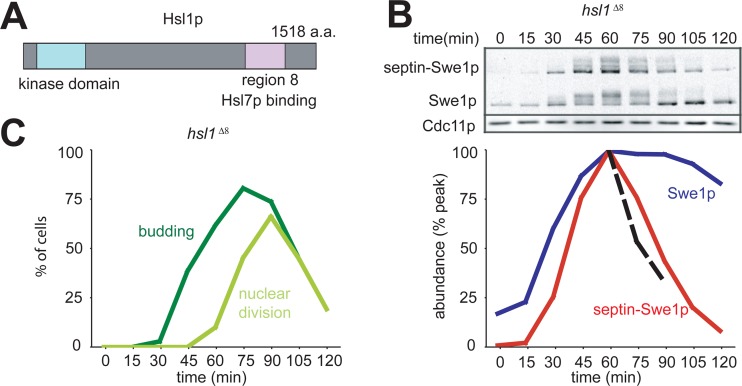
Hsl1p has an N-terminal kinase domain and a long C-terminal tail containing several regions with sequence motifs that are conserved among the Saccharomycetes. In particular, region 8 mediates Hsl1p-Hsl7p interactions (Fig. 3A) (10). In a strain where HSL1 is replaced with hsl1Δ8, neither Hsl7p nor Swe1p is recruited to the bud neck, and Swe1p is stable. However, we found that septin-Swe1p was degraded normally in this strain (Fig. 3B), showing that septin tethering bypasses the need for the Hsl1p-Hsl7p interaction in Swe1p degradation.
Fig 3.
Interaction between Hsl1p and Hsl7p is not required for degradation of septin-tethered Swe1p. (A) Schematic of Hsl1p. a.a., amino acids. (B) Swe1p is stabilized upon deletion of Hsl1p region 8, but septin-Swe1p is not. A single-cycle synchrony experiment, as described in the legend of Fig. 1C, was performed with hsl1Δ8 strain DLY15224. (C) Synchrony parameters (budding and nuclear division) for the experiment depicted in panel B were scored for >200 cells per sample.
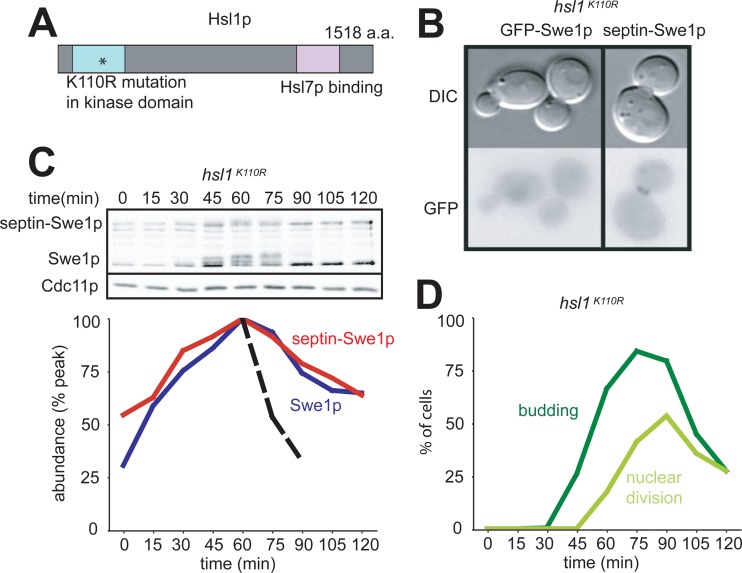
Like region 8, the kinase activity of Hsl1p is important for recruiting Hsl7p to the bud neck (28). In a strain where hsl1 is replaced with the kinase-dead hsl1K110R mutant, Swe1p is not detectably enriched at the bud neck, and it is stable. Although septin-Swe1p was effectively localized to the bud neck in an hsl1K110R strain, it was not degraded in G2/M phase (Fig. 4). Thus, the kinase activity of Hsl1p is needed not only for the effective recruitment of Swe1p to the bud neck but also for some subsequent step in Swe1p degradation.
Fig 4.
Hsl1p kinase activity is required for degradation of septin-tethered Swe1p. (A) Schematic of Hsl1p. The K110R point mutation in the kinase domain is noted by an asterisk. (B) Septin-Swe1p is localized to the bud neck in hsl1K110R cells, but GFP-Swe1p is not. Cells of strains DLY12973 and DLY15597 were grown at 30°C and photographed. (C) Swe1p and septin-Swe1p are both stabilized in a strain harboring the kinase-dead hsl1K110R mutant. A single-cycle synchrony experiment, as described in the legend of Fig. 1C, was performed with hsl1K110R strain DLY12973. (D) Synchrony parameters (budding and nuclear division) for the experiment depicted in panel C were scored for >200 cells per sample.
A previous screen for SWE1 mutants that caused a lethal G2 arrest in the absence of MIH1 identified many point mutants in a small conserved region in the Swe1p N-terminal regulatory domain (21). The deletion of that region (in a protein designated Swe1Δ1p) did not impair Swe1p function but greatly reduced its interaction with Hsl7p and eliminated detectable neck localization. The fusion of Swe1Δ1p to the septin Cdc3p restored neck localization, but septin-Swe1Δ1p was not degraded in G2/M phase (Fig. 5). The simplest interpretation of this result is that the Swe1p-Hsl7p interaction is required for Swe1p degradation, even when Swe1p is tethered to the neck.
Fig 5.
Interaction between Swe1p and Hsl7p is required for degradation of septin-tethered Swe1p. (A) Schematic of Swe1p. (B) Septin-tethered Swe1pΔ1 is localized to the bud neck. Cells of strain DLY13156 were grown at 30°C and photographed. (C) Swe1pΔ1 and septin-Swe1pΔ1 are both stable in G2/M phase. A single-cycle synchrony experiment, as described in the legend of Fig. 1C, was performed with strain DLY13156. (D) Synchrony parameters (budding and nuclear division) for the experiment in depicted panel C were scored for >200 cells per sample.
DISCUSSION
In this study, we have tested the hypothesis that the role of Hsl1p and Hsl7p in Swe1p degradation consists of tethering Swe1p to the septins at the bud neck. Our data indicate that even when Swe1p is artificially tethered to the neck by fusion to a septin, the degradation of the tethered septin-Swe1p still requires both Hsl1p and Hsl7p. Thus, Hsl1p and Hsl7p play roles in promoting Swe1p degradation beyond simply tethering it to the neck.
Cells with a septin-tethered Swe1p still required the Swe1p-Hsl7p interaction for degradation. As phosphorylation by Cdc5p is thought to be the trigger for Swe1p ubiquitination, one possibility would be that the Hsl7p-Swe1p interaction presents Swe1p to Cdc5p as a substrate. Hsl7p has also been reported to enhance Cdc5p neck localization, consistent with the idea that Hsl7p may interact with Cdc5p as well, potentially bridging a Cdc5p-Swe1p interaction.
Given that the interaction with Hsl7p is required for septin-Swe1p degradation, Hsl7p must presumably be present at the neck to promote degradation. How, then, is septin-Swe1p degraded in the hsl1Δ8 strain, where Hsl1p cannot recruit Hsl7p to the neck? It seems likely that in this case, septin-Swe1p itself recruits Hsl7p to the neck, reversing the normal tethering order.
Cells with a septin-tethered Swe1p also required Hsl1p kinase activity for degradation to occur. Hsl1p is homologous to the Nim1 kinase in Schizosaccharomyces pombe, which can directly phosphorylate the Swe1p homolog Wee1 in vitro (9, 22, 30). However, the direct phosphorylation of Swe1p by Hsl1p has been difficult to detect in vitro (7), and it remains unclear whether or not it occurs in vivo. Hsl1p is thought to phosphorylate Hsl7p in vivo (19), and it also catalyzes extensive autophosphorylation (3), but the functional relevance of these modifications is unknown. As Hsl1p kinase activity is required for optimal Hsl7p recruitment, it could be that the phosphorylation of either Hsl1p or Hsl7p enhances the interaction between those proteins. However, the degradation of septin-tethered Swe1p did not require an intact Hsl1p-Hsl7p interaction, so promoting that interaction cannot be the only role for the Hsl1p kinase activity.
The finding that the need for the Hsl1p-Hsl7p interaction in Swe1p degradation can be bypassed by tethering Swe1p to the septins provides strong evidence that tethering is indeed one important role for Hsl1p and Hsl7p. Thus, our findings in aggregate indicate that Hsl1p and Hsl7p play at least two separate roles in promoting Swe1p degradation: tethering Swe1p to the septins and a downstream role perhaps involving the presentation of Swe1p to other regulators for targeting to the degradation pathway.
Footnotes
Published ahead of print 5 October 2012
REFERENCES
- 1. Asano S, et al. 2005. Concerted mechanism of Swe1/Wee1 regulation by multiple kinases in budding yeast. EMBO J. 24:2194–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bailly E, Cabantous S, Sondaz D, Bernadac A, Simon MN. 2003. Differential cellular localization among mitotic cyclins from Saccharomyces cerevisiae: a new role for the axial budding protein Bud3 in targeting Clb2 to the mother-bud neck. J. Cell Sci. 116:4119–4130 [DOI] [PubMed] [Google Scholar]
- 3. Barral Y, Parra M, Bidlingmaier S, Snyder M. 1999. Nim1-related kinases coordinate cell cycle progression with the organization of the peripheral cytoskeleton in yeast. Genes Dev. 13:176–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Booher RN, Deshaies RJ, Kirschner MW. 1993. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 12:3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Caviston JP, Longtine M, Pringle JR, Bi E. 2003. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell 14:4051–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chowdhury S, Smith KW, Gustin MC. 1992. Osmotic stress and the yeast cytoskeleton: phenotype-specific suppression of an actin mutation. J. Cell Biol. 118:561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cid VJ, Shulewitz MJ, McDonald KL, Thorner J. 2001. Dynamic localization of the Swe1 regulator Hsl7 during the Saccharomyces cerevisiae cell cycle. Mol. Biol. Cell 12:1645–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Clotet J, et al. 2006. Phosphorylation of Hsl1 by Hog1 leads to a G(2) arrest essential for cell survival at high osmolarity. EMBO J. 25:2338–2346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Coleman TR, Tang Z, Dunphy WG. 1993. Negative regulation of the Wee1 protein kinase by direct action of the nim1/cdr1 mitotic inducer. Cell 72:919–929 [DOI] [PubMed] [Google Scholar]
- 10. Crutchley J, et al. 2009. Molecular dissection of the checkpoint kinase Hsl1p. Mol. Biol. Cell 20:1926–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Delley PA, Hall MN. 1999. Cell wall stress depolarizes cell growth via hyperactivation of RHO1. J. Cell Biol. 147:163–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harrison JC, Bardes ES, Ohya Y, Lew DJ. 2001. A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat. Cell Biol. 3:417–420 [DOI] [PubMed] [Google Scholar]
- 13. Keaton MA, et al. 2007. Differential susceptibility of yeast S and M phase CDK complexes to inhibitory tyrosine phosphorylation. Curr. Biol. 17:1181–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Keaton MA, et al. 2008. Nucleocytoplasmic trafficking of G2/M regulators in yeast. Mol. Biol. Cell 19:4006–4018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lew DJ. 2003. The morphogenesis checkpoint: how yeast cells watch their figures. Curr. Opin. Cell Biol. 15:648–653 [DOI] [PubMed] [Google Scholar]
- 16. Lew DJ, Reed SI. 1995. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J. Cell Biol. 129:739–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Longtine MS, et al. 2000. Septin-dependent assembly of a cell-cycle-regulatory module in Saccharomyces cerevisiae. Mol. Cell. Biol. 20:4049–4061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ma X-J, Lu Q, Grunstein M. 1996. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10:1327–1340 [DOI] [PubMed] [Google Scholar]
- 19. McMillan JN, et al. 1999. The morphogenesis checkpoint in Saccharomyces cerevisiae: cell cycle control of Swe1p degradation by Hsl1p and Hsl7p. Mol. Cell. Biol. 19:6929–6939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McMillan JN, Sia RAL, Bardes ESG, Lew DJ. 1999. Phosphorylation-independent inhibition of Cdc28p by the tyrosine kinase Swe1p in the morphogenesis checkpoint. Mol. Cell. Biol. 19:5981–5990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. McMillan JN, Theesfeld CL, Harrison JC, Bardes ES, Lew DJ. 2002. Determinants of Swe1p degradation in Saccharomyces cerevisiae. Mol. Biol. Cell 13:3560–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parker LL, Walter SA, Young PG, Piwnica-Worms H. 1993. Phosphorylation and inactivation of the mitotic inhibitor Wee1 by the nim1/cdr1 kinase. Nature 363:736–738 [DOI] [PubMed] [Google Scholar]
- 23. Raspelli E, Cassani C, Lucchini G, Fraschini R. 2011. Budding yeast Dma1 and Dma2 participate in regulation of Swe1 levels and localization. Mol. Biol. Cell 22:2185–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sakchaisri K, et al. 2004. Coupling morphogenesis to mitotic entry. Proc. Natl. Acad. Sci. U. S. A. 101:4124–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shulewitz MJ, Inouye CJ, Thorner J. 1999. Hsl7 localizes to a septin ring and serves as an adapter in a regulatory pathway that relieves tyrosine phosphorylation of Cdc28 protein kinase in Saccharomyces cerevisiae. Mol. Cell. Biol. 19:7123–7137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sia RAL, Bardes ESG, Lew DJ. 1998. Control of Swe1p degradation by the morphogenesis checkpoint. EMBO J. 17:6678–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sia RAL, Herald HA, Lew DJ. 1996. Cdc28 tyrosine phosphorylation and the morphogenesis checkpoint in budding yeast. Mol. Biol. Cell 7:1657–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Theesfeld CL, Zyla TR, Bardes EG, Lew DJ. 2003. A monitor for bud emergence in the yeast morphogenesis checkpoint. Mol. Biol. Cell 14:3280–3291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weirich CS, Erzberger JP, Barral Y. 2008. The septin family of GTPases: architecture and dynamics. Nat. Rev. Mol. Cell Biol. 9:478–489 [DOI] [PubMed] [Google Scholar]
- 30. Wu L, Russell P. 1993. Nim1 kinase promotes mitosis by inactivating Wee1 tyrosine kinase. Nature 363:738–741 [DOI] [PubMed] [Google Scholar]