Abstract
Erythroid Krüppel-like factor (EKLF or KLF1) is a transcriptional regulator that plays a critical role in lineage-restricted control of gene expression. KLF1 expression and activity are tightly controlled in a temporal and differentiation stage-specific manner. The mechanisms by which KLF1 is regulated encompass a range of biological processes, including control of KLF1 RNA transcription, protein stability, localization, and posttranslational modifications. Intact KLF1 regulation is essential to correctly regulate erythroid function by gene transcription and to maintain hematopoietic lineage homeostasis by ensuring a proper balance of erythroid/megakaryocytic differentiation. In turn, KLF1 regulates erythroid biology by a wide variety of mechanisms, including gene activation and repression by regulation of chromatin configuration, transcriptional initiation and elongation, and localization of gene loci to transcription factories in the nucleus. An extensive series of biochemical, molecular, and genetic analyses has uncovered some of the secrets of its success, and recent studies are highlighted here. These reveal a multilayered set of control mechanisms that enable efficient and specific integration of transcriptional and epigenetic controls and that pave the way for proper lineage commitment and differentiation.
INTRODUCTION
The correct balance of progenitor cell proliferation versus lineage-committed differentiation is central to the homeostasis of the hematopoietic system (1). The isolation and functional evaluation of transcription factors that direct expression of lineage-restricted target genes have led to significant advances in understanding how intracellular control of lineage commitment occurs at the molecular level (2). A major unresolved issue is the mechanism of how these proteins interact with chromatin-modifying factors and with the basal transcription machinery to direct the genotypic and phenotypic changes manifested during the process of lineage differentiation.
Erythroid Krüppel-like factor (EKLF) is a red cell-enriched DNA binding protein that interacts with its cognate 5′-CCMCRCCCN-3′ element at target promoters and enhancers (3) and is the founding member (now named KLF1) of the KLF family of transcription factors. This family is defined by the presence of three similar C2H2 zinc fingers at the C terminus that comprise its DNA binding domain (3–5). KLF1's role in β-like globin gene regulation has been the major focus of genetic, biochemical, and molecular studies since its discovery (6, 7). KLF1 is absolutely critical for the proper function and development of the erythroid lineage as supported by gene ablation studies in mice, of which the most obvious phenotypic effect is a profound, lethal β-thalassemia at the time of the switch to adult β-globin expression (8–10).
During the last 3 years, KLF1 has more fully emerged as one of the central players in establishing and maintaining global erythroid control (11–13). Recent studies utilizing chromatin immunoprecipitation (ChIP) followed by massively parallel sequencing (ChIP-seq) have demonstrated that KLF1 is physically bound to a large number of promoters, introns, and intergenic regions associated with erythroid cell-specific genes in both progenitors and terminally differentiating erythroid cells (13, 14). Together with expression profiling studies, KLF1's activation target repertoire has expanded beyond the classical β-globin gene to include protein-stabilizing, heme biosynthetic pathway, red cell membrane proteins, cell cycle, and transcription factor genes in both primitive and definitive erythroid cells (12–21). KLF1 activation targets have direct effects on erythroid biology, especially with respect to megakaryocyte-erythroid lineage decisions, control of cell cycle gene expression patterns (e.g., E2f2, E2f4, p18, and p21), and coordination of related events at late stages of erythroid differentiation (reviewed in reference 12).
The multiple defects in erythropoiesis and the erythroid lineage decisions that arise in KLF1-deficient embryos underline the importance of KLF1 in red cell biology. Its multifunctional roles in globin switching, lineage determination, terminal maturation, and human disease have been recently reviewed (12, 13). However, in addition to studies directed at understanding the role of KLF1 in the context of organismal biology, an extensive series of analyses has been performed to decipher the molecular mechanisms that govern KLF1 expression and function. These mechanistic aspects were last summarized more than 10 years ago (6, 7). In the present review, we update the latter studies and focus on recent exciting developments that elucidate KLF1's ability to directly integrate basal transcription and chromatin-modifying machineries, leading to temporal and lineage-restricted transcription of target genes.
GENERAL MECHANISMS BY WHICH KLF1 REGULATES TRANSCRIPTION, CHROMATIN ASSEMBLY, AND HIGHER-ORDER STRUCTURE IN ERYTHROID CELLS
Transcription initiation and promoter structure.
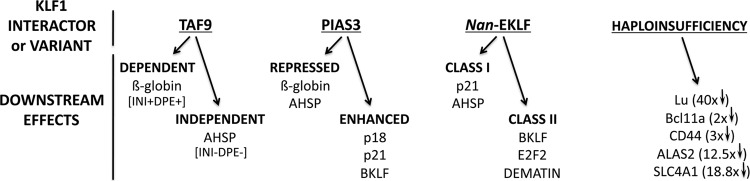
KLF1 interacts with the p62 subunit of TFIIH (22) and with TAF9 (23), each of which are components of the basal transcription machinery. These interactions are necessary for the optimal transactivation function of KLF1 and lead to two important observations that illuminate functional aspects of KLF1 initiation control. First, the interaction with p62/TFIIH imparts a helix-like structure to the normally unstructured region of the KLF1 activation domain, enabling the prediction and verification of a critical tryptophan amino acid within KLF1 (W73) required for its optimal activation function at the β-globin promoter (22). Since other proteins have been shown to interact with the structurally well characterized zinc finger region of KLF1 (see below), this result suggests that the whole of KLF1 may exist in a tightly structured conformation when complexed with transcription initiation components. Second, the discovery that association between KLF1 and TAF9 (a TATA-binding protein [TBP]-associated protein that is recruited by KLF1 to the β-globin promoter and is necessary for its activation) is not required at all KLF1 activation target promoters (23) suggests that interactions between KLF1 and the basal transcription machinery can be fine-tuned in a manner that may depend on specific promoter architectures and sequences (Fig. 1). In this instance, a conserved downstream promoter element (DPE) (24, 25), located at the β-globin promoter, interacts with TAF9. However, a TAF9-independent mode of KLF1 activation is seen at the alpha-hemoglobin stabilizing protein (AHSP) gene, which does not contain a discernible DPE. Together, these interactions with general transcription components are of critical importance for β-globin expression, since mutation within TFIIH leads to β-thalassemia (26), and some β-thalassemia mutations map to the β-globin DPE (27, 28).
Fig 1.
Nonequivalent KLF1 target gene regulatory mechanisms. KLF1 interaction with associated factors is either not required at all promoters (TAF9 [23]) or leads to differing transcriptional effects (PIAS3 [Siatecka and Bieker, unpublished]). Part of the TAF9 requirement may be related to whether INI and DPE promoter elements are present (23). Nan-EKLF contains a mutation within its second zinc finger (E339D) and recognizes a subset of normal KLF1 cognate elements: class I elements are recognized by both WT and Nan proteins, but class II elements are recognized only by the WT (29). Haploinsufficiency of KLF1 expression leads to a range of effects only on selected target genes (32, 126).
The nonequivalence of KLF1 gene targets (Fig. 1) is also manifested by analyses of the Nan-KLF1 (E339D mutant) interactions with DNA, where “class I” (p21 and AHSP) versus “class II” (KLF3, E2F2, and dematin) promoters are differentially activated compared to the wild type (WT) (12, 29). Additionally, some KLF1 activation targets are repressed (β-globin and AHSP) while others are enhanced (p18, p21, and BKLF/KLF3) by a SUMO-independent interaction with PIAS proteins (M. Siatecka and J. J. Bieker, unpublished). Finally, it must be noted in this context that some genes are uniquely sensitive to haploinsufficient levels of KLF1 (Lu and Bcl11a genes), leading to altered genetic expression patterns and hematologic parameters in humans [In(Lu) and hereditary persistence of fetal hemoglobin [HPFH] (11, 12, 30–33).
KLF1 displays selectivity in its interaction with histones and associates only with histone H3 (34). In this case, the KLF1 zinc finger region interacts with the H3 amino-terminal tail, but this interaction does not enable any discrimination between the H3.1 and H3.3 variants. As a result, the mechanism by which the H3.3 (but not the H3.1) variant is enriched at the β-globin promoter appears more complex than merely discrimination and recruitment by KLF1. However, interaction between KLF1 and H3.3 chaperones may provide a means to establish this enrichment (S. Soni and J. J. Bieker, unpublished).
GATA1 can physically interact with KLF1 and exhibits functional synergy with KLF1 at erythroid promoters as tested by cotransfection (35, 36). It is not known if the physical interaction between GATA1 and KLF1 is required for their synergism. One genome-wide KLF1 study has shown that most of its target sites are located a nonrandom distance away from a GATA binding motif (21). Another study that compared GATA1, SCL, and KLF1 ChIP-seq data suggests there are a significant number of target genes cooccupied by these three factors, forming a transcriptional core network (37). These observations have been supported and further buttressed by demonstrating their selective overlap with sites to which P300 is bound (38). On the other hand, another genome-wide study using the hemagglutinin (HA)-EKLF knock-in mouse (39) showed only a small overlap between KLF1 and GATA- or SCL-binding motifs (14), a difference possibly related to the use of the HA antibody and detection of a greater number of targets. Several recent studies, aimed at investigating interactions between erythroid cell-specific transcription factors and other proteins, have not shown KLF1 in complex with GATA1 in vivo (40–42). Nevertheless, identification of consensus sequences contained within KLF1-bound genomic regions implicates other transcription factors that may partner with KLF1 (21). Identifying these factors and deciphering the mode by which they interact with KLF1 to regulate target gene expression remains an open issue, one whose resolution will illuminate the transcription factor hierarchy (43) that operates to prime and maintain the erythroid program.
Chromatin protein interactions/higher-order structure.
KLF1 also interacts with chromatin-modifying and -remodeling factors, such as P/CAF, CBP/p300 (44), and the SWI/SNF complex (45, 46), the latter likely through its interaction with BRG1 (47–50) and possibly BAF47/BAF155 (51). Erythroid cells that lack KLF1 exhibit an aberrant chromatin configuration and altered components at KLF1 target promoters, resulting in histone hypoacetylation and loss of DNase hypersensitivity and an absence of CBP, BRG1, TBP, and RNA polymerase II (Pol II) (52).
KLF1 is necessary for transcription of AHSP mRNA and directly binds to a CACC site in the AHSP promoter (53, 54). In the absence of KLF1, a DNase I-hypersensitive site that is typically present in the promoter is lost, leading to a decrease but not complete loss of acetylated H3 and H4 across most of the AHSP gene, corresponding with loss of expression. KLF1 also directly regulates the erythroid cell-specific Alad(1b) promoter, enhancing the recruitment of other transcription factors, along with BRG1, P300/CBP, and RNA Pol II, that lead to induced changes in the local histone modification and chromatin structure (48). The absence of KLF1 disrupts hypersensitive-site formation at the β-globin (55) and E2F2 (19) genes, suggesting that in general, KLF1 plays a role in regulating chromatin conformation of its target loci. The critical role of KLF1 for these events has been most directly shown at the β-like globin locus, where KLF1 is necessary for formation of the active chromatin hub (ACH), a 3-dimensional nuclear interactome that brings distant DNA sequences into a spatially cooccupying complex (56–58). KLF1 not only binds the proximal adult β-globin promoter in vivo but also interacts with specific regions of the far-upstream locus control region (LCR) (59–61). It is noteworthy that an incomplete chromatin scaffold is formed in the absence of KLF1 (56). Although intergenic transcripts are normally generated throughout the β-locus, KLF1 binding and the three-dimensional (3D) structure of the locus are still retained when transcription initiation or elongation is inhibited (57, 62), suggesting that KLF1 directly drives its formation even in the absence of transcription.
Given the role of KLF1 in indirect regulation of embryonic and fetal globin genes via Bcl11a (30), it is perhaps surprising that KLF1 has been implicated in their direct control (61). In primitive erythroid cells isolated from embryonic day 10.5 (E10.5) murine embryos, the presence of KLF1 is required for enrichment of H3K9Ac (associated with an open chromatin configuration) at HS2 of the β-globin locus, as well as at the Ey and βh1 genes. It is also required for the enrichment of H3K4me3 (associated with active transcription) at HS2 and HS3, in addition to the Ey, βh1, and βmaj genes, despite the absence of βmaj transcription at this point (63). Part of KLF1's control mechanism in primitive cells appears to be coordinately regulated with KLF2 (64). For example, a global comparative analysis of transcripts expressed in WT, KLF1-null, or KLF1/KLF2-null primitive red cells led to the discovery that these two closely related transcription factors synergize to directly regulate c-Myc expression, leading to a network of genes that control the primitive red cell maturation program (65).
In erythroid progenitors, KLF1 is bound to an important enhancer element in the c-Myb promoter that, in conjunction with the LDB1 complex, is part of the ACH that forms at the c-Myb gene. As proposed for the β-like globin locus, the c-Myb ACH is suggested to be involved in transitioning the transcription machinery from initiation to elongation. However, this is a dynamic process: as erythroid cells terminally differentiate, KLF1 and the other proteins that form the ACH dissociate from the complex, and c-Myb expression decreases (66). Together with the effects at the globin locus, these data suggest that the presence of KLF1 is critical for transcription initiation in facilitating the formation of an open chromatin configuration at target loci. These data raise the possibility that in addition to transcription initiation, KLF1 also plays a role in regulating transcription elongation in erythroid cells (67), since the interaction between the upstream LCR and the proximal β-globin promoter, whose 3D chromatin structure requires KLF1 (56, 58), aids in establishing a postinitiation transcription elongation complex (68, 69). KLF1 is also recruited to the α-globin promoter and one of its upstream enhancer elements at late stages of differentiation (60, 70), possibly aiding in intrachromosomal interactions, preinitiation complex recruitment, and the transition from a poised to an elongating transcriptional state (70). However, it remains to be seen if stage-specific regulation of transcription elongation through chromatin looping is a generalized mechanism by which KLF1 functions. An additional detail in this context is the observation that KLF1 interacts with Ppm1b (42); although part of its role relates to KLF1 stability, a tantalizing layer arises from suggestions that Ppm1b dephosphorylates an important residue in the Cdk9 component of the pTEFb elongation complex (71, 72).
Collectively, these data show that KLF1 is an integrator of basic transcription and chromosomal remodeling signals that lead to modified histones, open chromatin, looped domains, and activated transcription. In this context, it is of interest that the majority of in vivo KLF1 binding sites are located at large distances (>10 kb) from transcription initiation sites (21), consistent with long-distance regulation. Use of the HA-tagged KLF1 mouse (39) has yielded slightly different estimates of their relative proportion (14). In any case, as observed with other erythroid factors, only ∼0.1 to 0.5% of theoretical target sites are actually bound in vivo. These interactions play a crucial role in establishing a limited number of transcription factories in vivo that enable efficient coordinate expression of select KLF1 target genes (73). KLF1-associated transcription factories tend to colocalize with loci that are highly transcribed in erythroid cells. For example, the globin genes associate with other active genes, revealing extensive and preferential intra- and interchromosomal transcription interactomes that are larger than transcription factories without KLF1 (74) and that may include PML bodies and Sc35 (75). Such interactions likely go hand in hand with the KLF1-, GATA1- and FOG1-dependent extrusion of the β-globin locus away from the nuclear periphery (76), an event that is necessary for its transcriptional activation (77). The KLF1 protein in erythroid cells from the HA-EKLF knock-in mouse is localized at the periphery in progenitor cells but then is dispersed throughout the nucleus in erythroblasts (14). These data suggest that KLF1 plays a global role in coordinating maximal transcription rates of select erythroid cell-specific genes by regulating their localization to specialized subnuclear compartments. The molecular mechanism by which these precise factories are established and mobilized within the intricate milieu of the nucleus remains a challenging cell biology and biochemical issue.
KLF1 POSTTRANSLATIONAL MODIFICATIONS AND EFFECTS ON PROTEIN-PROTEIN INTERACTIONS AND SUBSEQUENT FUNCTIONAL READOUT
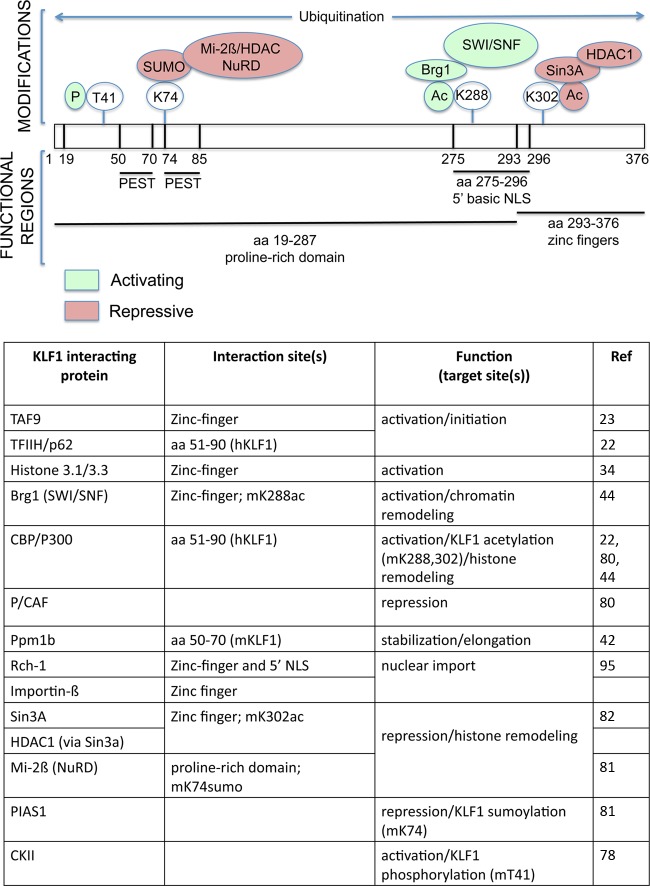
KLF1 activates a large number of genes that are broadly classified as important for erythroid function and development. It is highly expressed in the megakaryocyte-erythroid progenitor in addition to early differentiating primitive and definitive erythroid cells, and is especially critical at late stages of differentiation (e.g., erythroid lineage induction module 889 [2]). And yet its direct targets differ in different cell types, for example when comparing erythroid progenitors versus its erythroblast progeny (14) or primitive versus definitive red cells (61, 63). KLF1 is posttranslationally modified by phosphorylation (78), ubiquitylation (79), acetylation (80), and sumoylation (81). Posttranslational modifications of transcription factors can provide alternate protein interaction platforms that lead to varied downstream effects. As a result, these modifications may play a determining role in modulating KLF1's interactions with other proteins, fine-tuning its function in erythroid precursor, progenitor, and mature cells at different differentiation stages (Fig. 2).
Fig 2.
Summary of KLF1 posttranslational modifications and consequent protein-protein interactions. (Top) Schematic diagram of KLF1, showing the general locations of phosphorylation, sumoylation, acetylation, and ubiquitinylation modifications and their effects on subsequent protein-protein interactions and transcriptional readout. Those that are essential for optimal function as an activator are in green; those for repression are in pink (based on references 78, 79, 81, 82, and 44). Functional regions are based on mouse amino acid numbering (3, 79, 95, 127). (Bottom) Tabulation of KLF1 protein interactions, their interaction sites and any subsequent modification, and their downstream effects on transcription/chromatin. m, mouse; h, human.
Acetylation of KLF1 by CBP or P300 occurs at two sites, K288 and K302, and individually these play a critical role in subsequent protein-protein interactions that lead to different functional effects. Directed mutagenesis of K288 (but not K302) decreases the ability of KLF1 to transactivate the β-globin promoter and renders it unable to be superactivated by coexpressed p300 or CBP (44). Acetylated KLF1 has a higher affinity for the SWI/SNF chromatin remodeling complex and is a more potent transcriptional activator of chromatin-assembled templates in vitro (44). Using an in vivo rescue system (34), K288 acetylation has been shown to be critical for recruitment of CBP to the β-globin locus, modification of histone H3, occupancy by KLF1, opening of the chromatin structure, and transcription of adult β-globin (34).
On the other hand, the interaction of KLF1 with Sin3a is dependent on the acetylation status of KLF1 at K302. Mutagenesis of Lys-302 renders KLF1 no longer able to bind Sin3A, and it loses most of its repression activity (82). Endogenous KLF1 preferentially interacts with Sin3a in primitive erythroid cells and to a lesser extent in definitive erythroid cells (82, 83), suggesting that there are differentiation stage-dependent differences in KLF1 repression activity, possibly arising from variations in acetylation status (82). The biological significance of KLF1-mediated gene repression via this mechanism remains to be resolved and is further complicated by observations that an intact zinc finger region and thus target DNA specificity is not required for Sin3a interaction or repression.
KLF1 is also posttranslationally modified by SUMO (81) at a unique amino acid residue, K74, with the aid of PIAS1 as its E3 ligase. This leads to a more efficient interaction with the Mi2β subunit of the NuRD complex (an interaction that has been observed globally [84]) and transcriptional repression. Most intriguingly, a significant consequence of mutation in the KLF1 SUMO acceptor site is the loss of its inhibitory effect on megakaryocytic differentiation. The role of KLF1 in bipotential decisions emanating from the megakaryocyte/erythroid progenitor following gain- and loss-of-function experiments has been reviewed elsewhere (12). The importance of sumoylation in this process was shown in two ways. Using K562 cells, it was found that induction of megakaryopoiesis (with tetradecanoyl phorbol acetate [TPA]) was inhibited by the presence of wild-type KLF1 compared to that seen in the presence of KLF1 with the K74R mutation (K74R-KLF1), which exhibited little inhibition of the process. This was further supported by in vivo tests, where transgenic misexpression of WT KLF1 in megakaryocytes effectively decreased megakaryocyte cellularity and colony formation in the bone marrow, whereas transgenic bone marrow from the K74R-KLF1 line revealed no effect of the misexpression. Notably, erythroid colony formation by either of the transgenic lines, compared to the nontransgenic line, was not significantly affected. As a result, the availability of the SUMO modification site in KLF1 is absolutely critical for it to exert its normal inhibition of megakaryopoiesis prior to red cell onset (81). This KLF1 status may be directly relevant to cross-antagonistic models with Fli-1, a critical megakaryocytic transcription factor whose levels and activity are thought to be directly and/or indirectly repressed by KLF1 (reviewed in references 85 and 12).
At this point, the function of KLF1 as a repressor remains an area of speculation as its direct in vivo interaction with postulated repressed genes has not been extensively characterized. Comparison of expression profiling and ChIP experiments suggest that a majority of KLF1 targets are activated (14, 17, 21, 38); however, the smaller number of putative repressed targets may still be highly biologically significant. A potential complication is that BKLF/KLF3 and KLF8 are downstream targets of KLF1 and interact with a similar target DNA site (16, 86). KLF3 is a transcription factor that functions mainly as a repressor, and a recent model suggests that KLF3 acts as a feedback repressor to temper KLF1 activation of select targets (either promiscuous or specific) at late stages of erythropoiesis (87).
KLF1 is phosphorylated in vivo at serine and threonine residues, and casein kinase II (CKII)-dependent phosphorylation at T41 is essential for a GAL/EKLF(20–124) fusion to behave as an activator of transcription (78). Paradoxically, this study also showed that certain KLF1 kinases in MEL lysates decrease in activity when MEL cells are differentiated with hexamethylene bisacetimide (HMBA), raising the possibility that inhibition of KLF1 phosphorylation or an increase in phosphatase activity also plays a role in erythroid differentiation. Ppm1b, a serine/threonine phosphatase that interacts with KLF1, can inhibit the induction of erythropoiesis, while at the same time it has the ability to stabilize KLF1, augmenting its ability to activate transcription in committed erythroid cells (42). While exact mechanisms for how CKII and Ppm1b regulate erythroid homeostasis are not known and remain open areas of inquiry, these data suggest that both may play a role in part by regulating the transactivation capability and stability of the KLF1 protein. The functional coupling of phosphorylation to acetylation and to sumoylation (88) underscores the need to address whether there is any cross-regulation between these KLF1 modifications.
CRITICAL ROLE OF INTACT KLF1 ZINC FINGER SEQUENCES FOR NORMAL HEMATOLOGY
The mouse mutation Nan (neonatal anemia) was mapped to a single amino acid change (E339D) within the second zinc finger of KLF1 (29, 89, 90). The mutation alters the DNA binding specificity of KLF1 such that it no longer binds promoters of a subset of its DNA targets (29). Remarkably, even when mutant Nan and wild-type KLF1 alleles are expressed at equivalent levels, the mutant form selectively interferes with expression of KLF1 target genes whose promoter elements it no longer binds. This yields a distorted genetic output and selective protein deficiencies that differ from those seen in KLF1-heterozygous and KLF1-null red cells (29). This murine mutation has converged with human disease, particularly a subtype of congenital dyserythropoietic anemia (12, 91–94), where the same amino acid is altered (albeit to lysine; E325K) in human KLF1.
While it may not be surprising that zinc finger mutations that alter KLF1 interaction with its target site lead to functional changes, it should be kept in mind that this region is also responsible for protein interactions (Fig. 2). In some cases, an intact finger structure is not required (e.g., Sin3a [82] and importins [95]).
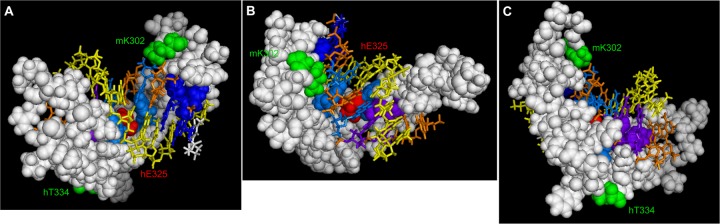
Although most human KLF1 mutations to date encode truncated proteins that ultimately lead to haploinsufficient expression levels, a significant number lead to missense mutations that result in expression of an altered protein (12, 33). It has been noted that most of these are localized to the DNA binding region, particularly the second zinc finger (31, 33, 94, 96, 97), and new mutations continue to support this initial observation (http://lovd.bx.psu.edu/variants.php?action=search_unique&select_db=KLF1). The structural implications of these alterations have been discussed elsewhere (32, 94), but it is interesting to note that, upon inspection of a model of KLF1 bound to its cognate DNA, almost all fall within the side of the protein that forms the interface with DNA (e.g., hE325) (Fig. 3). In most cases, these mutations alter important conserved and direct DNA-interacting amino acids, but a few are at sites that are predicted (from the model [98]) to be on a surface that faces away from DNA. As discussed above, numerous proteins interact with KLF1 via its zinc finger domain (Fig. 2). One can predict that mutations will be discovered that do not alter the DNA binding activity of KLF1 but disrupt protein-protein interactions, much like acetylated mK302 (hK288) enabling KLF1's interaction with Sin3a (82). In this context, human T334 (hT334) (mouse T348 [mT348]) (96) may fit with this idea and might be affecting an important protein interaction (Fig. 3). Alternatively, changes within amino acids that help orient KLF1 on its target DNA sites may alter the overall structure of dynamics of the fingers, thus indirectly disrupting KLF1's affinity for another associated protein(s). Future structural assessment of tripartite KLF1/protein/DNA complexes will help sort out how the zinc finger motif can assimilate such a mix.
Fig 3.
Location of selected amino acids within the KLF1 zinc fingers. The KLF1 structural model is based on the Zif268-DNA complex (98). Note that human (h) and mouse (m) numberings are not equivalent. KLF1/DNA structure is viewed at different angles (A, B, and C). Critical basic amino acids that interact with guanine residues on one strand of its double-stranded DNA (dsDNA) recognition element are selectively highlighted: blue for the first finger (mK306 and mH309), magenta for the second finger (mR336 and mR342), and purple for the third finger (mR364 and mH367). Note the two residues highlighted in green: mK302, known to be acetylated and critical for KLF1 interaction with Sin3a, and hT334, one of a collection of human KLF1 mutations. These are highlighted to demonstrate that they face away from the DNA interaction surface and are not directly involved in protein-DNA interactions. Most human KLF1 missense mutations are located at amino acids that are critical for its interaction/recognition of its cognate DNA element and face the DNA, such as hE325 (highlighted in red). Mutations are more fully discussed in the text. Images were generated using the MacPyMOL software program.
REGULATION OF KLF1 EXPRESSION
Transcriptional regulation of KLF1.
KLF1 expression is dramatically restricted during early mouse development, a property quite unlike findings for other members of the family but one that effectively circumscribes its functional effects (discussed in reference 99). KLF1 mRNA first appears at the neural plate stage (∼E7.5), where it is strictly localized to the earliest morphologically identified erythroid cells in the blood islands of the yolk sac, followed by a switch to the fetal liver by E10.5 (100). Expression of KLF1 mRNA in the adult mouse is restricted to the bone marrow and the red pulp of the spleen (3, 100). KLF1 transcript levels dramatically increase during hematopoietic differentiation, both in vivo and in cell culture assays (101–103). This is also reflected in the dynamic changes in histone marks (particularly H3K4/K9/K27/K36 and H4K20) and H2A.Z and RNA Pol II occupancy (101). A 950-bp region, located just upstream of the transcription initiation site, is sufficient to generate erythroid cell-specific expression in transient assays (104). This region exhibits the most significant homology upon a seven-species alignment of 30 kb of surrounding genomic DNA (105) and harbors erythroid-restricted DNase-hypersensitive sites. One of these sites (EHS1) behaves as a very strong enhancer, which in conjunction with the proximal promoter (106–108) accounts for KLF1's tissue-specific expression. The importance of this short region has been verified in vivo by the use of transgenic mice, where the 950-bp piece is sufficient to drive reporter (LacZ) expression specifically to the fetal liver and to the hematopoietic, but not endothelial or visceral endoderm, compartment of the yolk sac (109). Removal of the endogenous EHS1 enhancer decreases KLF1 levels by 50-fold (110).
Mouse embryonic stem (ES) cells differentiating in serum-free medium were used to show that BMP4 is necessary and sufficient to induce KLF1 expression as embryoid bodies (EBs) are being formed (111). The involvement of the Smad pathway in this process was directly implicated by showing that interference by constitutive expression of a dominant-negative BMP receptor or of the inhibitory Smad6 obviates KLF1 expression even in the presence of serum (111), results also seen after manipulation of Smad6 levels in human CD34+ cells (112). Related to this observation, conserved Gata and Smad binding motifs are located precisely within the previously mapped upstream enhancer and proximal promoter sequences, and their importance has been verified by mutational analyses using a KLF1 promoter/green fluorescent protein (GFP) transgene that faithfully recapitulates the onset of endogenous KLF1 expression during mouse EB differentiation (105). ChIP of GATA proteins reveal a switch in their occupancy (113, 114) when comparing early versus late times of EB differentiation, corresponding to GATA2 in the progenitor stage, followed by GATA1 after lineage commitment (105). In addition, use of a doxycycline-inducible short hairpin RNA (shRNA) line directed against Smad5 verified its critical importance for KLF1 expression. Together with the promoter analyses, these data led to the proposition of a two-tiered mechanism for transcriptional regulation of KLF1, with the GATA2 and SMAD5 proteins initially generating low transcript levels, followed by high quantities of KLF1 transcript after the GATA1 protein is produced (105). This partly explains the sustained level of KLF1 expression in GATA1-null erythroid cells (115). Both GATA1 and GATA2 have been shown to bind the KLF1 promoter in vivo (116, 117), and GATA2/SMAD4 (the partner of SMAD5) form part of a combinatorial interaction network (51). In addition, KLF1 retains a GATA1 “bookmark” that enables its rapid reactivation after mitosis (118).
As predicted from studies showing that KLF1 is highly expressed in the common megakaryocyte/erythroid progenitor (102), GFP+ fetal liver cells from a KLF1 promoter/GFP reporter mouse are enriched for erythroid and megakaryocyte progenitors as tested by colony-forming assays (105). This is also true when similar assays are performed with EB-derived cells.
Other proteins are also critical for KLF1 expression. Part of the mechanism by which c-Myb promotes erythropoiesis and represses megakaryopoiesis is by in vivo binding and transactivation of human KLF1, likely within the upstream enhancer region (119). It is probable that c-Myb plays a critical role in onset of KLF1 transcription specifically in progenitors. Analysis of the Gata1-binding region of the KLF1 promoter shows occupancy by the SCL, E2A, HEB, Zbtb7a/LRF/Pokemon, Fog1, and Mi2β proteins (117). How the proper protein complexes form at the upstream enhancer and proximal promoter regions that together are required for tissue-specific expression remains unanswered; its resolution will be relevant to understanding early events in the developing yolk sac as well as during hematopoiesis, when KLF1 is first detected in the multipotent and common myeloid progenitors (102).
Regulation of KLF1 stability.
The expression of the KLF1 protein is also regulated by proteosomal control of its stability (79). KLF1 can be ubiquitinated on multiple lysines that span its primary sequence, thus excluding sumoylation or acetylation from playing a controlling role. Although its two conserved PEST sequences are not required for ubiquitination, they may serve as docking sites for proteasome-mediated unfolding of the KLF1 protein prior to degradation (79). The Ppm1b protein phosphatase interacts with the KLF1 PEST1 sequence and increases the stability of KLF1 via its zinc finger domain, thereby regulating the availability of KLF1 to activate transcription (42).
The requirement for tight regulation of KLF1 levels is underlined by evidence that KLF1 haploinsufficiency perturbs regulation of erythropoiesis (reviewed in references 31, 12, and 33). Primitive erythroid cells in KLF1+/− mice possess ruffled membranes and exhibit an abnormal display of cell surface proteins, including an ectopic display of megakaryocytic cell surface markers (18). Haploinsufficiency of human KLF1 is linked to decreased levels of the Lu antigen in red cells (32) and to aberrant regulation of globin expression in adults; some patients that are borderline HbA2 or exhibit a hereditary persistence of fetal hemoglobin (HPFH) are heterozygous carriers of a nonsense mutation that ablates one KLF1 allele (12, 31, 33).
Last, KLF1 protein levels in definitive erythroid cells are 3-fold that of primitive erythroid cells (observed in references 63 and 61, although not in reference 120), possibly leading to altered binding patterns seen at different globin promoters. These observations highlight the idea that controlling the level of KLF1 gene transcription and stability of the protein likely play important roles in regulating its protein-protein interactions and subsequent involvement in downstream processes.
REGULATION OF KLF1 SUBCELLULAR LOCALIZATION
KLF1 possesses two nuclear localization signals (NLSs) near the carboxyl end that individually contribute to maximal nuclear localization (95, 121). The first NLS spans amino acids (aa) 275 to 296 of murine KLF1, a region adjacent to the second NLS that overlaps the KLF1 zinc fingers (aa 293 to 376). The acetylation status of K288 in NLS1 is irrelevant to its activity. KLF1 zinc fingers bind well in vitro to Rch-1 (importin α2) and importin β (95). Heterokaryon assays demonstrate that KLF1 is able to shuttle out of the nucleus, although its nuclear reentry is rapid (122). KLF1 does not contain an obvious nuclear export signal, and shuttling is relatively insensitive to leptomycin B inhibition of CRM1-mediated nuclear export.
In spite of the existence of two domains that behave as strong nuclear localization signals, a significant proportion of KLF1 quite unexpectedly resides in the cytoplasm (14, 73, 75, 122). Immunofluorescent and biochemical assays show that a substantial proportion of endogenous KLF1 is cytoplasmic at steady state in all erythroid cells examined (primary and MEL), that this pattern does not appear to change during either erythroid development or terminal differentiation, and that its nuclear/cytoplasmic ratio is not altered by a variety of treatments (122). Intriguingly, the murine KLF1 protein displays subtle yet distinct biochemical (migration on an SDS-PAGE gel) and functional (altered electrophoretic mobility shift assay [EMSA] complex) differences depending on which subcellular compartment it is isolated from (122), with PEST sequences (79) possibly playing a role in these differences.
The biological significance of KLF1 differential localization is unclear. Each KLF1 population may undergo different posttranslational modifications by virtue of being in different subcellular compartments, possibly enabling its differential retention. Such a mechanism may account for changes in DNA binding and transcription activity of KLF1 over the course of erythroid differentiation. Whether it is sequestered in a latent form or whether it maintains a totally separate function in the cytoplasm remains to be established.
FINAL REMARKS
Questions related to mechanistic details that remain outstanding have been alluded to throughout this review. In addition, other recent observations point to new unexplored areas of interest. For example, KLF1's critical role at late stages of erythropoiesis is further supported by the observation that it is expressed in the central macrophage of the erythroblastic island, where it activates DNase IIα (123), a protein that aids in the digestion of nuclei extruded in erythroblastic islands during erythroid enucleation (124). A totally unexpected observation is that an alternatively spliced form is found in the testis (125). Yet another interesting facet is that (paradoxically) both pro- and antiapoptosis pathway components targeted by KLF1 are enriched (13). Analysis of their activities in KLF1-null cells indicates that apoptosis is indeed perturbed but not in a straightforward way (38). Finally, it is likely that non-mRNA-coding genes, such as those encoding microRNAs (miRNAs), are also regulated by KLF1. In this regard, it is of interest that two recently identified long noncoding RNAs (lncRNAs) that exhibit erythroid cell-specific expression are directly regulated by KLF1 (38).
ACKNOWLEDGMENTS
Recent studies from our laboratory have been supported by PHS grants R01 DK046865, R01 DK077822, and RC1 DK086200 and by NYSTEM contract CO26435.
We thank Jim Omichinski and Shefali Soni for comments on the manuscript.
Footnotes
Published ahead of print 22 October 2012
REFERENCES
- 1. Orkin SH, Zon LI. 2008. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 132:631–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, Frampton GM, Drake AC, Leskov I, Nilsson B, Preffer F, Dombkowski D, Evans JW, Liefeld T, Smutko JS, Chen J, Friedman N, Young RA, Golub TR, Regev A, Ebert BL. 2011. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144:296–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Miller IJ, Bieker JJ. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol. Cell. Biol. 13:2776–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wolfe SA, Greisman HA, Ramm EI, Pabo CO. 1999. Analysis of zinc fingers optimized via phage display: evaluating the utility of a recognition code. J. Mol. Biol. 285:1917–1934 [DOI] [PubMed] [Google Scholar]
- 5. Wolfe SA, Nekludova L, Pabo CO. 2000. DNA recognition by Cys2His2 zinc finger proteins. Annu. Rev. Biophys. Biomol. Struct. 29:183–212 [DOI] [PubMed] [Google Scholar]
- 6. Bieker JJ. 2000. EKLF and the development of the erythroid lineage, p 71–84 In Ravid K, Licht JD. (ed), Transcription factors: normal and malignant development of blood cells. Wiley-Liss, New York, NY [Google Scholar]
- 7. Perkins A. 1999. Erythroid Kruppel like factor: from fishing expedition to gourmet meal. Int. J. Biochem. Cell Biol. 31:1175–1192 [DOI] [PubMed] [Google Scholar]
- 8. Lim SK, Bieker JJ, Lin CS, Costantini F. 1997. A shortened life span of EKLF−/− adult erythrocytes, due to a deficiency of β-globin chains, is ameliorated by human gamma-globin chains. Blood 90:1291–1299 [PubMed] [Google Scholar]
- 9. Nuez B, Michalovich D, Bygrave A, Ploemacher R, Grosveld F. 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375:316–318 [DOI] [PubMed] [Google Scholar]
- 10. Perkins AC, Sharpe AH, Orkin SH. 1995. Lethal β-thalassemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375:318–322 [DOI] [PubMed] [Google Scholar]
- 11. Bauer DE, Orkin SH. 2011. Update on fetal hemoglobin gene regulation in hemoglobinopathies. Curr. Opin. Pediatr. 23:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Siatecka M, Bieker JJ. 2011. The multifunctional role of EKLF/KLF1 during erythropoiesis. Blood 118:2044–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tallack MR, Perkins AC. 2010. KLF1 directly coordinates almost all aspects of terminal erythroid differentiation. IUBMB Life 62:886–890 [DOI] [PubMed] [Google Scholar]
- 14. Pilon AM, Ajay SS, Kumar SA, Steiner LA, Cherukuri PF, Wincovitch S, Anderson SM, Mullikin JC, Gallagher PG, Hardison RC, Margulies EH, Bodine DM. 2011. Genome-wide ChIP-Seq reveals a dramatic shift in the binding of the transcription factor erythroid Kruppel-like factor during erythrocyte differentiation. Blood 118:e139–e148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Drissen R, von Lindern M, Kolbus A, Driegen S, Steinlein P, Beug H, Grosveld F, Philipsen S. 2005. The erythroid phenotype of EKLF-null mice: defects in hemoglobin metabolism and membrane stability. Mol. Cell. Biol. 25:5205–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Funnell AP, Maloney CA, Thompson LJ, Keys J, Tallack M, Perkins AC, Crossley M. 2007. Erythroid Kruppel-like factor directly activates the basic Kruppel-like factor gene in erythroid cells. Mol. Cell. Biol. 27:2777–2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hodge D, Coghill E, Keys J, Maguire T, Hartmann B, McDowall A, Weiss M, Grimmond S, Perkins A. 2006. A global role for EKLF in definitive and primitive erythropoiesis. Blood 107:3359–3370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Isern J, Fraser ST, He Z, Zhang H, Baron MH. 2010. Dose-dependent regulation of primitive erythroid maturation and identity by the transcription factor Eklf. Blood 116:3972–3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pilon AM, Arcasoy MO, Dressman HK, Vayda SE, Maksimova YD, Sangerman JI, Gallagher PG, Bodine DM. 2008. Failure of terminal erythroid differentiation in EKLF-deficient mice is associated with cell cycle perturbation and reduced expression of E2F2. Mol. Cell. Biol. 28:7394–7401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Siatecka M, Lohmann F, Bao S, Bieker JJ. 2010. EKLF directly activates the p21WAF1/CIP1 gene by proximal promoter and novel intronic regulatory regions during erythroid differentiation. Mol. Cell. Biol. 30:2811–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tallack MR, Whitington T, Yuen WS, Wainwright EN, Keys JR, Gardiner BB, Nourbakhsh E, Cloonan N, Grimmond SM, Bailey TL, Perkins AC. 2010. A global role for KLF1 in erythropoiesis revealed by ChIP-seq in primary erythroid cells. Genome Res. 20:1052–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mas C, Lussier-Price M, Soni S, Morse T, Arseneault G, Di Lello P, Lafrance-Vanasse J, Bieker JJ, Omichinski JG. 2011. Structural and functional characterization of an atypical activation domain in erythroid Kruppel-like factor (EKLF). Proc. Natl. Acad. Sci. U. S. A. 108:10484–10489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sengupta T, Cohet N, Morle F, Bieker JJ. 2009. Distinct modes of gene regulation by a cell-specific transcriptional activator. Proc. Natl. Acad. Sci. U. S. A. 106:4213–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ohler U, Wassarman DA. 2010. Promoting developmental transcription. Development 137:15–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thomas MC, Chiang CM. 2006. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41:105–178 [DOI] [PubMed] [Google Scholar]
- 26. Viprakasit V, Gibbons RJ, Broughton BC, Tolmie JL, Brown D, Lunt P, Winter RM, Marinoni S, Stefanini M, Brueton L, Lehmann AR, Higgs DR. 2001. Mutations in the general transcription factor TFIIH result in beta-thalassaemia in individuals with trichothiodystrophy. Hum. Mol. Genet. 10:2797–2802 [DOI] [PubMed] [Google Scholar]
- 27. Cai SP, Eng B, Francombe WH, Olivieri NF, Kendall AG, Waye JS, Chui DH. 1992. Two novel beta-thalassemia mutations in the 5′ and 3′ noncoding regions of the beta-globin gene. Blood 79:1342–1346 [PubMed] [Google Scholar]
- 28. Oner R, Agarwal S, Dimovski AJ, Efremov GD, Petkov GH, Altay C, Gurgey A, Huisman TH. 1991. The G–A mutation at position +22 3′ to the Cap site of the beta-globin gene as a possible cause for a beta-thalassemia. Hemoglobin 15:67–76 [DOI] [PubMed] [Google Scholar]
- 29. Siatecka M, Sahr KE, Andersen SG, Mezei M, Bieker JJ, Peters LL. 2010. Severe anemia in the Nan mutant mouse caused by sequence-selective disruption of erythroid Kruppel-like factor. Proc. Natl. Acad. Sci. U. S. A. 107:15151–15156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bieker JJ. 2010. Putting a finger on the switch. Nat. Genet. 42:733–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Borg J, Patrinos GP, Felice AE, Philipsen S. 2011. Erythroid phenotypes associated with KLF1 mutations. Haematologica 96:635–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singleton BK, Burton NM, Green C, Brady RL, Anstee DJ. 2008. Mutations in EKLF/KLF1 form the molecular basis of the rare blood group In(Lu) phenotype. Blood 112:2081–2088 [DOI] [PubMed] [Google Scholar]
- 33. Singleton BK, Frayne J, Anstee DJ. 2012. Blood group phenotypes resulting from mutations in erythroid transcription factors. Curr. Opin. Hematol. 19:486–493 [DOI] [PubMed] [Google Scholar]
- 34. Sengupta T, Chen K, Milot E, Bieker JJ. 2008. Acetylation of EKLF is essential for epigenetic modification and transcriptional activation of the beta-globin locus. Mol. Cell. Biol. 28:6160–6170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gregory RC, Taxman DJ, Seshasayee D, Kensinger MH, Bieker JJ, Wojchowski DM. 1996. Functional interaction of GATA1 with erythroid Krüppel-like factor and SP1 at defined erythroid promoters. Blood 87:1793–1801 [PubMed] [Google Scholar]
- 36. Merika M, Orkin SH. 1995. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol. Cell. Biol. 15:2437–2447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wontakal SN, Guo X, Smith C, MacCarthy T, Bresnick EH, Bergman A, Snyder MP, Weissman SM, Zheng D, Skoultchi AI. 2012. A core erythroid transcriptional network is repressed by a master regulator of myelo-lymphoid differentiation. Proc. Natl. Acad. Sci. U. S. A. 109:3832–3837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tallack MR, Magor GW, Dartigues B, Sun L, Huang S, Fittock JM, Fry SV, Glazov EA, Bailey TL, Perkins AC. 26 July 2012. Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq. Genome Res. [Epub ahead of print.] doi:10.1101/gr.135707.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou D, Ren JX, Ryan TM, Higgins NP, Townes TM. 2004. Rapid tagging of endogenous mouse genes by recombineering and ES cell complementation of tetraploid blastocysts. Nucleic Acids Res. 32:e128 doi:10.1093/nar/gnh128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grosveld F, Rodriguez P, Meier N, Krpic S, Pourfarzad F, Papadopoulos P, Kolodziej K, Patrinos GP, Hostert A, Strouboulis J. 2005. Isolation and characterization of hematopoietic transcription factor complexes by in vivo biotinylation tagging and mass spectrometry. Ann. N. Y. Acad. Sci. 1054:55–67 [DOI] [PubMed] [Google Scholar]
- 41. Woo AJ, Moran TB, Schindler YL, Choe SK, Langer NB, Sullivan MR, Fujiwara Y, Paw BH, Cantor AB. 2008. Identification of ZBP-89 as a novel GATA-1-associated transcription factor involved in megakaryocytic and erythroid development. Mol. Cell. Biol. 28:2675–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yien YY, Bieker JJ. 2012. Functional interactions between erythroid Kruppel-like factor (EKLF/KLF1) and protein phosphatase PPM1B/PP2Cbeta. J. Biol. Chem. 287:15193–15204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garber M, Yosef N, Goren A, Raychowdhury R, Thielke A, Guttman M, Robinson J, Minie B, Chevrier N, Itzhaki Z, Blecher-Gonen R, Bornstein C, Amann-Zalcenstein D, Weiner A, Friedrich D, Meldrim J, Ram O, Cheng C, Gnirke A, Fisher S, Friedman N, Wong B, Bernstein BE, Nusbaum C, Hacohen N, Regev A, Amit I. 2012. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol. Cell 47:810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang W, Kadam S, Emerson BM, Bieker JJ. 2001. Site-specific acetylation by p300 or CREB binding protein regulates erythroid Kruppel-like factor transcriptional activity via its interaction with the SWI-SNF complex. Mol. Cell. Biol. 21:2413–2422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Armstrong JA, Bieker JJ, Emerson BM. 1998. A SWI/SNF-related chromatin remodeling complex, E-RC1, is required for tissue-specific transcriptional regulation by EKLF in vitro. Cell 95:93–104 [DOI] [PubMed] [Google Scholar]
- 46. Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, Emerson BM. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441–2451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Brown RC, Pattison S, van Ree J, Coghill E, Perkins A, Jane SM, Cunningham JM. 2002. Distinct domains of erythroid Kruppel-like factor modulate chromatin remodeling and transactivation at the endogenous beta-globin gene promoter. Mol. Cell. Biol. 22:161–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Desgardin AD, Abramova T, Rosanwo TO, Kartha S, Shim E-H, Jane SM, Cunningham JM. 2012. Regulation of delta-aminolevulinic acid dehydratase by Krüppel-like factor 1. PLoS One 7:e46482 doi:10.1371/journal.pone.0046482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kim SI, Bresnick EH, Bultman SJ. 2009. BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Res. 37:6019–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Perrine SP, Mankidy R, Boosalis MS, Bieker JJ, Faller DV. 2009. Erythroid Kruppel-like factor (EKLF) is recruited to the gamma-globin gene promoter as a co-activator and is required for gamma-globin gene induction by short-chain fatty acid derivatives. Eur. J. Haematol. 82:466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ravasi T, Suzuki H, Cannistraci CV, Katayama S, Bajic VB, Tan K, Akalin A, Schmeier S, Kanamori-Katayama M, Bertin N, Carninci P, Daub CO, Forrest AR, Gough J, Grimmond S, Han JH, Hashimoto T, Hide W, Hofmann O, Kamburov A, Kaur M, Kawaji H, Kubosaki A, Lassmann T, van Nimwegen E, MacPherson CR, Ogawa C, Radovanovic A, Schwartz A, Teasdale RD, Tegner J, Lenhard B, Teichmann SA, Arakawa T, Ninomiya N, Murakami K, Tagami M, Fukuda S, Imamura K, Kai C, Ishihara R, Kitazume Y, Kawai J, Hume DA, Ideker T, Hayashizaki Y. 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140:744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bottardi S, Ross J, Pierre-Charles N, Blank V, Milot E. 2006. Lineage-specific activators affect beta-globin locus chromatin in multipotent hematopoietic progenitors. EMBO J. 25:3586–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Keys JR, Tallack MR, Hodge DJ, Cridland SO, David R, Perkins AC. 2007. Genomic organisation and regulation of murine alpha haemoglobin stabilising protein by erythroid Kruppel-like factor. Br. J. Haematol. 136:150–157 [DOI] [PubMed] [Google Scholar]
- 54. Pilon A, Nilson D, Zhou D, Sangerman J, Townes T, Bodine D, Gallagher P. 2006. Alterations in expression and chromatin configuration of the alpha hemoglobin-stabilizing protein gene in erythroid Kruppel-like factor-deficient mice. Mol. Cell. Biol. 26:4368–4377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tewari R, Gillemans N, Wijgerde M, Nuez B, von Lindern M, Grosveld F, Philipsen S. 1998. Erythroid Kruppel-like factor (EKLF) is active in primitive and definitive erythroid cells and is required for the function of 5′HS3 of the beta-globin locus control region. EMBO J. 17:2334–2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. 2004. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 18:2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, de Laat W. 2008. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One 3:e1661 doi:10.1371/journal.pone.0001661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shimotsuma M, Matsuzaki H, Tanabe O, Campbell AD, Engel JD, Fukamizu A, Tanimoto K. 2007. Linear distance from the locus control region determines epsilon-globin transcriptional activity. Mol. Cell. Biol. 27:5664–5672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Im H, Grass JA, Johnson KD, Kim SI, Boyer ME, Imbalzano AN, Bieker JJ, Bresnick EH. 2005. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl. Acad. Sci. U. S. A. 102:17065–17070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shyu YC, Wen SC, Lee TL, Chen X, Hsu CT, Chen H, Chen RL, Hwang JL, Shen CK. 2006. Chromatin-binding in vivo of the erythroid kruppel-like factor, EKLF, in the murine globin loci. Cell Res. 16:347–355 [DOI] [PubMed] [Google Scholar]
- 61. Zhou D, Pawlik KM, Ren J, Sun CW, Townes TM. 2006. Differential binding of erythroid Krupple-like factor to embryonic/fetal globin gene promoters during development. J. Biol. Chem. 281:16052–16057 [DOI] [PubMed] [Google Scholar]
- 62. Mitchell JA, Fraser P. 2008. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 22:20–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Alhashem YN, Vinjamur DS, Basu M, Klingmuller U, Gaensler KM, Lloyd JA. 2011. Transcription factors KLF1 and KLF2 positively regulate embryonic and fetal beta-globin genes through direct promoter binding. J. Biol. Chem. 286:24819–24827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Basu P, Lung TK, Lemsaddek W, Sargent TG, Williams DC, Jr, Basu M, Redmond LC, Lingrel JB, Haar JL, Lloyd JA. 2007. EKLF and KLF2 have compensatory roles in embryonic beta-globin gene expression and primitive erythropoiesis. Blood 110:3417–3425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pang CJ, Lemsaddek W, Alhashem YN, Bondzi C, Redmond LC, Ah-Son N, Dumur CI, Archer KJ, Haar JL, Lloyd JA, Trudel M. 2012. Kruppel-like factor 1 (KLF1), KLF2, and Myc control a regulatory network essential for embryonic erythropoiesis. Mol. Cell. Biol. 32:2628–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stadhouders R, Thongjuea S, Andrieu-Soler C, Palstra RJ, Bryne JC, van den Heuvel A, Stevens M, de Boer E, Kockx C, van der Sloot A, van den Hout M, van Ijcken W, Eick D, Lenhard B, Grosveld F, Soler E. 2012. Dynamic long-range chromatin interactions control Myb proto-oncogene transcription during erythroid development. EMBO J. 31:986–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Stadhouders R, van den Heuvel A, Kolovos P, Jorna R, Leslie K, Grosveld F, Soler E. 1 July 2012. Transcription regulation by distal enhancers: who's in the loop? Transcription. [Epub ahead of print.] doi:10.4161/trns.20720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sawado T, Halow J, Bender MA, Groudine M. 2003. The beta-globin locus control region (LCR) functions primarily by enhancing the transition from transcription initiation to elongation. Genes Dev. 17:1009–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhou Z, Li X, Deng C, Ney PA, Huang S, Bungert J. 2010. USF and NF-E2 cooperate to regulate the recruitment and activity of RNA polymerase II in the beta-globin gene locus. J. Biol. Chem. 285:15894–15905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 26:2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cho S, Schroeder S, Ott M. 2010. CYCLINg through transcription: posttranslational modifications of P-TEFb regulate transcription elongation. Cell Cycle 9:1697–1705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wang Y, Dow EC, Liang YY, Ramakrishnan R, Liu H, Sung TL, Lin X, Rice AP. 2008. Phosphatase PPM1A regulates phosphorylation of Thr-186 in the Cdk9 T-loop. J. Biol. Chem. 283:33578–33584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, Eskiw CH, Luo Y, Wei CL, Ruan Y, Bieker JJ, Fraser P. 2010. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 42:53–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Eskiw CH, Fraser P. 2011. Ultrastructural study of transcription factories in mouse erythroblasts. J. Cell Sci. 124:3676–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Shyu YC, Lee TL, Wen SC, Chen H, Hsiao WY, Chen X, Hwang J, Shen CK. 2007. Subcellular transport of EKLF and switch-on of murine adult βmaj globin gene transcription. Mol. Cell. Biol. 27:2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lee HY, Johnson KD, Boyer ME, Bresnick EH. 2011. Relocalizing genetic loci into specific subnuclear neighborhoods. J. Biol. Chem. 286:18834–18844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Song SH, Kim A, Ragoczy T, Bender MA, Groudine M, Dean A. 2010. Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation. Blood 116:2356–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ouyang L, Chen X, Bieker JJ. 1998. Regulation of erythroid Kruppel-like factor (EKLF) transcriptional activity by phosphorylation of a protein kinase casein kinase II site within its interaction domain. J. Biol. Chem. 273:23019–23025 [DOI] [PubMed] [Google Scholar]
- 79. Quadrini KJ, Bieker JJ. 2006. EKLF/KLF1 is ubiquitinated in vivo and its stability is regulated by activation domain sequences through the 26S proteasome. FEBS Lett. 580:2285–2293 [DOI] [PubMed] [Google Scholar]
- 80. Zhang W, Bieker JJ. 1998. Acetylation and modulation of erythroid Kruppel-like factor (EKLF) activity by interaction with histone acetyltransferases. Proc. Natl. Acad. Sci. U. S. A. 95:9855–9860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Siatecka M, Xue L, Bieker JJ. 2007. Sumoylation of EKLF promotes transcriptional repression and is involved in inhibition of megakaryopoiesis. Mol. Cell. Biol. 27:8547–8560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chen X, Bieker JJ. 2004. Stage-specific repression by the EKLF transcriptional activator. Mol. Cell. Biol. 24:10416–10424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chen X, Bieker JJ. 2001. Unanticipated repression function linked to erythroid Kruppel-like factor. Mol. Cell. Biol. 21:3118–3125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Stielow B, Sapetschnig A, Kruger I, Kunert N, Brehm A, Boutros M, Suske G. 2008. Identification of SUMO-dependent chromatin-associated transcriptional repression components by a genome-wide RNAi screen. Mol. Cell 29:742–754 [DOI] [PubMed] [Google Scholar]
- 85. Dore LC, Crispino JD. 2011. Transcription factor networks in erythroid cell and megakaryocyte development. Blood 118:231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Eaton SA, Funnell AP, Sue N, Nicholas H, Pearson RC, Crossley M. 2008. A network of Kruppel-like factors (Klfs). Klf8 is repressed by Klf3 and activated by Klf1 in vivo. J. Biol. Chem. 283:26937–26947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Funnell AP, Norton LJ, Mak KS, Burdach J, Artuz CM, Twine NA, Wilkins MR, Power CA, Hung TT, Perdomo J, Koh P, Bell-Anderson KS, Orkin SH, Fraser ST, Perkins AC, Pearson RC, Crossley M. 2012. The CACCC-binding protein KLF3/BKLF represses a subset of KLF1/EKLF target genes and is required for proper erythroid maturation in vivo. Mol. Cell. Biol. 32:3281–3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, Lim WA, Fraser JS, Frydman J, Krogan NJ. 2012. Systematic functional prioritization of protein posttranslational modifications. Cell 150:413–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Heruth DP, Hawkins T, Logsdon DP, Gibson MI, Sokolovsky IV, Nsumu NN, Major SL, Fegley B, Woods GM, Lewing KB, Neville KA, Cornetta K, Peterson KR, White RA. 2010. Mutation in erythroid specific transcription factor KLF1 causes hereditary spherocytosis in the Nan hemolytic anemia mouse model. Genomics 96:303–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. White RA, Sokolovsky IV, Britt MI, Nsumu NN, Logsdon DP, McNulty SG, Wilmes LA, Brewer BP, Wirtz E, Joyce HR, Fegley B, Smith A, Heruth DP. 2009. Hematologic characterization and chromosomal localization of the novel dominantly inherited mouse hemolytic anemia, neonatal anemia (Nan). Blood Cells Mol. Dis. 43:141–148 [DOI] [PubMed] [Google Scholar]
- 91. Arnaud L, Saison C, Helias V, Lucien N, Steschenko D, Giarratana MC, Prehu C, Foliguet B, Montout L, de Brevern AG, Francina A, Ripoche P, Fenneteau O, Da Costa L, Peyrard T, Coghlan G, Illum N, Birgens H, Tamary H, Iolascon A, Delaunay J, Tchernia G, Cartron JP. 2010. A dominant mutation in the gene encoding the erythroid transcription factor KLF1 causes a congenital dyserythropoietic anemia. Am. J. Hum. Genet. 87:721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Mitchell WB, Gnanapragasam MN, Jaffray JA, Bieker JJ, Manwani D. 2011. Case report of erythroid transcription factor EKLF mutation causing a rare form of congenital dyserythropoietic anemia in a patient of Taiwanese origin. Blood 118:A2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ravindranath Y, Goyette G, Buck S, Gadgeel M, Dombkowski A, Boxer LA, Gallagher PG, Johnson RM. 2011. A new case of KLF1 G973A mutation and congenital dyserythropoeitic anemia (CDA)—further definition of emerging new syndrome and possible association with gonadal dysgenesis. Blood 118:A2101 [Google Scholar]
- 94. Singleton BK, Lau W, Fairweather VS, Burton NM, Wilson MC, Parsons SF, Richardson BM, Trakarnsanga K, Brady RL, Anstee DJ, Frayne J. 2011. Mutations in the second zinc finger of human EKLF reduce promoter affinity but give rise to benign and disease phenotypes. Blood 118:3137–3145 [DOI] [PubMed] [Google Scholar]
- 95. Quadrini KJ, Bieker JJ. 2002. Kruppel-like zinc fingers bind to nuclear import proteins and are required for efficient nuclear localization of EKLF. J. Biol. Chem. 277:32243–32252 [DOI] [PubMed] [Google Scholar]
- 96. Gallienne AE, Dreau HM, Schuh A, Old J, Henderson S. 2012. Ten novel mutations in the erythroid transcription factor KLF1 gene associated with increased fetal hemoglobin levels in adults. Haematologica 97:340–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Giardine B, Borg J, Higgs DR, Peterson KR, Philipsen S, Maglott D, Singleton BK, Anstee DJ, Basak AN, Clark B, Costa FC, Faustino P, Fedosyuk H, Felice AE, Francina A, Galanello R, Gallivan MV, Georgitsi M, Gibbons RJ, Giordano PC, Harteveld CL, Hoyer JD, Jarvis M, Joly P, Kanavakis E, Kollia P, Menzel S, Miller W, Moradkhani K, Old J, Papachatzopoulou A, Papadakis MN, Papadopoulos P, Pavlovic S, Perseu L, Radmilovic M, Riemer C, Satta S, Schrijver I, Stojiljkovic M, Thein SL, Traeger-Synodinos J, Tully R, Wada T, Waye JS, Wiemann C, Zukic B, Chui DH, Wajcman H, Hardison RC, Patrinos GP. 2011. Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat. Genet. 43:295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Feng WC, Southwood CM, Bieker JJ. 1994. Analyses of β-thalassemia mutant DNA interactions with erythroid Krüppel-like factor (EKLF), an erythroid cell-specific transcription factor. J. Biol. Chem. 269:1493–1500 [PubMed] [Google Scholar]
- 99. Bieker JJ. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 276:34355–34358 [DOI] [PubMed] [Google Scholar]
- 100. Southwood CM, Downs KM, Bieker JJ. 1996. Erythroid Kruppel-like Factor (EKLF) exhibits an early and sequentially localized pattern of expression during mammalian erythroid ontogeny. Dev. Dyn. 206:248–259 [DOI] [PubMed] [Google Scholar]
- 101. Cui K, Zang C, Roh TY, Schones DE, Childs RW, Peng W, Zhao K. 2009. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4:80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Frontelo P, Manwani D, Galdass M, Karsunky H, Lohmann F, Gallagher PG, Bieker JJ. 2007. Novel role for EKLF in megakaryocyte lineage commitment. Blood 110:3871–3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Li B, Ding L, Li W, Story MD, Pace BS. 2012. Characterization of the transcriptome profiles related to globin gene switching during in vitro erythroid maturation. BMC Genomics 13:153 doi:10.1186/1471-2164-13-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Chen X, Reitman M, Bieker JJ. 1998. Chromatin structure and transcriptional control elements of the erythroid Kruppel-like factor (EKLF) gene. J. Biol. Chem. 273:25031–25040 [DOI] [PubMed] [Google Scholar]
- 105. Lohmann F, Bieker JJ. 2008. Activation of Eklf expression during hematopoiesis by Gata2 and Smad5 prior to erythroid commitment. Development 135:2071–2082 [DOI] [PubMed] [Google Scholar]
- 106. Anderson KP, Crable SC, Lingrel JB. 2000. The GATA-E box-GATA motif in the EKLF promoter is required for in vivo expression. Blood 95:1652–1655 [PubMed] [Google Scholar]
- 107. Anderson KP, Crable SC, Lingrel JB. 1998. Multiple proteins binding to a GATA-E box-GATA motif regulate the erythroid Kruppel-like factor (EKLF) gene. J. Biol. Chem. 273:14347–14354 [DOI] [PubMed] [Google Scholar]
- 108. Crossley M, Tsang AP, Bieker JJ, Orkin SH. 1994. Regulation of the erythroid Kruppel-like factor (EKLF) gene promoter by the erythroid transcription factor GATA-1. J. Biol. Chem. 269:15440–15444 [PubMed] [Google Scholar]
- 109. Xue L, Chen X, Chang Y, Bieker JJ. 2004. Regulatory elements of the EKLF gene that direct erythroid cell-specific expression during mammalian development. Blood 103:4078–4083 [DOI] [PubMed] [Google Scholar]
- 110. Zhou D, Liu K, Sun CW, Pawlik KM, Townes TM. 2010. KLF1 regulates BCL11A expression and gamma- to beta-globin gene switching. Nat. Genet. 42:742–744 [DOI] [PubMed] [Google Scholar]
- 111. Adelman CA, Chattopadhyay S, Bieker JJ. 2002. The BMP/BMPR/Smad pathway directs expression of the erythroid-specific EKLF and GATA1 transcription factors during embryoid body differentiation in serum-free media. Development 129:539–549 [DOI] [PubMed] [Google Scholar]
- 112. Kang YJ, Shin JW, Yoon JH, Oh IH, Lee SP, Kim SY, Park SH, Mamura M. 2012. Inhibition of erythropoiesis by Smad6 in human cord blood hematopoietic stem cells. Biochem. Biophys. Res. Commun. 423:750–756 [DOI] [PubMed] [Google Scholar]
- 113. Bresnick EH, Lee HY, Fujiwara T, Johnson KD, Keles S. 2010. GATA switches as developmental drivers. J. Biol. Chem. 285:31087–31093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Dore LC, Chlon TM, Brown CD, White KP, Crispino JD. 2012. Chromatin occupancy analysis reveals genome-wide GATA factor switching during hematopoiesis. Blood 119:3724–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Weiss MJ, Keller G, Orkin SH. 1994. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1− embryonic stem cells. Genes Dev. 8:1184–1197 [DOI] [PubMed] [Google Scholar]
- 116. Fujiwara T, O'Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. 2009. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell 36:667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Yu M, Riva L, Xie H, Schindler Y, Moran TB, Cheng Y, Yu D, Hardison R, Weiss MJ, Orkin SH, Bernstein BE, Fraenkel E, Cantor AB. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell 36:682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Kadauke S, Udugama MI, Pawlicki JM, Achtman JC, Jain DP, Cheng Y, Hardison RC, Blobel GA. 2012. Tissue-specific mitotic bookmarking by hematopoietic transcription factor GATA1. Cell 150:725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Bianchi E, Zini R, Salati S, Tenedini E, Norfo R, Tagliafico E, Manfredini R, Ferrari S. 2010. c-myb supports erythropoiesis through the transactivation of KLF1 and LMO2 expression. Blood 116:e99–e110 [DOI] [PubMed] [Google Scholar]
- 120. McGrath KE, Frame JM, Fromm GJ, Koniski AD, Kingsley PD, Little J, Bulger M, Palis J. 2011. A transient definitive erythroid lineage with unique regulation of the beta-globin locus in the mammalian embryo. Blood 117:4600–4608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Pandya K, Townes TM. 2002. Basic residues within the Kruppel zinc finger DNA binding domains are the critical nuclear localization determinants of EKLF/KLF-1. J. Biol. Chem. 277:16304–16312 [DOI] [PubMed] [Google Scholar]
- 122. Quadrini KJ, Gruzglin E, Bieker JJ. 2008. Non-random subcellular distribution of variant EKLF in erythroid cells. Exp. Cell Res. 314:1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Porcu S, Manchinu MF, Marongiu MF, Sogos V, Poddie D, Asunis I, Porcu L, Marini MG, Moi P, Cao A, Grosveld F, Ristaldi MS. 2011. Klf1 affects DNase II-alpha expression in the central macrophage of a fetal liver erythroblastic island: a non-cell-autonomous role in definitive erythropoiesis. Mol. Cell. Biol. 31:4144–4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Kawane K, Fukuyama H, Kondoh G, Takeda J, Ohsawa Y, Uchiyama Y, Nagata S. 2001. Requirement of DNase II for definitive erythropoiesis in the mouse fetal liver. Science 292:1546–1549 [DOI] [PubMed] [Google Scholar]
- 125. Southwood CM, Lipovich L, Gow A. 29 August 2012. Tissue-restricted transcription from a conserved intragenic CpG island in the Klf1 gene in mice. Biol. Reprod. doi:10.1095/biolreprod.112.099879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Borg J, Papadopoulos P, Georgitsi M, Gutierrez L, Grech G, Fanis P, Phylactides M, Verkerk AJ, van der Spek PJ, Scerri CA, Cassar W, Galdies R, van Ijcken W, Ozgur Z, Gillemans N, Hou J, Bugeja M, Grosveld FG, von Lindern M, Felice AE, Patrinos GP, Philipsen S. 2010. Haploinsufficiency for the erythroid transcription factor KLF1 causes hereditary persistence of fetal hemoglobin. Nat. Genet. 42:801–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Bieker JJ, Southwood CM. 1995. The erythroid Krüppel-like factor (EKLF) transactivation domain is a critical component for cell-specific inducibility of a β-globin promoter. Mol. Cell. Biol. 15:852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]