Abstract
Cytoplasmic polyadenylation is a conserved mechanism that controls mRNA translation and stability. A key protein that promotes polyadenylation-induced translation of mRNAs in maturing Xenopus oocytes is the cytoplasmic polyadenylation element binding protein (CPEB). During this meiotic transition, CPEB is subjected to phosphorylation-dependent ubiquitination and partial destruction, which is necessary for successive waves of polyadenylation of distinct mRNAs. Here we identify the peptidyl-prolyl cis-trans isomerase Pin1 as an important factor mediating CPEB destruction. Pin1 interacts with CPEB in an unusual manner in which it occurs prior to CPEB phosphorylation and prior to Pin1 activation by serine 71 dephosphorylation. Upon induction of maturation, CPEB becomes phosphorylated, which occurs simultaneously with Pin1 dephosphorylation. At this time, the CPEB-Pin1 interaction requires cdk1-catalyzed CPEB phosphorylation on S/T-P motifs. Subsequent CPEB ubiquitination and destruction are mediated by a conformational change induced by Pin1 isomerization of CPEB. Similar to M phase progression in maturing Xenopus oocytes, the destruction of CPEB during the mammalian cell cycle requires Pin1 as well. These data identify Pin1 as a new and essential factor regulating CPEB degradation.
INTRODUCTION
Cytoplasmic polyadenylation is a widespread phenomenon that controls the translation and stability of mRNAs. Although some molecular details may differ according to species and/or cell type, much of the biochemistry of cytoplasmic polyadenylation that has been elucidated in Xenopus oocytes during meiotic progression serves to illustrate the general features of this regulatory process (1). A subset of RNAs that acquire a near-constitutive and typical long poly(A) tail in the nucleus undergo deadenylation in the cytoplasm where they are stored in a translational silent form (2). When the oocytes resume meiosis in response to hormonal (progesterone) stimulation, the activation of a kinase cascade culminates in poly(A) tail lengthening and the initiation of translation (3, 4). The 3′ untranslated region (UTR)-residing cytoplasmic polyadenylation element (CPE) dictates which mRNAs undergo these changes in poly(A). Cytoplasmic polyadenylation element binding protein (CPEB), an RNA recognition motif (RRM) and zinc finger-containing protein, associates with the CPE and a number of other factors, including Gld2, a noncanonical poly(A) polymerase; PARN, a deadenylating enzyme; symplekin, a ribonucleoprotein (RNP) assembly factor; and maskin, a eukaryotic translation initiation factor 4E (eIF4E) binding protein. Gld2 and PARN are constitutively active, but PARN activity is the more robust, and thus, cytoplasmic CPE-containing mRNAs are deadenylated and stored in an inactive form (5, 6). Maskin binding to eIF4E prevents the assembly of the initiation complex on the 5′ end of the mRNA, thus inhibiting translation (7, 8). Upon reentry into the meiotic divisions, CPEB is subjected to two waves of phosphorylation with two different outcomes. In the first wave, CPEB is phosphorylated by the kinase Aurora A on serine residue 174, which facilitates PARN expulsion from the RNP complex, resulting in Gld2-catalyzed polyadenylation of target mRNAs (5, 6, 9). In a second wave, CPEB undergoes six additional phosphorylation events that are catalyzed by cdk1; these phosphorylations are necessary for ubiquitin-mediated partial destruction of CPEB at the end of meiotic maturation (10).
Although several signaling events lead to ubiquitination and protein destruction, a prevalent one involves phosphorylation of S/T-P (Ser/Thr-Pro) motifs (11–13). For example, the SCF (Skp1–cullin–F-box) E3 complex directs phosphoproteins to be ubiquitinated and destroyed (14, 15). In many cases, phosphorylation-mediated protein destruction involves the peptidyl-prolyl cis-trans isomerase Pin1, which binds phosphorylated S/T-P motifs and enhances a normally slow cis-trans isomerization of the peptide bond. Pin1 is composed of an amino-terminal WW (Trp-Trp) domain that serves as a phosphoprotein binding domain and a carboxy-terminal peptidyl-prolyl cis-trans isomerase (PPIase) domain, which is essential for catalysis. Because of its broad range of target proteins, Pin1 functions in diverse cellular processes, such as cell cycle control, transcription, splicing regulation, DNA replication, and the DNA damage response (16). Not surprisingly, Pin1 activity is tightly regulated, mainly by phosphorylation, and its misregulation is associated with numerous human diseases, including cancer and neurologic disorders (17, 18).
The conformational changes induced by Pin1 affect the properties of a variety of target proteins. Nevertheless, changes in protein stability are the most common consequence (12). It has been suggested that the cis/trans isomerization and subsequent conformational change imposed by Pin1 establishes a proper three-dimensional architecture for E3 ligase (such as the SCF complex) recognition, resulting in the ubiquitination and destruction of target proteins (12, 14, 19).
We surmised that Pin1 might be involved in CPEB ubiquitination and degradation because it requires cdk1-catalyzed CPEB phosphorylation on S/T-P pairs (10). Here, we identify Pin1 as a major component of the CPEB destruction machinery. Surprisingly, we find that CPEB and Pin1 interact in arrested (immature) oocytes prior to any detectable CPEB phosphorylation. Following hormonal treatment to induce meiotic progression, Pin1 is dephosphorylated and, as a result, its activity is elevated. Coincidentally, CPEB is phosphorylated on 6 S/T-P pairs, which alters its interaction with Pin1 such that their association becomes CPEB phosphorylation dependent. By using a combination of neutralizing Pin1 antibody, Pin1 small interfering RNA (siRNA) depletion, and ectopic expression of wild-type (WT) and mutant Pin1 proteins, we find that it directly mediates CPEB ubiquitination and destruction in maturing Xenopus oocytes and cycling mammalian cells.
MATERIALS AND METHODS
Regents and antibodies.
The antibodies used in this study include those against Pin1 (H-123; Santa Cruz Biotechnology), CPEB (20), hemagglutinin (HA; Covance), tubulin (Sigma), green fluorescent protein (GFP; Abcam), and cyclin B1 (a kind gift of James Maller, University of Colorado Health Sciences Center). Recombinant Flag-ubiquitin was purchased from Boston Biochem, nontargeting siRNA from Dharmacon, and Pin1 targeting siRNA (Hs_PIN1_5 FlexiTube siRNA) from Qiagen.
Immunoprecipitation assays.
Fifty oocytes [some injected with 10 ng of mRNA synthesized with an Ambion mMESSAGE kit and polyadenylated in vitro with Escherichia coli poly(A) polymerase (New England BioLabs)] or HEK293T cells from one 10-cm dish were homogenized in lysis buffer containing 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% SDS, 50 mM Tris-HCl (pH 7.7), 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol, and protease inhibitor cocktail (Roche). The lysis buffer in some experiments also contained 10 μg/ml RNase A (Sigma). Clarified supernatants were incubated with Dynabeads protein A (Invitrogen) premixed with Pin1 or CPEB antibody or, as a control, IgG overnight at 4°C. The beads were washed extensively with lysis buffer before elution in SDS sample buffer.
GST pulldown assays.
Glutathione-Sepharose 4B (Amersham) coupled with glutathione S-transferase (GST) or GST-Pin1 and washed with 150 mM NaCl, 20 mM Tris-HCl (pH 8), 1 mM MgCl2, and 0.1% NP-40 was mixed with 1 mg of oocyte extract overnight at 4°C. The beads were then washed, and the protein was eluted with 20 mM reduced glutathione. The proteins were resolved by SDS-PAGE and visualized by Western analysis or autoradiography.
Pin1 phosphorylation and activity assays.
To examine Pin1 phosphorylation, oocytes, some of which were matured with progesterone, were homogenized in 80 mM β-glycerophosphate, 20 mM EGTA, 15 mM MgCl2, 50 mM NaVO4, and a protease inhibitor cocktail. The lysate was centrifuged for 5 min at 4°C, and the kinase assay performed at 37°C for 30 min in a 50-μl reaction volume containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 50 mM KCl, 1 mM dithiothreitol (DTT), 1 mM EGTA, 0.16 mCi/ml [γ-32P]ATP, and 20 μl oocyte extract. The beads were then washed, and the protein was eluted with reduced glutathione.
The method outlined by Nechama et al. (21) was used to investigate Pin1 activity. Briefly, oocytes were homogenized in reaction buffer containing100 mM NaCl, 50 mM HEPES (pH 7), 2 mM DTT, and 0.04 mg/ml bovine serum albumin (BSA) and the lysates were cleared by centrifugation at 12,000 × g for 10 min (4°C). PPIase activity was measured using equal amounts of oocyte cytoplasmic lysates and α-chymotrypsin (Sigma-Aldrich) using a synthetic tetrapeptide substrate Suc-Ala-Glu-Pro-Phe-pNa (Peptides International). Absorption at 390 nM was measured using an Ultraspec 2000 spectrophotometer. The results were expressed as the mean of three independent measurements at each time point.
Analysis of Pin1-CPEP interaction in mammalian cells.
Plasmid-transduced HEK293T cells were split into four 10-cm plates and cultured for 24 h. The medium was then replaced with Dulbecco's modified Eagle's medium (DMEM) lacking l-methionine and l-cysteine (Invitrogen) and then supplemented with [35S]methionine (6 μCi/ml) for 45 min. The cells were then washed with cold phosphate-buffered saline (PBS) and replaced with fresh DMEM supplement with 10% fetal calf serum (FCS). At the indicated times, the cells were washed with cold PBS, lysed, and processed for protein analysis.
For cell cycle analysis, HEK293T cells were arrested at S phase using double thymidine block as described previously (22). Briefly, 30% of confluent cells were incubated with DMEM supplemented with 2 mM thymidine for 18 h. The cells were washed and maintained in fresh DMEM for 9 h previous to a second block with 2 mM thymidine for another 17 h. The cells were then released by replacement of the culture medium. At the different time points after the release, the cells were washed and fixed using ice-cold 95% ethanol. The cells were sorted at the University of Massachusetts Medical Flow Cytometry Core Facility.
Statistics.
Values are reported as means ± standard errors of the means (SEM) unless otherwise stated; the data were analyzed by a Student's two-tailed t test with the significance set at a P value of <0.05.
RESULTS
Pin1 interacts with CPEB in a phosphorylation-independent manner.
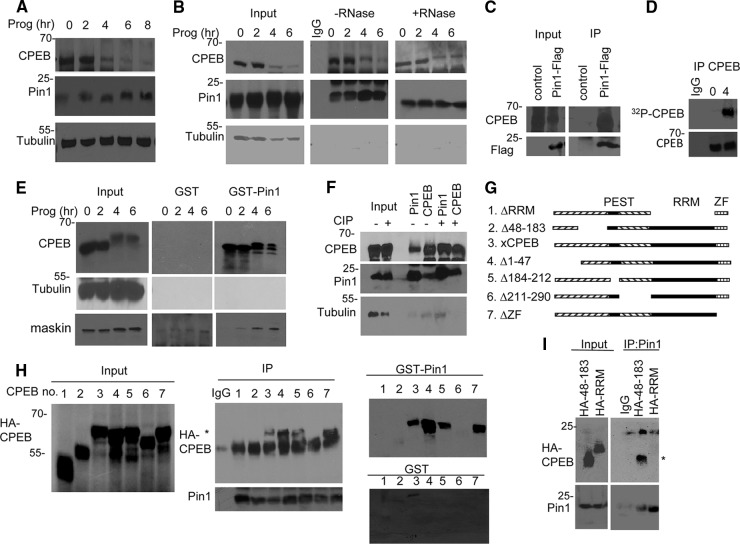
To investigate the possible involvement of Pin1 in CPEB destruction, extracts from Xenopus oocytes treated with progesterone to induce oocyte maturation were probed for Pin1, CPEB, and tubulin. The Western blots in Fig. 1A show that, as expected, CPEB decreased in amount as maturation progressed (10) while Pin1 levels were mostly unchanged. Because CPEB-mediated destruction requires cdk1-catalyzed, proline-directed serine/threonine phosphorylation (10), which also serves as recognition sites for Pin1, we examined a possible CPEB and Pin1 interaction. Extracts from oocytes treated with progesterone for varying times were used for immunoprecipitation of Pin1 and Western blotting for both Pin1 and CPEB (Fig. 1B). Pin1 and CPEB coimmunoprecipitated at 2, 4, and 6 h after progesterone treatment, which was coincident with CPEB phosphorylation (10). This interaction occurred in the presence or absence of RNase, indicating that RNA binding was not essential. Surprisingly, however, Pin1 and CPEB coimmunoprecipitated even from oocytes not exposed to progesterone when CPEB is not phosphorylated (3, 10). This unexpected and highly unusual result prompted us to examine this interaction further in a number of different ways. First, RNA encoding Flag-Pin1 was injected into oocytes, followed by an overnight incubation and Flag immunoprecipitation and Western blotting for Flag and CPEB. Figure 1C shows that CPEB was coimmunoprecipitated with ectopically expressed Pin1 from oocytes not exposed to progesterone. Second, to assess CPEB phosphorylation during maturation, some oocytes injected with [γ-32P]ATP and treated with progesterone for 4 h were used for CPEB immunoprecipitation and phosphorimaging; Fig. 1D shows that 32P-labeled CPEB was detected after, but not before progesterone application. Third, extracts from control or progesterone-treated oocytes were applied to GST or GST-Pin1 columns, followed by Western blotting for CPEB and tubulin as a negative control. We also blotted for maskin, a well-known CPEB-interacting protein (23) that is phosphorylated by cdk1 after progesterone treatment (8) and serves as a positive control for the GST-Pin1 pulldown (Fig. 1E). CPEB interacted with Pin1 irrespective of whether the oocytes were exposed to progesterone. On the other hand, maskin interaction with Pin1 was evident only after progesterone treatment when maskin is phosphorylated on S/T-P motifs. Fourth, to assess whether CPEB from immature oocytes might be phosphorylated, which previously went undetected, oocyte extracts were treated with calf intestinal alkaline phosphatase (CIP) and then subjected to CPEB-Pin1 coimmunoprecipitation. Figure 1F demonstrates a strong interaction between these proteins irrespective of CIP treatment. Finally, to identify the Pin1 interaction site(s) on CPEB prior to its phosphorylation, mRNA encoding HA-tagged CPEB deletion mutants (Fig. 1G) were injected into oocytes and, after overnight incubation, extracts were subjected to Pin1 immunoprecipitation and Western blotted for HA. Figure 1H (center panel) shows that the deletion of three regions, the RRMs, residues 48 to 183, and residues 211 to 290 abrogated the CPEB-Pin1 interaction. To further assess the need for these regions for Pin1 interaction, the different CPEB mutant proteins were ectopically expressed in oocytes as before. Extracts were then applied to GST or GST-Pin1 columns, followed by Western blotting for HA (Fig. 1H, right panel). As with the coimmunoprecipitation assay, deletion of the RRMs, residues 48 to 183, and residues 211 to 290 prevented the CPEB-Pin1 interaction. To identify the minimal region needed for Pin1 interaction, the three regions deleted in the experiments noted above were HA tagged and ectopically expressed by mRNA injection (Fig. 1I). Pin1 interacted with CPEB residues 48 to 183 but not the RRMs. We were unable to detect residues 211 to 290, probably because of the region's small size (∼7 kDa). We conclude that Pin1 associates with CPEB residues 48 to 183 in a phosphorylation-independent manner.
Fig 1.
CPEB and Pin1 interact in a phosphorylation-independent manner. (A) Western blots of CPEB and Pin1 during progesterone-induced oocyte maturation. Tubulin served as a loading control. (B) Extracts from oocytes exposed to progesterone for 0 to 6 h were subjected to Pin1 immunoprecipitation in the absence or presence of RNase A and Western blotting for Pin1, CPEB, and tubulin. The input represents 10% of the total lysate. (C) Oocytes were injected with mRNA encoding Flag-Pin1 and subjected to Flag immunoprecipitation and Western blot analysis for Flag and CPEB. Control refers to nonspecific IgG immunoprecipitation. (D) Oocytes were injected with [γ-32P]ATP, followed by treatment with progesterone for 0 or 4 h, immunoprecipitation of CPEB, and SDS-PAGE and autoradiography. (E) Extracts from oocytes exposed to progesterone for 0 to 6 h were applied to GST or GST-Pin1 columns, followed by Western blotting for CPEB, tubulin, and maskin. (F) Oocyte extracts were treated with calf intestinal alkaline phosphatase (CIP) prior to immunoprecipitation of Pin1 and CPEB and blotting for CPEB, Pin1, and tubulin. (G) Schematic of CPEB deletion mutants. (H) mRNA encoding HA-tagged CPEB deletion mutants (numbered in panel G) were injected into oocytes, followed by Pin1 immunoprecipitation and Western blotting for Pin1 and HA (center panel) or chromatography on a GST or GST-Pin1 column (right panels). The asterisk denotes the HA-CPEB proteins. (I) mRNA encoding HA-tagged CPEB proteins 48 to 183 and RRM were injected into oocytes, followed by Pin1 immunoprecipitation and Western blotting for HA and Pin1. The asterisk denotes CPEB HA 48–183.
Pin1 interacts with CPEB in two steps.
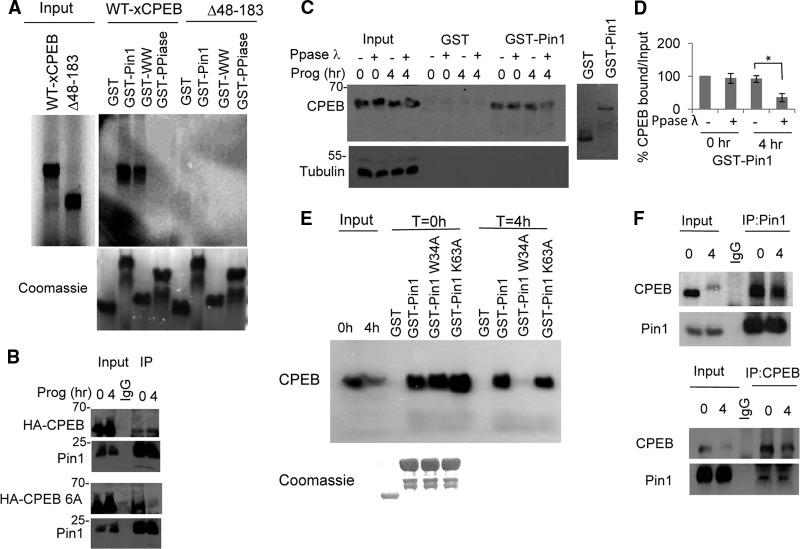
The data in Fig. 1 demonstrate that the interaction of CPEB with Pin1 is CPEB phosphorylation independent in immature (G2-arrested) oocytes. To evaluate the interaction of Pin1 and CPEB in vitro and the contribution of each of the Pin1 domains to this interaction, mRNAs encoding WT CPEB and the Δ48–183 CPEB mutant were translated in rabbit reticulocyte lysates in the presence of [35S]methionine. The lysates were then subjected to pulldown experiments using GST, GST-Pin1, and GST-WW and GST-PPIase proteins, which correspond to the Pin1 WW domain and Pin1 PPIase domains, respectively. As expected, GST-Pin1 interacted with full-length CPEB and not with the Δ48–183 CPEB mutant (Fig. 2A). Interestingly, full-length CPEB interacted with the GST-WW domain to the same extent as it did with GST-Pin1 but failed to interact with the GST-PPIase domain alone. As negative controls, neither of them interacted with the Δ48–183 CPEB mutant. These results indicate that the interaction between CPEB and Pin1, prior to detectable CPEB phosphorylation, requires the WW domain and the region spanning amino acids 48 to 183 of CPEB. Because CPEB has six known cdk1 S/T-P phosphorylation sites (10), we decided to investigate whether they might be involved in the Pin1-CPEB interaction after oocyte maturation. Consequently, mRNAs encoding WT HA-CPEB or a HA-CPEB mutant with alanine substitutions for the six phospho-S/T residues (referred to as CPEB-6A) were injected into oocytes that were then stimulated with progesterone. As demonstrated previously, Pin1 interacted with WT CPEB both before and after progesterone treatment (Fig. 2B). However, although CPEB-6A interacted with Pin1 prior to progesterone stimulation, it interacted only very weakly after stimulation (Fig. 2B). To examine this further, extracts from oocytes either before or after 4 h of progesterone treatment were mixed with bacteriophage λ phosphatase, which dephosphorylates S/T residues. The extracts were then subjected to chromatography on GST or GST-Pin1 columns followed by Western blotting for CPEB. As shown in Fig. 2C and D, the CPEB interaction with Pin1 before progesterone treatment was not affected by λ phosphatase treatment. Conversely, treatment of mature oocyte extracts with λ phosphatase reduced the CPEB-Pin1 interaction by more than 50%. These data suggest that CPEB interacts with Pin1 in two steps: step 1 occurs in immature oocytes and does not involve phosphorylation, while step 2 takes place after progesterone treatment and is mediated by the phosphorylation of CPEB on one or more S/T-P pairs.
Fig 2.
A two-step interaction between CPEB and Pin1. (A) mRNA encoding WT HA-tagged CPEB or the Δ48–183 CPEB mutant was in vitro translated in the presence of [35S]methionine and used for pulldown assays with GST-Pin1, the GST-WW domain, or the GST-PPIase domain as the bait. (B) mRNA encoding WT HA-tagged CPEB or a CPEB mutant with alanine substitutions for serine or threonine residues that are phosphorylated during maturation (CPEB 6A) was injected into oocytes that were then treated with progesterone for 0 or 4 h. Pin1 was then immunoprecipitated and Western blots probed for Pin1 or HA. A mock immunoprecipitation with nonspecific IgG was also performed. (C) Extracts from oocytes treated with progesterone for 0 or 4 h were treated with λ phosphatase and then applied to GST or GST-Pin1 columns. The eluted material was probed for CPEB and tubulin. At right is a Coomassie blue-stained gel showing the integrity of recombinant GST and GST-Pin1. (D) Quantification of the three experiments in panel B. The error bars refer to SEM, and the asterisk indicates statistical significance (P < 0.05). (E) Extracts from untreated or progesterone-treated oocytes were applied to GST, GST-Pin1, GST-Pin1 W34A, or GST-Pin1 K63A columns, followed by Western blotting for CPEB. (F) Control or progesterone-treated oocyte extracts were reciprocally immunoprecipitated for CPEB and Pin1 and blotted for Pin1 and CPEB, respectively.
To assess the contribution of each of the Pin1 domains to the interaction with CPEB either before or after progesterone treatment, we altered GST-Pin1 in either its WW domain (W34A) or its PPIase domain (K63A) and used these mutant proteins for GST-Pin1 pulldown experiments with oocyte extracts before or after progesterone treatment. As shown in Fig. 2E, GST-Pin1 and both mutant proteins interacted with CPEB from untreated oocyte extracts. In contrast, when progesterone-treated oocyte extracts were used, the GST-Pin1 W34A mutant protein failed to interact with CPEB. This result reinforces the notion that the Pin1-CPEB interaction is altered upon CPEB phosphorylation and becomes dependent on phospho-S/P sites. It is worth mentioning that the Pin1 K63A mutation had little effect on the Pin1-CPEB interaction compared to the W34A mutation, which emphasizes the importance of the WW domain in the second step of the interaction. These results suggest that there may be an enhanced Pin1 affinity for phosphorylated CPEB. Because CPEB is a highly insoluble protein, we could not determine the kinetics of CPEB-Pin1 interaction in vitro with purified components. Therefore, we performed a reciprocal immunoprecipitation of Pin1 or CPEB from either untreated or progesterone-treated oocytes and blotted for CPEB or Pin1. As shown in Fig. 2F, when Pin1 was immunoprecipitated, it interacted with CPEB from both untreated and progesterone-treated oocytes. As expected, this interaction was reduced after progesterone treatment because CPEB is partially destroyed in mature oocytes. However, although the reciprocal experiment shows that less CPEB was immunoprecipitated from mature versus immature oocyte extracts, nearly the same amount of Pin1 was coprecipitated from each extract. These results suggest that the elevated Pin1-CPEB interaction after progesterone treatment might be the result of an increased affinity of phospho-CPEB for Pin1.
Pin1 is phosphorylated and inactive prior to progesterone treatment.
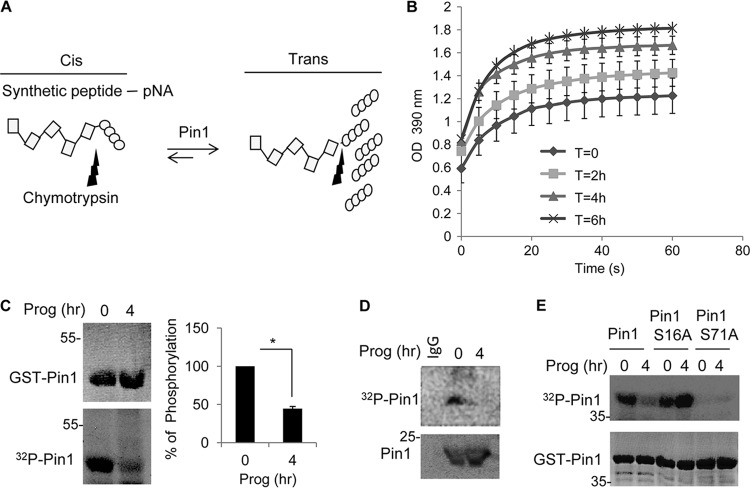
Pin1 catalytic activity is tightly regulated under normal physiological conditions and is mediated by at least two posttranslational events: protein kinase A (PKA)-mediated phosphorylation of serine 16, which affects its ability to interact with target proteins (24), and DAPK1-catalyzed phosphorylation of serine 71, which inhibits its isomerization activity (25). An additional phosphorylation on serine 65 is catalyzed by PLK (polo-like kinase) (26); however, this phosphorylation event seems to increase Pin1 protein stability rather than affect its activity. To assess whether Pin1 activity is regulated during oocyte maturation, extracts from control and progesterone-treated oocytes were incubated with a synthetic peptide containing a Pin1 interaction site in a cis orientation attached to p-nitroanilide; Pin1 catalyzes a relatively slow peptidyl-prolyl isomerization of the target peptide to the trans conformation, allowing for chymotrypsin cleavage and release of the p-nitroanilide moiety, which is monitored by an absorbance of 390 nm of light (Fig. 3A). Figure 3B shows that progesterone treatment significantly elevated Pin1 activity compared to untreated controls.
Fig 3.
Pin1 is activated during oocyte maturation. (A) Schematic to measure Pin1 activity (see the text for details). (B) Extracts from oocytes treated with progesterone for 0 to 6 h were used to assay for Pin1 activity as illustrated in panel A. (C) Extracts from oocytes treated with progesterone for 0 or 4 h were incubated with GST-Pin1 and [γ-32P]ATP; GST-Pin1 was then detected by Coomassie blue staining, and 32P-labeled GST-Pin1 was detected by autoradiography. (D) Oocytes were injected with [γ-32P]ATP, followed by progesterone treatment, immunoprecipitation of Pin1, and detection by Western blotting and autoradiography. (E) Oocytes and mature oocyte extracts were incubated with [γ-32P]ATP and GST-Pin1 or the GST-Pin1 S16A or GST-Pin1 S71A mutant proteins and analyzed by autoradiography. The lower panel shows Coomassie blue-stained GST-Pin1 isoforms.
To examine whether the activation of Pin1 was due to changes in phosphorylation, recombinant GST-Pin1 was incubated with [γ-32P]ATP and extracts from untreated or progesterone-treated oocytes. Figure 3C demonstrates that Pin1 was strongly phosphorylated when incubated with extracts from control oocytes, and this phosphorylation was reduced by ∼60% when incubated with extracts from progesterone-treated oocytes. To investigate whether Pin1 phosphorylation occurs in vivo, [γ-32P]ATP was injected into oocytes that were then incubated with progesterone for 4 h, followed by Pin1 immunoprecipitation. Figure 3D shows that Pin1 was phosphorylated in control oocytes but that this phosphorylation was reduced by almost 70% after progesterone treatment. To determine which residues on Pin1 were phosphorylated in oocytes, serine 16 or 71 was mutated to alanine and the same experiment as in Fig. 3C was performed. As before, GST-Pin1 was phosphorylated using extracts derived from untreated oocytes, which was reduced when extracts from progesterone-treated oocytes were used (Fig. 3E). There was no detectable phosphorylation of the GST-Pin1 S71A mutant protein irrespective of whether extracts from control or progesterone-treated oocytes were used. Interestingly, mutation of serine 16 did not abrogate Pin1 phosphorylation when extracts from control oocytes were used, indicating that serine 16 is not phosphorylated prior to maturation. However, in contrast to WT GST-Pin1, there was no reduction in phosphorylation of the GST-Pin1 S16A mutant protein when extracts from progesterone-treated oocyte extracts were used in the assay. This result might indicate that serine 16 is required for serine 71 dephosphorylation during maturation. Therefore, Pin1 is phosphorylated on serine 71 prior to oocyte maturation and is inactive. After progesterone-induced maturation, Pin1 is dephosphorylated and consequently activated.
Pin1 promotes CPEB destruction.
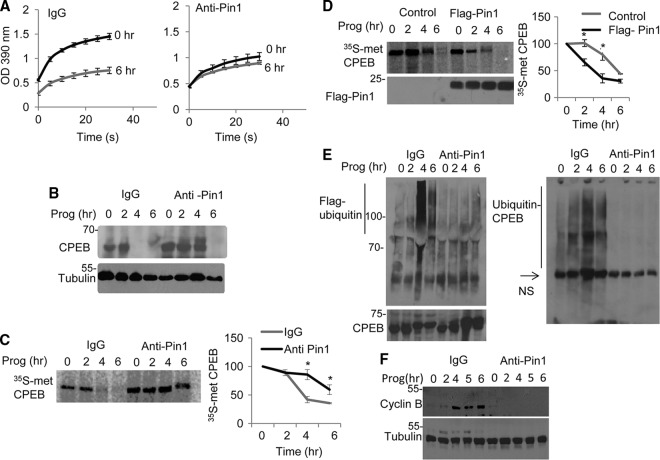
Nearly simultaneous with Pin1 activation are cdk1-catalyzed CPEB phosphorylation, ubiquitination, and partial destruction. We suspected that these events would be linked and began to test this hypothesis by inhibiting Pin1 activity via injection of neutralizing Pin1 antibody into oocytes. Although control IgG injection had no effect on Pin1 isomerase activity during maturation, Pin1 antibody significantly inhibited its activation (Fig. 4A). To assess whether Pin1 mediates CPEB destruction, oocytes were injected with nonspecific IgG or Pin1 antibody, followed by progesterone treatment. The Western blot in Fig. 4B shows that, compared to IgG, Pin1 antibody lowered the rate of CPEB destruction. This result was also confirmed in Fig. 4C; in this case, oocytes injected with IgG or Pin1 antibody were incubated with [35S]methionine for 3 h and then with radio-inert methionine, followed by progesterone treatment and immunoprecipitation of CPEB. Pin1 antibody lowered the rate of destruction of newly synthesized CPEB.
Fig 4.
Pin1 enhances ubiquitination and destruction of CPEB. (A) Extracts from oocytes, some of which were treated with progesterone, were injected with IgG or Pin1 antibody, and the cis-trans isomerization analysis as depicted in Fig. 3A was performed. (B) Oocytes were injected with IgG or Pin1 antibody, treated with progesterone, and immunoblotted for CPEB or tubulin. (C) Oocytes were injected with IgG or Pin1 antibody, followed by incubation in [35S]methionine-containing medium for 3 h and progesterone treatment. At the indicated times, the extracts were subjected to CPEB immunoprecipitation and analysis by SDS-PAGE and autoradiography. (D) Oocytes were injected with Flag-Pin1 or control RNA, followed by incubation in [35S]methionine-containing media for 3 h and then treated with progesterone. At the indicated times, the extracts were subjected to CPEB immunoprecipitation and analysis by SDS-PAGE and autoradiography. (E) Oocytes were injected with Flag-ubiquitin peptide together with IgG or Pin1 antibody. CPEB was then immunoprecipitated and probed for CPEB and Flag-ubiquitin (left) or Flag immunoprecipitated and probed for CPEB (right). NS refers to a nonspecific band. (F) Oocytes were injected with IgG or Pin1 antibody, treated with progesterone, and then probed in Western blots for cyclin B and tubulin.
We next performed a reciprocal experiment by examining whether CPEB destruction would be accelerated by the ectopic expression of Pin1. Oocytes were injected with Flag-Pin1 or control noncoding RNA, followed by metabolic labeling of protein with [35S]methionine for 3 h. The oocytes where then chased with radio-inert methionine and treated with progesterone for the indicated times. CPEB was then immunoprecipitated and analyzed by SDS-PAGE (Fig. 4D). Converse to the data shown with neutralizing Pin1 antibody, ectopic Pin1 expression enhanced CPEB destruction, reducing its half-life (t1/2) from 5 h in control oocytes to ∼3 h. These results demonstrate that Pin1 accelerates CPEB destruction.
The phosphorylation of CPEB leads to its ubiquitination and destruction (10, 27, 28). To assess whether Pin1 controls CPEB ubiquitination, oocytes were injected with Flag-ubiquitin peptide and IgG or Pin1 antibody, followed by a 1-h incubation with MG132 to prevent proteosome-mediated protein destruction. The oocytes were then incubated with progesterone, followed by CPEB or Flag immunoprecipitation and probing of Western blots for Flag or CPEB. Figure 4E (left panel) shows that CPEB antibody precipitated Flag-ubiquitin at 4 and 6 h after progesterone treatment when IgG was injected but not when Pin1 antibody was injected. Conversely, immunoprecipitation of Flag and probing with CPEB antibody again showed ubiquitinated CPEB when IgG was injected but not when Pin1 antibody was injected.
In Xenopus oocytes, CPEB destruction is vital for meiotic progression and cyclin B1 mRNA translation (10). To assess whether Pin1 control of CPEB destruction is necessary for cyclin B1 synthesis, oocytes were injected with IgG or Pin1 antibody as above, followed by progesterone treatment and Western blotting for cyclin B1. As demonstrated previously (10), cyclin B1 synthesis increased dramatically after progesterone treatment, which was abrogated by Pin1 antibody (Fig. 4F). These data show a close correlation for Pin1-mediated CPEB destruction, which is necessary for cyclin B1 mRNA translation.
Pin1 regulates CPEB destruction in mammalian cells.
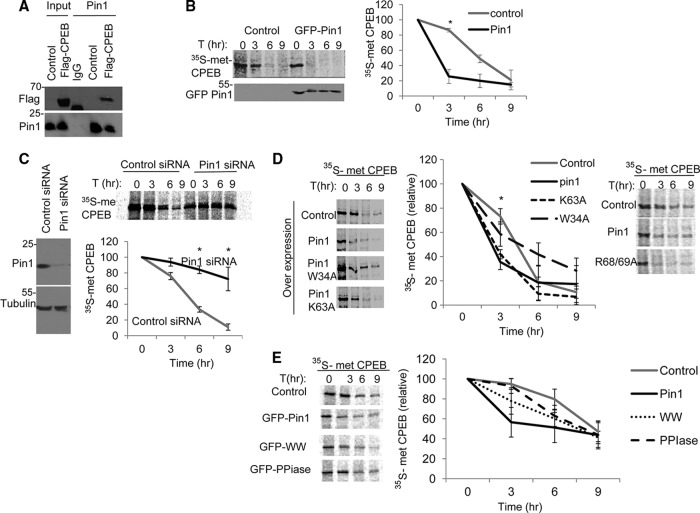
We next determined whether Pin1 regulates CPEB activity in mammalian cells. To do this, Flag-CPEB was ectopically expressed in HEK293 cells, followed by Pin1 immunoprecipitation and Western blotting for Flag. As shown in Fig. 5A, tagged CPEB was coimmunoprecipitated with Pin1. We also pulse-labeled cells coexpressing Flag-CPEB and GFP-Pin1, or GFP as a control, with [35S]methionine, followed by a chase with radio-inert methionine. CPEB was then immunoprecipitated and analyzed by SDS-PAGE. Ectopic Pin1 reduced the half-life of CPEB by >50%, from ∼6 h in control cells to less than 3 h in GFP-Pin1–expressing cells (Fig. 5B). Conversely, depletion of Pin1 with siRNA resulted in a 2-fold increase in the CPEB half-life, from 5 h to almost 9 h (Fig. 5C). Therefore, Pin1 regulates CPEB destruction in mammalian cells and in Xenopus oocytes.
Fig 5.
Pin1 controls CPEB destruction in mammalian cells. (A) HEK293T cells were transfected with Flag-CPEB, followed by IgG or Pin1 immunoprecipitation and Western analysis for Flag and Pin1. (B) Cells cotransfected with Flag-CPEB and either control GFP vector or GFP-Pin1 were pulse-labeled with [35S]methionine, followed by a chase of radio-inert methionine for the indicated times. The extracts were subjected to Flag immunoprecipitation and analyzed by SDS-PAGE and autoradiography. (C) Cells were cotransfected with Flag-CPEB and either control or Pin1 siRNA followed by pulse-labeling with 35S-methionine and chased with radio-inert methionine for the times indicated, immunoprecipitated for Flag and analyzed by SDS-PAGE and autoradiography. The quantification of 3 experiments is shown on the bottom panel. (D) Cells expressing ectopic WT or W34A or K34A mutant Pin1 proteins (left panel) or R68/69A double mutant Pin1 protein (right panel) were [35S]methionine labeled, followed by a chase of radio-inert methionine, and then immunoprecipitated for Flag-CPEB. (E) Cells expressing ectopic full-length Pin1, Pin1 WW domain, or Pin1 PPIase domain were [35S]methionine labeled, followed by a chase of radio-inert methionine, and then immunoprecipitated for Flag-CPEB. The quantification of three experiments is shown in the right panel.
To determine which of the Pin1 domains is required for regulating CPEB degradation, we repeated the pulse-chase experiment with the Pin1 W34A mutant protein in the WW domain, the Pin1 K63A mutant protein, and the Pin1 R68/69A double mutant protein in the PPIase domain (Fig. 5D). As expected, ectopic GFP-Pin1 expression decreased the CPEB half-life as above. However, in the presence of the Pin1 W34A mutant protein, the CPEB half-life remained mainly unchanged compared to the control ectopic GFP-Pin1-expressing cells. Interestingly, neither the K63A mutant protein nor the R68/69A double mutant protein had a significant effect compared to GFP-Pin1 in that they both significantly reduced the CPEB half-life. To examine whether the Pin1 WW domain is sufficient to induce CPEB destruction, we performed the [35S]methionine pulse-chase experiment as before using ectopic expression of the WW domain or the PPIase domain. In contrast to full Pin1 expression, ectopic expression of either domain had a smaller effect on the CPEB half-life, indicating that both of the domains are necessary for full Pin1 activity.
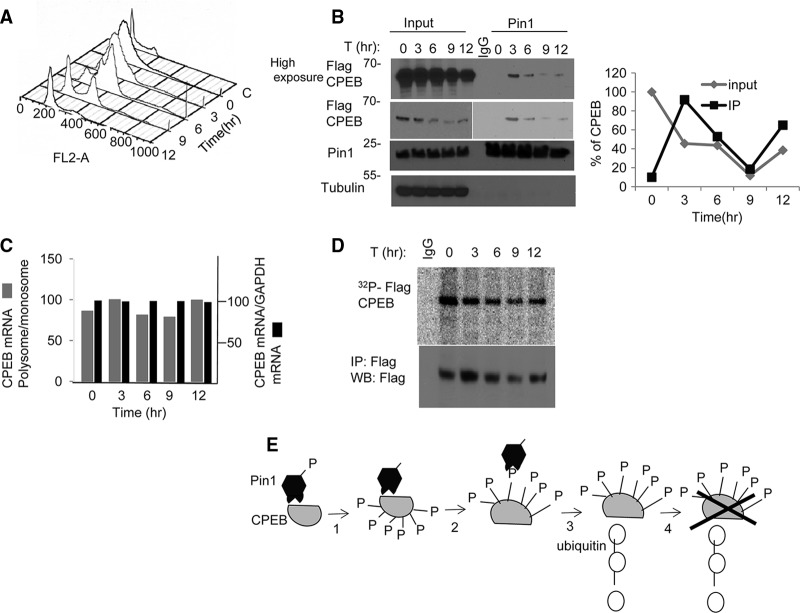
CPEB becomes phosphorylated and partially destroyed during oocyte maturation, a phase of meiosis that closely resembles the G2-to-M phase transition in mitotic cells. To assess whether CPEB is similarly phosphorylated and destroyed at M phase in the mammalian cell cycle, HEK293 cells were transduced with Flag-CPEB and then synchronized at S phase by a double thymidine block. The cells were then released from the block and monitored for cell cycle progression by flow cytometry (Fig. 6A) and for Flag-CPEB protein levels and CPEB-Pin1 interaction. The flow cytometry analysis shows that the G2-to-M transition occurred 6 to 9 h after entry into the cell cycle. Although there was no change in Pin1 levels at any phase, Flag-CPEB protein levels diminished by 9 h post-entry into the cell cycle (Fig. 6B). This reduction was not due to altered RNA levels, as measured by reverse transcriptase PCR (RT-PCR) (Fig. 6C), or a change in translation, as determined by the polysome/monosome ratio of Flag-CPEB mRNA (Fig. 6C).
Fig 6.
Cell cycle regulation of CPEB destruction by Pin1. (A) Fluorescence-activated cell sorter (FACS) analysis in HEK293T cells after release from double thymidine block. (B) Flag-CPEB transduced cells were synchronized by double thymidine block, released, and Pin1 immunoprecipitated at the times indicated and probed for Pin1 and Flag-CPEB. (C) Analysis of total CPEB RNA and the amount in polysomes (expressed as the polysome to monosome ratio), as quantified by quantitative reverse transcriptase PCR (qRT-PCR; average of two experiments) (D) As described for panel A, HEK293 cells were double thymidine blocked; 3 h prior to their cell cycle release, 10 μCi/ml [γ-32P]ATP was added to the culture media. At the indicated times, the cells were lysed and immunoprecipitated for Flag-CPEB. (E) Model for two-step interaction between CPEB and Pin1. In oocytes, unphosphorylated CPEB interacts with Pin1 that is phosphorylated on serine 71. Following progesterone stimulation, Pin1 is dephosphorylated and thereby activated; it then binds and isomerizes phospho-CPEB, resulting in ubiquitination of CPEB and its destruction.
To examine whether the Pin1-CPEB interaction correlates with CPEB destruction during cell cycle progression, Pin1 was immunoprecipitated at different phases of the cell cycle and Western blotted for Flag-CPEB. Figure 6B shows that, although there was very little interaction of Pin1 with CPEB at S phase (t = 0), a Pin1-CPEB association was clearly evident 3 h after release from the thymidine block. By 6 h and, especially, 9 h (G2 to M), when CPEB levels were low, there was a corresponding decrease in Pin1-CPEB interaction. At 12 h after the release, when the Flag-CPEB protein levels increased, there was a restoration of Pin1-CPEB association (Fig. 6B).
Because there is little information regarding CPEB phosphorylation during mitosis in mammalian cells, we double thymidine blocked the cells as before and labeled them in vivo by adding [γ-32P]ATP to the culture medium 3 h prior to release from the block. At the indicated times after the release, the cells were subjected to immunoprecipitation of Flag-CPEB, followed by autoradiography. As shown in Fig. 6D, CPEB was phosphorylated before the release, where the cells were S phase arrested. However, CPEB phosphorylation diminished by 9 h post-entry into the cell cycle, which corresponded to a reduced Pin1-CPEB interaction. These results indicate that, although CPEB is phosphorylated at S phase, it does not interact with Pin1. However, upon cell cycle progression from G2 to M, the CPEB-Pin1 interaction is evident, as was shown in Xenopus oocyte maturation upon hormonal induction. Taken together, we infer that, as shown for Xenopus oocytes, the interaction between CPEB and Pin1 leads to CPEB destruction in HEK293 cells.
Figure 6E summarizes the interaction of CPEB and Pin1. In immature oocytes, Pin1 is phosphorylated on serine 71 and therefore is enzymatically inactive. After progesterone treatment, Pin1 is dephosphorylated and then associates with CPEB that is phosphorylated on S/T-P residues. Pin1 then isomerizes one or more of these phosphoresidues, leading to ubiquitination and partial destruction of CPEB.
DISCUSSION
It may seem counterintuitive that CPEB destruction during oocyte maturation is necessary for stimulating the polyadenylation and translation of mRNAs. However, the polyadenylation of CPE-containing RNAs is hierarchical; that is, the polyadenylation of different mRNAs occurs in a temporal-specific manner that is governed by the levels of CPEB. For example, the single-CPE-containing c-mos mRNA is among the first to be polyadenylated while polyadenylation of the two-CPE-containing cyclin B1 mRNA takes place at a later time after CPEB has been partially destroyed (10, 29, 30). There are now thought to be a number of mRNAs whose time-dependent polyadenylation may be regulated by the number of CPEs they contain (31). Thus, the partial destruction of CPEB, which is necessary for oocyte development, is complex and involves multiple posttranslational modifications and recognition factors. Several phosphorylation events and two distinct pools of CPEB are involved in the regulation of its destruction. During oocyte maturation, CPEB becomes phosphorylated on 6 S/T-P pairs by cdk1, although the one on S210 is particularly important for protein destruction (10). CPEB also contains a PEST (proline, glutamic acid, serine, and threonine) domain, which often mediates the protein half-life. Within the PEST domain is a TSG (threonine, serine, and glycine) motif, which resembles the binding site for β-TrCP, the F-box protein of the SCF complex (28). Plx1, a polo-like kinase, recognizes a CPEB cdk1 phosphorylation site and in turn phosphorylates the TSG motif, which is recognized by β-TrCP that then ubiquitinates CPEB, resulting in proteasome-mediated destruction (28). Recent evidence also demonstrates that CPEB resides in two distinct pools in oocytes: one pool is bound to RNA while the second forms a homodimer through its RNA binding regions and thus does not bind RNA. The homodimers are destroyed very rapidly upon oocyte maturation, perhaps releasing essential factors to associate with the cytoplasmic polyadenylation complex (32).
In this study, we show that Pin1 is a new CPEB-interacting protein that mediates CPEB degradation. It was surprising to find that Pin1, which selectively recognizes and isomerizes phosphorylated S/T-P bonds, associates with CPEB prior to detectable CPEB phosphorylation. However, this finding is consistent with previous observations, indicating that Pin1 might have two different modes of protein interaction: a preferred one that is phosphorylation dependent and a secondary one that is phosphorylation independent (33, 34). Abrahamsen et al. (33) showed that, upon Pin1 interaction with and subsequent isomerization of protein kinase C (PKC), the conventional PKC isozymes are converted into species that are rapidly ubiquitinated following phorbol ester stimulation. Interestingly, Abrahamsen et al. also showed that the Pin1-PKC interaction is based on a hydrophobic motif in the C-terminal segment of the substrate and does not require phosphorylation, even though the interaction is strengthened when the Pin1 target sites are phosphorylated after phorbol ester treatment. This observation is consistent with our findings, indicating a two-step mode of Pin1-substrate interaction. Using a number of CPEB deletion mutants, we identified a site on CPEB corresponding to residues 48 to 183 that is necessary and sufficient for Pin1 interaction. It is worth noting that this domain (48 to 183) contains ∼30% hydrophobic amino acids, although they are not organized in any detectably conserved motif. Because the three-dimensional structure of CPEB is not known, we do not know which of these amino acids are solvent exposed and might serve as a platform for Pin1 interaction. An alternative hypothesis might be that several hydrophobic amino acids are needed to build a charged region for this interaction to occur. However, two additional regions of CPEB, the RRMs and residues 211 to 290, abrogate Pin1 interaction when deleted, which suggests that the tertiary structure of CPEB or multiple contact points are important for Pin1 binding prior to CPEB phosphorylation. Furthermore, we show that, at least in vitro, this first phase interaction is dependent on the WW domain of Pin1 (Fig. 2A). Although surprising, this first phosphorylation-independent interaction that does not induce substrate destruction may serve another purpose. Consider that Xenopus oocytes are exceptionally large (volume of 1 μl) and that CPEB must be rapidly destroyed after progesterone treatment. Therefore, to facilitate this rapid destruction, CPEB is “preloaded” with Pin1, where it may function as a molecular switch to coordinate subsequent cellular processes like maturation. Indeed, other proteins in the CPEB complex are subjected to cdk1-mediated phosphorylation upon progesterone stimulation and also might be subjected to Pin1-mediated regulation. Maskin, for example, is an eIF4E binding protein that prevents the assembly of the initiation complex (eIF4E-eIF4G) on the 5′ end of the mRNA, thus inhibiting translation (7, 8). Upon progesterone stimulation, maskin is phosphorylated by cdk1, inducing its dissociation from eIF4E, thereby ensuring the translation initiation complex assembly. CPEB destruction and maskin dissociation need to occur simultaneously to activate the translation of these mRNAs. Therefore, Pin1 assembly into this complex before progesterone treatment will ensure rapid isomerization of both CPEB and maskin for this manner.
Following the induction of oocyte maturation, nearly simultaneous CPEB phosphorylation on S/T-P pairs and Pin1 dephosphorylation of serine 71 cause the Pin1 interaction with CPEB to become CPEB phosphorylation dependent. We suspect that the affinity of Pin1 for CPEB might be higher when the substrate is phosphorylated because the ratio of immunoprecipitated CPEB to coprecipitated Pin1 is greater after maturation than before. However, this inference is somewhat speculative because we cannot perform an in vitro kinetic analysis of purified components due to the fact that it is difficult to isolate soluble recombinant CPEB.
This change in interaction sites and the isomerization of the phosphorylated residues results in CPEB ubiquitination and subsequent destruction. Accordingly, Pin1 dephosphorylation and subsequent enzymatic activation is a crucial step in CPEB ubiquitination. Although the inhibitory serine 71 phosphorylation is catalyzed by DAPK1 in mammalian cells (17), we do not know whether the same kinase acts in Xenopus oocytes. In addition, we find that serine 16 is not phosphorylated, but that its mutation abrogates serine 71 dephosphorylation during oocyte maturation. This result might indicate that serine 16 is necessary and acts as a docking site for recruitment of a phosphatase such as PP2A to Pin1 during maturation; such an enzyme could then dephosphorylate serine 71 for Pin1 activation. Irrespective of the kinase/phosphatase involved, an intriguing question is how does the Pin1-induced conformational change in CPEB control ubiquitination? Liou et al. (12) suggested that cis-trans isomerization of Pin1 substrates establishes a conformation needed for E3 ligase recognition, and possibly other regulatory proteins such as phosphatases (35, 36). Our data support this hypothesis and suggest that, upon Pin1 activation and simultaneously CPEB phosphorylation, Pin1 induces a CPEB conformational change that favors its interaction with the E3 ligase β-TrCP.
In maturing mouse oocytes, CPEB undergoes phosphorylation/destruction similar to that in Xenopus oocytes (37, 38). In mitotically cycling Xenopus embryonic cells, polyadenylation increases during M phase and decreases during interphase, which also occurs in cyclin mammalian cells (39). This observation prompted us to investigate whether CPEB undergoes an M phase destruction (like that which occurs during oocyte maturation, an M phase progression) and, if so, whether Pin1 might be involved. Indeed, Fig. 6 demonstrates that, in synchronized HEK293T cells, CPEB is partially destroyed at M phase and interacts with Pin1 prior to its destruction (as in Xenopus oocytes). However, it should not be assumed that maturing Xenopus oocytes and cycling mammalian cells are equivalent vis-à-vis cell cycle progression or by how mRNA translation is regulated. For example, oocytes are arrested at a G2-like phase and then enter M phase; there is no S phase in maturing oocytes. Xenopus CPEB is hyperphosphorylated only during the G2-to-M phase transition. At the same time, Pin1-CPEB interaction is evident before and after maturation. In mammalian cells, CPEB is phosphorylated at S phase and during other phases of the cell cycle. However, the CPEB-Pin1 interaction is only evident after cell cycle progression into the G2-to-M phase. The lack of Pin1-CPEB interaction in S phase likely indicates that the phospho-CPEB residues are not involved in recruiting Pin1. Despite the differences between these two systems, in both cases, the CPEB-Pin1 interaction is evident during the G2-to-M transition and precedes CPEB destruction. Moreover, ectopic Pin1 expression dramatically reduced the CPEB half-life while ectopic expression of a dominant negative Pin1 W34A mutant protein partially prevented CPEB destruction; mutations in the PPIase domain had no effect on Pin1's capability to promote CPEB destruction.
Fujimoto et al. (40) previously reported that Pin1 may affect the function and degradation of a target protein, peroxisome proliferator-activated receptor γ (PPARγ), solely by the interaction between the N-terminal activation function 1 (AF-1) domain of PPARγ and the WW domain in Pin1. In this case, the proline isomerization by the PPIase domain seemed to be dispensable. This prompted us to investigate the contribution of each Pin1 domain separately on CPEB destruction. We found that, in contrast to full-length Pin1, expression of either the WW or PPIase domain had minor effects on the CPEB half-life, indicating that both domains are needed for full Pin1 activity to promote CPEB destruction. These results may also indicate that other amino acids in the PPIase domain, such as C113, which was shown to be important for Pin1 isomerization activity (40), are needed for Pin1-mediated CPEB destruction.
Based on the functions of its interacting substrates, Liou et al. (12) suggest that Pin1 induces the destruction or inactivation of tumor suppressors but enhances the stability of proteins involved in malignant transformation. Although we have focused most of this present work on Xenopus oocytes, it is important to note that CPEB might be considered to be a tumor suppressor, and thus Pin1 could have important implications for oncogenic transformation via translation. That is, CPEB knockout (KO) mice, while not developing cancer spontaneously, contract papillomas much more readily than wild-type animals when challenged with a DNA damaging agent (41). Moreover, mouse embryo fibroblasts (MEFs) lacking CPEB do not senesce as do wild-type MEFS, but instead are immortal (42, 43). Primary human cells depleted of CPEB also bypass senescence (41, 44). Because the lack of CPEB leads to reduced translation of p53 mRNA, and because reduced p53 immortalizes primary cells, it may be inferred that CPEB can also act as a tumor suppressor. Although we do not know whether Pin1 control of CPEB destruction leads to a bypass of senescence, we speculate that it might and are presently investigating this phenomenon.
ACKNOWLEDGMENTS
We thank Kun Ping Lu (Beth Israel Deaconess Medical Center and Harvard University) for kindly providing GFP-Pin1 and the GFP-Pin1 W34A and K63A mutant proteins, as well as the GFP-WW- and GFP-PPIase-encoding plasmids. We also thank Frank Eckerdt (Department of Gynecology and Obstetrics, J. W. Goethe University) for kindly providing the GST-Pin1- and GST-Pin1 WW domain-encoding plasmids.
This work was supported by NIH grant GM46779. Additional core support from the Diabetes Endocrinology Research Center (grant P30 DK32520) is gratefully acknowledged.
M.N. performed all experiments, C.-L.L. generated the CPEB deletion plasmids, and M.N. and J.D.R. designed the experiments and wrote the manuscript.
We declare that we have no conflicts of interest.
Footnotes
Published ahead of print 22 October 2012
REFERENCES
- 1. Richter JD. 2007. CPEB: a life in translation. Trends Biochem. Sci. 32: 279–285 [DOI] [PubMed] [Google Scholar]
- 2. Lin CL, Evans V, Shen S, Xing Y, Richter JD. 2010. The nuclear experience of CPEB: implications for RNA processing and translational control. RNA 16: 338–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mendez R, Hake LE, Andresson T, Littlepage LE, Ruderman JV, Richter JD. 2000. Phosphorylation of CPE binding factor by Eg2 regulates translation of c-mos mRNA. Nature 404: 302–307 [DOI] [PubMed] [Google Scholar]
- 4. Sarkissian M, Mendez R, Richter JD. 2004. Progesterone and insulin stimulation of CPEB-dependent polyadenylation is regulated by Aurora A and glycogen synthase kinase-3. Genes Dev. 18: 48–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barnard DC, Ryan K, Manley JL, Richter JD. 2004. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119: 641–651 [DOI] [PubMed] [Google Scholar]
- 6. Kim JH, Richter JD. 2006. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell 24: 173–183 [DOI] [PubMed] [Google Scholar]
- 7. Cao Q, Richter JD. 2002. Dissolution of the maskin-eIF4E complex by cytoplasmic polyadenylation and poly(A)-binding protein controls cyclin B1 mRNA translation and oocyte maturation. EMBO J. 21: 3852–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao Q, Kim JH, Richter JD. 2006. CDK1 and calcineurin regulate Maskin association with eIF4E and translational control of cell cycle progression. Nat. Struct. Mol. Biol. 13: 1128–1134 [DOI] [PubMed] [Google Scholar]
- 9. Mendez R, Murthy KG, Ryan K, Manley JL, Richter JD. 2000. Phosphorylation of CPEB by Eg2 mediates the recruitment of CPSF into an active cytoplasmic polyadenylation complex. Mol. Cell 6: 1253–1259 [DOI] [PubMed] [Google Scholar]
- 10. Mendez R, Barnard D, Richter JD. 2002. Differential mRNA translation and meiotic progression require Cdc2-mediated CPEB destruction. EMBO J. 21: 1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hunter T. 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell 28: 730–738 [DOI] [PubMed] [Google Scholar]
- 12. Liou YC, Zhou XZ, Lu KP. 2011. Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem. Sci. 36: 501–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weissman AM, Shabek N, Ciechanover A. 2011. The predator becomes the prey: regulating the ubiquitin system by ubiquitylation and degradation. Nat. Rev. Mol. Cell Biol. 12: 605–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gutierrez GJ, Ronai Z. 2006. Ubiquitin and SUMO systems in the regulation of mitotic checkpoints. Trends Biochem. Sci. 31: 324–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Silverman JS, Skaar JR, Pagano M. 2012. SCF ubiquitin ligases in the maintenance of genome stability. Trends Biochem. Sci. 37: 66–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Berger M, Stahl N, Del Sal G, Haupt Y. 2005. Mutations in proline 82 of p53 impair its activation by Pin1 and Chk2 in response to DNA damage. Mol. Cell. Biol. 25: 5380–5388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee TH, Pastorino L, Lu KP. 2011. Peptidyl-prolyl cis-trans isomerase Pin1 in ageing, cancer and Alzheimer disease. Expert Rev. Mol. Med. 13: e21 doi:http://dx.doi.org/10.1017/S1462399411001906 [DOI] [PubMed] [Google Scholar]
- 18. Nakamura K, Greenwood A, Binder L, Bigio EH, Denial S, Nicholson L, Zhou XZ, Lu KP. 2012. Proline isomer-specific antibodies reveal the early pathogenic tau conformation in Alzheimer's disease. Cell 149: 232–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. 2003. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell 112: 243–256 [DOI] [PubMed] [Google Scholar]
- 20. Hake LE, Richter JD. 1994. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79: 617–627 [DOI] [PubMed] [Google Scholar]
- 21. Nechama M, Uchida T, Mor Yosef-Levi I, Silver J, Naveh-Many T. 2009. The peptidyl-prolyl isomerase Pin1 determines parathyroid hormone mRNA levels and stability in rat models of secondary hyperparathyroidism. J. Clin. Invest. 119: 3102–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, Botstein D. 2002. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell 13: 1977–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stebbins-Boaz B, Cao Q, de Moor CH, Mendez R, Richter JD. 1999. Maskin is a CPEB-associated factor that transiently interacts with elF-4E. Mol. Cell 4: 1017–1027 [DOI] [PubMed] [Google Scholar]
- 24. Lu PJ, Zhou XZ, Liou YC, Noel JP, Lu KP. 2002. Critical role of WW domain phosphorylation in regulating phosphoserine binding activity and Pin1 function. J. Biol. Chem. 277: 2381–2384 [DOI] [PubMed] [Google Scholar]
- 25. Lee TH, Chen CH, Suizu F, Huang P, Schiene-Fischer C, Daum S, Zhang YJ, Goate A, Chen RH, Zhou XZ, Lu KP. 2011. Death-associated protein kinase 1 phosphorylates Pin1 and inhibits its prolyl isomerase activity and cellular function. Mol. Cell 42: 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eckerdt F, Yuan J, Saxena K, Martin B, Kappel S, Lindenau C, Kramer A, Naumann S, Daum S, Fischer G, Dikic I, Kaufmann M, Strebhardt K. 2005. Polo-like kinase 1-mediated phosphorylation stabilizes Pin1 by inhibiting its ubiquitination in human cells. J. Biol. Chem. 280: 36575–36583 [DOI] [PubMed] [Google Scholar]
- 27. Reverte CG, Ahearn MD, Hake LE. 2001. CPEB degradation during Xenopus oocyte maturation requires a PEST domain and the 26S proteasome. Dev. Biol. 231: 447–458 [DOI] [PubMed] [Google Scholar]
- 28. Setoyama D, Yamashita M, Sagata N. 2007. Mechanism of degradation of CPEB during Xenopus oocyte maturation. Proc. Natl. Acad. Sci. U. S. A. 104: 18001–18006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ballantyne S, Daniel DL, Jr, Wickens M. 1997. A dependent pathway of cytoplasmic polyadenylation reactions linked to cell cycle control by c-mos and CDK1 activation. Mol. Biol. Cell 8: 1633–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. de Moor CH, Richter JD. 1997. The Mos pathway regulates cytoplasmic polyadenylation in Xenopus oocytes. Mol. Cell. Biol. 17: 6419–6426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Piqué M, López JM, Foissac S, Guigó R, Méndez R. 2008. A combinatorial code for CPE mediated translational control. Cell 132: 434–448 [DOI] [PubMed] [Google Scholar]
- 32. Lin CL, Huang YT, Richter JD. 2012. Transient CPEB dimerization and translational control. RNA 18: 1050–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abrahamsen H, O'Neill AK, Kannan N, Kruse N, Taylor SS, Jennings PA, Newton AC. 2012. Peptidyl-prolyl isomerase Pin1 controls down-regulation of conventional protein kinase C isozymes. J. Biol. Chem. 287: 13262–13278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Crenshaw DG, Yang J, Means AR, Kornbluth S. 1998. The mitotic peptidyl-prolyl isomerase, Pin1, interacts with Cdc25 and Plx1. EMBO J. 17: 1315–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Werner-Allen JW, Lee CJ, Liu P, Nicely NI, Wang S, Greenleaf AL, Zhou P. 2011. cis-Proline-mediated Ser(P)5 dephosphorylation by the RNA polymerase II C-terminal domain phosphatase Ssu72. J. Biol. Chem. 286: 5717–5726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhou XZ, Kops O, Werner A, Lu PJ, Shen M, Stoller G, Küllertz G, Stark M, Fischer G, Lu KP. 2000. Pin1-dependent prolyl isomerization regulates dephosphorylation of Cdc25C and tau proteins. Mol. Cell 6: 873–883 [DOI] [PubMed] [Google Scholar]
- 37. Hodgman R, Tay J, Mendez R, Richter JD. 2001. CPEB phosphorylation and cytoplasmic polyadenylation are catalyzed by the kinase IAK1/Eg2 in maturing mouse oocytes. Development 128: 2815–2822 [DOI] [PubMed] [Google Scholar]
- 38. Tay J, Hodgman R, Richter JD. 2000. The control of cyclin B1 mRNA translation during mouse oocyte maturation. Dev. Biol. 221: 1–9 [DOI] [PubMed] [Google Scholar]
- 39. Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD. 2002. Translational control of the embryonic cell cycle. Cell 109: 473–483 [DOI] [PubMed] [Google Scholar]
- 40. Fujimoto Y, Shiraki T, Horiuchi Y, Waku T, Shigenaga A, Otaka A, Ikura T, Igarashi K, Aimoto S, Tate S, Morikawa K. 2010. Proline cis/trans-isomerase Pin1 regulates peroxisome proliferator-activated receptor gamma activity through the direct binding to the activation function-1 domain. J. Biol. Chem. 285: 3126–3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Burns DM, Richter JD. 2008. CPEB regulation of human cellular senescence, energy metabolism, and p53 mRNA translation. Genes Dev. 22: 3449–3460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Groisman I, Ivshina M, Marin V, Kennedy NJ, Davis RJ, Richter JD. 2006. Control of cellular senescence by CPEB. Genes Dev. 20: 2701–2712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Groppo R, Richter JD. 2011. CPEB control of NF-kappaB nuclear localization and interleukin-6 production mediates cellular senescence. Mol. Cell. Biol. 31: 2707–2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Burns DM, D'Ambrogio A, Nottrott S, Richter JD. 2011. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature 473: 105–108 [DOI] [PMC free article] [PubMed] [Google Scholar]