Abstract
Understanding the mechanisms that drive the differentiation of dopaminergic (DA) neurons is crucial for successful development of novel therapies for Parkinson's disease, in which DA neurons progressively degenerate. However, the mechanisms underlying the differentiation-promoting effects of Wnt5a on DA precursors are poorly understood. Here, we present the molecular and functional characterization of a signaling pathway downstream of Wnt5a, the Wnt/Dvl/Rac1 pathway. First, we characterize the interaction between Rac1 and Dvl and identify the N-terminal part of Dvl3 as necessary for Rac1 binding. Next, we show that Tiam1, a Rac1 guanosine exchange factor (GEF), is expressed in the ventral midbrain, interacts with Dvl, facilitates Dvl-Rac1 interaction, and is required for Dvl- or Wnt5a-induced activation of Rac1. Moreover, we show that Wnt5a promotes whereas casein kinase 1 (CK1), a negative regulator of the Wnt/Dvl/Rac1 pathway, abolishes the interactions between Dvl and Tiam1. Finally, using ventral midbrain neurosphere cultures, we demonstrate that the generation of DA neurons in culture is impaired after Tiam1 knockdown, indicating that Tiam1 is required for midbrain DA differentiation. In summary, our data identify Tiam1 as a novel regulator of DA neuron development and as a Dvl-associated and Rac1-specific GEF acting in the Wnt/Dvl/Rac1 pathway.
INTRODUCTION
Wnt ligands are known to activate a number of highly evolutionary conserved pathways that regulate various essential aspects of embryo development and adult tissue homeostasis. The Wnt/β-catenin pathway, also referred to as the canonical Wnt signaling pathway, is the most intensively studied and best defined pathway (1, 2). In this study, we focus on Wnt signaling pathways independent of β-catenin, in particular on Wnt-mediated activation of small GTPases (3, 4). These pathways are also involved in many aspects of embryogenesis, such as proper morphogenesis and differentiation of neuronal tissue (5), including the ventral midbrain (6, 7), the area where dopaminergic (DA) neurons are born. We and others have previously shown that Wnt5a activates a β-catenin-independent signaling in DA cells (8) and promotes DA differentiation of primary ventral midbrain precursor cells, as well as of mouse neural and embryonic stem cells (9–12).
To date, the precise molecular mechanisms underlying the prodifferentiation effects of Wnt5a and the signaling pathways activated by Wnt5a in DA cells, as well as in other cell types, are only beginning to emerge. We recently demonstrated that the prodifferentiation effects of Wnt5a require the activity of the Rac1 GTPase (6). Rac1 belongs to the Rho family of small GTPases, and its activity is controlled by guanosine exchange factors (GEFs), GTPase activity-activating proteins (GAPs), and guanine nucleotide exchange inhibitors (GDIs) (13, 14). While Wnt ligands are known to trigger activation of various small GTPases from the Rho and Ras families in various cell types (4), evidence for the employment of specific GEFs in Wnt5a/Rac1 signaling is missing, and the mechanism of small GTPase activation in context of Wnt signaling is poorly understood.
In the present study, we aimed to elucidate the mechanism of Rac1 activation in the Wnt5a/Rac1 pathway. Earlier work proposed that Rac1 interacts with Dishevelled (Dvl) (15, 16), a cytosolic protein with three homologs in mammals. Dvl is a critical component of Wnt-driven signaling, acts as a scaffolding protein associated with many different intracellular proteins (17, 18), and is phosphorylated by several kinases, such as casein kinase 1 (CK1) (9, 19).
In our study, we first characterize the Dvl-Rac1 interaction and show that the N-terminal part of Dvl3 is required for complex formation between Rac1 and Dvl3, as well as for the activation of Rac1. We next searched for Dvl interactors and found that the Rac1 GEF T-cell lymphoma invasion and metastasis 1 (Tiam1) (20) is a novel binding partner of Dvl. Moreover, using loss-of-function (LOF) approaches, we demonstrated that Tiam1 is functionally required for the activation of Rac1 by Wnt5a or Dvl, as well as for generation of DA neurons from DA progenitors in neurosphere cultures. In sum, we hereby provide biochemical and functional evidence that Tiam1 is a novel regulator of Wnt/Dvl/Rac1 signaling that is required for midbrain DA neuron differentiation.
MATERIALS AND METHODS
Cell culture, transfection, and treatments.
The mouse SN4741 DA cell line was propagated as described before (21). For the purpose of transient gene overexpression, cells were transfected with Superfect (Qiagen) according to the manufacturer's instructions. The following constructs were used: MYC-Dvl2 (22), FLAG-Dvl3, hemagglutinin (HA)-Dvl3 (23), Dvl2-enhanced green fluorescent protein (EGFP) (24), HA-Dvl2 (wild type [wt] and M1, M2, and M4 mutants) (25), FLAG-Tiam1 (26), and MYC-Rac1 (27). For experiments involving small interfering RNA (siRNA), the SN4741 cells were transfected with 50 to 100 nM siGENOME Tiam1 siRNA (SMARTpool M-047-08-01) or nontargeting siRNA (both from Dharmacon) by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. The efficiency of the silencing was assessed by Western blotting or quantitative PCR (qPCR). For analysis of cellular signaling, the cells were stimulated with 100 ng/ml of mouse Wnt5a (R&D Systems) for 2.5 h. Control stimulations were done with vehicle (0.1% bovine serum albumin [BSA], 0.05% CHAPS in phosphate-buffered saline [PBS]). HEK293A cells, T98G cells, U78MG cells, or mouse embryonic fibroblasts (MEFs) were grown in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal calf serum (FCS), 2 mM l-glutamine, 50 units/ml penicillin, and 50 units/ml streptomycin. Transient transfections of HEK293A cells were carried out with polyethylimin (PEI) with a ratio of 1 μg DNA per 3 μl of PEI in 100 ul serum-free medium, incubating the mixture for 10 min at room temperature, and adding the mixture dropwise to HEK293A cells in their culture medium. The following constructs, in addition to those listed above, were used: FLAG-Dvl3 deletion mutants (23). Inhibitors of CK1 (D4476, Calbiochem; PF-670462, Tocris Bioscience) were used as described previously (28, 29), where indicated.
Immunoprecipitation, Western blotting, and densitometry analyses.
HEK293A cells (24 to 36 h after transfection), SN4741 cells, T98G cells, or U78MG cells (endogenous immunoprecipitation [IP]) were harvested for total cell extract in RIPA buffer (150 mM NaCl, 50 mM Tris-Cl [pH 7.4], 1 mM EDTA, 0.5% NP-40, 1× protease inhibitor cocktail [Roche]) or RIPA plus 0.05% SDS buffer (for Rac1 immunoprecipitations). Whole adult brain extract has been prepared by mechanical dissociation of brain tissue and subsequent lysis in complete RIPA buffer. Cell extracts were cleared from cell debris by centrifugation (16,000 × g for 15 min at +4°C), and supernatants were incubated with protein G-Sepharose (GE Healthcare) (1 h at +4°C in an orbital shaker) to pull down nonspecific interactors. Subsequently, precleared extracts obtained after centrifugation (200 × g for 5 min at +4°C) were incubated with 1 μg (10 μg in case of IPs for mass spectrometry [MS] analyses) of the following antibodies: mouse monoclonal anti-Rac1 (clone 23A8; Millipore), rabbit polyclonal anti-Tiam1 (sc-872), mouse monoclonal anti-MYC (sc-40), mouse monoclonal anti-Dvl2 (sc-8026), mouse monoclonal anti-Dvl3 (sc-8027) (Santa Cruz Biotechnology), rabbit polyclonal anti-FLAG (F7425; Sigma) antibodies, for 30 min at +4°C in an orbital shaker and subsequently with protein G-Sepharose. Following overnight incubation, samples were rigorously washed with RIPA buffer and subsequently analyzed by Western blotting. Sample preparation and Western blotting was done as described before (30). Luminescence was detected by either film exposure (Agfa), in the case of Fig. 3G and H and 4B to E, or the ChemiDoc XRS system (Bio-Rad). Nonsaturated images, within dynamic range of the charge-coupled-device (CCD) camera, were chosen as representative and used for densitometric analyses. The following antibodies were used, in addition to those described before: mouse monoclonal anti-FLAG antibody (M2; Sigma), rabbit polyclonal anti-Dvl2 antibody (sc-13974; Santa Cruz Biotechnology), horseradish peroxidase (HRP)-conjugated anti-mouse secondary antibody (GE Healthcare), and HRP-conjugated anti-rabbit secondary antibody (Sigma).
GTPase pulldown assay.
Cells were harvested in GTPase lysis buffer (150 mM NaCl, 10 mM Tris-Cl [pH 7.4], 10 mM MgCl2, 1% Triton X-100, 0.1% SDS, 1 mM dithiothreitol [DTT], 1× protease inhibitor cocktail [Roche]). The pulldowns were performed with recombinant GST-PAK-CRIB as described before (31). For experiments with Tiam1 siRNA, the SN4741 cells were seeded out (750,000 cells/6-well plate), transfected first with either control or Tiam1 siRNA 24 h after seeding, and then transfected with the indicated expression constructs after an additional 24 h, processed 24 h after the last transfection, and subsequently analyzed by Western blotting. In experiments involving Dvl overexpression, MYC-Rac1 was cotransfected, and changes in Rac1 activity were subsequently measured for MYC-Rac1. A densitometry analysis was performed using Image J software, where indicated. The level of activated Rac1 (GTP-Rac1) was normalized to the level of Rac1 in total cell lysate (TCL).
LC-MS/MS and data mining.
Following the coimmunoprecipitation (extracts from 5- by 15-cm plates of 80 to 90% confluent MEFs were used per one condition), samples were washed in detergent-free RIPA buffer and subjected to proteomics grade trypsin digest (10 ng/μl at 37°C for 12 h) (Sigma). Liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed on a NanoAcquity ultraperformance liquid chromatograph (UPLC; Waters) coupled to an ESI Q-Tof Premier (Waters) mass spectrometer. Digested peptides were first desalted using a ZipTip C18 column (Millipore), diluted in MQ water, and loaded onto a nanoAcquity UPLC symmetry trap column (Waters) packed with 5 μm BEH C18 beads. Peptides were eluted through a nanoAcquity (Waters) analytical column packed with 1.7 μm BEH C18 beads at a flow rate of 400 nl/min using a gradient of 3 to 40% acetonitrile with 0.1% formic acid for 35 min. Effluent was directly fed into the ESI source of the MS instrument. Raw data were acquired in MSE Identity mode and later subjected to a search using UniProt and NCBI mouse protein database by the PLGS2.3 software (Waters). Acetyl N terminus, deamidation N and Q, carbamidomethyl C, and oxidation M were set as variable modifications. Peptide accuracy and MS/MS fragment mass accuracy was set to less than 20 ppm. Generated hit lists were manually curated to remove common contaminants and hits found in IgG IP samples.
Immunocytochemistry and confocal microscopy.
SN4741 cells (20,000 to 40,000 cells/well of a 24-well plate) were grown overnight on glass coverslips and transfected with the indicated plasmids. Twenty-four hours posttransfection, cells were fixed in 4% paraformaldehyde (15 min at +4°C), serum blocked, and incubated in the appropriate primary and, subsequently, secondary antibodies as previously described (32). The following antibodies were used: mouse monoclonal anti-Rac1 (1:100; Millipore), rabbit polyclonal anti-FLAG (1:1,000; Sigma), rabbit polyclonal anti-MYC, mouse monoclonal anti-MYC (both 1:1,000; Santa Cruz Biotechnology), Alexa Fluor 488–goat anti-mouse or anti-rabbit, and Alexa Fluor 555–donkey anti-mouse or anti-rabbit (all 1:1,000; Molecular Probes, Invitrogen) antibodies. Fluorescent labeling was examined using a Zeiss LSM5 exciter inverted confocal scanning laser microscope.
Precursor cultures.
Ventral midbrain (VM) precursor cultures were grown as neurospheres, as previously described (12). Briefly, VMs were dissected from E10.5 mouse embryos, dissociated with collagenase/dispase (30 min at 37°C on a rocking platform), followed by mechanical trituration. Next, cells were plated at a density of 100,000 cells/cm2 in 1 ml of N2 medium (supplemented with 250 ng/nl Shh, 25 ng/ml fibroblast growth factor 8 [FGF8], and 20 ng/ml FGF2) and grown as neurospheres for 7 days. A total of 0.5 ml of fresh medium was added every second day. At day 7, spheres were collected and broken into small cell clusters (collagenase/dispase treatment plus mechanical trituration), which were then seeded at a density of 100,000 cells/cm2 on poly-d-lysine–laminin-precoated plates in N2 medium (no growth factors) and transfected with appropriate siRNA (50 to 100 nM) using Lipofectamine 2000. Fresh N2 medium was added 24 h after transfection (supplemented with brain-derived neurotrophic factor (BDNF) and glial cell-derived neurotrophic factor (GDNF) to a final concentration 20 ng/ml of each), and cells were harvested 3 days after the transfection.
For experiments with lentiviral particles, the VM tissue was dissected and plated as described above. The cells were transduced immediately after dissection in N2 medium containing Shh, FGF2, FGF8, and polybrene (2 μg/ml). After 24 h, the medium was replaced with a fresh one. Cells were grown as neurospheres for 5 days, with the addition of fresh medium every 2 to 3 days. At day 5, spheres were collected and seeded at a density of 50 spheres/cm2 on PDL-laminin-precoated plates in N2 medium (with BDNF and GDNF, 20 ng/ml each) and differentiated for 3 days.
The efficiency of the silencing was determined by qPCR. For immunocytochemistry experiments, the following antibodies were used: rabbit polyclonal anti-tyrosine hydroxylase (anti-TH) (1:500; Pel-Freeze), mouse monoclonal anti-βIII tubulin (Tuj, 1:1,000; Promega), mouse monoclonal anti-TH (1:400; Sigma), mouse anti-GFP (1:75; Millipore), and rabbit anti-GFP (1:800; Invitrogen) antibodies. Two or three wells per condition were analyzed in each experiment. The DA neuron numbers were counted as either the number of TH+ cells per area of Tuj+ cell clusters (10 randomly selected observation fields per well, siRNA experiments) or the number of TH+ cells per area of DAPI+ (4′,6-diamidino-2-phenylindole) clusters (all spheres within the well analyzed, short hairpin RNA [shRNA] experiments). The area of DAPI+ clusters was measured in square pixels and subsequently converted into square micrometers (1 square pixel = 0.6209 square μm, for 10× objective and 2× zoom).
CD1 mice (Charles River) were housed, fed, and sacrificed according to Karolinska Institute guidance for animal experiments and the ethical permits to E.A.
Production of lentiviral shRNA particles.
Third-generation replication-incompetent lentiviral vectors (pLKO.1-CMV-tGFP) encoding either nontargeting shRNA or Tiam1 shRNA (TRCN0000042594 and TRCN0000042595) were obtained from Sigma. The viral particles, vesicular stomatitis virus glycoprotein (VSVG) pseudotyped, were produced using the four-plasmid system as previously described (33). Forty-eight hours later, after transfection of HEK293T cells, the supernatant was collected and filtered. High-titer stocks were obtained by ultracentrifugation (50,000 × g for 2 h), and the pellets were resuspended in PBS with 1% BSA and stored at −80°C until further use. The titers used for primary cell transduction were in the range of 1 × 107 to 1.5 × 107 transducing units per ml, and the multiplicity of infection was 1 to 2.
Reverse transcription (RT)-PCR and quantitative PCR.
Total RNA from either SN4741 or differentiated VM cultures was extracted using the RNeasy extraction kit (Qiagen) according to the manufacturer's instructions. Preparation of cDNA, quantitative PCR (qPCR), and primers used in this study were described previously (10), with the exception of the following: Tiam1 primers (forward, GAG CCA GAG GGA GGC GTG GA; reverse, TGG CAT CCT GAG GGG ACG GG), Rac1 primers (forward, CGT CCC CTC TCC TAC CCG CA; reverse, CTT TCG CCA TGG CCA GCC CC). 18S was used as an internal control. Relative mRNA expression was calculated as a fold change versus the control.
Statistical analyses.
Statistical analyses (Student's t test, Mann-Whitney test, one-way analysis of variance [ANOVA] with Neumann-Keuls multiple comparison test, and Kolmogorov-Smirnov test) were performed using Prism 4 (GraphPad Software). P values of <0.05 were considered statistically significant differences. Results are presented as means ± standard errors of the means (SEM).
RESULTS
Characterization of the interaction between Rac1 and Dvl.
We previously found that Wnt5a treatment activates the small GTPase Rac1 in DA cells (6). Indeed, stimulation of SN4741 dopaminergic (DA) cells with recombinant Wnt5a increased the level of Rac1-GTP (activated Rac1) compared to vehicle-treated cells (Fig. 1A and B). It has been shown that Dishevelled (Dvl), a critical component of Wnt signaling machinery, is sufficient to increase Rac1 activity (15) and that it coimmunoprecipitates with Rac1 in hippocampal neurons (16). We thus decided to examine and characterize the possible interaction between Rac1 and Dvl in DA cells.
Fig 1.
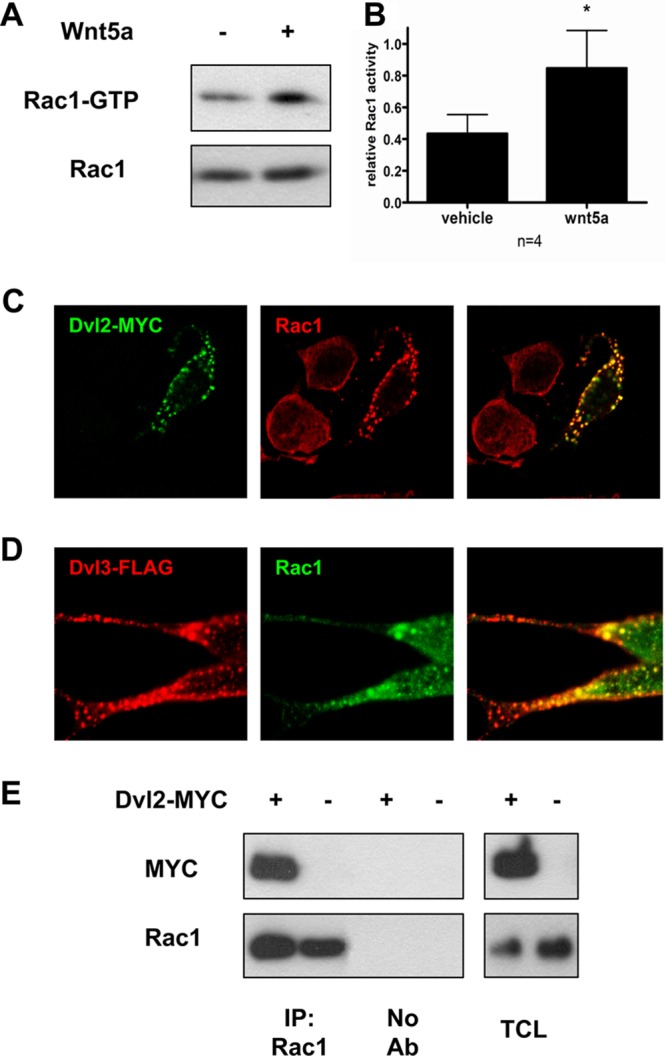
Dvl and Rac1 form a complex. (A) Wnt5a treatment (100 ng/ml; 2.5 h) triggers the activation of Rac1 (increase in Rac1-GTP) in SN4741 DA cells. (B) Quantification of the effect of Wnt5a on Rac1 activity, compared to vehicle treatment. The levels of Rac1-GTP were normalized to the total amount of Rac1 under each condition. (C) Colocalization of Rac1 (Alexa 555, red) and Dvl2-MYC (Alexa 488, green). Note the recruitment of endogenous Rac1 into immunoreactive puncta by expression of Dvl2-MYC. (D) Colocalization of Dvl3-FLAG (Alexa 555, red) and Rac1 (Alexa 488, green). (E) Dvl2-MYC coimmunoprecipitates with endogenous Rac1 in HEK293A cells. The right panel shows total cell lysate (TCL). *, P < 0.05 by Student t test.
As Dvl2 and Dvl3 are both involved in Wnt5a-mediated signaling in DA cells (9), we first analyzed the cellular distribution of Dvl2-MYC or Dvl3-FLAG after overexpression in SN4741 cells. Dvl, as a scaffolding protein, undergoes highly dynamic polymerization. This was reflected by formation of dynamic protein complexes in the cytosol (34), referred to as Dvl dots/puncta (35, 36). Interestingly, Dvl2-MYC/Dvl3-FLAG immmunoreactive puncta colocalized with endogenous Rac1 in cells where either Dvl2-MYC or Dvl3-FLAG was overexpressed (Fig. 1C and D), suggesting that endogenous Rac1 is recruited to the Dvl-containing protein complexes.
We next performed coimmunoprecipitation experiments in HEK293A cells and observed that endogenous Rac1 coimmunoprecipitates with both Dvl2-MYC (Fig. 1E) and Dvl3-FLAG (Fig. 2A) with similar efficiency. To get further insight into the interaction between Dvl and Rac1 and its possible functional consequences for Wnt signaling, the region of Dvl that mediates binding to Rac1 was determined using the FLAG-tagged deletion constructs of Dvl3 (23). The full-length Dvl3-FLAG and the three C-terminal deletion Dvl3 mutants coimmunoprecipitated with endogenous Rac1 (Fig. 2A and B). Importantly, however, deletion mutants lacking the N-terminal part, including the DIX domain, failed to be detected after Rac1 immunoprecipitation (see lanes 2 to 4 in Fig. 2A). Note that Dvl3 mutant 5, which lacks the C-terminal part, including the PDZ and DEP domains, showed somewhat weaker coimmunoprecipitation with Rac1 than the remaining Dvl mutants. These results indicated that the DIX domain of Dvl is required for its interaction with Rac1 and that this complex is possibly stabilized by interaction(s) with the C-terminal part of Dvl.
Fig 2.
The N-terminal part of Dvl is required to interact with Rac1. (A) Deletion mapping of the Dvl3 domain mediating the interaction with Rac1 in HEK293A cells. The FLAG-Dvl3 expression constructs used in coimmunoprecipitation experiments are showed in panel B (total cell lysates [TCL] shown to the right). Note that FLAG-Dvl3 pulldown after Rac1 immunoprecipitation (IP) was lost in Dvl3 mutants lacking the N-terminal part, including the DIX domain. Arrows point to the Dvl3-FLAG variants coimmunoprecipitating with endogenous Rac1. (B) Schematic representation of the constructs used in panel A and the results obtained. + or − symbols indicate the efficiency of coimmunoprecipitation for the different mutants. (C) HEK293A cells were transfected with the indicated expression constructs of Dvl2 harboring point mutations in the DIX domain. Subcellular localization of individual Dvl2 mutants has been determined by immunocytochemical staining using anti-HA antibody. (D) Coimmunoprecipitation of all Dvl2 constructs was detected after Rac1 immunoprecipitation but not if control IgG was used instead of an anti-Rac1 antibody. (E) HEK293A cells were transfected with the indicated Dvl constructs and Rac1-MYC. Rac1 activity was determined by the level of Rac1-GTP pulldown. Note that Rac1 was not activated by the Dvl3 ΔN-FLAG construct. (F) Summary of experiments shown in panel E. *, P < 0.05; **, P < 0.01 by Student t test (compared to mock).
Since the DIX domain of Dvl is required for the formation of Dvl-containing polymers (25), we tested whether the failure to interact with Rac1 could be due to impaired polymerization of Dvl. Several amino acids (Y27, F43, and V67 together with K68) within the DIX domain have been demonstrated as critical for Dvl polymerization and Wnt/β-catenin signaling (25). While Dvl2 DIX domain mutant M4 (Y27D) retained some of the capacity of Dvl2 to form large cytosolic complexes in SN4741 cells, mutants M1 (F43S) and M2 (V67A and K68A) completely failed to form such complexes (Fig. 2C) (25). Interestingly, M1 and M2 mutants retained both (i) the ability to coimmunoprecipitate with Rac1 (Fig. 2D), even if the M1 mutant was somewhat less efficient in coimmunoprecipitation, and (ii) the potential to induce Rac1 activation, as shown by the increased levels of Rac1-GTP (Fig. 2E and F). On the other hand, Dvl3-ΔN (construct 2 in Fig. 2B), which lacks the whole DIX domain, failed to activate Rac1 (Fig. 2E and F). These results indicate that the N terminus of Dvl, including the DIX domain (amino acids 1 to 81), is required for the Dvl-Rac1 interaction and for the Dvl-mediated Rac1 activation. In contrast, structural integrity of the DIX domain, which is absolutely crucial for DIX-mediated polymerization and Dvl function in Wnt/β-catenin signaling, is dispensable for activation of Rac1. These findings prompted us to examine alternative mechanisms of Rac1 activation by Dvl.
Tiam1, a Rac1 GEF, interacts with Dvl.
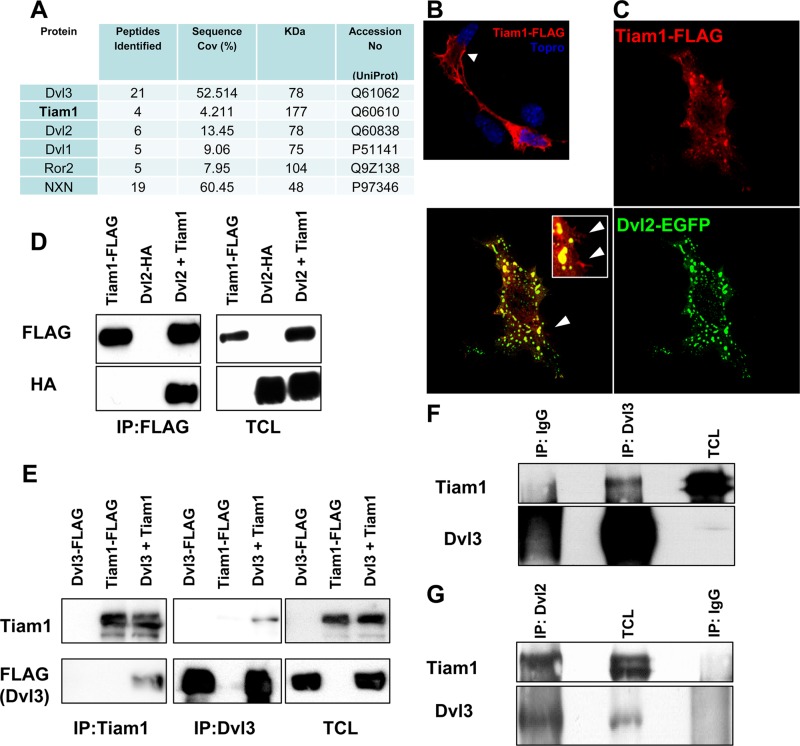
We next used a proteomic approach to identify endogenous interactors of Dvl that could be involved in the activation of Rac1. The PDZ domain-containing proteins have been previously found to act as critical scaffolds for the activation of GTPases from the Rho family (37, 38). In our experiments, we performed immunoprecipitation of Dvl3 in mouse embryonic fibroblasts (MEFs) and subsequent LS-MS/MS. In addition to previously described Dvl binding partners such as nucleoredoxin (NXN) (17) or Ror2 (17), we found T-cell lymphoma invasion and metastasis-inducing protein 1 (Tiam1) (Fig. 3A), which is a Rac1 GEF known to activate Rac1 (20).
Fig 3.
Tiam1 is a novel Dvl binding partner. (A) Tiam1 is present in Dvl3-associated complexes, identified by LC-MS/MS after endogenous Dvl3 coimmunoprecipitation. (B) Tiam1-FLAG protein (red) localizes both in the cytosol and at the membrane (arrow) upon overexpression in SN4741. Topro3 was used to counterstain the cell nucleus. (C) Coexpression of Dvl2-EGFP (green) and Tiam1-FLAG (red) in SN4741 cells leads to a cytoplasmic colocalization of Dvl2-EGFP and Tiam1-FLAG in puncta. Arrowheads show that a small pool of Tiam1-FLAG that did not colocalize with Dvl2-EGFP remained in the membrane. (D) Dvl2-HA was found to form a complex with Tiam1-FLAG, as assessed by immunoprecipitation with Tiam1-FLAG in HEK293A cells (first panel, last lane). The next panel shows total cell lysates (TCL). (E) Dvl3-FLAG was also immunoprecipitated by Tiam1-FLAG in HEK293A cells. The complex formed by Tiam1-FLAG and Dvl3-FLAG was detected by immunoprecipitation with either anti-Dvl3 or anti-Tiam1 antibodies. (F and G) Endogenous Dvl3 (F) or endogenous Dvl2 (G) were immunoprecipitated from whole adult mouse brain lysates, and the presence of Tiam1 has been detected with an anti-Tiam1 antibody. IgG, control antibody.
In order to verify the Dvl-Tiam1 interaction, we first overexpressed Tiam1-FLAG and Dvl2-EGFP in SN4741 DA cells and subsequently analyzed their subcellular localization by confocal microscopy. In cells transfected with Tiam1-FLAG only, Tiam1-FLAG was localized both in cytosol and at the membrane (Fig. 3B). Upon Dvl2-EGFP coexpression, the membrane-localized pool of Tiam1-FLAG was not significantly mobilized, but the cytosolic pool was efficiently relocated and colocalized with Dvl2-EGFP in cytosolic puncta. These findings thus supported the possibility of a Dvl-Tiam1 protein-protein interaction in the cytosolic puncta (Fig. 3C).
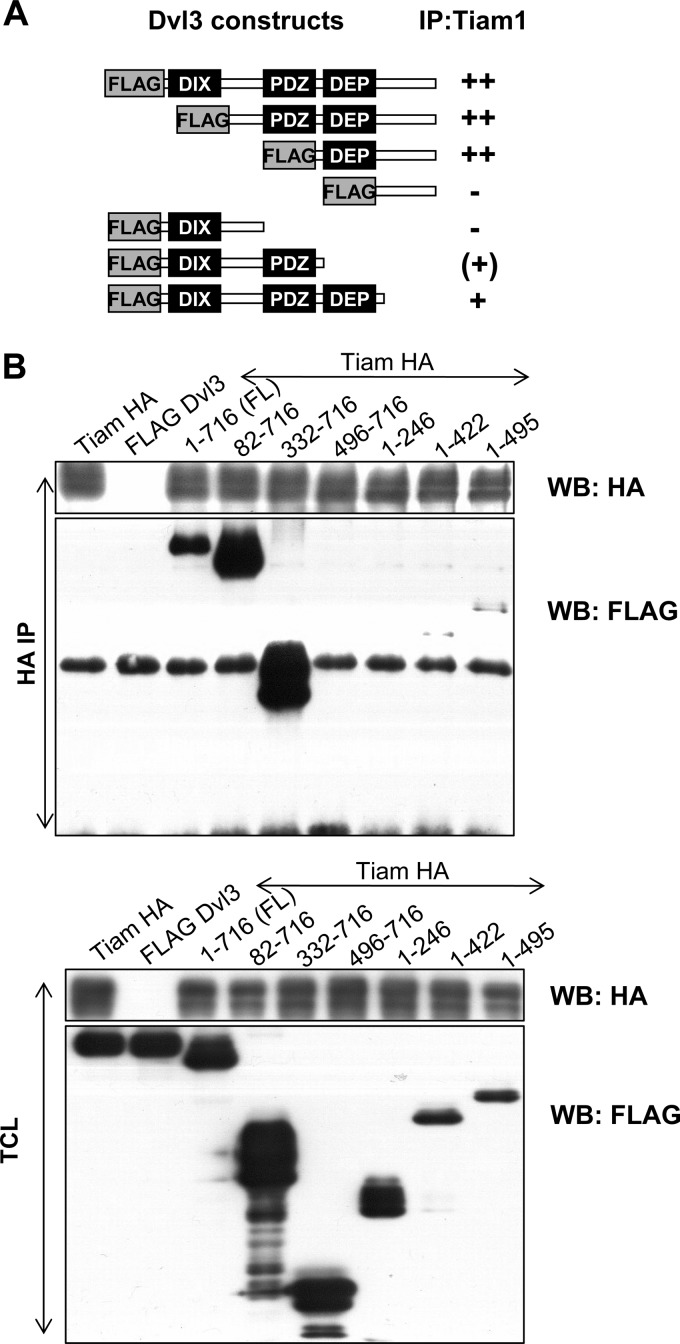
To further confirm that Dvl and Tiam1 are part of the same protein complex, we performed coimmunoprecipitations in HEK293A cells. We found that Tiam1 interacted with both Dvl2-HA (Fig. 3D) and Dvl3-FLAG (Fig. 3E), as detected by their coimmunoprecipitation with Tiam1-FLAG. Importantly, analysis of adult mouse brain lysates showed that endogenous Tiam1 could be pulled down both with Dvl3 (Fig. 3F) and Dvl2 antibodies (Fig. 3G). These results thus show that both endogenous Dvl2 and Dvl3 form complexes with endogenous Tiam1. Detailed mapping of the interaction domains using Dvl3 deletion constructs described in Fig. 2B showed that the proline-rich domain between PDZ and DEP is essential for Tiam1 binding (Fig. 4A and B). Our domain mapping of Dvl suggests that while Rac1 binds at the N-terminal DIX domain, Tiam1 binds at the C-terminal proline-rich domain of Dvl, and hence Dvl can serve as a scaffold for the Rac1-Tiam1 interaction.
Fig 4.
Proline-rich region of Dvl3 is required for interaction with Tiam1. (A) Schematic representation of the domain mapping of the interaction between Tiam1-HA and Dvl3-FLAG deletion mutants. + or − symbols indicate the efficiency of coimmunoprecipitation for the different mutants used in panel B. The top panel shows detection of Dvl3-FLAG mutants after immunoprecipitation of Tiam1-HA by anti-HA antibody. Note the lack of signal from FLAG antibody in lanes with Dvl3 mutants 496-716 and 1-246. Bottom panel shows total cell lysate (TCL).
Tiam1 interaction with Dvl is promoted by Wnt5a and CK1 inhibition.
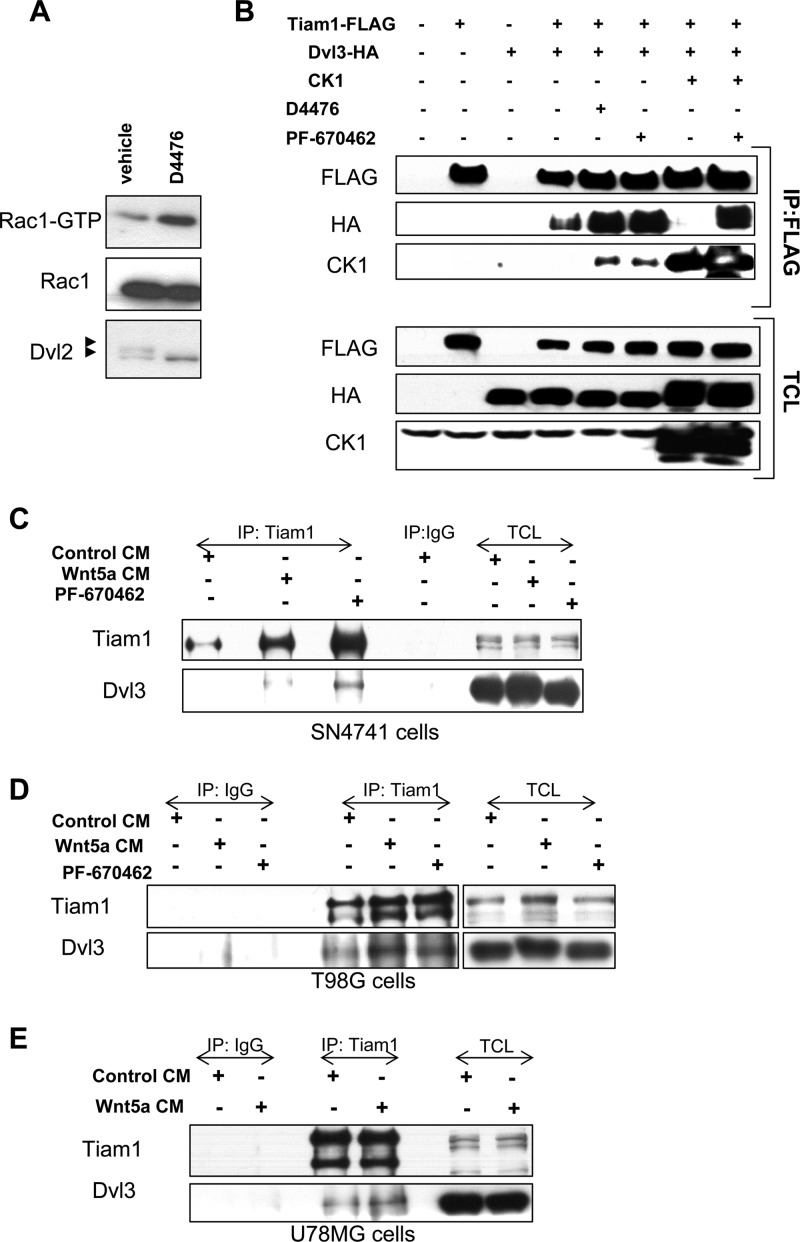
We next examined whether the interaction between Dvl and Tiam1 could be modulated by casein kinase 1 (CK1), a critical regulator known to phosphorylate several components of the Wnt pathway, including Dvl (9, 19). Importantly, the proline-rich domain of Dvl, which we found to be required for Tiam1 binding, contains many residues phosphorylated by CK1 (39, 40). We previously identified CK1ε as a negative regulator of Dvl-induced Rac1 activation and found that pharmacological inhibition of CK1 by D4476 increases the level of Rac1-GTP in mouse embryonic fibroblasts (31). As shown in Fig. 5A, D4476 (50 μM, 2 h) led to increased Rac1 activity also in SN4741 cells. We therefore examined whether the interaction between Dvl and Tiam1 is modified by CK1ε overexpression or pharmacological inhibition of CK1. Overexpression of CK1ε completely abrogated the interaction between Tiam1-FLAG and Dvl3-HA (compare lanes 4 and 7 in Fig. 5B). Moreover, both endogenous and overexpressed CK1 was found to coimmunoprecipitate with Tiam1-FLAG (lanes 5 to 8 in Fig. 5B). On the other hand, the inhibition of CK1 by either D4476 (50 μM) or PF-670462 (5 μM) for 2 to 3 h led to a robust increase in the amount of Dvl3-HA that coimmunoprecipitated with Tiam1-FLAG. Importantly, the negative effect of CK1 overexpression was rescued by CK1 inhibition (compare the last two lanes in Fig. 5B). These results demonstrate that the interaction between Dvl and Tiam1 is under the dynamic control of CK1 and suggest that CK1 may also regulate the interaction between Dvl and Tiam1 in the context of endogenous Wnt signaling. In order to test this possibility, we have analyzed the effects of Wnt5a treatment and CK1 inhibition on the interaction of endogenous Dvl3 and Tiam1 in dopaminergic SN4741 cells. Our results show that while the interaction between Tiam1 and Dvl3 is almost negligible in control conditions, it can be clearly induced by Wnt5a treatment or by the CK1 inhibition by PF-670462 (Fig. 5C). These findings were also confirmed in other neural cell lines, such as U78MG (Fig. 5D) and T98G (Fig. 5E). Thus, our results indicate that Wnt5a and CK1 inhibition promote the interaction between endogenous Dvl and Tiam1 in different neural types, including midbrain DA neurons.
Fig 5.
CK1 and Wnt5a regulate Tiam1-Dvl3 interaction. (A) Inhibition of CK1 with 50 μM D4476 (for 2 h) induced the activation of Rac1 in SN4741 cells. Activation was determined by pulldown of Rac1-GTP and subsequent Western blotting. The efficiency of the inhibition of CK1 was monitored by the disappearance of the hyperphosphorylated form of Dvl2 after D4476 treatment. (B) CK1 negatively regulates the interaction between Tiam1-FLAG and Dvl3-HA as assessed by coimmunoprecipitation of Dvl3-HA by FLAG antibodies and subsequent Western blotting. Note that the coexpression of CK1ε abrogated the Tiam1-Dvl3 interaction (compare lanes 4 and 7), while pharmacological inhibition of CK1 increased the coimmunoprecipitation of Dvl3-HA (lanes 5 and 6) and rescued the effect of CK1ε overexpression (lane 8). CK1 inhibitors D4476 (50 μM) and PF-670462 (5 μM) were added to the medium 2 to 3 h before cells were harvested for immunoprecipitation. (C to E) Wnt5a and CK1 inhibitor (PF-670462) promoted the interaction between endogenous Dvl3 and Tiam1 in dopaminergic SN4741 (C), glioma T98G (D), and U78G (E) cell lines. Immunoprecipitation with anti-Tiam1 antibodies was followed by Western blot detection of Dvl3 interacting with Tiam1.
Does Tiam1 regulate Rac1 function and the formation of Dvl-Rac1 complexes?
The results obtained until this point indicated that Dvl2 and Dvl3 interact and colocalize not only with Rac1 but also with Tiam1 and that Tiam1-Dvl interaction is under tight control of CK1 and Wnt5a. We thus decided to examine whether Tiam1 can regulate the formation of the Dvl-Rac1 complex and the activity of Rac1. We found that the coimmunoprecipitation of Dvl2-MYC with Rac1 was enhanced when Tiam1-FLAG was overexpressed (Fig. 6A). Quantification of these results (n = 5) indicated that the expression of Tiam1 increased the Dvl2-Rac1 coimmunoprecipitation by 46%, compared to the condition without Tiam1 expression (Fig. 6B). Thus, our results suggest that the presence of Tiam1 facilitates the Dvl-Rac1 interaction.
Fig 6.
Tiam1 functionally interacts with Dvl to regulate Rac1 activation. (A) Dvl2-MYC was pulled down by Rac1 (first lane), and this coimmunoprecipitation was enhanced by coexpression of Tiam1-FLAG (third lane). (B) Densitometric quantification of the relative amount of Dvl2-MYC coimmunoprecipitating with Rac1 in control or Tiam1 cotransfected cells. Values were normalized to the total level of Dvl-MYC in total cell lysate (TCL). (C) SN4741 and HEK293A cells were transfected with a Tiam1-FLAG or mock plasmid and analyzed for the level of GTP-Rac1 24 h after transfection. Tiam1-FLAG was sufficient to increase GTP-Rac1 levels, compared to that of the control, in both cell lines. (D) SN4741 cells were transfected with control or Tiam1 siRNAs and subsequently with Rac1-MYC and Dvl3-FLAG or mock vector. Dvl3-FLAG increased the levels of MYC-tagged Rac1-GTP compared to those of mock-transfected SN4741 cells, but not in cells where Tiam1 expression was knocked down by siRNA. (E) Densitometric quantification of the relative level of Rac1-GTP (normalized to total MYC-tagged Rac1) induced by Dvl3-FLAG in control or Tiam1 siRNA conditions. (F) SN4741 cells transfected with control or Tiam1 siRNA and 24 h after they were stimulated with Wnt5a (100 ng/ml) or vehicle (PBS-CHAPS) for 2.5 h and subsequently analyzed for Rac1 activation (level of Rac1-GTP). (G) Densitometric quantification of the relative level of Rac1-GTP (normalized to total Rac1) induced by Wnt5a stimulation in control siRNA versus Tiam1 siRNA (second versus fourth lane). *, P < 0.05 by Mann-Whitney test.
We next examined whether Tiam1 also acts as a Rac1 GEF and regulates the activity of Rac1. As expected, Tiam1 overexpression was sufficient to induce Rac1 activation in both HEK293A and SN4741 cells (Fig. 6C). In light of these data, we found it plausible that Tiam1 could be functionally required for Rac1 activation in context of the Wnt/Dvl/Rac1 pathway. In order to test this possibility, we knocked down Tiam1 protein in SN4741 DA cells using a Tiam1 siRNA. While Dvl2-MYC or Dvl3-FLAG overexpression increased the level of activated MYC-tagged Rac1-GTP in control siRNA cells, they did not activate Rac1 when Tiam1 expression was knocked down (Fig. 6D and data not shown). Moreover, the relative increases in Rac1-GTP levels after Dvl2-MYC or Dvl3-FLAG overexpression were significantly decreased to almost basal levels after Tiam1 knockdown (Fig. 6E and data not shown). Next, we tested whether Tiam1 was similarly required for Wnt5a ligand-dependent activation of Rac1 in SN4741 DA cells. We found that the ability of Wnt5a to increase Rac1-GTP levels was impaired when Tiam1 expression was knocked down by siRNA (Fig. 6F and G). In sum, our results show that Tiam1 is sufficient for Rac1 activation in DA cells and, more importantly, that Tiam1 is required for the activation of Rac1 by either Dvl overexpression or acute stimulation by Wnt5a. Thus, our results identify Tiam1 as a novel regulator of the noncanonical Wnt/Dvl/Rac1 signaling pathway.
Tiam1 is required for the differentiation of midbrain DA precursors into neurons.
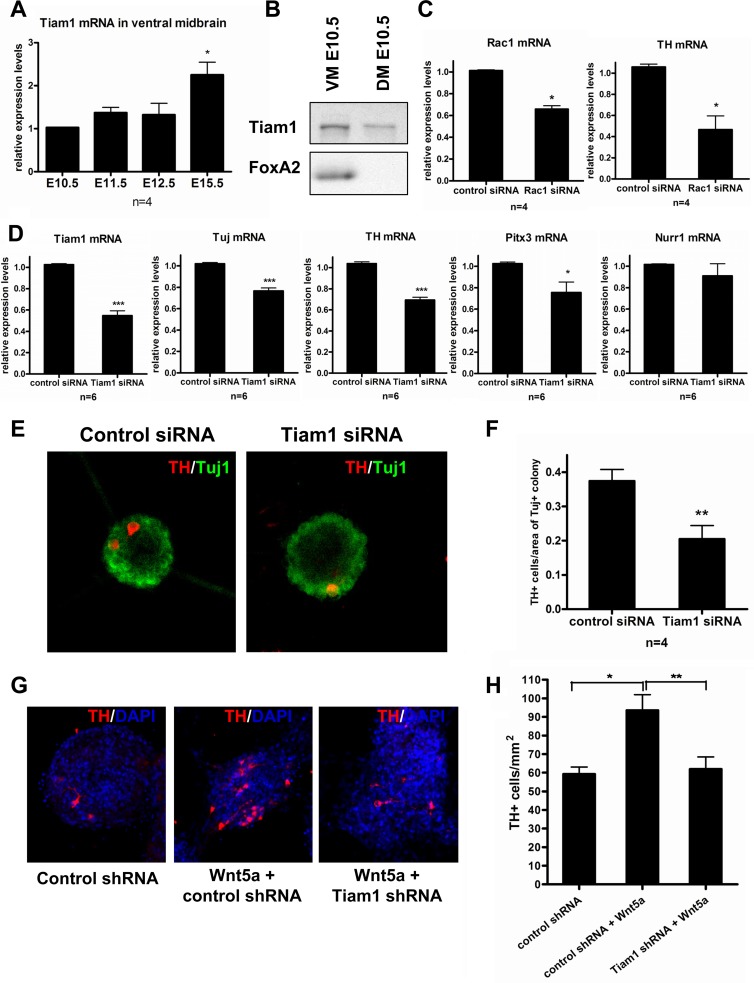
As we hereby report that Tiam1 regulates the function of Rac1 in the context of the Wnt/Dvl/Rac1 pathway and we have previously reported that Wnt5a and Rac1 are involved in differentiation of primary ventral midbrain DA neurons (6), we decided to examine the possible contribution of Tiam1 to this process. First, we analyzed the expression of Tiam1 mRNA in mouse ventral midbrain (VM) at different embryonic (E) stages, from E10.5 to E15.5, by qPCR. We found that the expression of Tiam1 mRNA progressively increased during embryonic development, reaching maximal levels at E15.5 (Fig. 7A). Moreover, when we examined the levels of Tiam1 protein in the developing VM, we found that Tiam1 is present as early as at E10.5 (Fig. 7B). These results indicated a possible function of Tiam1 in midbrain development, at the indicated developmental stages.
Fig 7.
Tiam1 is expressed in the developing VM and is required for Wnt5a-induced DA neuron differentiation. (A) qPCR analysis showed that Tiam1 mRNA is expressed between E10.5 and E15.5 in mouse ventral midbrain (VM). *, P < 0.05 by Mann-Whitney test (compared to E10.5). (B) Western blot analyses confirmed the presence of Tiam1 protein in both ventral midbrain and dorsal midbrain (DM) samples at E10.5. The floor plate marker Foxa2 was used to confirm the identity of the VM sample. (C) Rac1 siRNA of Rac1 and TH mRNA in differentiating ventral midbrain precursors, 3 days after transfection. (D) Tiam1 siRNA decreased the expression of Tiam1 mRNA, as well as Tuj, Th, and Pitx3 mRNAs but not Nurr1 mRNA in differentiating ventral midbrain precursors 3 days after transfection. These results suggested an effect of Tiam1 in DA differentiation, similar to that of Wnt5a. (E) Immunostaining of control or Tiam1 siRNA VM neurospheres showed similar numbers of Tuj1+ cells (green) but lower numbers of TH+ dopamine neurons (red) after 3 days of differentiation. (F) Quantification of the number of TH+ cells per area of Tuj+ neurospheres revealed that Tiam1 siRNA significantly reduced the number of dopamine neurons. (G) Treatment with Wnt5a (100 ng/ml, 3 days) increased the number of TH+ neurons in neurosphere cultures treated with control shRNA lentiviruses but not in Tiam1 shRNA lentiviruses. DAPI was used for nuclei counterstaining. (H) Quantification of the number of TH+ cells per area (mm2) of sphere, summary of 4 independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 by Student t test or one-way analysis of variance (ANOVA) (Fig. 7H).
We therefore tested whether Tiam1 mRNA interference impaired the generation of midbrain DA neurons in midbrain neural stem/progenitor cells expanded as neurospheres in the presence of Shh, basic FGF, and FGF8 for 7 days (12). Primary neurospheres were gently dissociated to smaller cell clusters, transfected with either control siRNA or Tiam1 siRNA, and differentiated for 3 days. Transfection of differentiated progenitor cultures with Tiam1 siRNA reduced the expression of Tiam1 mRNA by 50% (Fig. 7D) at day 3 of differentiation. Tiam1 gene knockdown led to significant decreases in the mRNA expression of beta III tubulin (Tuj), a neuronal marker, and tyrosine hydroxylase (TH), the rate-limiting enzyme in dopamine synthesis and marker of DA neurons (Fig. 7D). Similarly, Rac1 siRNA, which efficiency reduced Rac1 mRNA levels in SN4741 DA cells by 50 to 75% (data not shown), caused a decrease in the expression of TH mRNA (and Rac1 mRNA) in VM neurospheres differentiated for 3 days (Fig. 7C). Interestingly, Tiam1 gene knockdown also decreased the mRNA expression of Pitx3 (paired-like homeodomain transcription factor 3), a specific marker of VM DA neurons, but did not affect the expression of Nurr1, a transcription factor expressed both in DA neurons and postmitotic neuroblasts (Fig. 7D). These results suggested a role of Tiam1 in the last stages of differentiation, in the acquisition of two DA neuron markers, Pitx3 and TH, by Nurr1+ neuroblasts. Indeed, immunocytochemical analysis of differentiated ventral midbrain neurospheres showed that Tiam1 gene knockdown reduced by 50% the number of TH+ DA neurons in Tuj+ clusters normalized by area (Fig. 7E and F). These results indicated that Tiam1 is required for the generation of midbrain DA neurons from neural stem/progenitor cell preparations. Finally, since our biochemical data identified Tiam1 as a downstream component of Wnt5a-mediated signaling, we examined whether Tiam1 knockdown can also block the Wnt5a-induced DA differentiation in neurosphere cultures. In our experiments, Wnt5a treatment (100 ng/ml) increased the number of TH+ DA neurons per area by 58%, from 59.2 ± 3.8 TH+ cells/mm2 to 93.6 ± 8.3 TH+ cells/mm2 (Fig. 7G and H). These effects were efficiently ablated by lentiviral delivery of Tiam1 shRNA (61.9 ± 6.5 TH+ cells/mm2). In sum, our data suggest that Rac1 and its upstream regulator Tiam1 are involved in the differentiation of midbrain DA neurons and that Tiam1 works downstream of Wnt5a in DA differentiation.
DISCUSSION
In this study, we investigated the mechanisms of activation of the small GTPase, Rac1, in the context of Wnt signaling in DA cells and found that the Rac1 GEF Tiam1 regulates the Wnt/Dvl/Rac1 signaling pathway and the differentiation of midbrain dopaminergic (DA) neurons. We previously reported that Wnt5a promotes the differentiation of DA neurons via a Wnt/β-catenin-independent pathway (6, 8, 9) that involves Rac1 (31). The activation of Rac1 by Wnt/β-catenin-independent signaling has been reported in various cellular systems (4), and Rac1 has been found to play a role in cytoskeleton rearrangements, cell polarity, and cell migration (13, 41), as well as in neuronal development and differentiation (42–44). Surprisingly, however, little is known about upstream mechanisms involved in the regulation of small GTPases in the context of the Wnt/β-catenin independent signaling. Previous studies have shown that Dvl is both required and sufficient for the activation of Rac1 (15, 31, 45). Our results further suggest that the activation of Rac1 by Dvl requires the formation of protein complexes containing Dvl and Rac1, as Dvl mutants lacking the ability to interact with Rac1 also fail to induce Rac1 activation. We identified the N-terminal region of Dvl3 (1 to 81 amino acids) as critical for mediating the Dvl3-Rac1 interaction and the activation of Rac1 by Dvl3. Our results differ from the study by Habas and colleagues (15), who reported that the DEP domain of Dvl2 is sufficient to bind and activate Rac1 and failed to detect activation of Rac1 by Dvl3. In our study, we found that both Dvl2 and Dvl3 are sufficient to induce Rac1 activation and that the DEP domain of Dvl3 is not sufficient for Rac1 interaction. It is possible that methodological and cell-type-specific factors account for the discrepancy. However, data in the literature indirectly support an interaction of Rac1 with the N-terminal part of Dvl, as the N-terminal part of Dvl1 is sufficient for association with PAK, a direct effector and binding partner of Rac1 (46).
Evidence for the employment of specific GEFs in Wnt/β-catenin-independent signaling has only recently begun to emerge. WGEF, p114-RhoGEF, and GEF-H1 have been proposed as GEFs for RhoA (47, 48). However, GEFs involved in the Wnt5a/Dvl-mediated activation of Rac1 have not been described until now. Our data show first evidence for the involvement of a specific Rac1 GEF, Tiam1, in the noncanonical Wnt/Dvl/Rac1 pathway. This conclusion is based on the following lines of evidence. First, we found that Tiam1 interacts with Dvl and enhances the Dvl-Rac1 interaction. Second, we showed that the Dvl-Tiam1 interaction is regulated by CK1. Third, siRNA knockdown experiments demonstrated that Tiam1 is functionally required for the activation of Rac1 by Wnt5a or Dvl. These findings are in full agreement with the known negative role of CK1 in Wnt/Dvl/Rac1 signaling and provide a possible molecular mechanism by which CK1 regulates DA differentiation, namely by regulating both the interaction between Dvl and Tiam1 and the activation of Rac1. We previously reported that inhibition of CK1 or overexpression of dominant negative CK1ε increases the level of Rac1-GTP, in a Dvl-dependent manner, while overexpression of CK1 blocks the activation of Rac1 mediated by Dvl (31). Here, we show that inhibition of CK1 facilitates, whereas CK1 overexpression blocks, the interaction between Dvl and Tiam1. Moreover, we could detect CK1 present in Tiam1 immunoprecipitates. Interestingly, a recent proteomic study also found CK1 as a Tiam1-associated protein (49). Combined, our results suggest that CK1 controls changes in protein-protein interactions between Dvl and Tiam1, which result in the activation of Rac1 by Tiam1. Surprisingly, however, we were unable to detect the interaction between Tiam1 and Rac1. One possible explanation for this is the instability of the trimeric complex formed by Tiam1-Rac1-GTP (50). Evidence in the literature supports the view that the activity of small GTPases is regulated by protein-protein interactions between GEFs and scaffolding proteins (37, 38, 51). This possibility is in agreement with our observation of a Dvl-Tiam1 interaction and a negative effect of CK1 on Dvl-Tiam1 binding and Wnt/Dvl/Rac1 signaling. We thus propose a model where CK1 activity determines the amount of Tiam1 associated with the scaffolding protein Dvl, which is hence available to bind and activate Dvl-bound Rac1. In this model, the Wnt-induced activation of CK1 (9, 52) may thus represent a negative feedback mechanism to fine-tune the level of Rac1-GTP.
Future experiments should aim at addressing whether the activation of Rac1 by Wnt5a/Dvl is exclusively controlled by Tiam1 or is redundantly controlled by other GEFs. Another pending issue is the identity of the signaling component(s) downstream of the Wnt5a/Dvl/Rac1 pathway. It has been previously shown that cJun acts downstream of Tiam1/Rac1 (53), opening the intriguing possibility that the Wnt5a/Dvl/Rac1 pathway directly regulates transcription via cJun. However, we found that Wnt5a did not activate a cJun-luciferase reporter in DA cells (data not shown). Thus, our results suggest that (i) cJun may not be downstream of Rac1 in the context of Wnt5a-mediated signaling and that (ii) an alternative factor(s) mediates the Wnt5a/Dvl/Rac1 pathway in DA neurons.
At a functional level, Tiam1 has been previously shown to regulate neurite and axon outgrowth in neuroblastoma cells (54, 55) as well as in hippocampal neurons (56), migration of cortical neurons (57), and the formation of dendritic spines (26). Previous studies have also shown that the attenuation of Tiam1 and/or Rac1 activity impairs cytoskeleton remodeling and leads to loss of cell polarity, which in context of neuronal differentiation can be reflected by impaired neurogenic cell division, migration, and/or neuritogenesis (43, 58–60). Moreover, Wnt5a/Dvl-mediated signaling events have been implicated in the regulation of polarity in differentiating neurons (4, 6, 61). Thus, it is plausible that cell polarity underlies the requirement of Wnt5a/Dvl/Rac1 signaling for neuronal differentiation. Importantly, the role of Tiam1 in the ventral midbrain (VM) and its possible implication in the differentiation/development of DA neurons has not been addressed before. Here, we show that Tiam1 is expressed in the VM at the time when DA neurons are born. Moreover, we found that Tiam1 knockdown impairs the generation of DA neurons in VM progenitor cultures. Thus, it is plausible to expect that Tiam1 regulates a signaling pathway(s) relevant to DA development and differentiation (62). Interestingly, Wnt5a and Rac1 activation had previously been shown to promote the differentiation of Nurr1+ TH− postmitotic neuroblasts into Nurr1+ TH+ Pitx3+ DA neurons (6, 10). In this regard, our biochemical and functional data suggest that Tiam1 mediates Wnt5a-induced DA neuron differentiation. Furthermore, our results indicating that Tiam1 knockdown decreases TH, Pitx3, and Tuj mRNA levels but does not affect the expression of Nurr1 are fully in agreement with the idea that Wnt5a promotes the transition of Nurr1+ DA neuroblasts into mature DA neurons. Thus, combined, our results suggest that Tiam1 plays a role in the differentiation of Nurr1+ DA precursors and is a key modulator of the Wnt/Dvl/Rac1 pathway.
In summary, we hereby show that Rac1 interacts with its upstream regulator Dvl. We identified Tiam1, a Rac1-specific GEF, as a binding partner of Dvl and a critical regulator of Rac1 activation in the context of the Wnt5a/Dvl/Rac1 pathway. Finally, we provide functional evidence that Tiam1 is required for the Wnt5a-mediated generation of DA neurons in ventral midbrain progenitor cultures. Hence, our data highlight the important role of the Wnt5a/Dvl/Rac1 signaling pathway in DA neuron differentiation and pinpoint Tiam1, the first Rac1 GEF identified in noncanonical Wnt signaling.
ACKNOWLEDGMENTS
We thank R. J. Lefkowitz (HHMI, Duke University, Durham, NC) for Dvl2-EGFP, S. Yanagawa (Kyoto University) for Dvl2-MYC, R. T. Moon (HHMI, University of Washington, Seattle, WA) for Dvl3 constructs, Alan Hall (Memorial Sloan-Kettering Cancer Center, New York, NY) for Rac1-MYC, M. Bienz (LMB, MRC, Cambridge, United Kingdom) for Dvl DIX mutants (M1 to M4), Pontus Aspenström (KI, Stockholm, Germany) for GST-PAK CRIB, K. Tolias (Baylor College of Medicine, Houston, TX) for Tiam1-FLAG constructs, and Ondřej Slabý for U78MG and T98G cells. We acknowledge Johnny Söderlund, Nad'a Bílá, Jakub Harnoš, and Alessandra Nanni for excellent assistance.
This work was supported by grants from the European Union (Neurostemcell), Swedish Foundation for Strategic Research (SRL Program and CEDB project), Swedish Research Council (VR2008:2811, VR2011:3116, and DBRM), Norwegian Research Council, and Karolinska Institute to E.A., as well as grants from the Ministry of Education, Youth, and Sports of the Czech Republic (MSM0021622430, CZ.1.07/2.3.00/20.0180), EMBO Installation Grant, and Czech Science Foundation (204/09/0498; 204/09/H058) to V.B. L.C. was supported by a KID Ph.D. student fellowship (6110/06-225) from Karolinska Institute. S.T. was supported by a VR fellowship for visiting scientists and short-term fellowship from the Onassis Foundation.
We declare no competing interests.
Footnotes
Published ahead of print 29 October 2012
REFERENCES
- 1. Angers S, Moon RT. 2009. Proximal events in Wnt signal transduction. Nat. Rev. Mol. Cell Biol. 10: 468–477 [DOI] [PubMed] [Google Scholar]
- 2. MacDonald BT, Tamai K, He X. 2009. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev. Cell 17: 9–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lai SL, Chien AJ, Moon RT. 2009. Wnt/Fz signaling and the cytoskeleton: potential roles in tumorigenesis. Cell Res. 19: 532–545 [DOI] [PubMed] [Google Scholar]
- 4. Schlessinger K, Hall A, Tolwinski N. 2009. Wnt signaling pathways meet Rho GTPases. Genes Dev. 23: 265–277 [DOI] [PubMed] [Google Scholar]
- 5. Montcouquiol M, Crenshaw EB, III, Kelley MW. 2006. Noncanonical Wnt signaling and neural polarity. Annu. Rev. Neurosci. 29: 363–386 [DOI] [PubMed] [Google Scholar]
- 6. Andersson ER, Prakash N, Cajanek L, Minina E, Bryja V, Bryjova L, Yamaguchi TP, Hall AC, Wurst W, Arenas E. 2008. Wnt5a regulates ventral midbrain morphogenesis and the development of A9-A10 dopaminergic cells in vivo. PLoS One 3: e3517 doi:10.1371/journal.pone.0003517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inestrosa NC, Arenas E. 2010. Emerging roles of Wnts in the adult nervous system. Nat. Rev. Neurosci. 11: 77–86 [DOI] [PubMed] [Google Scholar]
- 8. Schulte G, Bryja V, Rawal N, Castelo-Branco G, Sousa KM, Arenas E. 2005. Purified Wnt-5a increases differentiation of midbrain dopaminergic cells and dishevelled phosphorylation. J. Neurochem. 92: 1550–1553 [DOI] [PubMed] [Google Scholar]
- 9. Bryja V, Schulte G, Rawal N, Grahn A, Arenas E. 2007. Wnt-5a induces Dishevelled phosphorylation and dopaminergic differentiation via a CK1-dependent mechanism. J. Cell Sci. 120: 586–595 [DOI] [PubMed] [Google Scholar]
- 10. Castelo-Branco G, Wagner J, Rodriguez FJ, Kele J, Sousa K, Rawal N, Pasolli HA, Fuchs E, Kitajewski J, Arenas E. 2003. Differential regulation of midbrain dopaminergic neuron development by Wnt-1, Wnt-3a, and Wnt-5a. Proc. Natl. Acad. Sci. U. S. A. 100: 12747–12752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hayashi H, Morizane A, Koyanagi M, Ono Y, Sasai Y, Hashimoto N, Takahashi J. 2008. Meningeal cells induce dopaminergic neurons from embryonic stem cells. Eur. J. Neurosci. 27: 261–268 [DOI] [PubMed] [Google Scholar]
- 12. Parish CL, Castelo-Branco G, Rawal N, Tonnesen J, Sorensen AT, Salto C, Kokaia M, Lindvall O, Arenas E. 2008. Wnt5a-treated midbrain neural stem cells improve dopamine cell replacement therapy in parkinsonian mice. J. Clin. Invest. 118: 149–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jaffe AB, Hall A. 2005. Rho GTPases: biochemistry and biology. Annu. Rev. Cell Dev. Biol. 21: 247–269 [DOI] [PubMed] [Google Scholar]
- 14. Rossman KL, Der CJ, Sondek J. 2005. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 6: 167–180 [DOI] [PubMed] [Google Scholar]
- 15. Habas R, Dawid IB, He X. 2003. Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes Dev. 17: 295–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. 2005. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat. Neurosci. 8: 34–42 [DOI] [PubMed] [Google Scholar]
- 17. Gao C, Chen YG. 2010. Dishevelled: the hub of Wnt signaling. Cell Signal. 22: 717–727 [DOI] [PubMed] [Google Scholar]
- 18. Wallingford JB, Habas R. 2005. The developmental biology of Dishevelled: an enigmatic protein governing cell fate and cell polarity. Development 132: 4421–4436 [DOI] [PubMed] [Google Scholar]
- 19. Cong F, Schweizer L, Varmus H. 2004. Casein kinase Iepsilon modulates the signaling specificities of Dishevelled. Mol. Cell. Biol. 24: 2000–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Habets GG, Scholtes EH, Zuydgeest D, van der Kammen RA, Stam JC, Berns A, Collard JG. 1994. Identification of an invasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell 77: 537–549 [DOI] [PubMed] [Google Scholar]
- 21. Bryja V, Cajanek L, Grahn A, Schulte G. 2007. Inhibition of endocytosis blocks Wnt signalling to beta-catenin by promoting Dishevelled degradation. Acta Physiol. (Oxf.) 190: 55–61 [DOI] [PubMed] [Google Scholar]
- 22. Lee JS, Ishimoto A, Yanagawa S. 1999. Characterization of mouse Dishevelled (Dvl) proteins in Wnt/Wingless signaling pathway. J. Biol. Chem. 274: 21464–21470 [DOI] [PubMed] [Google Scholar]
- 23. Angers S, Thorpe CJ, Biechele TL, Goldenberg SJ, Zheng N, Maccoss MJ, Moon RT. 2006. The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat. Cell Biol. 8: 348–357 [DOI] [PubMed] [Google Scholar]
- 24. Chen W, ten Berge D, Brown J, Ahn S, Hu LA, Miller WE, Caron MG, Barak LS, Nusse R, Lefkowitz RJ. 2003. Dishevelled 2 recruits beta-arrestin 2 to mediate Wnt5A-stimulated endocytosis of Frizzled 4. Science 301: 1391–1394 [DOI] [PubMed] [Google Scholar]
- 25. Schwarz-Romond T, Fiedler M, Shibata N, Butler PJ, Kikuchi A, Higuchi Y, Bienz M. 2007. The DIX domain of Dishevelled confers Wnt signaling by dynamic polymerization. Nat. Struct. Mol. Biol. 14: 484–492 [DOI] [PubMed] [Google Scholar]
- 26. Tolias KF, Bikoff JB, Burette A, Paradis S, Harrar D, Tavazoie S, Weinberg RJ, Greenberg ME. 2005. The Rac1-GEF Tiam1 couples the NMDA receptor to the activity-dependent development of dendritic arbors and spines. Neuron 45: 525–538 [DOI] [PubMed] [Google Scholar]
- 27. Ridley AJ, Paterson HF, Johnston CL, Diekmann D, Hall A. 1992. The small GTP-binding protein Rac regulates growth factor-induced membrane ruffling. Cell 70: 401–410 [DOI] [PubMed] [Google Scholar]
- 28. Badura L, Swanson T, Adamowicz W, Adams J, Cianfrogna J, Fisher K, Holland J, Kleiman R, Nelson F, Reynolds L, St Germain K, Schaeffer E, Tate B, Sprouse J. 2007. An inhibitor of casein kinase I epsilon induces phase delays in circadian rhythms under free-running and entrained conditions. J. Pharmacol. Exp. Ther. 322: 730–738 [DOI] [PubMed] [Google Scholar]
- 29. Rena G, Bain J, Elliott M, Cohen P. 2004. D4476, a cell-permeant inhibitor of CK1, suppresses the site-specific phosphorylation and nuclear exclusion of FOXO1a. EMBO Rep. 5: 60–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cajanek L, Ribeiro D, Liste I, Parish CL, Bryja V, Arenas E. 2009. Wnt/beta-catenin signaling blockade promotes neuronal induction and dopaminergic differentiation in embryonic stem cells. Stem Cells 27: 2917–2927 [DOI] [PubMed] [Google Scholar]
- 31. Bryja V, Schambony A, Cajanek L, Dominguez I, Arenas E, Schulte G. 2008. Beta-arrestin and casein kinase 1/2 define distinct branches of noncanonical WNT signalling pathways. EMBO Rep. 9: 1244–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Parish CL, Parisi S, Persico MG, Arenas E, Minchiotti G. 2005. Cripto as a target for improving embryonic stem cell-based therapy in Parkinson's disease. Stem Cells 23: 471–476 [DOI] [PubMed] [Google Scholar]
- 33. Dull T, Zufferey R, Kelly M, Mandel RJ, Nquyen M, Trono D, Naldini L. 1998. A third-generation lentivirus vector with a conditional packaging system. J. Virol. 72: 8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yokoyama N, Golebiewska U, Wang HY, Malbon CC. 2010. Wnt-dependent assembly of supermolecular Dishevelled-3-based complexes. J. Cell Sci. 123: 3693–3702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwarz-Romond T, Merrifield C, Nichols BJ, Bienz M. 2005. The Wnt signalling effector Dishevelled forms dynamic protein assemblies rather than stable associations with cytoplasmic vesicles. J. Cell Sci. 118: 5269–5277 [DOI] [PubMed] [Google Scholar]
- 36. Smalley MJ, Signoret N, Robertson D, Tilley A, Hann A, Ewan K, Ding Y, Paterson H, Dale TC. 2005. Dishevelled (Dvl-2) activates canonical Wnt signalling in the absence of cytoplasmic puncta. J. Cell Sci. 118: 5279–5289 [DOI] [PubMed] [Google Scholar]
- 37. Garcia-Mata R, Burridge K. 2007. Catching a GEF by its tail. Trends Cell Biol. 17: 36–43 [DOI] [PubMed] [Google Scholar]
- 38. Marinissen MJ, Gutkind JS. 2005. Scaffold proteins dictate Rho GTPase-signaling specificity. Trends Biochem. Sci. 30: 423–426 [DOI] [PubMed] [Google Scholar]
- 39. Bernatik O, Ganji R, Dijksterhuis J, Konik P, Cervenka I, Polonio T, Krejci P, Schulte G, Bryja V. 2011. Sequential activation and inactivation of Dishevelled in the Wnt/beta-catenin pathway by casein kinases. J. Biol. Chem. 286: 10396–10410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Klein TJ, Jenny A, Djiane A, Mlodzik M. 2006. CKIepsilon/discs overgrown promotes both Wnt-Fz/beta-catenin and Fz/PCP signaling in Drosophila. Curr. Biol. 16: 1337–1343 [DOI] [PubMed] [Google Scholar]
- 41. Etienne-Manneville S, Hall A. 2002. Rho GTPases in cell biology. Nature 420: 629–635 [DOI] [PubMed] [Google Scholar]
- 42. de Curtis I. 2008. Functions of Rac GTPases during neuronal development. Dev. Neurosci. 30: 47–58 [DOI] [PubMed] [Google Scholar]
- 43. Govek EE, Newey SE, Van Aelst L. 2005. The role of the Rho GTPases in neuronal development. Genes Dev. 19: 1–49 [DOI] [PubMed] [Google Scholar]
- 44. Tahirovic S, Hellal F, Neukirchen D, Hindges R, Garvalov BK, Flynn KC, Stradal TE, Chrostek-Grashoff A, Brakebusch C, Bradke F. 2010. Rac1 regulates neuronal polarization through the WAVE complex. J. Neurosci. 30: 6930–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ishida-Takagishi M, Enomoto A, Asai N, Ushida K, Watanabe T, Hashimoto T, Kato T, Weng L, Matsumoto S, Asai M, Murakumo Y, Kikuchi A, Takahashi M. 2012. The Dishevelled-associated protein Daple controls the noncanonical Wnt/Rac pathway and cell motility. Nat. Commun. 3: 89. [DOI] [PubMed] [Google Scholar]
- 46. Luo ZG, Wang Q, Zhou JZ, Wang J, Luo Z, Liu M, He X, Wynshaw-Boris A, Xiong WC, Lu B, Mei L. 2002. Regulation of AChR clustering by Dishevelled interacting with MuSK and PAK1. Neuron 35: 489–505 [DOI] [PubMed] [Google Scholar]
- 47. Tanegashima K, Zhao H, Dawid IB. 2008. WGEF activates Rho in the Wnt-PCP pathway and controls convergent extension in Xenopus gastrulation. EMBO J. 27: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tsuji T, Ohta Y, Kanno Y, Hirose K, Ohashi K, Mizuno K. 2010. Involvement of p114-RhoGEF and Lfc in Wnt-3a- and Dishevelled-induced RhoA activation and neurite retraction in N1E-115 mouse neuroblastoma cells. Mol. Biol. Cell 21: 3590–3600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Woodcock SA, Jones RC, Edmondson RD, Malliri A. 2009. A modified tandem affinity purification technique identifies that 14-3-3 proteins interact with Tiam1, an interaction which controls Tiam1 stability. J. Proteome Res. 8: 5629–5641 [DOI] [PubMed] [Google Scholar]
- 50. Worthylake DK, Rossman KL, Sondek J. 2000. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature 408: 682–688 [DOI] [PubMed] [Google Scholar]
- 51. Buchsbaum RJ, Connolly BA, Feig LA. 2003. Regulation of p70 S6 kinase by complex formation between the Rac guanine nucleotide exchange factor (Rac-GEF) Tiam1 and the scaffold spinophilin. J. Biol. Chem. 278: 18833–18841 [DOI] [PubMed] [Google Scholar]
- 52. Swiatek W, Tsai IC, Klimowski L, Pepler A, Barnette J, Yost HJ, Virshup DM. 2004. Regulation of casein kinase I epsilon activity by Wnt signaling. J. Biol. Chem. 279: 13011–13017 [DOI] [PubMed] [Google Scholar]
- 53. Fanto M, Weber U, Strutt DI, Mlodzik M. 2000. Nuclear signaling by Rac and Rho GTPases is required in the establishment of epithelial planar polarity in the Drosophila eye. Curr. Biol. 10: 979–988 [DOI] [PubMed] [Google Scholar]
- 54. Leeuwen FN, Kain HE, Kammen RA, Michiels F, Kranenburg OW, Collard JG. 1997. The guanine nucleotide exchange factor Tiam1 affects neuronal morphology; opposing roles for the small GTPases Rac and Rho. J. Cell Biol. 139: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tanaka M, Ohashi R, Nakamura R, Shinmura K, Kamo T, Sakai R, Sugimura H. 2004. Tiam1 mediates neurite outgrowth induced by ephrin-B1 and EphA2. EMBO J. 23: 1075–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kunda P, Paglini G, Quiroga S, Kosik K, Caceres A. 2001. Evidence for the involvement of Tiam1 in axon formation. J. Neurosci. 21: 2361–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kawauchi T, Chihama K, Nabeshima Y, Hoshino M. 2003. The in vivo roles of STEF/Tiam1, Rac1 and JNK in cortical neuronal migration. EMBO J. 22: 4190–4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mertens AE, Pegtel DM, Collard JG. 2006. Tiam1 takes PARt in cell polarity. Trends Cell Biol. 16: 308–316 [DOI] [PubMed] [Google Scholar]
- 59. Minobe S, Sakakibara A, Ohdachi T, Kanda R, Kimura M, Nakatani S, Tadokoro R, Ochiai W, Nishizawa Y, Mizoguchi A, Kawauchi T, Miyata T. 2009. Rac is involved in the interkinetic nuclear migration of cortical progenitor cells. Neurosci. Res. 63: 294–301 [DOI] [PubMed] [Google Scholar]
- 60. Schlessinger K, McManus EJ, Hall A. 2007. Cdc42 and noncanonical Wnt signal transduction pathways cooperate to promote cell polarity. J. Cell Biol. 178: 355–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang X, Zhu J, Yang GY, Wang QJ, Qian L, Chen YM, Chen F, Tao Y, Hu HS, Wang T, Luo ZG. 2007. Dishevelled promotes axon differentiation by regulating atypical protein kinase C. Nat. Cell Biol. 9: 743–754 [DOI] [PubMed] [Google Scholar]
- 62. Sasaki N, Kurisu J, Kengaku M. 2010. Sonic hedgehog signaling regulates actin cytoskeleton via Tiam1-Rac1 cascade during spine formation. Mol. Cell. Neurosci. 45: 335–344 [DOI] [PubMed] [Google Scholar]