Abstract
The histone variant H2AX is a principal component of chromatin involved in the detection, signaling, and repair of DNA double-strand breaks (DSBs). H2AX is thought to operate primarily through its C-terminal S139 phosphorylation, which mediates the recruitment of DNA damage response (DDR) factors to chromatin at DSB sites. Here, we describe a comprehensive screen of 67 residues in H2AX to determine their contributions to H2AX functions. Our analysis revealed that H2AX is both sumoylated and ubiquitylated. Individual residues defective for sumoylation, ubiquitylation, and S139 phosphorylation in untreated and damaged cells were identified. Specifically, we identified an acidic triad region in both H2A and H2AX that is required in cis for their ubiquitylation. We also report the characterization of a human H2AX knockout cell line, which exhibits DDR defects, including p53 activation, following DNA damage. Collectively, this work constitutes the first genetic complementation system for a histone in human cells. Finally, our data reveal new roles for several residues in H2AX and define distinct functions for H2AX in human cells.
INTRODUCTION
Nuclear DNA is bound by histones within nucleosomes to form chromatin (1). Core nucleosomes consist of two copies each of four canonical histones (H2A, H2B, H3, and H4) in an octamer that contains ∼146 bp of DNA wrapped around the histone protein core. In mammalian genomes, several histone variants resembling core histones exist, such as the histone variant H2AX, which is nearly identical to H2A except for a divergent and extended C terminus. Histones can be modified on specific amino acid residues by various posttranslational chemical modifications (PTMs), including methylation, acetylation, and phosphorylation (2–4). In addition, lysine residues can be modified by the covalent attachment of small polypeptides such as ubiquitin (Ub) and SUMO (small ubiquitin-like modifier) (5). These various PTMs are catalyzed by “writer” enzymes and are removed by additional enzymes that act to “erase” these marks (3). Together, these enzymes and chromatin binding proteins dynamically regulate the structure and functions of chromatin, which in turn regulates fundamental nuclear processes, such as chromosome replication and segregation, transcription, and DNA repair.
The protection of our genetic material is paramount for averting various human diseases, and chromatin plays an important role in coordinating the repair of nuclear DNA (6, 7). Cells have evolved a complex network of diverse cellular pathways, termed the DNA damage response (DDR), which detects damaged DNA, signals its presence, and promotes DNA repair (6, 7). DNA double-strand breaks (DSBs) represent a particularly challenging and cytotoxic form of DNA damage. DSBs create discontinuities in chromosomal DNA that, if not repaired or repaired incorrectly, result in mutations, chromosome loss, and/or ongoing genome instability. DSBs are predominantly repaired by either homologous recombination (HR) or nonhomologous end joining (NHEJ) (8). DSB repair by HR is initiated by the process of DNA end resection that facilitates the utilization of a homologous DNA molecule for the accurate copying and repair of a DSB, whereas NHEJ joins DSBs with minimal DNA end processing. It is becoming clear that chromatin and histone modifications, as well as chromatin binding and modifying enzymes, are important regulators of both HR and NHEJ (9).
The histone variant H2AX represents the quintessential example of how chromatin is involved in the DDR. Upon DSB formation, H2AX is phosphorylated on Ser-139 within its C-terminal tail by the DDR kinases ATM, ATR, and DNA-dependent protein kinase (DNA-PK), to yield γH2AX (10, 11). γH2AX generation can be propagated for over a megabase of chromatin surrounding the DSB site, thus creating microscopically visible ionizing radiation-induced nuclear foci (IRIF) (10, 12–14). γH2AX does this, at least in part, by creating a binding site for the DDR protein MDC1, which then helps mediate the DSB localization of the ubiquitin E3 ligases RNF4, RNF8, RNF168, HERC2, and BRCA1, which also colocalize at DSB sites with the SUMO E3 ligases PIAS1 and PIAS4 (15–22). Ubiquitylated histones H2AX and H2A mediate the chromatin association of BRCA1 through their recognition by the ubiquitin-interaction motif (UIM) domains of the BRCA1-interacting protein RAP80 (23). Notably, in the absence of γH2AX, many DDR proteins, including the DDR mediator proteins MDC1 and 53BP1, fail to form foci effectively at DSB sites (24–27). Accordingly, H2AX−/− deletion in mice results in genome instability, hypersensitivity to DNA damage, and elevated cancer predisposition, potentially due to defective IRIF formation and activation of alternative error-prone DNA repair pathways (25, 28–30, 52). Whether these functions for H2AX are entirely conserved in humans, however, is currently unknown due to the unavailability of a H2AX knockout (KO) model in human cells.
As is the case for core histones, H2AX PTMs have been described, and several of these have been linked to the DDR. For example, H2AX is acetylated on lysine 5 by the histone acetyltransferase (HAT) TIP60, which regulates H2AX ubiquitylation that, in turn, affects H2AX chromatin dynamics in response to DNA damage (31). Furthermore, H2AX is phosphorylated on tyrosine 142 by the kinase WSTF (32), and upon DNA damage, this site is dephosphorylated by the phosphatase EYA (33), with impairment of either of these two enzymes resulting in a defective DDR (32, 33). By using reconstituted H2AX−/− mouse embryonic stem (ES) cells, additional IR-induced H2AX modifications, including acetylation of lysine 36 and phosphorylation of threonine 101, have been identified (34). Collectively, these studies have shown that the engagement of H2AX in the DDR is not solely governed by γH2AX and have suggested that H2AX is decorated by multiple PTMs that may contribute combinatorially to a chromatin-mediated DDR.
As described herein, in an effort to comprehensively analyze H2AX PTMs, we performed an extensive screen of 67 individual alanine substitutions of key H2AX residues involved in PTM formation or DNA damage tolerance. By using this library, we have screened each mutant for its ability to affect H2AX ubiquitylation and Ser-139 phosphorylation (γH2AX generation) upon DSB induction, as well as sumoylation, a histone mark previously unidentified in H2AX. We find that several residues support the ubiquitylation and phosphorylation of H2AX and furthermore, identify an acidic triad motif required in cis for H2AX and H2A ubiquitylation. Additionally, we present the first analysis of an H2AX gene knockout in human cells. We find that deletion of H2AX results in increased DSB formation, p53 activation, DNA damage focus formation, and defective cell cycle progression. Upon DNA damage, DDR signaling is defective in the absence of H2AX, including defects in both ATM-dependent phosphorylations and p53 induction. Collectively, our results establish a map of key H2AX residues and establish a powerful genetic complementation system to comprehensively dissect the function of individual, as well as combinatorial, PTMs of H2AX. The differences between mouse and human cells for the requirement of H2AX in the DDR highlight the importance of creating and analyzing gene knockouts, such as H2AX, in multiple human cell types.
MATERIALS AND METHODS
Cells, cell culture, reagents, and treatments.
U2OS cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin, 100 mg/ml streptomycin, and 2 mM l-glutamine. The MCF10A H2AX−/− knockout cell line was obtained by zinc finger nuclease (ZFN)-mediated deletion (Sigma). MCF10A H2AX+/+ and H2AX−/− cell lines were grown in DMEM–Ham's nutrient mixture F-12 (1:1) (Sigma) with 2.5 mM l-glutamine, 5% horse serum, 10 μg/ml human insulin, 0.5 μg/ml hydrocortisone, 10 ng/ml epidermal growth factor (EGF), and 100 ng/ml cholera toxin. A 3×Flag tag was added to the N terminus of H2AX by PCR, and this construct (fH2AX) was cloned into pENTR11. 3×Flag-H2AX was Gateway cloned into pcDNA 6.2V5/DEST. A stop codon in the C terminus coding sequence was introduced to inhibit V5 tagging of H2AX. Indicated point mutants in H2AX were introduced using the QuikChange mutagenesis kit (Stratagene). 3×Flag-H2AX-9K to R was custom synthesized and subsequently cloned in the same manner as 3×Flag-H2AX into the pcDNA 6.2V5/DEST vector. Single arginine-to-lysine revertants in 3×Flag-H2AX-9K to R were introduced using the QuikChange mutagenesis kit (Stratagene). All constructs were fully sequenced following mutagenesis. To analyze the expression of proteins from vectors, cells were transfected with FugeneHD in antibiotic-free DMEM with the indicated plasmid DNA. After 12 h, normal medium was added and cells were allowed to grow for 48 to 72 h before being analyzed. All vector constructs used in this study are listed in Table S1 in the supplemental material. DNA damage treatments were as follows. Cells were exposed to 10 Gy ionizing radiation (IR) using a Faxitron X-ray unit and subsequently placed in a 37°C incubator for recovery for 1 h unless otherwise indicated. UV treatments were at 20 J/m2 followed by recovery for 1 h. Drug treatments were with methyl methanesulfonate (MMS) (Sigma) (3 mM, 60 min), camptothecin (CPT) (Sigma) (1 μM, 60 min), H2O2 (500 μM, 30 min), etoposide (25 μM, 60 min) or phleomycin (60 μg/ml, 2 h unless stated otherwise). ATM inhibitor (KU-55933; Tocris) (20 μM, 2 h) and MG132 (Tocris) (10 μM, 1 h) were used to inhibit ATM and the proteosome, respectively. Small interfering RNA (siRNA) experiments were performed as previously described (17). The antibodies used in this study are listed in Table S2 in the supplemental material.
Protein extracts and Western blotting.
For whole-cell extracts (WCEs), cells were washed once with phosphate-buffered saline (PBS), collected by adding Laemmli buffer (4% [vol/vol] SDS, 20% [vol/vol] glycerol, and 120 mM Tris [pH 6.8]), sonicated in a Diagenode Bioruptor 300 for 10 min, and boiled for 5 min at 95°C before loading. For immunoprecipitations (IPs), cells were washed once with PBS and scraped into radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1.0% IGEPAL CA-630 [octylphenoxypolyethoxyethanol] [NP-40], 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris [pH 8.0], 2 mM EDTA) containing 1× protease inhibitor without EDTA tablet (Roche) and 2.5 mM NEM (N-ethylmaleimide). Extracts were sonicated in a Diagenode Bioruptor 300 for 10 min, followed by clearing of the extract by centrifugation. Flag-tagged proteins were immunoprecipitated with EZview Red anti-Flag M2 affinity gel beads (Sigma) and eluted from the beads with the 3×Flag peptide (Sigma) following the manufacturer's suggestions. Samples from whole-cell extracts or immunoprecipitations (IPs) were resolved by SDS-PAGE and analyzed by standard Western blotting techniques. Antigens were detected by standard chemiluminescence (GE Healthcare, Amersham ECL Prime system) using a Bio-Rad molecular imager ChemiDoc XRS+ system. Secondary antibodies used for enhanced chemiluminescence (ECL) were goat anti-rabbit IgG conjugated with horseradish peroxidase (HRP) (Cell Signaling) and horse anti-mouse IgG-HRP (Cell Signaling). Quantification of chemiluminescence signals was performed using the Bio-Rad molecular imager ChemiDoc XRS+ system with Image Lab software.
RNA analysis.
Total RNA was purified from each sample using the RNeasy purification kit from Qiagen and treated with Turbo DNase from Ambion following the manufacturer's protocol. cDNA synthesis of 500 ng of total RNA was performed with the Superscript III first-strand synthesis system (Invitrogen) using oligo(dT) and random hexamer primers. For quantitative real-time PCR (qRT-PCR) analyses, 1/50 of each reaction was used. The following gene-specific Quantitect primer assays were used in our analysis: ALAS1, B2M, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), p53, and p21. To analyze mRNA from the H2AX locus in MCF10A H2AX+/+ and H2AX−/− cells, we designed unique primers specific for 3 individual regions predicted to be located within the H2AX mRNA sequence. The sequences of these 3 primers are as follows: A, 5′ primer CGGGCGTCTGTTCTAGTGTT and 3′ primer GGTGTACACGGCCCACTG; B, 5′ primer ACGAGGAGCTCAACAAGCTG and 3′ primer CGGGCCCTCTTAGTACTCCT; and C, 5′ primer GGTGCTTAGCCCAGGACTTT and 3′ primer CCCAGCGCAGACCTATGAAT. RT-PCR analysis was performed using SYBR green (Applied Biosystems) for detection with an Applied Biosystems StepOne Plus system.
MTT and FACS analysis.
MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] assays were performed with MCF10A H2AX+/+ and H2AX−/− cell lines using the Vybrant MTT cell proliferation assay kit (Invitrogen) following the manufacturer's protocol. Several dilutions of cells were analyzed, and data obtained from dilutions that were within the linear range of detection are shown. Data obtained from 3 independent experiments were analyzed and graphed using Graphpad Prism software. Error bars represent standard errors of the mean (SEM). For fluorescence-activated cell sorter (FACS) analysis, MCF10A H2AX+/+ and H2AX−/− cell lines were trypsinized and washed in ice-cold PBS followed by resuspension with a small pipette tip to get a single-cell suspension. Cells were added dropwise into a slowly vortexed conical tube containing ice-cold 70% ethanol to fix. Cells were fixed overnight at 4°C. Cells were pelleted and resuspended in PBS containing propidium iodide (40 μg/ml) and RNase A (100 μg/ml) followed by incubation for 1 h at 37 C. Cells were filtered through a 70-μm-mesh tube and analyzed on a BD LSRFortessa cell analyzer. FlowJo software was used to analyze flow cytometry data.
Neutral comet assays.
Comet assays were carried out with the Trevigen standardized CometAssay system (Trevigen) and performed as previously described (35). Images were captured with an EVOSfl fluorescence microscope (AMG) containing a green fluorescent protein (GFP) filter. Images were analyzed with CometScore to determine the percentage of DNA in the tail. For each sample, more than 100 cells were counted from five independent experiments. Values were plotted in Prism. (Error bars on figures represent the SEM.) P values between samples were calculated with the paired t test using Graphpad software.
Immunofluorescence analyses.
MCF10A cells were grown in Nunc Lab-Tek II chamber slides. After the indicated treatments, slide chambers were washed once with PBS at room temperature. Cells were washed once in cold PBS and fixed with 2% (vol/vol) paraformaldehyde (PFA) for 15 min at room temperature followed by three washes with PBS. Cells were permeabilized with 0.2% Triton X-100 for 10 min followed by three washes with PBS. Slides were blocked for 15 min at room temperature in blocking buffer (PBS containing 3% bovine serum albumin [BSA]). Primary antibodies were incubated for 1 h at room temperature in the same buffer. Cells were then washed three times in PBS before incubation in the dark with Alexa Fluor-conjugated secondary antibodies (Invitrogen) in PBS for 45 min at room temperature. Cells were again washed three times in PBS. The slides were then mounted in Vectashield containing DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories). Cells were imaged with an inverted IX71 fluorescence microscope (Olympus), and data were collected and analyzed with CellSens software.
Cell survival assays.
MCF10A H2AX+/+ and H2AX−/− cells were treated with phleomycin (for 2 h followed by washing). Cells were left to form colonies for 10 to 14 days at 37°C. Colonies were stained with crystal violet, washed, and counted. Results were normalized to plating efficiencies of untreated cells for each cell line.
RESULTS
H2AX is ubiquitylated and sumoylated.
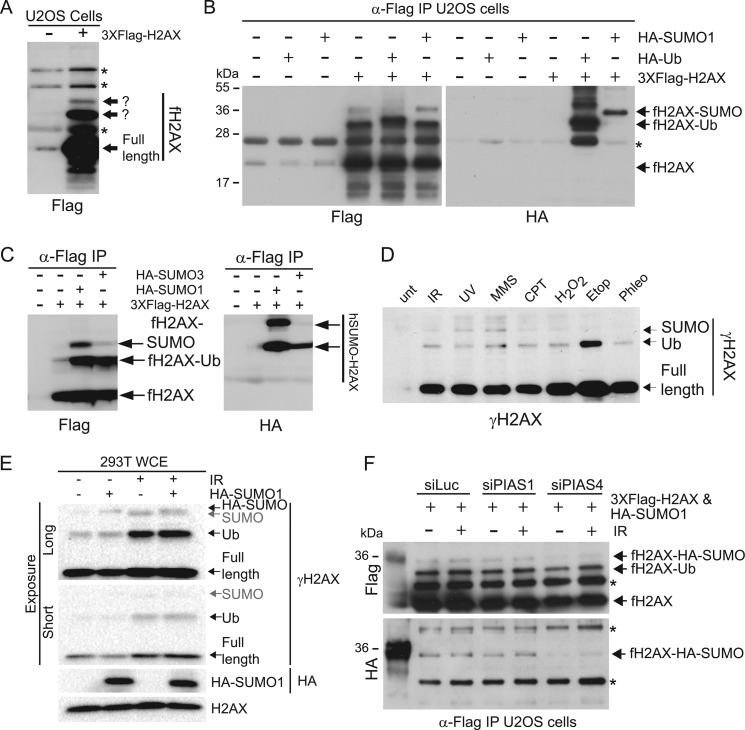
While many studies have focused on how H2AX phosphorylation affects the DDR, we reasoned that additional, as-yet-unidentified H2AX PTMs, as well as regulatory regions, could contribute to H2AX functions. To provide sensitive and specific detection of H2AX, we cloned a triple-Flag affinity tag on the H2AX N terminus (fH2AX) and verified expression of 3×Flag-H2AX in human U2OS cancer cells by transfection followed by immunoprecipitation (IP) and Western blotting (Fig. 1A). Notably, while confirming expression of this fusion protein, this analysis detected multiple protein bands, suggesting that H2AX was modified by additional PTMs that retarded its electrophoretic mobility (asterisks indicate cross-reacting protein species). As previous studies have shown that both H2A and H2AX are ubiquitylated (11, 18, 36), we predicted that one H2AX species represented ubiquitylated H2AX and hypothesized that one or more of the other bands could reflect H2AX sumoylation (ubiquitin [Ub] and SUMO have molecular masses of 8.5 and 12 kDa, respectively, which is consistent with electrophoretic mobilities of the two additional forms of H2AX [arrows in Fig. 1A and B]). To test this, we performed cotransfection experiments with fH2AX and hemagglutinin (HA)-tagged forms of ubiquitin (HA-Ub) and SUMO1 (HA-SUMO1). We observed decreased mobility of the suspected H2AX-ubiquitin band when overexpressing HA-Ub, while overexpression of HA-SUMO1 decreased the mobility of the predicted H2AX-SUMO band (Fig. 1B). Furthermore, Western blotting of anti-Flag IP samples with an HA-specific antibody confirmed the identification of fH2AX-ubiquitin and fH2AX-SUMO1 (Fig. 1B) (These H2AX modifications also occurred in 293T cells [data not shown].) In addition to detecting sumoylation of H2AX, we detected sumoylated core histone H4, as has been previously reported (37), as well as sumoylated H2A and H3 (data not shown). Human cells express SUMO1 and two highly related and functionally overlapping SUMO isoforms, SUMO2 and SUMO3. By performing experiments similar to those in Fig. 1B but with either HA-SUMO1 or HA-SUMO3, we observed that H2AX was preferentially sumoylated by SUMO1 (Fig. 1C). In addition, by analyzing Ser-139 H2AX phosphorylation with a γH2AX-specific antibody, we found that the phospho-form of endogenous H2AX (γH2AX), produced in response to various DNA-damaging agents exhibited a similar staining pattern to fH2AX (Fig. 1D). The ratios of γH2AX of full-length, ubiquitylated and sumoylated forms H2AX were not identical upon treatment with different DNA-damaging agents, however, suggesting that certain types of DNA damage might regulate these H2AX PTMs differently. To unambiguously assign this band as being that of endogenous sumoylated H2AX, we reasoned that expression of HA-tagged SUMO1 would result in a mobility shift of this band due to the increased size of the tagged SUMO. Indeed, as in Fig. 1D, we observed a band corresponding to sumoylated endogenous H2AX in 293T cells treated with DNA damage (Fig. 1E; SUMO-γH2AX). Expression of HA-SUMO1 resulted in a mobility shift of this band while not affecting the mobility of other H2AX species (Fig. 1E). Therefore, we conclude that we can detect endogenous H2AX that is sumoylated. We next sought to identify the SUMO E3 ligase that is responsible for H2AX sumoylation. Knockdown of PIAS4, but not PIAS1, resulted in the reduction of fH2AX-SUMO (Fig. 1F). Coexpression of HA-SUMO, like in Fig. 1B, allowed us to assign the fH2AX-SUMO band in these experiments. As PIAS4 is involved in the DNA damage response (17), these experiments suggest that H2AX sumoylation could play a role in this process as well, although we were unable to detect a DNA damage induction of this modification under these experimental conditions. Collectively, these results identify sumoylation as a new modification on H2AX that is mediated by the PIAS4 SUMO E3 ligase.
Fig 1.
H2AX is ubiquitylated and sumoylated. (A) H2AX is posttranslationally modified. Empty vector (−) or vector expressing 3×Flag-tagged H2AX (fH2AX) (+) was transfected into U2OS cells, and then H2AX was immunoprecipitated with anti-Flag antibody and detected by Western blotting with this antibody. Arrows indicate full-length fH2AX as well as two, slower-migrating fH2AX species. (B) H2AX is ubiquitylated and sumoylated. As indicated, U2OS cells were transfected with fH2AX or empty vector, either together with (+) or in the absence of (−) vectors encoding HA-tagged ubiquitin (HA-Ub) or SUMO1. Cells were treated as in panel A and then analyzed with the indicated antibodies. α-Flag, anti-Flag. Arrows indicate H2AX species. (C) H2AX is preferentially sumoylated by SUMO1. fH2AX coimmunoprecipitations with either HA-SUMO1 or HA-SUMO3 were performed as in panel B. (D and E) Endogenous H2AX is sumoylated. For panel D, U2OS cells were untreated (unt) or were treated with the indicated DNA-damaging agents and then analyzed and detected by Western blotting with a γH2AX-specific antibody. Etop, etoposide. For panel E, 293T cells were transfected with either an empty vector or a vector encoding HA-tagged SUMO1. Cells were untreated or treated with 5 Gy IR and then analyzed and detected by Western blotting with the specified antibodies. Arrows indicate the various H2AX species. Note the shift of mobility of the H2AX sumoylation band in cells expressing HA-tagged SUMO1. WCE, whole-cell extracts. (F) H2AX sumoylation is dependent on PIAS4. U2OS cells were transfected with control (siLuc), PIAS1, or PIAS4 siRNAs followed by transfection with both fH2AX and HA-tagged SUMO1. Cells were analyzed as in panel B. Asterisks indicate background bands.
We next sought to identify the sumoylation and ubiquitylation sites in H2AX. Because ubiquitylation and sumoylation occur on lysine residues, we generated a synthetic H2AX cDNA containing a triple-Flag epitope on its N terminus (termed “fH2AX-9K to R”) in which nine lysine residues were mutated to arginines, which conserve the positive charge but cannot be ubiquitylated or sumoylated (Fig. 2A). The choice of these nine lysine residues was based on modeling of H2AX onto the nucleosome crystal structure of histone H2A, which predicted that these residues would be surface exposed (data not shown). In accord with our predictions, Western blot analysis of fH2AX-9K to R, unlike wild-type (wt) fH2AX, showed virtually no ubiquitylation or sumoylation (Fig. 2B). For modification site mapping of these PTMs, we next made nine additional fH2AX constructs in which each arginine was individually mutated back to a modifiable lysine. (Each construct thus only contained one acceptor lysine within the nine possible sites.) While fH2AX-9K to R exhibited neither ubiquitylation nor sumoylation, specific arginine-to-lysine reversion mutations restored the ability of fH2AX to be modified (Fig. 2C). For ubiquitylation, both fH2AX-9K to R-R118K and -R119K restored near wild-type levels of ubiquitylation (Fig. 2C), and we confirmed ubiquitylation of these H2AX mutants by the use of tagged ubiquitin (data not shown). In contrast, restoration of fH2AX-SUMO was observed, although to various degrees, for all single arginine-to-lysine revertants and sumoylated H2AX was confirmed using HA-tagged SUMO1 (Fig. 2C). H2AX Lys-5 and Lys-127 are predicted to be sumoylation sites by the SUMOplot program. H2AX-9K to R lacks detectable sumoylation, suggesting that the lysine acceptor site is among these lysine residues. Collectively, our results have mapped the ubiquitin receptor sites of H2AX to Lys-118 and Lys-119 for monoubiquitylation and suggest that multiple lysine residues found in H2AX-9K to R can act as an H2AX sumoylation site.
Fig 2.
Ubiquitylation and sumoylation site mapping in H2AX by fH2AX-9K to R analysis. (A) Diagram of the fH2AX-9K to R construct. All lysine (K)-to-arginine (R) substitutions in fH2AX-9K to R are indicated. (B) fH2AX-9K to R blocks ubiquitylation and sumoylation. The indicated constructs were transfected into U2OS cells and analyzed as in Fig. 1A. Vec, empty vector. (C) Lysines 118 and 119 of H2AX are the preferred ubiquitylation sites. Cells were transfected with various Flag-H2AX constructs and HA-tagged SUMO1 as indicated. Experiments were analyzed as in panel B with the indicated antibodies, except that anti-Flag immunoprecipitants were eluted with Flag peptide as described in Materials and Methods. Exp., exposure.
Comprehensive screen for H2AX PTMs and functional residues.
After establishing that H2AX is phosphorylated, ubiquitylated, and sumoylated, we sought to systematically analyze the contribution of individual H2AX residues to the regulation of these PTMs both under normal conditions and in response to DNA damage. By using fH2AX, we were able to assess its ubiquitylation, sumoylation, and S139 phosphorylation (γH2AX) status by Western blotting (Fig. 1 and 2). To screen for functional amino acid residues, we mutated individual residues in fH2AX to alanine to disrupt their function (Fig. 3A). Residues targeted for mutagenesis included serines, threonines, tyrosines, and histidines that can be phosphorylated, lysines that can be modified by ubiquitylation, sumoylation, acetylation, and methylation, as well as arginines, which can be methylated. In addition to this group of 45 H2AX mutations (residues coded green in Fig. 3A), we also mutated conserved residues that, when mutated in yeast (Saccharomyces cerevisiae) H2A, resulted in DNA damage hypersensitivity. (H2AX constitutes the core H2A histone in yeast, and mutational analysis has revealed DNA damage hypersensitivity for 15 individual residues, all of which are conserved in mammalian H2AX [38].) We reasoned that the latter group of mutations (residues coded red in Fig. 3A) could affect H2AX PTMs. We also mutated additional residues corresponding to amino acids (with the exception of glycines and alanines) that are present in the H2AX C-terminal extension but not in H2A (10, 11) (colored purple in Fig. 3A).
Fig 3.
Comprehensive screen for residues in H2AX that affect its ubiquitylation, sumoylation, and/or S139 phosphorylation. (A) Schematic outline of the H2AX screen. The amino acid sequence of H2AX is shown with individually mutated residues highlighted in one of 4 colors to indicate its category (see the text and legend for details). The diagram on the right graphically represents the protocol for the screen. (B) Complete H2AX screen in untreated and DNA damage conditions. The indicated constructs containing a single-amino-acid substitution to alanine in fH2AX, along with either vector or a wild-type fH2AX control, were transfected into U2OS cells. Cells were either untreated or treated with 60 μg/ml of phleomycin (Phleo) for 2 h. Whole-cell extracts were analyzed by Western blotting with Flag- and γH2AX-specific antibodies. Phospho S139, SUMO, and ubiquitylated (Ub) and unmodified (full-length) species of fH2AX are indicated. (C) Confirmation of positive hits from the H2AX screen. Mutants from panel B that affected any of the 3 PTMs analyzed in the screen compared to the wild type were reanalyzed as in panel B. (D) Quantification of γH2AX levels. The signal from full-length fH2AX from panel C was quantified. The signal from γH2AX was quantified, normalized to full-length levels, and plotted. Samples showing a greater than 25% difference in DNA damage-induced γH2AX compared to wild-type fH2AX were considered positive and are shown in red. (E) Quantification of H2AX ubiquitin levels. H2AX-Ub signals from panel C were quantified, normalized to full length, and compared to wild-type H2AX-Ub levels. Mutants whose Ub levels were greater than 25% different from wild-type H2AX-Ub levels were considered positive and are labeled in red. (F) Results of the H2AX screen. Mutants reproducibly affecting either of three PTMs of H2AX in both experiments from panels B and C are listed (see the text for details).
Each of the resulting 67 mutant H2AX derivatives was then transfected into human U2OS cells, the cells were mock treated or treated with phleomycin to induce DSBs, and cell extracts were analyzed by Western blotting with antibodies specific to Flag or γH2AX to determine ubiquitylation, sumoylation, and γH2AX levels (Fig. 3A). As expected, fH2AX expression was detected as three prominent bands representing the full-length, ubiquitylated and sumoylated forms of H2AX (Fig. 3B). Of the 67 mutant H2AX derivatives, 52 exhibited apparently normal ubiquitylation, sumoylation, and γH2AX levels (Fig. 3B). No DNA damage-dependent changes in either H2AX ubiquitylation or sumoylation were detected in our screen, suggesting that these modifications are not DNA damage responsive under our conditions. As anticipated, fH2AX S139A did not yield a γH2AX signal, confirming the specificity of the antibody. Notably, several mutations surrounding S139 reduced the extent of γH2AX detection (category 1A mutants). These mutations likely disrupted the γH2AX epitope, so a direct comparison of phosphorylation levels could not be performed. However, Q137A, Q140A, and Y142A did show some, albeit low, levels of γH2AX induction after DNA damage, indicating that these residues are not absolutely required for γH2AX formation and/or detection. After our initial screen, we repeated these experiments with all potential hits from the first screen, excluding the mutations that were in proximity to S139 (Fig. 3C). We note that expression of several H2AX mutants resulted in spurious bands detected by both γH2AX and Flag antibodies (for example, see fH2AX H38A, R77A, or E92A). These bands are background bands that arise in these samples due to increased extract loading used to normalize the loading of full-length H2AX, which was required for our analysis for quantification. To overcome this issue, we quantified full-length H2AX for the wild type and all mutants and used these values to normalize γH2AX levels upon DNA damage (Fig. 3D) and H2AX-Ub levels (Fig. 3E). This analysis revealed mutants that displayed reduced γH2AX formation and mutants defective in H2AX ubiquitylation compared to wild-type H2AX (listed in Fig. 3F). This analysis identified 3 mutations that were not in the proximity of S139 that reduced γH2AX levels upon DNA damage (category 1B) (Fig. 3B to D). Strikingly, eight amino acid residues were identified that affected H2AX ubiquitylation (category 2) (Fig. 3B, C, E, and F). Interestingly, some but not all mutants in this category reduced γH2AX formation, suggesting that normal ubiquitylation levels were not absolutely required for this phosphorylation, as has been suggested previously (39, 40). In contrast, no mutated single amino acid residue in fH2AX resulted in a clear reduction in fH2AX-SUMO (Fig. 3B, C, and F). These data are consistent with our results from fH2AX-9K to R, which suggested that SUMO can independently modify multiple H2AX lysine residues.
Mapping of these mutations onto the nucleosome crystal structure revealed that several positive hits, including I111A and L116A, are located in close proximity to the prominent ubiquitylated Lys-119 residue (data not shown). Another interesting mutation, E92A, occurs within the region of H2AX corresponding to the conserved acidic patch region of H2A that acts as an interaction domain for several proteins, including histone H4 (41–43). Furthermore, some mutations that reduced H2AX ubiquitylation, including L65A and L93A, mapped near E92 on the histone H2A(X) surface (data not shown). Collectively, this work provided a comprehensive analysis of amino acid residues in H2AX that impact its modification states, thus providing a framework for future analyses to decipher their functions.
An acidic triad motif, containing E92, promotes H2A(X) ubiquitylation.
We next focused on the E92A mutation due to its location within a known interaction domain in H2A. We confirmed that fH2AX E92A displayed reduced γH2AX formation and ubiquitylation compared to wild-type fH2AX (Fig. 4A; graph showing quantification of Western blotting signals), and immunoprecipitation experiments with H2AX E92A and HA-ubiquitin confirmed that the reduced H2AX species corresponded to monoubiquitylated H2AX (Fig. 4B). Notably, HA-Ub expression increased the ubiquitylation of fH2AX E92, suggesting that HA-ubiquitin overexpression could partially alleviate the requirement of fH2AX E92A for this modification (Fig. 1B and 4B). We also noted that HA-ubiquitin overexpression reduced fH2AX sumoylation, suggesting an interplay/competition between H2AX ubiquitylation and sumoylation. These results established that H2AX E92 was required for effective fH2AX ubiquitylation. Published crystal structures of two H2A-interacting proteins, LANA and RCC1, revealed an interaction between E61 and D90, which together with E92, created an acidic triad structure on H2A (41, 43). Consistent with this acidic triad being required for H2AX ubiquitylation, as for E92A, mutation of either E61 or D90 impaired fH2AX monoubiquitylation (Fig. 4C) (Note that mutation of H2AX E91, another acidic residue adjacent to E92, caused only a minor reduction in fH2AX-ubiquitin formation.) In parallel studies, we observed that analogous mutations in histone H2A impaired its ubiquitylation (Fig. 4D), suggesting that histone H2AX and H2A are ubiquitylated by a similar mechanism. Further analysis of these mutations with coexpression of HA-SUMO1 followed by an anti-Flag IP revealed that sumoylation of H2AX could occur independently from its acidic triad-dependent ubiquitylation (Fig. 4E). In accord with Fig. 1E, we could observe a mobility shift of sumoylated H2AX by the coexpression of HA-tagged SUMO, which allows us to unambiguously assign this band as being that of sumoylated H2AX. These results thereby uncovered a novel interaction domain, the acidic triad, which is required in cis for both H2A and H2AX ubiquitylation (Fig. 4F). Our structural mapping analysis revealed that the acidic triad is located on the same surface and side as Lys-118 and Lys-119 on the H2A(X) structure (Fig. 4F). Notably, several other residues that, when mutated, affected H2AX ubiquitylation (including I111A and L116A) are located between the ubiquitylation site(s) and the acidic triad on the surface of H2A(X), while L65 and L93, whose mutation also impaired H2AX ubiquitylation, are located adjacent to the acidic triad (data not shown). Thus, our results are consistent with these residues collectively defining an interaction interface that allows H2A(X) ubiquitylation.
Fig 4.
Identification of acidic residues in H2AX and H2A that are required for ubiquitylation in cis. (A) fH2AX E92A exhibits reduced γH2AX formation upon DNA damage. Cells were transfected with the indicated vectors as in Fig. 3B and C. For DNA damage, transfected cells were treated with 60 μg/ml of phleomycin for 2 h and samples were collected at the indicated times post-phleomycin treatment. Samples were analyzed as in Fig. 3B and quantified as in Fig. 3D. Note that fH2AX E92A shows defective fH2AX-Ub and γH2AX formation. (B) Ubiquitylation of H2AX is reduced in H2AX E92A. Experiments were performed with the indicated constructs as in Fig. 1B. Cells were damaged as in panel A. (C) Mutations of specific acidic amino acids in H2AX affect its monoubiquitylation. The indicated fH2AX mutants were analyzed as in Fig. 2C. Cells were damaged as in panel A. (D) Mutation of specific acidic amino acids in H2A affects its monoubiquitylation. The indicated fH2A mutants were analyzed as in Fig. 2C. Cells were damaged as in panel A. (E) Acidic triad mutants of H2AX are sumoylated. Experiments were performed as in Fig. 1B by cotransfections with HA-SUMO1 and fH2AX mutants. Arrows identify H2AX and its modified forms as labeled. Note the shift of mobility of the H2AX sumoylation band in cells expressing HA-tagged SUMO1. (F) Molecular modeling of acidic triad motif mutants and Lys-119 ubiquitylation of H2A(X). Nucleosomes were modeled from the X-ray crystal structure (1.9 A) of a Xenopus nucleosome associated with human DNA (42). The structure was retrieved from a protein data bank (accession no. 1KX5). Modeling and coloring were performed in PyMol. The acidic triad motif refers to E61, D90, and E92 of H2A(X). Individual components of the model were colored to facilitate visualization as indicated in the legend.
H2AX-deficient human cells exhibit growth and repair defects.
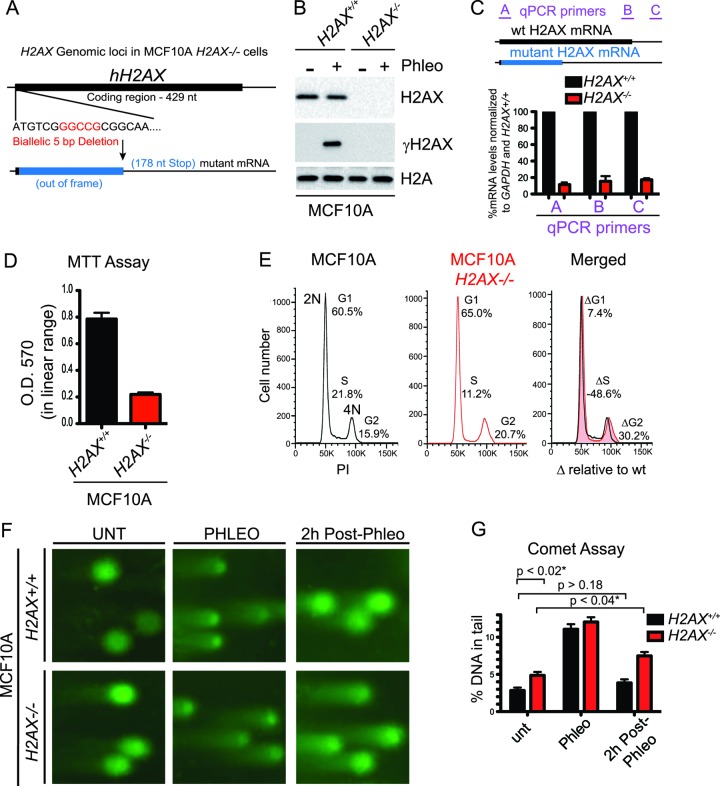
We next sought to create a genetic complementation system to analyze H2AX mutations in human cells. To this end, we obtained a human breast epithelial cell line, MCF10A, that contained a full gene knockout of H2AX that was targeted by zinc finger nuclease (ZFN)-mediated deletion (Fig. 5A). Western blot analysis confirmed the loss of H2AX in the MCF10A H2AX−/− cell line compared to MCF10A H2AX+/+ cells (Fig. 5B). Furthermore, DNA-damage induced γH2AX formation was undetectable in this cell line (Fig. 5B). Real-time quantitative PCR (RT-qPCR) analysis of mRNA levels using 3 qPCR primers targeting different regions of the H2AX transcript revealed that the mutant RNA expressed in MCF10A H2AX−/− cells is reduced by over 80% compared to H2AX mRNA levels in wild-type MCF10A cells (Fig. 5C). Taken together, these data confirm that these cells lack H2AX at both the protein and mRNA levels. While culturing the MCF10A H2AX−/− cell line, we observed a slow-growth phenotype, which was confirmed by MTT growth assays, revealing ∼4-fold decreased growth of MCF10A H2AX−/− cells compared to MCF10A H2AX+/+ cells (Fig. 5D). FACS analysis of MCF10A H2AX−/− cells showed an approximate 50% reduction in S-phase cells, as well as an increase in cells in the G2/M phase of the cell cycle, compared to their H2AX-containing counterparts (Fig. 5E). Consistent with these phenotypes and the known functions of H2AX, MCF10A H2AX−/− cells exhibited higher levels of spontaneously arising DSBs, as detected by neutral comet assays (Fig. 5F and G). Moreover, we observed that while MCF10A H2AX−/− cells were able to repair phleomycin-induced DSBs, the rate at which they did so was reduced compared to that of MCF10A H2AX+/+ cells (Fig. 5F and G). These results indicate that, as in mouse cells (25, 28, 29), H2AX promotes DSB repair in human cells.
Fig 5.
Deletion of H2AX in human cells results in cell cycle and growth defects. (A) Schematic of H2AX genomic loci in human MCF10A H2AX−/− cells. ZFN-mediated knockout of H2AX resulted in a biallelic 5-bp deletion resulting in a truncated and out-of-frame mRNA transcript. (B) Human MCF10A H2AX−/− cells lack H2AX. Loss of H2AX was confirmed by Western blotting with H2AX and γH2AX antibodies in wild-type and H2AX knockout MCF10A cells. H2A is a loading control. Cells were damaged as in Fig. 3B. (C) mRNA from the H2AX locus is reduced in MCF10A H2AX−/− cells. RNA was isolated from both MCF10A H2AX+/+ and H2AX−/− cells and analyzed by RT-qPCR with primers specific for the H2AX locus and GAPDH. The locations of H2AX locus-specific qPCR primers A, B, and C are indicated. Data were normalized to GAPDH and MCF10A H2AX+/+ samples. The graph represents data from 3 independent experiments, and error bars represent SEM. (D) Reduced proliferation in MCF10A H2AX−/− cells. MTT assays were performed on the indicated cell lines as described in Materials and Methods. O.D., optical density. Error bars represent SEM. (E) Cell cycle analysis by flow cytometry (FACS). Wild-type and H2AX-deleted MCF10A cells were analyzed by FACS with propidium iodide (PI) staining. DNA contents (2N and 4N) are indicated, and the DNA profiles are compared (overlaid right panel). The Δ cell cycle values represent percentages of the indicated cell cycle phase in MCF10A H2AX−/− cells relative to MCF10A cells. (F) MCF10A H2AX−/− cells accumulate DNA double-strand breaks (DSBs). Wild-type and MCF10A H2AX−/− cells were either untreated (UNT) or were treated with 60 μg/ml of phleomycin for 2 h followed by 2 h of recovery. Samples were analyzed by a neutral comet assay to detect DSBs. (G) Quantification of experiments from panel F. Data were quantified in Cometscore and are represented as the percentage of DNA in the tail. Error bars represent SEM of five independent experiments. P values are indicated for some comparisons. *, statistically significant at P < 0.05.
Analysis of 53BP1 localization in the absence of H2AX.
In addition to it being recruited to IRIF, the DDR mediator protein 53BP1 also localizes to nuclear bodies, termed 53BP1-OPT domains, in G1 cells that mark sites of endogenous replicative stress encountered during the preceding S phase (44, 45). We found that 53BP1 nuclear bodies were present in MCF10A cells, regardless of their H2AX status (Fig. 6A). Furthermore, as described previously (44, 45), these structures were restricted to cyclin A-negative, G1 cells, and their maintenance was decreased by inhibiting the activity of ATM or the proteosome (44, 45; data not shown). Notably, consistent with H2AX−/− cells exhibiting higher levels of DNA damage than control cells, the proportion of cells displaying 53BP1-OPT domains was enhanced in the absence of H2AX (Fig. 6A and B). (Note that H2AX status did not affect the cell cycle restriction of 53BP1 foci to G1 cells.) Previous work has shown that mouse H2AX−/− cells exhibit defective MDC1 and 53BP1 IRIF formation (25, 27). In accord with these reports, we found that MCF10A H2AX−/− cells were impaired in accumulating both of these factors in IRIF (Fig. 6C). H2AX−/− cells also showed impaired 53BP1 accumulation at DNA damage sites induced by phleomycin and doxorubicin (data not shown). In mouse H2AX−/− cells, initial recruitment of 53BP1 to IRIF was reported to be essentially normal, while by 30 min, the maintenance of 53BP1 foci was impaired (24). In contrast, in human cells lacking H2AX, we observed impaired 53BP1 IRIF recruitment as early as 15 min after irradiation (Fig. 6D). At 30 and 60 min after irradiation, small nuclear 53BP1 foci started to accumulate in H2AX−/− cells, but these did not form foci comparable to those in H2AX+/+ cells (Fig. 6D). Thus, in human MCF10A epithelial cells, initial detection, as well as retention, of 53BP1 at DNA damage sites induced by several different DNA-damaging agents is largely H2AX dependent.
Fig 6.
H2AX loss affects 53BP1 focus formation in unperturbed and DNA damage-treated MCF10A cells. (A) Untreated MCF10A H2AX−/− cells accumulate 53BP1 foci. Wild-type and H2AX-deleted MCF10A cells were cultured, fixed, and analyzed by immunofluorescence (IF) with the indicated antibodies. The percentages of total cells staining positive for 53BP1 foci are shown in white. Over 100 cells from each cell line were quantified for 53BP1 foci and graphed. The numbers shown represent the numbers of 53BP1 foci, and error bars represent SEM (n = 3). Note that large 53BP1 foci in MCF10A cells contain γH2AX. (B) 53BP1 foci in untreated cells are enriched in the G1 phase of the cell cycle. Asynchronous cells were analyzed by IF with the indicated antibodies. Cyclin A staining was performed to detect S/G2-phase cells. Over 100 cells were counted for 53BP1 foci and cyclin A staining in each cell line, and error bars represent SEM (n = 3). We note that MCF10A H2AX−/− cells exhibit reduced cyclin A-positive cells and an increase in 53BP1 foci relative to MCF10A cells. (C) Impaired recruitment of MDC1 to DNA damage in MCF10A H2AX−/− cells. Cells were treated with 3 Gy IR and analyzed 60 min post-IR with the indicated antibodies. (D) DNA-damage-induced 53BP1 foci are defective in MCF10A H2AX−/− cells. Cells were irradiated and analyzed with the indicated antibodies at 15, 30, and 60 min after 3 Gy IR. Scale bars, 10 μm.
H2AX ubiquitylation is dispensable for DNA damage recruitment of 53BP1.
The establishment and characterization of a human cell line lacking H2AX allowed us to carry out complementation experiments with mutated H2AX derivatives. Importantly, expression of wild-type fH2AX in MCF10A H2AX−/− cells restored IR-induced 53BP1 focus formation (Fig. 7A). In contrast, and in accord with our other data, expression of fH2AX S139A did not rescue 53BP1 IRIF formation, thus confirming the requirement for H2AX Ser-139 phosphorylation in this process (Fig. 7A). Some studies have implicated H2AX ubiquitylation in the formation of γH2AX (39, 40), while another study showed that γH2AX occurred independently from its ubiquitylation (46). Nevertheless, despite our finding that fH2AX E92A was markedly defective in its ability to be ubiquitylated, expression of this mutated H2AX derivative, in MCF10A H2AX−/− cells rescued the ability of these cells to effectively mediate 53BP1 IRIF formation (Fig. 7A; quantified in panel B). In addition, reintroduction of fH2AX-9K to R, as well as all of the single R-to-K reversions, also restored to near-wild-type levels the ability of 53BP1 to form IRIF (Fig. 7A [data not shown]; quantified in panel B). We also sought to determine if BRCA1, a protein involved in homologous recombination, required ubiquitylated H2AX, as has been suggested. In agreement with mouse H2AX null cells, BRCA1 IRIFs were defective in MCF10A H2AX−/− cells (Fig. 7C). Reintroduction of either wild-type H2AX or H2AX-9K to R restored both 53BP1 and BRCA1 IRIF in complemented MCF10A H2AX−/− cells (Fig. 7C; compare GFP-positive to -negative cells to see differences in IRIF formation). Thus, in our human cell-based complementation system, γH2AX, but not H2AX ubiquitylation or sumoylation, is required for 53BP1 and BRCA1 accrual at DNA damage sites.
Fig 7.
Complementation of 53BP1 focus formation in MCF10A H2AX−/− cells. (A) Wild type, fH2AX E92A, and fH2AX-9K to R, but not fH2AX S139A, rescued 53BP1 IR-induced foci in MCF10A H2AX−/− cells. MCF10A H2AX−/− cells were transfected with either empty vector or vectors containing the indicated fH2AX constructs followed by 48 h of recovery. Cells were then treated and analyzed as in Fig. 6C at 15 min posttreatment with 3 Gy IR. Flag IF identified H2AX-expressing cells. (B) Quantification of panel A. More than 50 cells from both Flag-positive and Flag-negative cells were scored from wild-type, fH2AX E92A-, fH2AX-9K to R-, and fH2AX S139A-transfected MCF10A H2AX−/− cells for 53BP1 focus formation. Cells were scored as having either 0 to 10 53BP1 foci or more than 10 53BP1 foci. Cells staining negative for Flag from each of the 4 fH2AX samples were scored and combined to represent the Flag-negative sample. (C) Impaired BRCA1 recruitment to DNA damage sites in MCF10A H2AX−/− cells is rescued by H2AX-9K to R. Cells were treated and analyzed as in panel A, except using GFP-tagged H2AX and H2AX-9K. Dotted circles indicate cell nuclei. Scale bars, 10 μm.
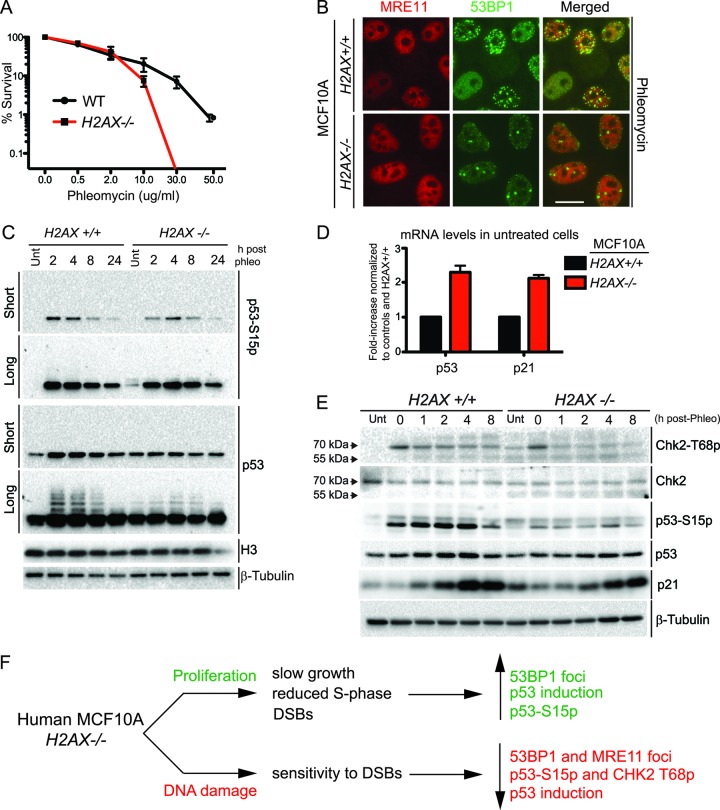
H2AX promotes human cell survival following DNA damage induction.
In line with data published on the phenotypes of H2AX−/− cells (25, 28), we found that H2AX loss caused human MCF10A cells to be hypersensitive to DNA-damaging agents (Fig. 8A). Before γH2AX is generated, the MRE11-RAD50-NBS1 (MRN) complex is recruited to DSBs. This complex then mediates the recruitment of the protein kinase ATM (ataxia-telangiectasia mutated), which phosphorylates various proteins, including H2AX (reviewed in reference 47). Interestingly, unlike the situation in MCF10A H2AX+/+ cells, MRE11 did not form IRIF effectively in MCF10A H2AX−/− cells (Fig. 8B). Given that MRN foci were defective, we reasoned that ATM activation could also be impaired in the absence of H2AX in these cells, which could help explain their hypersensitivity to DSB-inducing agents. To address this idea, we first analyzed the phosphorylation of p53 on S15, a largely ATM-dependent site that is induced upon DNA damage (48). When cells were grown in the absence of a DNA-damaging agent, p53 S15 phosphorylation was higher in H2AX−/− cells than in H2AX+/+ cells (Fig. 8C). We also detected an increase in total p53 protein levels under these conditions, and real-time quantitative PCR (RT-qPCR) analysis of mRNA levels showed an induction of both p53 and p21, a direct p53 target, in MCF10A H2AX−/− cells (Fig. 8D). These results are consistent with MCF10A H2AX−/− cells accumulating DSBs with a concomitant reduction in proliferation under normal growth conditions (Fig. 5D, F, and G). Furthermore, while H2AX+/+ cells displayed marked inductions of p53 phosphorylation and total protein levels after phleomycin treatment, these inductions were less pronounced in H2AX−/− cells, suggesting that H2AX loss impairs the ability of MCF10A cells to mount a robust DDR (Fig. 8C; compare 4-h time points between H2AX+/+ and H2AX−/− cells). Longer exposures of the blots revealed that DNA damage also led to increased levels of slower-migrating forms of p53, consistent with numerous modifications that occur on p53 in the presence of DNA damage, which were induced less dramatically in H2AX−/− cells than in H2AX+/+ cells (Fig. 8C). Similar analyses with shorter time points confirmed the above findings and also showed that H2AX−/− cells were defective in the induction of the p53 target, p21, in response to DNA damage (Fig. 8E). Such analyses also revealed that, while H2AX−/− cells were able to execute apparently normal phosphorylation of another direct ATM target, CHK2 T68, they were unable to sustain this phosphorylation compared to H2AX+/+ cells (Fig. 8E). Collectively, these data revealed that human cells lacking H2AX display DDR defects, including defective recruitment of the mediator proteins MDC1 and 53BP1 as well as the MRN complex to DNA damage sites and impaired ATM-dependent signaling and p53 regulation following DNA damage.
Fig 8.
H2AX is required for the DNA damage response in human MCF10A cells. (A) H2AX promotes cellular resistance to DNA damage. Clonogenic survival assays of MCF10A H2AX+/+ and H2AX−/− cells were performed with various phleomycin concentrations as shown. n = 3 ± SEM. (B) DNA damage-induced MRE11 foci require H2AX. Cells were treated with phleomycin (60 μg/ml) for 1 h and analyzed by IF with the indicated antibodies. (C) DNA damage-dependent p53 activation requires H2AX. MCF10A H2AX+/+ and H2AX−/− cells were either untreated or were treated with phleomycin (30 μg/ml) for 2 h. After 2 h, fresh medium was used to wash out phleomycin for postrepair analysis. Samples were taken at the indicated times and analyzed by Western blotting with the antibodies as shown with both short and long exposures. Note that the long exposure of p53-S15p shows activation in MCF10A H2AX−/− cells under undamaged conditions. (D) p53 and p21 mRNA levels are increased in MCF10A H2AX−/− cells. RNA was isolated from both MCF10A H2AX+/+ and H2AX−/− cells and analyzed by RT-qPCR with primers specific for p53 and p21 and with control primers (GAPDH). Experiments were analyzed as in Fig. 5C. The graph represents data from 3 independent experiments, and error bars represent SEM. (E) DNA damage signaling is defective in MCF10A H2AX−/− cells. Experiments were performed and analyzed as in panel C with the indicated antibodies. (F) Summary of defects occurring in untreated and DNA damage-treated human MCF10A H2AX−/− cells.
DISCUSSION
Studies of the histone variant H2AX have provided important insights into how chromatin impacts cellular responses to DNA damage. Our present work has comprehensively surveyed residues in H2AX that regulate its ubiquitylation and sumoylation as well as γH2AX formation in response to DNA damage. This analysis has identified a key acidic triad motif in both H2A and H2AX that is required for their ubiquitylation. Furthermore, we have characterized the first reported knockout of H2AX in human cells. Notably, these cells exhibit phenotypes that are apparently somewhat distinct from those of mouse cells that lack H2AX. Our analysis indicates that H2AX is critical for maintaining genomic integrity during normal growth in human cells and in response to DNA damage and aids in the activation and orchestration of a fully functional DDR. Together, we have combined these results to create a powerful genetic complementation system for analysis of specific PTMs and regulatory regions of H2AX to understand how they contribute to the functions of H2AX in human cells.
Previous studies have focused mainly on the DNA damage-induced phosphorylation of H2AX S139 to form γH2AX, an early chromatin mark for DSBs that regulates the DDR. Recent studies have implicated ubiquitylation of various factors, including H2AX, as being critical for an effective DDR, as exemplified by the recruitment and activity of numerous ubiquitin E3 ligases that modify H2A and H2AX to DNA damage sites (18–20, 39, 40, 49). Our work has revealed that any mutation within the acidic triad motif in either H2A or H2AX virtually abolishes their monoubiquitylation. Although the H2AX E92A mutant also displayed defective γH2AX production following DNA damage (Fig. 4), this was not sufficient to appreciably alter 53BP1 accrual at DNA damage sites, a process dependent on H2AX phosphorylation (Fig. 7). We note, however, that these cells still express the core histone H2A, which contains a functional acidic triad motif that could compensate for the loss of this region in H2AX. However, we cannot rule out that γH2AX could promote DNA damage-induced processes that mediate 53BP1 IRIF that are distinct from histone ubiquitylation. Regardless, by using our methodology with both H2AX E92A and H2AX-9K to R, we were able to unambiguously determine the requirement of H2AX PTMs, including ubiquitylation, for these functions. Moreover, we were able to definitively show that monoubiquitylation of H2AX is dispensable for 53BP1 focus formation after DNA damage in human cells, which is consistent with similar data obtained in mouse ES H2AX−/− cells (34). Recently, H2A and H2AX were shown to be ubiquitylated on K13 and K15 by RNF168 in a DNA damage-dependent manner (50). In support of our findings, this study was also unable to observe any defects in either 53BP1 or BRCA1 IRIF in knockout (KO) mouse embryonic fibroblast (MEF) cells expressing lysine mutant H2AX at K13, K15, K118, and K119 (50). Collectively, these observations raise the question of what are the functions of H2AX ubiquitylation at each specific lysine acceptor site, and how does the acidic triad motif promote ubiquitin modification(s)? Several complexes, including the polycomb complex PRC1, promote H2A monoubiquitylation, although their potential link with H2AX ubiquitylation is unclear (51) and RNF168 catalyzes K63-linked ubiquitin chains on both H2A and H2AX (50). Since the acidic patch of H2A interacts with at least three different proteins, histone H4, LANA, and RCC1, we propose that this region also mediates an interaction with a protein that promotes H2A and H2AX monoubiquitylation and potentially K63 ubiquitylation (41–43) (Fig. 4). Future work will focus on the identification of acidic triad-interacting factors, which should help determine the mechanisms regulating H2A and H2AX ubiquitylation and their associated functions.
The deletion of H2AX in mice has been a powerful system for determining how H2AX promotes DNA repair, genome stability, and tumor suppressor functions (24, 25, 28, 29, 52). Our study now provides the first analysis of a human cell line deleted for H2AX. We find that human cells lacking H2AX share several phenotypes with H2AX−/− mouse cells, including slow growth, accumulation of DSBs, and altered cell cycle profiles. Both mouse and human cells lacking H2AX exhibit defective DNA damage recruitment of several DDR factors, including the MRN complex, MDC1, and 53BP1, resulting in aberrant ATM-dependent signaling and hypersensitivity to DNA damage. However, we observed several key differences between human and mouse cells lacking H2AX. Unlike mouse H2AX−/− cells, and what was inferred to be occurring in some human cells, OPT domain-containing 53BP1 foci arising in G1 cells still formed in MCF10A H2AX−/− cells (44, 45) (Fig. 6A and B). These 53BP1 foci accumulated at a higher frequency in MCF10A H2AX−/− cells compared to wild-type MCF10A cells. Our findings are consistent with this type of 53BP1 foci arising as a consequence of replicative stress because MCF10A H2AX−/− cells accumulated DSBs and displayed a reduced S-phase index, suggesting that H2AX in part functions to prevent DNA damage, especially endogenous damage occurring in S phase. Consistent with H2AX-independent recruitment of 53BP1, telomere fusions resulting from telomere dysfunction occur at normal frequencies in wild-type versus H2AX−/− MEFs (53) and telomere fusions arising from telomere uncapping are dependent on 53BP1 (54). Upon DNA damage, the initial recruitment of 53BP1 to DSBs was defective in MCF10A H2AX−/− cells (Fig. 6D), which is in contrast to observations in mouse H2AX KO cells that have suggested that the initial recognition of DNA damage by 53BP1 is H2AX independent in this system (24). Taken together, the available evidence implies that the dependencies that regulate the recruitment of 53BP1 to DNA lesions appear to differ, depending on the type of DNA damage and the origin of cells being analyzed. This work highlights the benefits of studying H2AX in different cellular contexts, including human cells, which will aid in assigning functional significance to these observed differences.
DDR signaling pathways have been suggested to act as tumor suppressor barriers to cancer development (55, 56), and mutations in DDR factors have been observed in various human cancers (57). Indeed, H2AX has been suggested to act as a tumor suppressor and has been linked to several human cancers (reviewed in reference10). The human H2AX gene maps to a region commonly deleted in several tumor types, and this loss of H2AX could promote tumorigenesis through its role in maintaining genome stability, as has been seen in H2AX knockout mouse models (25, 28, 29, 52). Our study has revealed that human cells lacking H2AX exhibit defective ATM signaling and p53 responses after DNA damage. While wild-type cells phosphorylated p53 on S15, resulting in increased p53 protein levels after DNA damage, human H2AX-deficient cells were defective in these aspects of DDR signaling. In contrast, mouse cells lacking H2AX do not exhibit these phenotypes (58). Thus, while deletion of H2AX could help drive tumorigenesis through increased genome instability, our results suggest that defects in p53 activation observed in these cells could also contribute by impairing downstream apoptotic pathways. Such a model could help explain why chromosomal regions containing H2AX are deleted in several human cancer types since this event could provide a selective advantage for these cells. Why mice lacking H2AX have not been reported to exhibit defects in p53 induction upon DNA damage when immortalized human epithelial cells lacking H2AX do is still an open question. One possibility is that H2AX functions in a cell-type-specific manner. In addition, MCF10A cells are spontaneously immortalized breast epithelial cells whose genetic analysis has shown MYC amplifications and deletion of CDKN2A (59). Thus, H2AX could acquire novel genetic interactions, depending on the genetic background of the cells being analyzed. Both of these possibilities could explain the observed differences between mouse H2AX KO cells and our model. In conclusion, our work has revealed the drastic consequences of deleting H2AX in human cells (Fig. 8F). With our extensive H2AX mutational library and human knockout cell line for H2AX, we have created the framework for obtaining additional mechanistic insights into how H2AX maintains genome integrity during normal cell growth, DDR activation, and tumor suppression.
Supplementary Material
ACKNOWLEDGMENTS
We thank Yaron Galanty for tagged ubiquitin and SUMO reagents. We are grateful to Blerta Xhemalce who provided assistance with culturing MCF10A cells and performed growth and sensitivity assays.
Research in the S.P.J. laboratory is supported by the European Community (EU Projects DNA Repair LSHG-CT-2005-512113 and GENICA), ERC Advanced Researcher Grant, DNA-Damage Responses: Regulation and Mechanisms (DDREAM), grant agreement no. 268536, and core infrastructure provided by Cancer Research UK and the Wellcome Trust. K.M.M. was funded by a Wellcome Trust project grant (086861/Z/08/Z) while in the S.P.J. laboratory. Research in the K.M.M. laboratory is supported by start-up funds from the University of Texas at Austin and from the Cancer Prevention Research Institute of Texas (CPRIT). K.M.M. is a CPRIT scholar.
K.M.M., W.C., and A.A. designed, analyzed, and conducted experiments. C.L. and F.G. conducted experiments. S.J. designed and analyzed experiments and assisted K.M.M. in writing the manuscript. K.M.M. conceived the study and wrote the manuscript. All authors commented on the manuscript before submission.
Footnotes
Published ahead of print 29 October 2012
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01024-12.
REFERENCES
- 1.Margueron R, Reinberg D. 2010. Chromatin structure and the inheritance of epigenetic information. Nat. Rev. Genet. 11:285–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campos EI, Reinberg D. 2009. Histones: annotating chromatin. Annu. Rev. Genet. 43:559–599 [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- 4.Suganuma T, Workman JL. 2011. Signals and combinatorial functions of histone modifications. Annu. Rev. Biochem. 80:473–499 [DOI] [PubMed] [Google Scholar]
- 5.Weake VM, Workman JL. 2008. Histone ubiquitination: triggering gene activity. Mol. Cell 29:653–663 [DOI] [PubMed] [Google Scholar]
- 6.Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol. Cell 40:179–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson SP, Bartek J. 2009. The DNA-damage response in human biology and disease. Nature 461:1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huertas P. 2010. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 17:11–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miller KM, Jackson SP. 2012. Histone marks: repairing DNA breaks within the context of chromatin. Biochem. Soc. Trans. 40:370–376 [DOI] [PubMed] [Google Scholar]
- 10.Bonner WM, Redon CE, Dickey JS, Nakamura AJ, Sedelnikova OA, Solier S, Pommier Y. 2008. GammaH2AX and cancer. Nat. Rev. Cancer 8:957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 [DOI] [PubMed] [Google Scholar]
- 12.Bekker-Jensen S, Lukas C, Kitagawa R, Melander F, Kastan MB, Bartek J, Lukas J. 2006. Spatial organization of the mammalian genome surveillance machinery in response to DNA strand breaks. J. Cell Biol. 173:195–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. 2010. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 29:1446–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogakou EP, Boon C, Redon C, Bonner WM. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bekker-Jensen S, Rendtlew Danielsen J, Fugger K, Gromova I, Nerstedt A, Lukas C, Bartek J, Lukas J, Mailand N. 2010. HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 12:80–86 (Erratum, 12:412.) [DOI] [PubMed] [Google Scholar]
- 16.Galanty Y, Belotserkovskaya R, Coates J, Jackson SP. 2012. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26:1179–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galanty Y, Belotserkovskaya R, Coates J, Polo S, Miller KM, Jackson SP. 2009. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462:935–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131:901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolas NK, Chapman JR, Nakada S, Ylanko J, Chahwan R, Sweeney FD, Panier S, Mendez M, Wildenhain J, Thomson TM, Pelletier L, Jackson SP, Durocher D. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318:1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131:887–900 [DOI] [PubMed] [Google Scholar]
- 21.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, Ng T, Solomon E. 2009. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462:886–890 [DOI] [PubMed] [Google Scholar]
- 22.Stucki M, Clapperton JA, Mohammad D, Yaffe MB, Smerdon SJ, Jackson SP. 2005. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123:1213–1226 [DOI] [PubMed] [Google Scholar]
- 23.Greenberg RA. 2011. Histone tails: directing the chromatin response to DNA damage. FEBS Lett. 585:2883–2890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A. 2003. Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat. Cell Biol. 5:675–679 [DOI] [PubMed] [Google Scholar]
- 25.Celeste A, Petersen S, Romanienko PJ, Fernandez-Capetillo O, Chen HT, Sedelnikova OA, Reina-San-Martin B, Coppola V, Meffre E, Difilippantonio MJ, Redon C, Pilch DR, Olaru A, Eckhaus M, Camerini-Otero RD, Tessarollo L, Livak F, Manova K, Bonner WM, Nussenzweig MC, Nussenzweig A. 2002. Genomic instability in mice lacking histone H2AX. Science 296:922–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. 2000. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr. Biol. 10:886–895 [DOI] [PubMed] [Google Scholar]
- 27.Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. 2003. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421:961–966 [DOI] [PubMed] [Google Scholar]
- 28.Bassing CH, Chua KF, Sekiguchi J, Suh H, Whitlow SR, Fleming JC, Monroe BC, Ciccone DN, Yan C, Vlasakova K, Livingston DM, Ferguson DO, Scully R, Alt FW. 2002. Increased ionizing radiation sensitivity and genomic instability in the absence of histone H2AX. Proc. Natl. Acad. Sci. U. S. A. 99:8173–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW. 2003. Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114:359–370 [DOI] [PubMed] [Google Scholar]
- 30.Xie A, Puget N, Shim I, Odate S, Jarzyna I, Bassing CH, Alt FW, Scully R. 2004. Control of sister chromatid recombination by histone H2AX. Mol. Cell 16:1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ikura T, Tashiro S, Kakino A, Shima H, Jacob N, Amunugama R, Yoder K, Izumi S, Kuraoka I, Tanaka K, Kimura H, Ikura M, Nishikubo S, Ito T, Muto A, Miyagawa K, Takeda S, Fishel R, Igarashi K, Kamiya K. 2007. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 27:7028–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, Patel DJ, Elledge SJ, Allis CD. 2009. WSTF regulates the H2A. X DNA damage response via a novel tyrosine kinase activity.Nature 457:57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cook PJ, Ju BG, Telese F, Wang X, Glass CK, Rosenfeld MG. 2009. Tyrosine dephosphorylation of H2AX modulates apoptosis and survival decisions. Nature 458:591–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xie A, Odate S, Chandramouly G, Scully R. 2010. H2AX post-translational modifications in the ionizing radiation response and homologous recombination. Cell Cycle 9:3602–3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. 2010. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat. Struct. Mol. Biol. 17:1144–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goldknopf IL, Taylor CW, Baum RM, Yeoman LC, Olson MO, Prestayko AW, Busch H. 1975. Isolation and characterization of protein A24, a “histone-like” non-histone chromosomal protein. J. Biol. Chem. 250:7182–7187 [PubMed] [Google Scholar]
- 37.Shiio Y, Eisenman RN. 2003. Histone sumoylation is associated with transcriptional repression. Proc. Natl. Acad. Sci. U. S. A. 100:13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsubara K, Sano N, Umehara T, Horikoshi M. 2007. Global analysis of functional surfaces of core histones with comprehensive point mutants. Genes Cells 12:13–33 [DOI] [PubMed] [Google Scholar]
- 39.Pan MR, Peng G, Hung WC, Lin SY. 2011. Monoubiquitination of H2AX protein regulates DNA damage response signaling. J. Biol. Chem. 286:28599–28607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu CY, Kang HY, Yang WL, Wu J, Jeong YS, Wang J, Chan CH, Lee SW, Zhang X, Lamothe B, Campos AD, Darnay BG, Lin HK. 2011. Critical role of monoubiquitination of histone H2AX protein in histone H2AX phosphorylation and DNA damage response. J. Biol. Chem. 286:30806–30815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbera AJ, Chodaparambil JV, Kelley-Clarke B, Joukov V, Walter JC, Luger K, Kaye KM. 2006. The nucleosomal surface as a docking station for Kaposi's sarcoma herpesvirus LANA. Science 311:856–861 [DOI] [PubMed] [Google Scholar]
- 42.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260 [DOI] [PubMed] [Google Scholar]
- 43.Makde RD, England JR, Yennawar HP, Tan S. 2010. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature 467:562–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harrigan JA, Belotserkovskaya R, Coates J, Dimitrova DS, Polo SE, Bradshaw CR, Fraser P, Jackson SP. 2011. Replication stress induces 53BP1-containing OPT domains in G1 cells. J. Cell Biol. 193:97–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, Lukas J. 2011. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 13:243–253 [DOI] [PubMed] [Google Scholar]
- 46.Wu J, Huen MS, Lu LY, Ye L, Dou Y, Ljungman M, Chen J, Yu X. 2009. Histone ubiquitination associates with BRCA1-dependent DNA damage response. Mol. Cell. Biol. 29:849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams RS, Williams JS, Tainer JA. 2007. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85:509–520 [DOI] [PubMed] [Google Scholar]
- 48.Giaccia AJ, Kastan MB. 1998. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 12:2973–2983 [DOI] [PubMed] [Google Scholar]
- 49.Panier S, Durocher D. 2009. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair 8:436–443 [DOI] [PubMed] [Google Scholar]
- 50.Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, Marteijn JA, Sixma TK. 2012. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150:1182–1195 [DOI] [PubMed] [Google Scholar]
- 51.Zhou W, Wang X, Rosenfeld MG. 2009. Histone H2A ubiquitination in transcriptional regulation and DNA damage repair. Int. J. Biochem. Cell Biol. 41:12–15 [DOI] [PubMed] [Google Scholar]
- 52.Celeste A, Difilippantonio S, Difilippantonio MJ, Fernandez-Capetillo O, Pilch DR, Sedelnikova OA, Eckhaus M, Ried T, Bonner WM, Nussenzweig A. 2003. H2AX haploinsufficiency modifies genomic stability and tumor susceptibility. Cell 114:371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fernandez-Capetillo O, Liebe B, Scherthan H, Nussenzweig A. 2003. H2AX regulates meiotic telomere clustering. J. Cell Biol. 163:15–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dimitrova N, Chen YC, Spector DL, de Lange T. 2008. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 456:524–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, Orntoft T, Lukas J, Bartek J. 2005. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434:864–870 [DOI] [PubMed] [Google Scholar]
- 56.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, Kletsas D, Yoneta A, Herlyn M, Kittas C, Halazonetis TD. 2005. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434:907–913 [DOI] [PubMed] [Google Scholar]
- 57.Halazonetis TD, Gorgoulis VG, Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science 319:1352–1355 [DOI] [PubMed] [Google Scholar]
- 58.Kang J, Ferguson D, Song H, Bassing C, Eckersdorff M, Alt FW, Xu Y. 2005. Functional interaction of H2AX, NBS1, and p53 in ATM-dependent DNA damage responses and tumor suppression. Mol. Cell. Biol. 25:661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kadota M, Yang HH, Gomez B, Sato M, Clifford RJ, Meerzaman D, Dunn BK, Wakefield LM, Lee MP. 2010. Delineating genetic alterations for tumor progression in the MCF10A series of breast cancer cell lines. PLoS One 5:e9201 doi:10.1371/journal.pone.0009201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.