Abstract
Multicellular organisms have evolved processes to prevent abnormal proliferation or inappropriate tissue infiltration of cells, and these tumor suppressive mechanisms serve to prevent tissue hyperplasia, tumor development, and metastatic spread of tumors. These include potentially reversible processes such as cell cycle arrest and cellular senescence, as well as apoptotic cell death, which in contrast eliminates dangerous cells that may initiate tumor development. Tumor suppressive processes are organized as complex, extensive signaling networks, controlled by central “nodes.” These “nodes” are prominent tumor suppressors, such as P53 or PTEN, whose loss is responsible for the development of the majority of human cancers. In this review we discuss the processes by which some of these prominent tumor suppressors trigger apoptotic cell death and how this process protects us from cancer development.
Tumor suppressors can trigger apoptosis and protect against cancer. P53 does this by up-regulating the expression of several proapoptotic genes (e.g., PUMA and NOXA).
A malignant tumor is characterized by the ability to expand in an uncontrolled manner, destroy normal tissue architecture, and ultimately undergo metastatic spread (Hanahan and Weinberg 2000). Although the number of mutations required for neoplastic transformation may vary, all tumors are reliant on two critical mechanisms for their development; the activation of oncogenes that promote proliferation and survival of cancer cells, as well as the inactivation of tumor suppressor genes that normally repress development and growth of tumors (Hanahan and Weinberg 2000).
Oncogenes can be activated via multiple mechanisms, including chromosomal translocations, deletions or insertions, as well as point mutations. One such example is the translocation between chromosomes 9 and 22 that is present in most cases of chronic myeloid leukemia. The juxtaposition of the BCR and c-ABL genes results in the production of an abnormal BCR-ABL fusion protein with constitutive kinase activity (Deininger et al. 2005). However, in other cancer-causing chromosomal translocations, such as the t[8;14] translocation in Burkitt’s lymphoma, the coding sequence of the oncogene, c-MYC, is unchanged; rather its activation results from deregulated expression in B lymphoid cells as a consequence of its proximity to the IGH gene enhancer (Cory et al. 1987). Tumorigenesis promoted by deregulated kinase activity frequently results from the acquisition of point mutations. In this context, a single amino acid substitution can dramatically enhance kinase activity by preventing binding of negative regulators or “locking” the catalytic domain in the active conformation. This is exemplified by the BRAF(V600E) mutation frequently observed in melanoma or colon carcinoma (Poulikakos and Rosen 2011) and the activating mutations in EGF-R observed in lung adenocarcinoma (Sharma et al. 2007).
Analogous to the activation of oncogenes, tumor suppressor genes can be inactivated through multiple mechanisms, including large-scale chromosomal alterations or point mutations. However, in most cases both alleles of the gene must be compromised to abolish gene function, unless the mutated protein can act in a dominant-negative fashion to block the activity of its wild-type counterpart.
Multicellular organisms have evolved a plethora of mechanisms to restrain the growth or even eliminate aberrant cells—these processes can all function as tumor suppressors. Notably, of the attributes that cells must acquire to become cancerous (“hallmarks of cancer”) discussed by Hanahan and Weinberg (2000), several relate to escape from regulatory processes that would normally suppress tumor growth. They include cell cycle arrest, cellular senescence, and cell death; of these only cell death is irreversible, all others can (at least potentially) be reversed. In this review, we describe the mechanisms by which tumor suppressors that are disabled in a broad range and large fraction of cancers trigger cell death, and how components of the apoptotic machinery can themselves act as tumor suppressors.
APOPTOSIS AS A MEDIATOR OF TUMOR SUPPRESSION
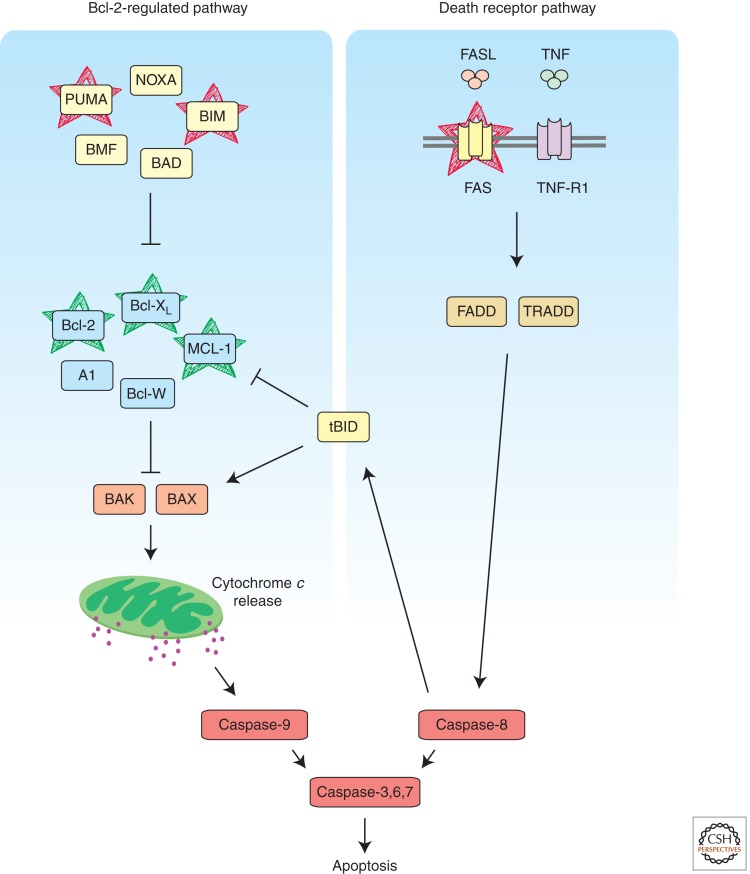
Apoptosis, also known as programmed cell death, is a highly regulated program of ordered cellular destruction that facilitates the removal of damaged or superfluous cells. This process is critical for many physiological processes, including embryonic development and tissue homeostasis in adulthood (Strasser et al. 2000; Hotchkiss et al. 2009). In vertebrates, apoptosis can be initiated by two distinct, albeit ultimately converging, signaling pathways (Strasser et al. 1995), termed “BCL-2-regulated” (“intrinsic,” “mitochondrial,” “stress-induced”) (Chipuk and Green 2008) and “death receptor-” (Strasser et al. 2009) induced apoptosis (Fig. 1). In both pathways, cell demolition is mediated by aspartate-specific cysteine proteases (caspases) that proteolyze hundreds of cellular proteins (Timmer and Salvesen 2007). Cell surface “death receptors” (members of the TNF-R family with an intracellular “death domain,” e.g., FAS, TNF-R1) can trigger apoptosis by direct activation of caspases, through adaptor protein (FADD, TRADD)-mediated activation (via conformational change) of initiator caspase-8, which then proteolytically activates effector caspases (caspase-3, -6, and -7) (Strasser et al. 2009) (Fig. 1). Mutations in FAS or its ligand, FASL, perturb peripheral lymphoid homeostasis, ultimately leading to severe lymphadenopathy, a systemic autoimmune disease and a predisposition to hematopoietic malignancy in both mice (Watanabe-Fukunaga et al. 1992; O’Reilly et al. 2009) and humans (Rieux-Laucat et al. 1995; Drappa et al. 1996).
Figure 1.
Apoptosis can be initiated by activation of two distinct, albeit ultimately converging, pathways, the “Bcl-2-regulated pathway” (left; also known as the “stress” or “mitochondrial” pathway) and the “death receptor pathway” (right). As the name suggests, protein–protein interactions between members of the Bcl-2 family govern activation of the Bcl-2-regulated pathway, whereas binding of their cognate ligands activates the death receptors (e.g., FAS, TNF-R1). Cellular demolition is performed by the effector caspases that act downstream of both pathways. Proteins that have been identified as suppressors or oncogenes are indicated in red or green, respectively.
The “BCL-2-regulated” apoptotic pathway can be activated by developmental cues and a broad range of cytotoxic insults, including cytokine deprivation or DNA damage. This pathway is regulated by the BCL-2 protein family and relies (at least in part) on the initiator caspase-9 (and its adaptor APAF-1) to activate the effector caspases (Marsden et al. 2002; Shi 2002; Riedl and Shi 2004) (Fig. 1). The BCL-2 protein family is composed of one antiapoptotic and two proapoptotic subgroups that regulate commitment to cell death through complex protein–protein interactions (Youle and Strasser 2008). The antiapoptotic members (BCL-2, BCL-XL, BCL-W, MCL-1, and A1) share four BH (BCL-2 Homology) domains and are essential for cell survival, functioning in a cell type-specific manner, albeit with significant functional overlap (Youle and Strasser 2008). Proapoptotic BAX and BAK share remarkable structural similarity with their prosurvival relatives, but they have largely overlapping functions during the execution of apoptosis and are essential for mitochondrial outer membrane permeabilization (MOMP) with consequent release of apoptogenic molecules (e.g., cytochrome c, DIABLO/Smac) and activation of caspase-9 (Lindsten et al. 2000; Chipuk and Green 2008). The proapoptotic BH3-only proteins (BIM, PUMA, BID, BAD, BMF, HRK, BIK, and NOXA) share with each other and the BCL-2 family overall only the BH3 domain and are required for initiation of apoptosis (Huang and Strasser 2000). These proteins are activated transcriptionally and/or post-translationally and exert stimulus-specific as well as cell type-specific actions. For example, BIM is critical for cytokine deprivation-induced apoptosis (Bouillet et al. 1999), whereas PUMA and, to a lesser extent, NOXA trigger the apoptosis activated by the tumor suppressor P53 (Jeffers et al. 2003; Villunger et al. 2003; Michalak et al. 2008). BH3-only proteins are thought to activate BAX/BAK either through direct binding and/or indirectly by binding to their repressors, the prosurvival BCL-2 proteins (Chipuk and Green 2008; Merino et al. 2009).
Deregulated expression of prosurvival BCL-2 proteins (e.g., because of the t[14;18], BCL2;IGH chromosomal translocation in human follicular center lymphoma) promotes tumorigenesis by sustaining the viability of cells undergoing neoplastic transformation, thereby facilitating the acquisition of additional oncogenic mutations (Vaux et al. 1988; Strasser et al. 1990, 1993). Similarly, loss of proapoptotic BCL-2 family members in isolation is not potently transforming, but it is consistent with the notion that cancer cells must acquire the ability to evade apoptosis; such defects promote tumorigenesis when they occur in concert with additional oncogenic mutations. For example, homozygous deletion of the BIM gene was found in ∼20% of human mantle cell lymphoma cases and, accordingly, loss of Bim accelerated Eμ-MYC-driven lymphomagenesis in mice (Egle et al. 2004). Moreover, ∼40% of human Burkitt’s lymphomas fail to express PUMA (Garrison et al. 2008), and loss of PUMA can also (like loss of BIM) accelerate lymphoma development in Eμ-MYC transgenic mice (Garrison et al. 2008; Michalak et al. 2009). In addition, loss of the BH3-only proteins NOXA (Michalak et al. 2010) and BMF (Labi et al. 2008) as well as loss of BAX (Eischen et al. 2001) were found to promote development of lymphoma in different experimental mouse models, but so far deregulation of these genes has not been detected in human cancers. As mentioned above, defects in apoptosis by themselves are not potently transforming. Therefore, mutations of oncogenes or tumor suppressor genes that encode master regulators of multiple pathways (including apoptosis) are frequently observed in human cancers because one (or two) oncogenic lesions will simultaneously activate several tumorigenic processes. In the following sections, we discuss the mechanisms by which tumor suppressors trigger apoptosis and their importance for prevention of neoplastic disease.
TUMOR SUPPRESSORS AND THEIR MECHANISMS FOR INDUCING APOPTOSIS
P53, Orchestrator of the Cellular Response to DNA Damage, Hypoxia, and Oncogenic Stress
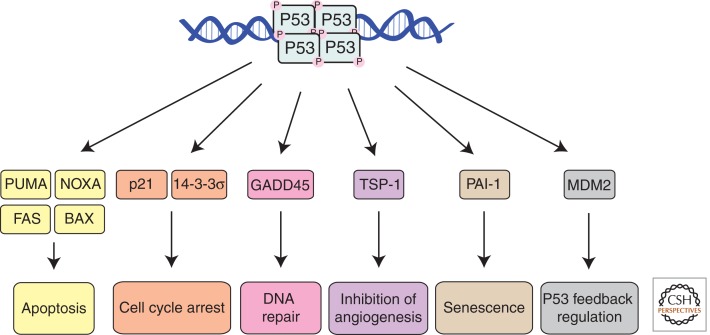
The tumor suppressor P53, a transcriptional regulator, is mutated in ∼50% of human cancers and in those cancers that lack P53 mutations, P53 signaling is often defective because of the acquisition of some other mutation(s) (Vousden and Lane 2007). P53 can be activated by a broad range of cytotoxic stress signals, including DNA damage, hypoxia, and activation of certain oncogenes (e.g., c-MYC) (Vousden and Lane 2007). In the majority of unstressed cells, P53 mRNA is expressed constitutively, but P53 protein levels are low. This is mainly attributable to the action of MDM2, an E3 ubiquitin ligase, which targets P53 for K48 linkage-mediated ubiquitination and proteasomal degradation (Haupt et al. 1997; Kubbutat et al. 1997). Stress-induced P53 activation involves stabilization of the P53 protein. This is mediated primarily through inhibition of MDM2 by the tumor suppressor ARF (Vousden and Lane 2007). However, in addition, multiple posttranslational modifications, including phosphorylation, acetylation, and neddylation, also affect P53 stability, its binding to target genes, and/or its transcriptional activity (Vousden and Lane 2007). Upon activation, P53 binds as a homotetramer to specific sequences within the regulatory regions of a broad range of target genes (Riley et al. 2008) and thereby triggers a multitude of effector pathways, including cell cycle arrest (Livingstone et al. 1992), cellular senescence (Metz et al. 1995), coordination of DNA repair, and apoptosis (Vousden and Lane 2007) (Fig. 2). In addition, P53 triggers a process that regulates its own activity that operates mainly through direct transcriptional activation of MDM2 (Barak et al. 1993; Wu et al. 1993). The critical importance of appropriate P53/MDM2 feedback regulation was revealed by the discovery that loss of MDM2 causes embryonic lethality in mice and that this can be prevented by concomitant loss of P53 (Jones et al. 1995; Montes de Oca Luna et al. 1995).
Figure 2.
The tumor suppressor P53 acts as a transcriptional regulator. It has the capacity to activate diverse cellular processes. Stimulus and cell type-specific effects determine which particular effector pathway(s) will dominate.
It remains unclear why a specific effector pathway will dominate in a particular cell in response to P53 activation. For example, why does low-dose γ-irradiation elicit cell G1/S cycle arrest and DNA repair in fibroblasts but apoptosis in thymocytes? It is possible that parallel signaling pathways, active in some cells but not others, can modulate the overall outcome of P53 activation. In addition, the various post-translational modifications may affect the preference of P53 for different target genes, thereby determining which effector pathway will predominate. To induce apoptosis, P53 directly transcriptionally up-regulates the expression of several proapoptotic BCL-2 family members, the BH3-only proteins PUMA (Nakano and Vousden 2001; Yu et al. 2001) and NOXA (Oda et al. 2000) as well as the multi-BH domain proapoptotic protein BAX (Miyashita and Reed 1995). Experiments with gene-targeted mice have shown that P53-induced apoptosis in a broad range of cell types is mediated predominantly by PUMA and to lesser extent by NOXA (Jeffers et al. 2003; Villunger et al. 2003; Erlacher et al. 2005; Naik et al. 2007; Michalak et al. 2008). BAX is expressed in P53-deficient cells, and many P53-independent apoptotic stimuli such as glucocorticoids rely on BAX (plus BAK) for cell killing (Lindsten et al. 2000). Thus, the P53-mediated induction of BAX probably serves to increase the efficiency of apoptosis signaling but does not determine whether a cell will live or die. As mentioned, P53 activates diverse effector pathways and it is not yet clear which one(s) is/are critical for tumor suppression. Loss of P53’s apoptotic pathway, through loss of PUMA (Garrison et al. 2008; Michalak et al. 2009) and/or NOXA (Michalak et al. 2010), can accelerate lymphoma development elicited by MYC overexpression or low-dose γ-irradiation, respectively. However, in contrast to P53-deficient mice (Donehower et al. 1992), animals lacking both PUMA and NOXA are not tumor prone, although their cells display comparable resistance to P53-dependent apoptotic stimuli (e.g., γ-irradiation) as those lacking P53 itself (Michalak et al. 2008). Thus, apoptosis induction does not account for all of the tumor suppressive action of P53. Loss of P21, the major effector of P53-mediated G1/S cell cycle arrest, also does not constitute a potently transforming oncogenic lesion (Deng et al. 1995). It therefore appears that it is the composite loss of several effector pathways (coordination of DNA repair, cell cycle arrest, apoptosis, and/or senescence) and not the loss of a single process that accounts for the high incidence and rapid onset of tumors when P53 is mutated in mice (Donehower et al. 1992) and in humans (Li-Fraumeni syndrome [Srivastava et al. 1990]). Recent evidence has emerged that suggests that signaling resulting from acute P53 activation is distinct from its ability to effect tumor suppression (Brady et al. 2011). Furthermore, there is now evidence that even combined defects in P53-mediated induction of apoptosis, cell cycle arrest, and cell senescence are all dispensable for P53-mediated tumor suppression (Li et al. 2012). This may indicate that regulation of cellular metabolism and perhaps coordination of DNA repair may be critical for the tumor suppressive action of P53.
Retinoblastoma Protein, a Negative Regulator of Cell Cycle Entry
The retinoblastoma protein (RB) is the central element in a tumor suppression network that is interconnected with that of P53 through the regulation of cell cycle arrest and also through activation by oncogenic stress (Lee et al. 1987). Rb was the first tumor suppressor identified in human cancer following the realization that inheritance of one mutated copy of Rb followed by mutation or loss of the wild-type allele resulted in the development of familial retinoblastoma in early childhood (Lee et al. 1987). Accordingly, heterozygous loss of RB in mice results in the development of pituitary tumors (that have lost the wt Rb allele), but curiously not retinoblastoma (Jacks et al. 1992). As for P53, the critical importance of the RB protein in the regulation of cell proliferation came from the discovery that DNA tumor viruses, such as the human papillomaviruses (HPV), promote host cell survival and proliferation and thereby viral replication by encoding specific inhibitors of these two tumor suppressors (Levine 2009). At that time, it was thought that most cancers were of viral origin; although this has not proven to be the case, the studies that have followed have illuminated RB (and P53) as critical components of the tumor suppression network.
RB controls cell proliferation through its regulation of the E2F family of transcription factors. In its active form, RB binds and sequesters the E2F proteins, which play critical roles in S phase entry. Upon phosphorylation, RB releases the E2F proteins allowing them to regulate transcription of their target genes. In addition to its role in cell cycle regulation, RB is also critical for the differentiation programs of certain tissues; however, the relevance, if any, of these activities for tumor suppression is currently unclear. This division of labor is achieved through the presence of distinct subsets within the E2F family. E2F1-3 promote cell cycle progression and are regulated by RB (Lees et al. 1993), whereas E2F4-8 act as transcriptional repressors and promote cell cycle exit and differentiation. In addition to its role in controlling P53 protein levels, MDM2 can also regulate the process by which E2F1 and its cofactor, DP1, acts to promote cell division (Martin et al. 1995; Xiao et al. 1995). The ability of MDM2 to potentiate E2F1 activity relies on inhibition of SKP2-mediated degradation of E2F1 (Zhang et al. 2005). The critical role that the activator E2Fs play in the control of proliferation is underscored by the finding that the loss of their negative regulator, RB, and its homologs abrogates the G1/S checkpoint and thereby promotes cellular immortalization (Dannenberg et al. 2000; Sage et al. 2000).
In addition to their role in promoting proliferation, paradoxically, the activator E2Fs can also promote apoptosis, at least in certain settings. This is particularly clear in the context of DNA damage when E2F1 is stabilized post-translationally through ATM-, ATR-, CHK1-, and CHK2-mediated phosphorylation, as well as by acetylation (Lin et al. 2001; Pediconi et al. 2003; Stevens et al. 2003; Urist et al. 2004). These modifications appear to drive E2F1 to up-regulate proapoptotic genes (Hershko and Ginsberg 2004), such as the BH3-only gene Bim (O’Connor et al. 1998). This duality of function is thought to represent a fail-safe mechanism whereby cells that have sustained DNA damage are predisposed to undergo apoptosis following E2F1 activation rather than DNA synthesis and proliferation. Indeed, E2F-driven proliferation can impose an “oncogene activation”-like stress on cells stimulating P53 in an ARF-dependent manner (Bates et al. 1998). The relevance of this signaling network for tumorigenesis is exemplified by studies of an E2F1-driven mouse model of skin cancer. Overexpression of E2F1 resulted in hyperplasia in the epidermis but this was held in check by a concomitant increase in apoptosis (Pierce et al. 1998a). Loss of P53 prevented this apoptosis and caused progression to skin carcinoma (Pierce et al. 1998b).
The realization that loss of RB, through consequent activation of the E2Fs, can have proapoptotic effects may at least in part explain why loss of RB activity commonly occurs late in tumor development and is associated with progression rather than initiation (Polager and Ginsberg 2009). Thus, apoptotic signaling may first need to be compromised (e.g., by loss of P53) so that the oncogenic stress imposed by RB inactivation does not eliminate the cells undergoing transformation.
Phosphatase with Tensin Homology, the Critical Safety Catch of the PI3K/AKT Signaling Pathway
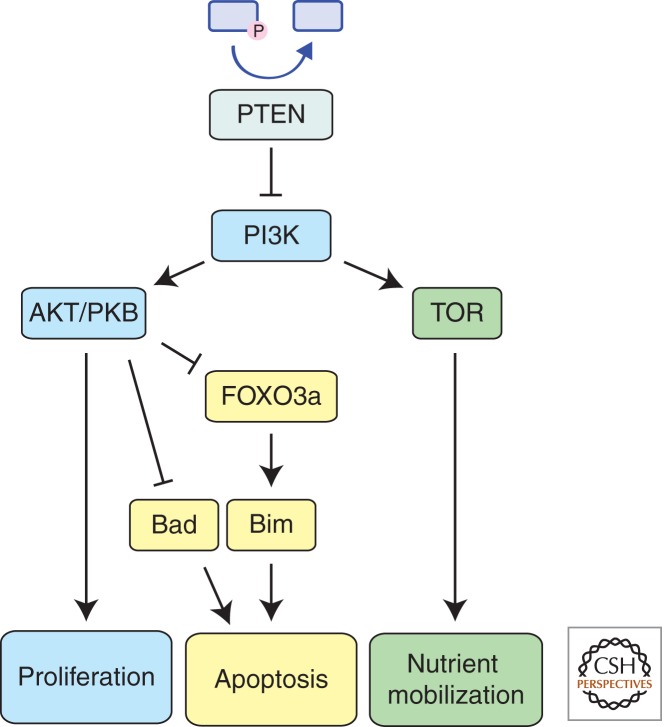
PTEN (phosphatase with tensin homology), a lipid phosphatase, is the key negative regulator of the phosphatidylinositol 3-kinase (PI3K) signaling pathway that promotes cell survival and proliferation and is frequently deregulated in various human cancers (Stambolic et al. 1998) (Fig. 3). Whereas the PI3K pathway is extensive and can direct diverse cellular processes, PTEN inhibits this pathway by breaking down PI3K’s active second messenger molecule, PIP3. It is also noteworthy that PTEN is a direct P53 target and thus represents a constituent of the wider P53 tumor suppression network (Stambolic et al. 2001). PI3K can be activated by receptor tyrosine kinases (RTKs) as well as RAS. PI3K in turn activates AKT (also known as PKB), which regulates a broad range of cellular processes, including proliferation, nutrient mobilization, and cell survival.
Figure 3.
PTEN acts as the endogenous antagonist of the PI3K-regulated growth pathway. PTEN breaks down the key signaling intermediate PIP3, thereby inhibiting downstream signaling mediated by AKT and TOR. Thus, PTEN negates PI3K’s ability to drive proliferation, facilitate nutrient mobilization, and inhibit apoptosis.
AKT was reported to directly inhibit apoptosis by phosphorylating and thereby causing sequestration of the BH3-only protein BAD (del Peso et al. 1997). However, given that loss of BAD (Ranger et al. 2003) or even combined loss of BAD and BIM (Kelly et al. 2010) had only minimal effects on cell survival indicates that this process is not critical for sustaining survival of cells undergoing transformation. AKT has also been shown to promote cell survival by phosphorylating FOXO3a, thereby preventing this transcription factor from activating its target genes, such as the proapoptotic BH3-only gene Bim (O’Connor et al. 1998; Dijkers et al. 2000). Because BIM deletions are found in human mantle cell lymphomas (Tagawa et al. 2005) and BIM loss promotes lymphomagenesis in mice (Egle et al. 2004), this pathway is more likely to be critical for tumor suppression.
AKT also activates TOR signaling by inhibiting its negative regulators TSC1 and TSC2, thereby promoting RHEB activity (Potter et al. 2002). RHEB allows TOR to act as a nutrient sensor and liberate additional glucose and amino acids to promote cell growth and proliferation (Garami et al. 2003). This metabolic process, inhibited by PTEN, is likely to intersect with the control of cell survival.
The inappropriate activation of TOR signaling has particular relevance to patients with Tuberous Sclerosis Complex disease, which is caused by inherited germline mutations in either TSC1 or TSC2 that act in an autosomal dominant manner. This disease is characterized by skin, brain, kidney, and heart abnormalities, with brain tumors accounting for most of the morbidity and mortality. Inherited mutations in PTEN are the cause of the PTEN Hamartoma Tumor Syndrome. These patients are predisposed to the development of cancers of the breast, thyroid, and endometrium, of which breast tumors are the most common with a lifetime risk of 25%–50%. Accordingly, in mice loss of one Pten allele resulted in hyperplasia in multiple organs, such as the skin and prostate, which progressed to colon adenocarcinoma, gonadostromal tumors, teratomas, thyroid carcinoma, and lymphoma (Di Cristofano et al. 1998; Suzuki et al. 1998).
In conclusion, acting as a safety catch at the apex of the PI3K/AKT signaling pathway, PTEN plays a critical role in tumor suppression by preventing inappropriate activation of cellular metabolism (via effects on TOR), proliferation and survival (via effects on proapoptotic BH3-only proteins).
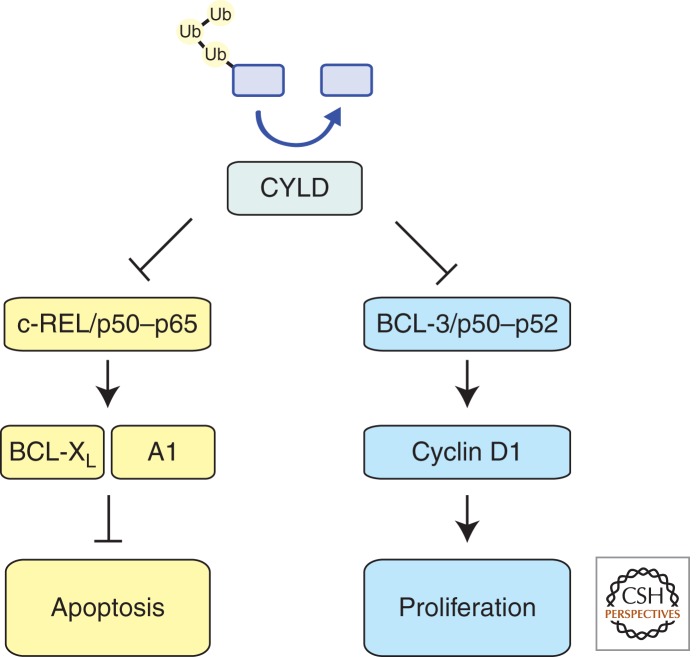
CYLD, a Negative Regulator of the NF-κB Signaling Pathway
CYLD is a member of the USP subfamily of deubiquitinases (DUBs), first identified as the gene mutated in the inherited condition familial cylindromatosis (Bignell et al. 2000) that is characterized by the development of benign skin tumors. In these patients, one mutated copy of CYLD is inherited and the wild-type allele is commonly lost during neoplastic progression.
Although ubiquitination was first identified as a process for targeting protein substrates for proteasome-mediated degradation, conjugation of ubiquitin moieties to substrates by linkages other than K48 (particularly K63 linkages) can activate signal transduction pathways (Wertz and Dixit 2010). Like many other cell signaling processes, ubiquitination is a reversible process, and whereas ubiquitin chains are assembled on target proteins by E3 ligases, they can also be hydrolyzed by DUBs. The existence of ∼100 DUBs in humans presumably allows highly specific regulation of a broad range of signaling pathways.
CYLD acts as a specific negative regulator of NF-κB signaling through its interaction with NEMO, the regulatory subunit of the IKK proteins, and TRAF2, an adaptor for several TNF-R family members (Brummelkamp et al. 2003; Kovalenko et al. 2003; Trompouki et al. 2003) (Fig. 4). CYLD antagonizes the conjugation of K63 ubiquitin chains on TRAF2 and thereby prevents IKK activation, which is required for nuclear import and functional activation of NF-κB complexes (e.g., REL/P50). Transcriptional targets of NF-κB can modulate the duration and intensity of signaling forming either feed-forward or negative feedback loops. Whereas IκBα induction constitutes a negative feedback loop, BCL3 acts to potentiate signaling through the noncanonical NF-κB pathway. This process is of particular relevance to tumorigenesis, because cyclin D1, which requires BCL3 and NF-κB for induction and is repressed by CYLD, has been shown to promote tumor cell proliferation (Massoumi et al. 2006).
Figure 4.
CYLD acts proximal to the membrane as a negative regulator of TNF-induced NF-κB signaling. The deubiquitinase activity of CYLD allows it to hydrolyze K63-linked ubiquitin chains, which are critical for the activity of multiple components within the NF-κB signaling network, and thereby acts as a suppressor by inhibiting proliferation and promoting apoptosis.
In addition to driving cellular proliferation, NF-κB also promotes cell survival, at least in part via transcriptional induction of antiapoptotic Bcl-2 family members (BCL-2, BCL-XL, and A1) (Grumont et al. 1999; Lee et al. 1999; Zong et al. 1999; Chen et al. 2000; Grossmann et al. 2000) as well as several IAP (inhibitor of apoptosis) proteins (Chu et al. 1997; Stehlik et al. 1998; Wang et al. 1998). These NF-κB targets are known to be essential for the survival of various cell types in response to diverse stress stimuli. Collectively, these data indicate that CYLD suppresses tumorigenesis by reducing cell cycling (e.g., via repression of CYCLIN D1 expression) and by reducing expression of antiapoptotic regulators. Indeed, CYLD is frequently deleted in cases of multiple myeloma showing hyperactive NF-κB and abnormalities in cell cycling and apoptosis induction (Annunziata et al. 2007).
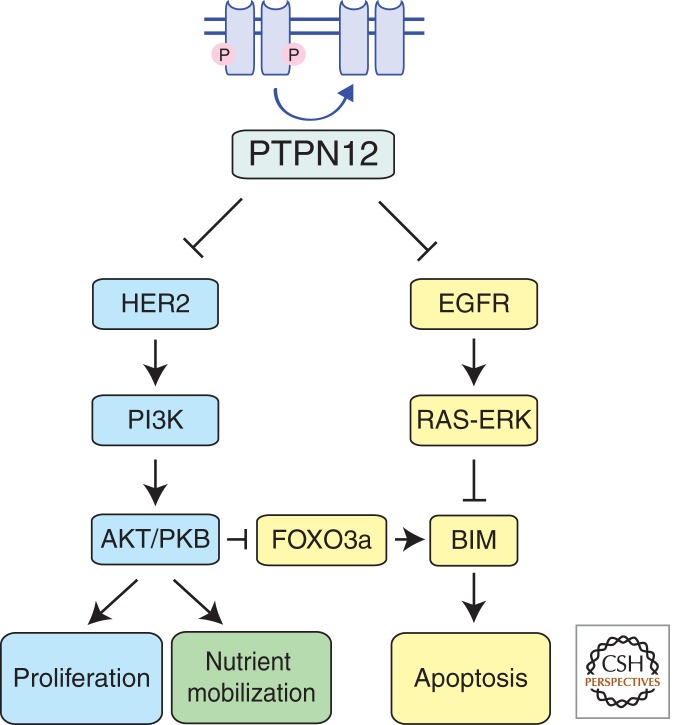
PTPN12, a Safety Catch for Several Oncogenic Receptor Kinases
PTPN12 has recently been described as a tumor suppressor in HER2, estrogen receptor and progesterone receptor negative (so-called “triple negative”) breast cancer (TNBC) (Sun et al. 2011). PTPN12 expression was found to be low or undetectable in primary breast cancer samples, and the locus from which it is expressed is more frequently lost in TNBC compared to other breast cancer subtypes (Sun et al. 2011). PTPN12 encodes a receptor tyrosine phosphatase that can dephosphorylate, and thereby inactivate, several kinase-containing surface receptors (e.g., ER, HER2, EGF-R) that promote breast cancer development when abnormally activated or overexpressed. Knock-down of the PTPN12 phosphatase by RNAi enhanced anchorage-independent growth of breast epithelial cells in culture and the formation of abnormal acini in 3D culture conditions (Sun et al. 2011). Interestingly, the enhanced proliferation and acini formation of MCF10A breast epithelial derived cells resulting from reduced PTPN12 function was not accompanied by increased apoptosis (Sun et al. 2011). Mutants of PTPN12 deficient in phosphatase activity could not inhibit cell growth and acini formation, indicating that PTPN12’s catalytic activity is required for tumor suppression. Consistent with the hypothesis that PTPN12 acts as a safety catch for HER2 and EGFR signaling for cell growth, pharmacological or shRNA-mediated inhibition of these receptors reduced the efficiency of transformation achieved through PTPN12 knockdown (Sun et al. 2011).
Loss of PTPN12 is thought to promote breast cancer development by allowing unrestrained signaling from the HER2, EGF-R, and PDGF-R receptors (Sun et al. 2011). These receptor tyrosine kinases all stimulate pathways for cell proliferation and, through activation of AKT as well as ERK, they probably also promote cell survival by repressing expression and/or function of the BH3-only proteins BIM and BAD (Fig. 5). Interestingly, it has been shown that repression of BIM (and to a lesser extent BAD) is critical for the sustained survival of certain other cancers that are driven by other oncogenic kinases—for example, BCR-ABL in CML (Kuroda et al. 2006), mutant EGF-R in lung cancer (Costa et al. 2007; Cragg et al. 2007), mutant B-RAF in melanoma, and colon carcinoma (Cragg et al. 2008).
Figure 5.
PTPN12 is a recently described tumor suppressor with phosphatase activity. It negatively regulates signaling through multiple oncogenic receptor kinases, such as EGFR and HER2. By limiting the activity of these receptors, PTPN12 blocks their ability to promote proliferation, nutrient mobilization, and inhibit apoptosis.
These data show that oncogenic hyperactivation of several receptor tyrosine kinases that promote abnormal cell proliferation and survival can be achieved more readily through loss of a single common “safety catch” than through coordinate activation of multiple cell surface receptors.
CLOSING REMARKS
Multicellular organisms require mechanisms to ensure appropriate cellularity of their tissues and, consequently, these processes impose barriers against tumor development. Some tumor suppressive mechanisms, such as cell cycle arrest or cellular senescence, facilitate repair and recovery of damaged cells. However, because of their reversible nature, these mechanisms can be subverted allowing outgrowth of clonal offspring. In contrast, apoptotic cell death eliminates cells, and although this unequivocally prevents clonal outgrowth of mutated cells, it comes at a cost to the organism. Some cells once lost cannot be readily replaced, and if the threshold for apoptosis initiation is set too low, cells may be eliminated unnecessarily and tissue degeneration may ensue. So, in tissues that are critical for function of the organism but have only low regenerative capacity (e.g., brain), processes such as cell cycle arrest, senescence, and differentiation are more appropriate mechanisms for tumor suppression than apoptosis. Conversely, in tissues with considerable regenerative capacity, such as the immune system, apoptosis would be the preferred mechanism for tumor suppression.
Although apoptosis is clearly a potent mechanism for tumor suppression and must be evaded to facilitate neoplastic transformation and sustained tumor expansion, other mechanisms are also critical to protect us from cancer. Thus, if escape from cell cycle arrest, senescence, apoptosis, and the mobilization of nutrients are all required for tumorigenesis to proceed, then mutational activation or inactivation of oncogenes or tumor suppressor genes, respectively, that regulate a number of these processes will enable cells to circumvent homeostatic controls in a more efficient manner than if each of these processes must be deregulated independently through multiple oncogenic events. The prevalence of mutations in certain tumor suppressors, particularly P53 and PTEN, in diverse human cancers identifies them as “Achilles heels” in the cell’s tumor suppression network, in which loss or inactivation of these targets can confer on cells undergoing neoplastic transformation multiple advantages rather than enhanced survival or increased cycling alone. Deciphering the complex networks for tumor suppression is not only critical for understanding the development and growth of cancer, but also has ramifications for cancer therapy because many tumor suppressors, and their processes for apoptosis induction in particular, also regulate the responses of tumor cells to anticancer therapy (Johnstone et al. 2002).
ACKNOWLEDGMENTS
The authors thank all present and past members of the apoptosis research programs at WEHI, particularly Drs. J. Adams, S. Cory, D. Vaux, D. Huang, P. Colman, P. Bouillet, A. Harris, R. Kluck, J. Silke, and C. Scott, for their outstanding contributions to cell death research and for stimulating discussions. Research in the authors’ laboratories is supported by fellowships and grants from the Australian NHMRC (257502, 461299), Cancer Council of Victoria, NIH (CA 043540), Leukemia and Lymphoma Society (LLS SCOR 7413), and the JDRF/NHMRC (466658).
Footnotes
Editors: Eric H. Baehrecke, Douglas R. Green, Sally Kornbluth, and Guy S. Salvesen
Additional Perspectives on Cell Survival and Cell Death available at www.cshperspectives.org
REFERENCES
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, et al. 2007. Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12: 115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak Y, Juven T, Haffner R, Oren M 1993. mdm2 expression is induced by wild type p53 activity. EMBO J 12: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates S, Phillips AC, Clark PA, Stott F, Peters G, Ludwig RL, Vousden KH 1998. p14ARF links the tumour suppressors RB and p53. Nature 395 [DOI] [PubMed] [Google Scholar]
- Bignell GR, Warren W, Seal S, Takahashi M, Rapley E, Barfoot R, Green H, Brown C, Biggs PJ, Lakhani SR, et al. 2000. Identification of the familial cylindromatosis tumour-suppressor gene. Nat Genet 25: 160–165 [DOI] [PubMed] [Google Scholar]
- Bouillet P, Metcalf D, Huang DCS, Tarlinton DM, Kay TWH, Köntgen F, Adams JM, Strasser A 1999. Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286: 1735–1738 [DOI] [PubMed] [Google Scholar]
- Brady CA, Jiang D, Mello SS, Johnson TM, Jarvis LA, Kozak MM, Broz DK, Basak S, Park EJ, McLaughlin ME, et al. 2011. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumour suppression. Cell 145: 571–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R 2003. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-κB. Nature 424: 797–801 [DOI] [PubMed] [Google Scholar]
- Chen C, Edelstein LC, Gelinas C 2000. The Rel/NF-κB family directly activates expression of the apoptosis inhibitor Bcl-xL. Mol Cell Biol 20: 2687–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk JE, Green DR 2008. How do BCL-2 proteins induce mitochondrial outer membrane permeabilization? Trends Cell Biol 18: 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Z-L, McKinsey TA, Liu L, Gentry JJ, Malim MH, Ballard DW 1997. Suppression of tumour necrosis factor-induced cell death by inhibitor of apoptosis c-IAP2 is under NF-κB control. Proc Natl Acad Sci 94: 10057–10062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory S, Harris AW, Langdon WY, Alexander WS, Corcoran LM, Palmiter RD, Pinkert CA, Brinster RL, Adams JM 1987. The myc oncogene and lymphoid neoplasia: From translocations to transgenic mice. Modern Trends Human Leuk VII: 248–251 [DOI] [PubMed] [Google Scholar]
- Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, Tenen DG, Kobayashi S 2007. BIM mediates EGFR tyrosine kinase inhibitor-induced apoptosis in lung cancers with oncogenic EGFR mutations. PLoS Med 4: e315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg MS, Kuroda J, Puthalakath H, Huang DCS, Strasser A 2007. Gefitinib-induced killing of NSCLC cell lines expressing mutant EGFR requires Bim and can be enhanced by BH3 mimetics. PLoS Med 4: 1681–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg MS, Jansen ES, Cook M, Harris C, Strasser A, Scott CL 2008. Treatment of B-RAF mutant human tumour cells with a MEK inhibitor requires Bim and is enhanced by a BH3 mimetic. J Clin Invest 118: 3651–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannenberg JH, van Rossum A, Schuijff L, te Riele H 2000. Ablation of the retinoblastoma gene family deregulates G1 control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev 14: 3051–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger M, Buchdunger E, Druker BJ 2005. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood 105: 2640–2653 [DOI] [PubMed] [Google Scholar]
- del Peso L, González-Garcia M, Page C, Herrera R, Nuñez G 1997. Interleukin-3–induced phosphorylation of BAD through the protein kinase Akt. Science 278: 687–689 [DOI] [PubMed] [Google Scholar]
- Deng C, Zhang P, Harper JW, Elledge SJ, Leder P 1995. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell 82: 675–684 [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP 1998. Pten is essential for embryonic development and tumour suppression. Nat Genet 19: 348–355 [DOI] [PubMed] [Google Scholar]
- Dijkers PF, Medema RH, Lammers JJ, Koenderman L, Coffer PJ 2000. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol 10: 1201–1204 [DOI] [PubMed] [Google Scholar]
- Donehower LA, Harvey M, Slagle BL, McArthur MJ, Montgomery CAJ, Butel JS, Bradley A 1992. Mice deficient for p53 are developmentally normal but are susceptible to spontaneous tumours. Nature 356: 215–221 [DOI] [PubMed] [Google Scholar]
- Drappa J, Vaishnaw AK, Sullivan KE, Chu JL, Elkon KB 1996. Fas gene mutations in the Canale–Smith syndrome, an inherited lymphoproliferative disorder associated with autoimmunity. N Engl J Med 335: 1643–1649 [DOI] [PubMed] [Google Scholar]
- Egle A, Harris AW, Bouillet P, Cory S 2004. Bim is a suppressor of Myc-induced mouse B cell leukemia. Proc Natl Acad Sci 101: 6164–6169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eischen CM, Roussel MF, Korsmeyer SJ, Cleveland JL 2001. Bax loss impairs Myc-induced apoptosis and circumvents the selection of p53 mutations during Myc-mediated lymphomagenesis. Mol Cell Biol 21: 7653–7662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlacher M, Michalak EM, Kelly PN, Labi V, Niederegger H, Coultas L, Adams JM, Strasser A, Villunger A 2005. BH3-only proteins Puma and Bim are rate-limiting for γ-radiation and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood 106: 4131–4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garami A, Zwartkruis FJ, Nobukuni T, Joaquin M, Roccio M, Stocker H, Kozma SC, Hafen E, Bos JL, Thomas G 2003. Insulin activation of Rheb, a mediator of mTOR/S6K/4E-BP signaling, is inhibited by TSC1 and 2. Mol Cell 11: 1457–1466 [DOI] [PubMed] [Google Scholar]
- Garrison SP, Jeffers JR, Yang C, Nilsson JA, Hall MA, Rehg JE, Yue W, Yu J, Zhang L, Onciu M, et al. 2008. Selection against PUMA gene expression in Myc-driven B-cell lymphomagenesis. Mol Cell Biol 28: 5391–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann M, O’Reilly LA, Gugasyan R, Strasser A, Adams JM, Gerondakis S 2000. The anti-apoptotic activities of rel and RelA required during B-cell maturation involve the regulation of Bcl-2 expression. EMBO J 19: 6351–6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumont RJ, Rourke IJ, Gerondakis S 1999. Rel-dependent induction of A1 transcription is required to protect B cells from antigen receptor ligation-induced apoptosis. Gen Dev 13: 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA 2000. The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M 1997. Mdm2 promotes the rapid degradation of p53. Nature 387: 296–299 [DOI] [PubMed] [Google Scholar]
- Hershko T, Ginsberg D 2004. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem 279: 8627–8634 [DOI] [PubMed] [Google Scholar]
- Hotchkiss RS, Strasser A, McDunn JE, Swanson PE 2009. Cell death. N Engl J Med 361: 1570–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DCS, Strasser A 2000. BH3-only proteins—Essential initiators of apoptotic cell death. Cell 103: 839–842 [DOI] [PubMed] [Google Scholar]
- Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA 1992. Effects of an Rb mutation in the mouse. Nature 359: 295–300 [DOI] [PubMed] [Google Scholar]
- Jeffers JR, Parganas E, Lee Y, Yang C, Wang J, Brennan J, MacLean KH, Han J, Chittenden T, Ihle JN, et al. 2003. Puma is an essential mediator of p53–dependent and -independent apoptotic pathways. Cancer Cell 4: 321–328 [DOI] [PubMed] [Google Scholar]
- Johnstone RW, Ruefli AA, Lowe SW 2002. Apoptosis: A link between cancer genetics and chemotherapy. Cell 108: 153–164 [DOI] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A 1995. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378: 206–208 [DOI] [PubMed] [Google Scholar]
- Kelly PN, White MJ, Goschnick MW, Fairfax KA, Tarlinton DM, Kinkel SA, Bouillet P, Adams JM, Kile BT, Strasser A 2010. Individual and overlapping roles of BH3-only proteins Bim and Bad in apoptosis of lymphocytes and platelets and in suppression of thymic lymphoma development. Cell Death Differ 17: 1655–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G 2003. The tumour suppressor CYLD negatively regulates NF-κB signalling by deubiquitination. Nature 424: 801–805 [DOI] [PubMed] [Google Scholar]
- Kubbutat MH, Jones SN, Vousden KH 1997. Regulation of p53 stability by Mdm2. Nature 387: 299–303 [DOI] [PubMed] [Google Scholar]
- Kuroda J, Puthalakath H, Cragg MS, Kelly PN, Bouillet P, Huang DC, Kimura S, Ottmann OG, Druker BJ, Villunger A, et al. 2006. Bim and Bad mediate imatinib-induced killing of Bcr/Abl+ leukemic cells, and resistance due to their loss is overcome by a BH3 mimetic. Proc Natl Acad Sci 103: 14907–14912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labi V, Erlacher M, Kiessling S, Manzl C, Frenzel A, O'Reilly L, Strasser A, Villunger A 2008. Loss of the BH3-only protein Bmf impairs B cell homeostasis and accelerates γ irradiation-induced thymic lymphoma development. J Exp Med 205: 641–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W-H, Bookstein R, Hong F, Young L-J, Shew J-Y, Lee EY-HP 1987. Human retinoblastoma susceptibility gene: cloning, identification and sequence. Science 235: 1394–1399 [DOI] [PubMed] [Google Scholar]
- Lee HH, Dadgostar H, Cheng Q, Shu J, Cheng G 1999. NF-κB-mediated up-regulation of Bcl-x and Bfl-1/A1 is required for CD40 survival signaling in B lymphocytes. Proc Natl Acad Sci 96: 9136–9141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees JA, Saito M, Vidal M, Valentine M, Look T, Harlow E, Dyson N, Helin K 1993. The retinoblastoma protein binds to a family of E2F transcription factors. Mol Cell Biol 13: 7813–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ 2009. The common mechanisms of transformation by the small DNA tumour viruses: The inactivation of tumour suppressor gene products: p53. Virology 384: 285–293 [DOI] [PubMed] [Google Scholar]
- Li T, Kon N, Jiang L, Tan M, Ludwig T, Zhao Y, Baer R, Gu W 2012. Tumour suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149: 1269–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WC, Lin FT, Nevins JR 2001. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev 15: 1833–1844 [PMC free article] [PubMed] [Google Scholar]
- Lindsten T, Ross AJ, King A, Zong W, Rathmell JC, Shiels HA, Ulrich E, Waymire KG, Mahar P, Frauwirth K, et al. 2000. The combined functions of proapoptotic Bcl-2 family members Bak and Bax are essential for normal development of multiple tissues. Mol Cell 6: 1389–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livingstone LR, White A, Sprouse J, Livanos E, Jacks T, Tlsty TD 1992. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell 70: 923–935 [DOI] [PubMed] [Google Scholar]
- Marsden V, O'Connor L, O'Reilly LA, Silke J, Metcalf D, Ekert P, Huang DCS, Cecconi F, Kuida K, Tomaselli KJ, et al. 2002. Apoptosis initiated by Bcl-2-regulated caspase activation independently of the cytochrome c/Apaf–1/caspase–9 apoptosome. Nature 419: 634–637 [DOI] [PubMed] [Google Scholar]
- Martin K, Trouche D, Hagemeier C, Sorensen TS, La Thangue NB, Kouzarides T 1995. Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature 375: 691–694 [DOI] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R 2006. Cyld inhibits tumour cell proliferation by blocking Bcl-3-dependent NF-κB signaling. Cell 125: 665–677 [DOI] [PubMed] [Google Scholar]
- Merino D, Giam M, Hughes PD, Siggs OM, Heger K, O’Reilly LA, Adams JM, Strasser A, Lee EF, Fairlie WD, et al. 2009. The role of BH3-only protein Bim extends beyond inhibiting Bcl-2-like prosurvival proteins. J Cell Biol 186: 355–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz T, Harris AW, Adams JM 1995. Absence of p53 allows direct immortalization of hematopoietic cells by the myc and raf oncogenes. Cell 82: 29–36 [DOI] [PubMed] [Google Scholar]
- Michalak EM, Villunger A, Adams JM, Strasser A 2008. In several cell types the tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ 15: 1019–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Jansen ES, Happo L, Cragg MS, Tai L, Smyth GK, Strasser A, Adams JM, Scott CL 2009. Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ 16: 684–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak EM, Vandenberg CJ, Delbridge ARD, Wu L, Scott CL, Adams JM, Strasser A 2010. Apoptosis-promoted tumorigenesis: γ-Irradiation-induced thymic lymphomagenesis requires Puma-driven leukocyte death. Genes Dev 24: 1608–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita T, Reed JC 1995. Tumour suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 80: 293–299 [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G 1995. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378: 203–206 [DOI] [PubMed] [Google Scholar]
- Naik E, Michalak EM, Villunger A, Adams JM, Strasser A 2007. UV-radiation triggers apoptosis of fibroblasts and skin keratinocytes mainly via the BH3-only protein Noxa. J Cell Biol 176: 415–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K, Vousden KH 2001. PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell 7: 683–694 [DOI] [PubMed] [Google Scholar]
- O’Connor L, Strasser A, O’Reilly LA, Hausmann G, Adams JM, Cory S, Huang DCS 1998. Bim: A novel member of the Bcl-2 family that promotes apoptosis. EMBO J 17: 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda E, Ohki R, Murasawa H, Nemoto J, Shibue T, Yamashita T, Tokino T, Taniguchi T, Tanaka N 2000. Noxa, a BH3-only member of the bcl-2 family and candidate mediator of p53–induced apoptosis. Science 288: 1053–1058 [DOI] [PubMed] [Google Scholar]
- O’Reilly LA, Tai L, Lee L, Kruse EA, Grabow S, Fairlie WD, Haynes NM, Tarlinton DM, Zhang JG, Belz GT, et al. 2009. Membrane-bound Fas ligand only is essential for Fas-induced apoptosis. Nature 461: 659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, Porcellini A, Screpanti I, Balsano C, Alesse E, et al. 2003. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat Cell Biol 5: 552–558 [DOI] [PubMed] [Google Scholar]
- Pierce AM, Fisher SM, Conti CJ, Johnson DG 1998a. Deregulated expression of E2F1 induces hyperplasia and cooperates with ras in skin tumour development. Oncogene 16: 1267–1276 [DOI] [PubMed] [Google Scholar]
- Pierce AM, Gimenez-Conti IB, Schneider-Broussard R, Martinez LA, Conti CJ, Johnson DG 1998b. Increased E2F1 activity induces skin tumours in mice heterozygous and nullizygous for p53. Proc Natl Acad Sci 95: 8858–8863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polager S, Ginsberg D 2009. p53 and E2f: Partners in life and death. Nat Rev Cancer 9: 738–748 [DOI] [PubMed] [Google Scholar]
- Potter CJ, Pedraza LG, Xu T 2002. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol 4: 658–665 [DOI] [PubMed] [Google Scholar]
- Poulikakos PI, Rosen N 2011. Mutant BRAF melanomas—Dependence and resistance. Cancer Cell 19: 11–15 [DOI] [PubMed] [Google Scholar]
- Ranger AM, Zha J, Harada H, Datta SR, Danial NN, Gilmore AP, Kutok JL, Le Beau MM, Greenberg ME, Korsmeyer SJ 2003. Bad-deficient mice develop diffuse large B cell lymphoma. Proc Natl Acad Sci 100: 9324–9329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl SJ, Shi Y 2004. Molecular mechanisms of caspase regulation during apoptosis. Nat Rev Mol Cell Biol 5: 897–907 [DOI] [PubMed] [Google Scholar]
- Rieux-Laucat F, Le Deist F, Hivroz C, Roberts IAG, Debatin KM, Fischer A, de Villartay JP 1995. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science 268: 1347–1349 [DOI] [PubMed] [Google Scholar]
- Riley T, Sontag E, Chen P, Levine A 2008. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol 9: 402–412 [DOI] [PubMed] [Google Scholar]
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, Theodorou E, Jacks T 2000. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev 14: 3037–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Bell DW, Settleman J, Haber DA 2007. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 7: 169–181 [DOI] [PubMed] [Google Scholar]
- Shi Y 2002. Mechanisms of caspase activation and inhibition during apoptosis. Mol Cell 9: 459–470 [DOI] [PubMed] [Google Scholar]
- Srivastava S, Zou ZQ, Pirollo K, Plattner W, Chang EH 1990. Germ-line transmission of a mutated p53 gene in a cancer-prone family with Li-Fraumeni syndrome. Nature 348: 747–749 [DOI] [PubMed] [Google Scholar]
- Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW 1998. Negative regulation of PKB/Akt-dependent cell survival by the tumour suppressor PTEN. Cell 95: 29–39 [DOI] [PubMed] [Google Scholar]
- Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW 2001. Regulation of PTEN transcription by p53. Mol Cell 8: 317–325 [DOI] [PubMed] [Google Scholar]
- Stehlik C, de Martin R, Kumabashiri I, Schmid JA, Binder BR, Lipp J 1998. Nuclear factor NF-κB-regulated X-chromosome-linked iap gene expression protects endothelial cells from tumour necrosis factor alpha-induced apoptosis. J Exp Med 188: 211–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C, Smith L, La Thangue NB 2003. Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol 5: 401–409 [DOI] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Bath ML, Cory S 1990. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature 348: 331–333 [DOI] [PubMed] [Google Scholar]
- Strasser A, Harris AW, Cory S 1993. Em-bcl-2 transgene facilitates spontaneous transformation of early pre-B and immunoglobulin-secreting cells but not T cells. Oncogene 8: 1–9 [PubMed] [Google Scholar]
- Strasser A, Harris AW, Huang DCS, Krammer PH, Cory S 1995. Bcl-2 and Fas/APO-1 regulate distinct pathways to lymphocyte apoptosis. EMBO J 14: 6136–6147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A, O’Connor L, Dixit VM 2000. Apoptosis signaling. Ann Rev Biochem 69: 217–245 [DOI] [PubMed] [Google Scholar]
- Strasser A, Jost PJ, Nagata S 2009. The many roles of FAS receptor signaling in the immune system. Immunity 30: 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Aceto N, Meerbrey KL, Kessler JD, Zhou C, Migliaccio I, Nguyen DX, Pavlova NN, Botero M, Huang J, et al. 2011. activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell 144: 703–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, et al. 1998. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumour suppressor gene in mice. Curr Biol 8: 1169–1178 [DOI] [PubMed] [Google Scholar]
- Tagawa H, Karnan S, Suzuki R, Matsuo K, Zhang X, Ota A, Morishima Y, Nakamura S, Seto M 2005. Genome-wide array-based CGH for mantle cell lymphoma: Identification of homozygous deletions of the proapoptotic gene BIM. Oncogene 24: 1348–1358 [DOI] [PubMed] [Google Scholar]
- Timmer JC, Salvesen GS 2007. Caspase substrates. Cell Death Differ 14: 66–72 [DOI] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G 2003. CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature 424: 793–796 [DOI] [PubMed] [Google Scholar]
- Urist M, Tanaka T, Poyurovsky MV, Prives C 2004. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev 18: 3041–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaux DL, Cory S, Adams JM 1988. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 335: 440–442 [DOI] [PubMed] [Google Scholar]
- Villunger A, Michalak EM, Coultas L, Mullauer F, Bock G, Ausserlechner MJ, Adams JM, Strasser A 2003. p53- and drug-induced apoptotic responses mediated by BH3-only proteins Puma and Noxa. Science 302: 1036–1038 [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP 2007. p53 in health and disease. Nat Rev Mol Cell Biol 8: 275–283 [DOI] [PubMed] [Google Scholar]
- Wang C-Y, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS Jr 1998. NF-κB antiapoptosis: Induction of TRAF1 and TRAF2 and c-IAP1 and c- IAP2 to suppress caspase-8 activation. Science 281: 1680–1683 [DOI] [PubMed] [Google Scholar]
- Watanabe-Fukunaga R, Brannan CI, Copeland NG, Jenkins NA, Nagata S 1992. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature 356: 314–317 [DOI] [PubMed] [Google Scholar]
- Wertz IE, Dixit VM 2010. Regulation of death receptor signaling by the ubiquitin system. Cell Death Differ 17: 14–24 [DOI] [PubMed] [Google Scholar]
- Wu X, Bayle JH, Olson D, Levine AJ 1993. The p53–mdm-2 autoregulatory feedback loop. Genes Dev 7: 1126–1132 [DOI] [PubMed] [Google Scholar]
- Xiao ZX, Chen J, Levine AJ, Modjtahedi N, Xing J, Sellers WR, Livingston DM 1995. Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature 375: 694–698 [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A 2008. The BCL-2 protein family: Opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9: 47–59 [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B 2001. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell 7: 673–682 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang H, Li M, Rayburn ER, Agrawal S, Zhang R 2005. Stabilization of E2F1 protein by MDM2 through the E2F1 ubiquitination pathway. Oncogene 24: 7238–7247 [DOI] [PubMed] [Google Scholar]
- Zong WX, Edelstein LC, Chen C, Bash J, Gelinas C 1999. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-κB that blocks TNFα-induced apoptosis. Genes Dev 13: 382–387 [DOI] [PMC free article] [PubMed] [Google Scholar]